ABSTRACT
Rabies virus (RABV) P gene mRNA encodes five in-frame start codons, resulting in expression of full-length P protein (P1) and N-terminally truncated P proteins (tPs), designated P2, P3, P4, and P5. Despite the fact that some tPs are known as interferon (IFN) antagonists, the importance of tPs in the pathogenesis of RABV is still unclear. In this study, to examine whether tPs contribute to pathogenesis, we exploited a reverse genetics approach to generate CE(NiP)ΔP2-5, a mutant of pathogenic CE(NiP) in which the P gene was mutated by replacing all of the start codons (AUG) for tPs with AUA. We confirmed that while CE(NiP) expresses detectable levels of P2 and P3, CE(NiP)ΔP2-5 has an impaired ability to express these tPs. After intramuscular inoculation, CE(NiP)ΔP2-5 caused significantly lower morbidity and mortality rates in mice than did CE(NiP), indicating that tPs play a critical role in RABV neuroinvasiveness. Further examinations revealed that this less neuroinvasive phenotype of CE(NiP)ΔP2-5 correlates with its impaired ability to replicate in muscle cells, indicative of the importance of tPs in viral replication in muscle cells. We also demonstrated that CE(NiP)ΔP2-5 infection induced a higher level of Ifn-β gene expression in muscle cells than did CE(NiP) infection, consistent with the results of an IFN-β promoter reporter assay suggesting that all tPs function to antagonize IFN induction in muscle cells. Taken together, our findings strongly suggest that tPs promote viral replication in muscle cells through their IFN antagonist activities and thereby support infection of peripheral nerves.
IMPORTANCE Despite the fact that previous studies have demonstrated that P2 and P3 of RABV have IFN antagonist activities, the actual importance of tPs in pathogenesis has remained unclear. Here, we provide the first evidence that tPs contribute to the pathogenesis of RABV, especially its neuroinvasiveness. Our results also show the mechanism underlying the neuroinvasiveness driven by tPs, highlighting the importance of their IFN antagonist activities, which support viral replication in muscle cells.
INTRODUCTION
Rabies virus (RABV), a member of the genus Lyssavirus of the family Rhabdoviridae, is a zoonotic agent that causes a lethal neurological disease in various mammal species, including humans. After transmission via a bite wound caused by an infected animal, RABV infects peripheral nerves and then spreads to and in the central nervous system (CNS), resulting in severe neurological symptoms with a high case fatality rate of almost 100% (reviewed in reference 1). Due to the absence of an effective cure and insufficient provision of postexposure prophylaxis, approximately 59,000 people die from rabies every year, mainly in developing countries (2). To establish an effective cure and also to develop a novel prophylaxis approach for rabies, it is necessary to understand the molecular mechanisms of the pathogenesis, including immune evasion, of RABV.
The phosphoprotein (P protein) of RABV is a multifunctional protein that is indispensable not only for viral replication but also for evasion of host innate immunity. Specifically, this protein plays an essential role in viral RNA synthesis as a cofactor of viral RNA-dependent RNA polymerase (L protein) by bridging nucleoprotein (N protein), which directly binds to viral genomic RNA, and L protein in the ribonucleoprotein complex (reviewed in reference 3). In addition, P protein functions to antagonize the type I interferon (IFN)-mediated antiviral responses by inhibiting both signaling pathways for IFN induction and response (4–12). P protein suppresses activation of interferon regulatory factor 3 (IRF-3), which is an important transcription factor for IFN induction (5, 8). Also, P protein binds to the transcriptional factors signal transducers and activator of transcription 1 (STAT1) and STAT2, which play a key role in the IFN response by activating expression of IFN-stimulated genes (ISGs), and inhibits their nuclear translocation and DNA binding (6, 10, 11).
In RABV-infected cells, mRNA of the P gene is translated from five in-frame start codons by a ribosomal leaky scanning mechanism, resulting in expression of full-length P protein (P1; 297 amino acids) and also less abundant expression of N-terminally truncated P proteins (tPs), designated P2, P3, P4, and P5, the amino acid sequences of which correspond to those of P1 at positions 20 to 297, 53 to 297, 69 to 297, and 83 to 297, respectively (Fig. 1A) (13). While P1 physically interacts with L protein by its N-terminal domain (amino acids 1 to 19) (14) to act as a cofactor of viral RNA polymerase, tPs lack the L protein-binding domain, indicating that tPs do not have cofactor activity. Importantly, start codons for the translation of tPs are highly conserved among RABV strains (15, 16), strongly suggesting a critical role of tPs in RABV infection.
FIG 1.
Schematic diagrams of the expression patterns and primary structures of P protein isoforms from P gene mRNA of CE(NiP) (A) and that of CE(NiP)ΔP2-5 (B). Mutations introduced into the P gene of CE(NiP)ΔP2-5 are indicated in boldface. (C) P protein isoforms in NA cells infected with CE(NiP) and CE(NiP)ΔP2-5 together with the respective isoforms expressed in transfected cells were analyzed by Western blotting. Tubulin in each sample was also detected as a loading control.
Notably, all of the RABV P protein isoforms (P1 and tPs) retain a functionally important domain for inhibition of IRF-3 activation (amino acids 176 to 181 in P1) (8) as well as the STAT1-binding domain (amino acids 267 to 297 in P1) (10), implying that tPs have activity to antagonize the host IFN system. In fact, by using a recombinant protein expression system, it was previously demonstrated that P3 inhibits nuclear translocation and DNA binding of STATs (11, 17). Further, Marschalek et al. (18) showed by using an RABV strain genetically modified to overexpress P2 that this isoform plays an important role in the IFN resistance of the virus. In addition, Blondel et al. (4) reported that P3 directly interacts with an ISG product, promyelocytic leukemia protein, that has antiviral activity (19, 20), implying that P3 inhibits the antiviral function of this host protein. Although the overall functions of tPs in IFN antagonism remain to be elucidated, these findings suggest that tPs contribute to evasion of IFN-mediated innate immunity and thereby also to the pathogenesis of RABV. However, the actual contribution of tPs to the pathogenesis has not been elucidated.
The pathogenesis of RABV depends on the ability of the virus to infect peripheral nerves and to spread to the CNS (neuroinvasiveness) and also the ability to spread in the CNS and to cause neurological disease (neurovirulence). We previously reported that RABV P protein has a key role in both neuroinvasiveness and neurovirulence (21, 22). This is supported by experimental data showing that chimeric mutant CE(NiP), which has the P gene from virulent Nishigahara (Ni) in the genome of the attenuated variant Ni-CE, caused lethal infection in mice after both intramuscular (i.m.) and intracerebral (i.c.) inoculations, whereas Ni-CE caused asymptomatic infection and nonlethal, mild disease after the respective inoculations. Therefore, we consider that CE(NiP) provides a good model to evaluate the contribution of tPs to the pathogenesis of RABV, including neuroinvasiveness and neurovirulence.
In the present study, to determine whether tPs play a significant role in the pathogenesis of RABV, we generated a CE(NiP) mutant [CE(NiP)ΔP2-5] in which the P gene was mutated by replacing all of the start codons (AUG) for tPs with AUA (Fig. 1B), and we examined its virulence in mice. The results indicated that CE(NiP)ΔP2-5 was less pathogenic than the parental CE(NiP) in mice, especially in infection via i.m. inoculation, indicating that tPs have a critical role in neuroinvasiveness. The findings obtained from further examinations strongly suggest that tPs support viral propagation in muscle cells by suppressing IFN induction, consequently facilitating infection of peripheral nerves. To our knowledge, this study provides the first substantial evidence that tPs play a critical role in the pathogenesis of RABV.
MATERIALS AND METHODS
Cells.
Mouse neuroblastoma NA cells and human neuroblastoma SYM-I cells (kindly provided by Akihiko Kawai) were maintained in Eagle's minimal essential medium (MEM) supplemented with 10% fetal calf serum (FCS). A baby hamster kidney (BHK) cell clone, BHK/T7-9 cells (23), which constitutively express T7 RNA polymerase, were maintained in Eagle's MEM supplemented with 10% tryptose phosphate broth and 5% FCS. Mouse muscle myoblast G-8 cells (American Type Culture Collection [ATCC] no. CRL-1456) were grown in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% FCS and 10% horse serum (HS). Before being used for the experiments, G-8 cells were differentiated by reducing the FCS and HS to 2% each.
Viruses.
CE(NiP) was previously generated by a reverse genetics approach (21). Recombinant CE(NiP) expressing firefly luciferase [CE(NiP)-Luc] was also generated in our previous study (22).
To generate CE(NiP)ΔP2-5, all of the start codons (AUG; shown in positive sense) for P2, P3, P4, and P5 in the full-length genome plasmid of CE(NiP) (Fig. 1A) (21) were replaced with AUA (positive sense) (Fig. 1B) by using conventional molecular cloning techniques. To rescue CE(NiP)ΔP2-5 from the resulting genome plasmid, we transfected BHK/T7-9 cells with this plasmid together with pT7IRES-RN, -RP, and -RL, expressing viral N, P, and L proteins, respectively, as previously reported (23). By using the same methods, we also obtained a recombinant CE(NiP)ΔP2-5 expressing firefly luciferase [CE(NiP)ΔP2-5-Luc] after inserting a PCR-amplified cDNA fragment of the luciferase gene into the G-L intergenic region of the above-described genome plasmid of CE(NiP)ΔP2-5 as previously reported (22, 23). Details of the construction of the above-described genome plasmids will be provided by the authors on request.
Working stocks of all viruses were prepared in NA cells and stored at −80°C. The infectious viruses in these stocks were titrated by focus assays on confluent NA cells as previously reported (22).
Construction of plasmids.
To construct plasmids expressing the respective P protein isoforms (P1, P2, P3, P4, and P5) of Ni and P1 of CE(NiP)ΔP2-5 (P1ΔP2-5), cDNA fragments containing the genetic regions from a start codon for each isoform to a stop codon were amplified by PCR using the genome plasmid of CE(NiP) (21) and CE(NiP)ΔP2-5 (described above) as a template. The cDNA fragments then were cloned into an RNA polymerase II-based mammalian expression plasmid, pCAGGS/MCS (kindly provided by Yoshihiro Kawaoka). The resulting plasmids were designated pCAGGS-P1, -P2, -P3, -P4, -P5, and -P1ΔP2-5, respectively. To obtain plasmids expressing N-terminally green fluorescent protein (GFP)-tagged P1s of CE(NiP) (GFP-P1) and the tagged P1 of CE(NiP)ΔP2-5 (GFP-P1ΔP2-5), cDNA fragments containing the full-length P1 coding region of the respective viruses were amplified by PCR and then cloned into pEGFP-C1 vector (Clontech, Mountain View, CA). We named the resulting plasmids pEGFP-P1 and pEGFP-P1ΔP2-5, respectively.
To establish a minigenome assay system (see below), we constructed a plasmid expressing luciferase-encoding minigenome RNA (named pCAGGS-RVDI-Luc). Briefly, we amplified a cDNA fragment consisting of a hammerhead ribozyme, the 5′ trailer region of RABV, a firefly luciferase gene, the 3′ leader region of RABV, and a hepatitis delta virus antigenomic ribozyme by PCR using pRVDI-luc (12) as a template and then cloned the fragment into the pCAGGS/MCS vector. We also constructed a pCAGGS-based plasmid expressing L protein of Ni-CE (pCAGGS-CEL) by stepwise cloning of cDNA fragments, which had been amplified by PCR or purified from the genome plasmid of Ni-CE (21). Details of the construction of these plasmids are available from the authors on request.
Western blotting.
NA cells grown in a 24-well tissue culture plate were transfected with 0.8 μg of pCAGGS-P1 to -P5 by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Apart from this procedure, NA cells were infected with CE(NiP) or CE(NiP)ΔP2-5 at a multiplicity of infection (MOI) of 2. After 2 days, the transfected or infected cells were lysed with 2× sample buffer solution containing 2-mercaptoethanol (Wako, Japan). The cell lysate samples were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis before being transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). The membranes then were blocked with phosphate-buffered saline containing 0.1% Tween 20 and 5% nonfat dry milk and treated with the following antibodies to visualize the blots: anti-P protein peptide rabbit antibody recognizing the region of Ni P1 at positions 187 to 197 (SATNEEDDLSV, from N to C terminus) (produced by Wako), which is included in all P protein isoforms (Fig. 1A), anti-N protein mouse monoclonal antibody 13-27 (24), and anti-α-tubulin antibody (Sigma-Aldrich, St. Louis, MO). Antibody signals on the membranes were detected as previously reported (25). In other experiments, lysates of transfected SYM-I, NA, and G-8 cells prepared for the promoter reporter assays (see below) were analyzed by Western blotting under the same conditions except for the use of anti-P protein rabbit polyclonal antibody (kindly provided by A. Kawai) to circumvent nonspecific reaction in SYM-I cells.
Viral replication in cultured cells.
NA and G-8 cells were inoculated with each virus at an MOI of 0.01 and MOI of 1, respectively. After collecting the culture media at 1, 3, and 5 days postinoculation (dpi), viral titers in the supernatant (calculated as focus-forming units [FFU] per milliliter) were determined by focus assays on confluent NA cells as previously reported (22).
Minigenome assay.
NA cells grown in a 24-well tissue culture plate were transfected by using transfection reagent with 0.12 μg of pEGFP-P1, pEGFP-P1ΔP2-5, or an empty vector (pEGFP-C1) (described above) together with 0.8 μg of pCAGGS-RVDI-Luc, 0.4 μg of pCAGGS-CEL (described above), and 1.2 μg of pCAGGS-CEN, which was previously constructed to express N protein of Ni-CE (25). After 48 h, the cells were lysed to measure firefly luciferase activity, which reflects efficiency of the replication/transcription of minigenome RNA, by using the luciferase assay system (Promega, Madison, WI).
G-8 cells grown in a 24-well tissue culture plate were transfected by using transfection reagent with 0.2 μg of pCAGGS-P1, pCAGGS-P1ΔP2-5, or an empty vector (pCAGGS/MCS) (described above), together with 0.8 μg of pCAGGS-RVDI-Luc, 0.4 μg of pCAGGS-CEL (described above), and 0.4 μg of pCAGGS-CEN (25). After 48 h, the cells were lysed to measure firefly luciferase activities by using the luciferase assay system (Promega).
IFN-β promoter reporter assay.
SYM-I cells grown in a 24-well tissue culture plate were transfected by using Lipofectamine 2000 (Invitrogen) with 0.25 μg of pEGFP-P1, pEGFP-P1ΔP2-5, or empty vector pEGFP-C1 (described above) together with 0.04 μg of pRL-TK (Promega), which expresses the Renilla luciferase, and 0.25 μg of IFNB-pGL3 plasmid (kindly provided by Rongtuan Lin), which has an IFN-β promoter upstream of the firefly luciferase gene. After 24 h, the cells were transfected with 5 μg of poly(I·C), a double-stranded RNA analog, to stimulate the IFN-β promoter. Twenty-four hours later, lysates of the cells were prepared and used to measure the activities of firefly and Renilla luciferases by using a dual-luciferase reporter assay system (Promega). The IFN-β promoter activity was calculated by dividing the firefly luciferase activity by the Renilla luciferase activity. In some experiments, pCAGGS-P1, -P1ΔP2-5, and empty vector pCAGGS/MCS were used instead of pEGFP-C1-based plasmids.
The same experiments were carried out in G-8 cells with the following minor modifications: TransIT-LT1 transfection reagent (Mirus, Madison, WI) instead of Lipofectamine 2000 was used, and the above-described pEGFP-C1-based plasmids were replaced with pCAGGS-P1 to -P5 and -P1ΔP2-5 (described above), pCAGGS-NiN, previously constructed to express N protein of Ni (25), or empty vector pCAGGS/MCS. After 24 h posttransfection, the cells were transfected with 0.5 μg of poly(I·C). Six hours later, lysates of the cells were prepared and used.
ISRE reporter assay.
NA cells grown in a 24-well tissue culture plate were transfected with 0.25 μg of pEGFP-P1, pEGFP-P1ΔP2-5, or empty vector pEGFP-C1 together with 0.04 μg of pRL-TK and 0.25 μg of pISRE-luc plasmid (Stratagene, La Jolla, CA), which encodes the firefly luciferase gene downstream of an IFN-stimulated response element (ISRE)-containing promoter, by using Lipofectamine 2000. After 24 h, the transfected cells were treated with 1,000 U/ml of universal IFN-α (PBL Assay Science, Piscataway, NJ) for 6 h. Subsequently, the cells were lysed to measure the activities of firefly and Renilla luciferases as mentioned above. The ISRE activity was determined as firefly luciferase activity normalized to Renilla luciferase activity.
Pathogenicity of each virus in mice.
Four-week-old male ddY mice (20 mice/group; Japan SLC, Inc., Shizuoka, Japan) were inoculated via the i.c. route with 0.03 ml of 104 FFU or via the i.m. route (into the left thigh muscle) with 0.1 ml of 106 FFU of each virus and then observed for 14 or 24 days, respectively. The symptoms in mice were classified into 5 grades as reported previously (22): (i) normal, (ii) body weight loss (5% reduction from maximum body weight), (iii) mild neurological symptoms (such as stagger or gait abnormality of a unilateral hind limb), (iv) severe neurological symptoms (such as gait abnormality of bilateral hind limbs), and (v) death. Mice were euthanized when they showed a lack of righting reflex (mice unable to right themselves within 10 s after being placed on their side). All animal experiments in this study were conducted in accordance with the Regulations for Animal Experiments at Gifu University; the protocols were approved by the Committee for Animal Research and Welfare of Gifu University (approval no. 13069).
In vivo examination of viral replication and propagation in the brain.
Four-week-old male ddY mice (three mice/group; Japan SLC, Inc.) were inoculated via the i.c. route with 0.03 ml of 104 FFU of CE(NiP)-Luc or CE(NiP)ΔP2-5-Luc. Brains of infected mice were collected at 1, 3, and 5 dpi and then homogenized before being lysed by using 1 ml of passive lysis buffer (Promega). After one cycle of freezing and thawing, the lysate samples were centrifuged at 20,000 × g for 10 min. Ten microliters of each of the supernatants then was used to measure luciferase activities (calculated as relative light units [RLU] per second per gram of brain weight) with the Promega luciferase assay systems.
Four-week-old male ddY mice (five mice/group; Japan SLC, Inc., Japan) were inoculated with 104 FFU of CE(NiP) or CE(NiP)ΔP2-5 by the i.c. route. Their brains were collected and homogenized at 5 dpi. The virus titers in homogenates (calculated as FFU per gram) were determined by focus assays on confluent NA cells as previously reported (22).
Biodistribution of each virus in mice.
Four-week-old male ddY mice (five mice/group; Japan SLC, Inc.) were inoculated via the i.m. route with 0.1 ml of 106 FFU of each virus. Mice were euthanized at 5 dpi, and then their brains, spinal cords, sciatic nerves, and thigh muscles were collected. All of those tissues were frozen in liquid nitrogen and stored at −80°C before examination for the presence of viral genomic RNA by reverse transcription (RT)-nested PCR as previously reported (22).
In vivo examination of viral replication in muscle.
Four-week-old male ddY mice (three mice/group; Japan SLC, Inc.) were inoculated via the i.m. route with 0.1 ml of 106 FFU of CE(NiP)-Luc or CE(NiP)ΔP2-5-Luc. Thigh muscles of infected mice were collected at 0, 24, 48, and 72 h postinoculation (hpi). The luciferase activities in these muscles were measured as previously reported (22).
In vitro examination of viral replication in muscle cells.
Differentiated G-8 cells grown in a 24-well tissue culture plate were infected with CE(NiP)-Luc or CE(NiP)ΔP2-5-Luc at an MOI of 1. At 1, 3, and 5 dpi, cell lysates were prepared to measure luciferase activities as previously reported (22).
Real-time RT-PCR.
Differentiated G-8 cells grown in a 24-well tissue culture plate were infected with each virus at an MOI of 1. At 24 hpi, the expression levels of Ifn-β, Mx1, Oas1, and Gapdh (glyceraldehyde-3-phosphate dehydrogenase) genes in the cells were examined with the ABI StepOnePlus real-time PCR system (Applied Biosystems, Carlsbad, CA) as previously reported (22).
Statistical analyses.
Student's t test and Fisher's exact test were used to determine statistical significance. One-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test was also conducted. P values of <0.05 were considered statistically significant.
RESULTS
Expression of tPs in CE(NiP)-infected cells.
To confirm the expression of tPs via a ribosomal leaky scanning mechanism in CE(NiP)-infected cells, we analyzed by Western blotting a lysate sample of mouse neuroblastoma NA cells infected with CE(NiP) together with lysates of NA cells transfected with pCAGGS-P1 to -P5 to express each isoform (P1 to P5). The results obtained with the recombinant proteins revealed that mobilities of the P protein isoforms did not correspond to their actual molecular sizes for an unknown reason (Fig. 1C): the full-length P1 migrated slightly faster than the shorter P2 did (Fig. 1C, lanes 2 and 3), and P4 moved slower than the longer P3 did (lanes 4 and 5). Notably, only the largest isoform was detected in the lysate of cells transfected to express each isoform (lanes 2 to 6), indicating that translation of tPs by the ribosomal leaky scanning mechanism does not occur efficiently in the transfected cells.
Based on the mobilities of these recombinant proteins, we sought to identify each isoform produced in CE(NiP)-infected NA cells. In addition to a band corresponding to P1 and P2, a band of P3 was observed in the sample of the infected cells (lane 8). Expression of P4 and P5 was under the detectable level, consistent with previous observations of other RABV strains (5, 13). We repeated the Western blot analysis under conditions with a higher level of exposure, but we were not able to detect the expression of P4 and P5 (data not shown). However, the fact that P3 was detected in CE(NiP)-infected NA cells strongly suggests that translation of tPs via the ribosomal leaky scanning mechanism occurs in infected cells.
Generation of CE(NiP)ΔP2-5, which has impaired ability to express tPs.
By using a reverse genetics system for CE(NiP), we successfully generated CE(NiP)ΔP2-5. Sequence analysis confirmed that CE(NiP)ΔP2-5 has the P gene, in which the start codons (AUG, in positive sense) for all of the tPs are changed to AUA (data not shown). To examine whether the introduced AUA codons impair the ability of CE(NiP)ΔP2-5 to express tPs, we analyzed a lysate sample of NA cells infected with CE(NiP)ΔP2-5 together with that of CE(NiP)-infected NA cells. Expression of P3 was detected in CE(NiP)-infected cells but not in CE(NiP)ΔP2-5-infected cells (Fig. 1C, lane 9). Notably, a strong band detected in the sample of CE(NiP)ΔP2-5-infected cells migrated slightly faster than did a band corresponding to P1 and P2 observed in CE(NiP)-infected cells, confirming that P2 is expressed in CE(NiP)-infected cells and also indicating that expression of P2 is significantly diminished in CE(NiP)ΔP2-5-infected cells. These results indicated that the AUA codons introduced into the P gene of CE(NiP)ΔP2-5 significantly impaired the expression of tPs in infected cells.
Replication of CE(NiP)ΔP2-5 in NA cells.
To assess whether the above-described gene manipulation affects the viral viability of CE(NiP)ΔP2-5, we compared growth curves of CE(NiP)ΔP2-5 and CE(NiP) in NA cells. The two viruses showed similar growth curves, and their titers in the culture supernatant reached over 108 FFU/ml at 5 dpi (Fig. 2A). There was no statistically significant difference between the titers of CE(NiP)ΔP2-5 and CE(NiP) in the supernatant at 5 dpi (P ≧ 0.05). These results demonstrated that the growth ability of CE(NiP)ΔP2-5 in NA cells was comparable to that of CE(NiP), leading to the conclusion that the replacement of all start codons for tPs with AUA codons had no significant impact on the viral viability of CE(NiP)ΔP2-5.
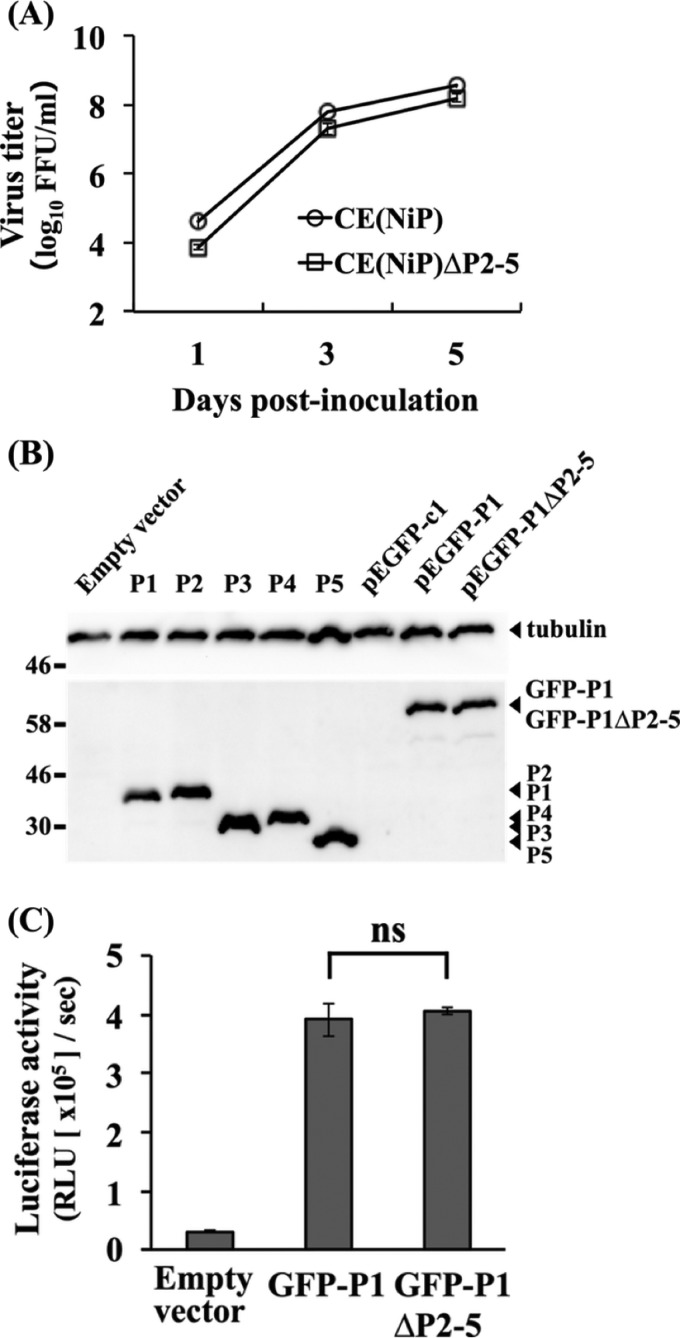
FIG 2.
Effects of mutations introduced into CE(NiP)ΔP2-5 on viral replication and activity of P1 as a cofactor of viral RNA polymerase. (A) Growth curves of CE(NiP) and CE(NiP)ΔP2-5 in NA cells. Each virus was inoculated into NA cells at an MOI of 0.01. Viral titers in culture supernatants collected at 1, 3, and 5 dpi were determined by focus assays. (B) GFP-tagged P1s in NA cells transfected with pEGFP-P1 and pEGFP-P1ΔP2-5 together with the respective isoforms expressed in transfected cells were analyzed by Western blotting. Tubulin in each sample was also detected as a loading control. (C) Minigenome assay to compare polymerase cofactor activities of P1s of CE(NiP) and CE(NiP)ΔP2-5. NA cells were transfected with empty plasmid, pEGFP-P1, or pEGFP-P1ΔP2-5, together with plasmids expressing luciferase-based minigenome RNA and viral N and L proteins. At 48 h posttransfection, luciferase activities in cell lysates were measured. All assays were carried out in triplicate, and the values in the graph are shown as means ± standard errors of the means. ns, not significant (P ≧ 0.05).
Function of P1 of CE(NiP)ΔP2-5 as a cofactor of viral RNA polymerase.
The above-described gene manipulation resulted in introduction of four Met-to-Ile mutations into P1 of CE(NiP)ΔP2-5 at positions 20, 53, 69, and 83 (Fig. 1B). This raised the possibility that these mutations affected not only the expression of tPs but also P1 functions of CE(NiP)ΔP2-5. Although the similar growth efficiencies of CE(NiP) and CE(NiP)ΔP2-5 (Fig. 2A) strongly suggested that these mutations in P1 of CE(NiP)ΔP2-5 did not affect its function as a cofactor of viral RNA polymerase, we sought to confirm this point directly by a luciferase-based minigenome assay, which enables evaluation of the efficiency of replication/transcription of artificial minigenome RNA driven by recombinant P protein together with recombinant N and L proteins. In this experiment, to suppress translation of tPs initiated from downstream in-frame AUG codons and investigate the function of only P1, we used pEGFP-P1ΔP2-5 or pEGFP-P1 to express GFP-P1ΔP2-5 and GFP-P1, the recombinant P1s of CE(NiP)ΔP2-5 and CE(NiP), respectively, the N terminus of which was fused with a GFP tag. Western blot analysis demonstrated that pEGFP-P1ΔP2-5 and pEGFP-P1 did not express detectable levels of tPs in NA cells transfected with these plasmids (Fig. 2B). We found that expression of GFP-P1ΔP2-5 and GFP-P1 resulted in comparable levels of luciferase expression from minigenome RNA (Fig. 2C). These results confirmed that P1 of CE(NiP)ΔP2-5 retained functional integrity as a cofactor of viral RNA polymerase.
Function of P1 of CE(NiP)ΔP2-5 as a viral IFN antagonist.
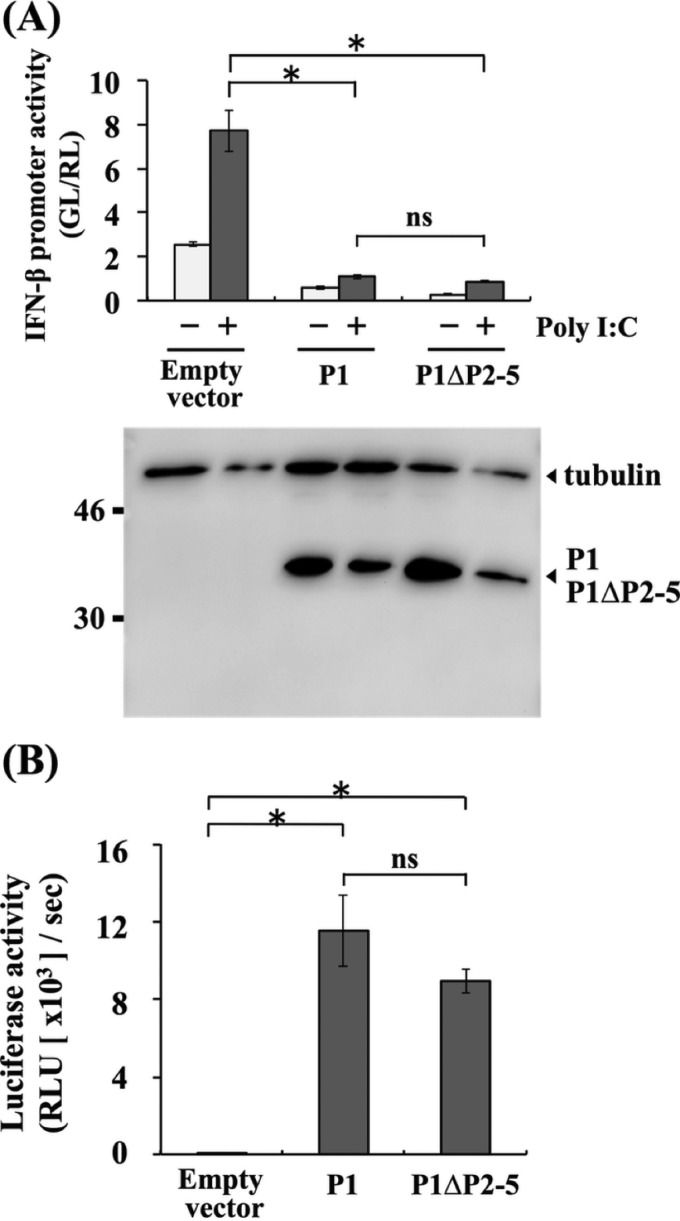
To examine whether the above-described Met-to-Ile mutations in P1 of CE(NiP)ΔP2-5 affect its activity to antagonize IFN induction, we conducted luciferase-based IFN-β promoter-reporter assays by using SYM-I cells transfected with pEGFP-P1ΔP2-5 or pEGFP-P1. We found that GFP-P1ΔP2-5 and GFP-P1 moderately, but statistically significantly (P < 0.05), inhibited activity of the IFN-β promoter, which was activated by treatment with poly(I·C), to almost the same extents (Fig. 3A, top). We observed that the moderate suppression of the promoter activity by GFP-P1ΔP2-5 and GFP-P1 was concomitant with the low expression levels of these proteins (Fig. 3A, bottom). Therefore, to exclude the possible influence of the GFP tag on protein expression, we repeated the experiments with pCAGGS-P1ΔP2-5 and pCAGGS-P1, expressing P1ΔP2-5 and P1, respectively, which are non-GFP-tagged forms of the P1s. In the cells transfected with the two plasmids, poly(I·C)-induced IFN-β promoter activity was significantly inhibited to equivalent levels (P < 0.01) (Fig. 3B, top). Importantly, neither pCAGGS-P1ΔP2-5 nor pCAGGS-P1 expressed detectable levels of tPs, while these plasmids expressed high levels of P1s in the SYM-I cells (Fig. 3B, bottom). These results indicated that P1s of both CE(NiP) and CE(NiP)ΔP2-5 had activity to antagonize IFN induction.
FIG 3.
Effects of mutations introduced into CE(NiP)ΔP2-5 on IFN antagonist activities of P1. IFN-β promoter-reporter assay to compare the activities of P1s of CE(NiP) and CE(NiP)ΔP2-5 to suppress IFN induction. (A) SYM-I cells were transfected with empty plasmid, pEGFP-P1, or pEGFP-P1ΔP2-5, together with the reporter plasmid IFNB-pGL3. (B) SYM-I cells were transfected with an empty plasmid, pCAGGS-P1, or pCAGGS-P1ΔP2-5, together with the reporter plasmid IFNB-pGL3. The cells were transfected with poly(I·C) at 24 h posttransfection and incubated for 24 h. Cell lysates then were prepared and used to measure luciferase activities. (Bottom) P1s and tubulin in the cell lysates were detected by Western blotting. (C) ISRE reporter assay to compare the activities of P1s of CE(NiP) and CE(NiP)ΔP2-5 to suppress IFN response. NA cells were transfected with empty plasmid, pEGFP-P1, or pEGFP-P1ΔP2-5, together with the reporter plasmid pISRE-luc. The cells were treated with IFN-α at 24 h posttransfection and incubated for 6 h. Cell lysates then were prepared and used to measure luciferase activities. (Bottom) P1s and tubulin in the cell lysates were detected by Western blotting. GL, firefly luciferase activity; RL, Renilla luciferase activity. All assays were carried out in triplicate, and the values in the graph are shown as means ± standard errors of the means. *, Significant difference at a P value of <0.05; **, significant difference at a P value of < 0.01; ns, not significant (P ≧ 0.05).
We next examined whether the Met-to-Ile mutations in P1 of CE(NiP)ΔP2-5 affect activity to antagonize IFN responses by ISRE-reporter assay in NA cells transfected with pEGFP-P1ΔP2-5 or pEGFP-P1. We found that both of the GFP-tagged P1s significantly blocked the ISRE activity induced by IFN-α treatment with similar efficiencies (P < 0.01) (Fig. 3C, top), under the condition that expression levels of the two proteins were comparable (Fig. 3C, bottom). These results indicated that P1s of both CE(NiP) and CE(NiP)ΔP2-5 maintained activity to antagonize IFN responses. Taken together, we concluded that the P1 of CE(NiP)ΔP2-5 retained functional integrity as a viral IFN antagonist, at least in neural cells.
Examination of the pathogenicity of CE(NiP)ΔP2-5 in mice by i.c. inoculation.
Based on the finding that the mutations introduced into CE(NiP)ΔP2-5 impair its ability to express tPs without affecting the functions of P1 for viral RNA synthesis and IFN antagonism, we considered that CE(NiP)ΔP2-5 and CE(NiP) would be useful tools to examine whether tPs contribute to the pathogenesis of RABV in vivo. To compare pathogenicities of CE(NiP)ΔP2-5 and CE(NiP) in mice, first we inoculated mice with each virus via the i.c. route. After the inoculation, both viruses caused lethal infection in 100% of the inoculated mice, most of which showed severe neurological symptoms before death (Fig. 4A and Table 1).
FIG 4.
Progression of symptoms in mice inoculated via the i.c. route with 104 FFU (A) or via the i.m. route with 106 FFU (B) of CE(NiP) or CE(NiP)ΔP2-5. The mice infected via the i.c. route and those infected via the i.m. route were observed daily for 14 and 24 days, respectively. The symptoms in mice were classified into 5 grades: (i) normal, (ii) body weight loss, (iii) mild neurological symptoms, (iv) severe neurological symptoms, and (v) death.
TABLE 1.
Morbidity and mortality rates of mice inoculated with each virus via the i.c. or i.m. route
| Strain | % Morbidity (i.c.; no. sick/inoculated) | % Mortality (i.c.; no. dead/inoculated) | % Morbidity (i.m.; no. sick/inoculated) | % Mortality (i.m.; no. dead/inoculated) |
|---|---|---|---|---|
| Mocka | 0 (0/5) | 0 (0/5) | 0 (0/5) | 0 (0/5) |
| CE(NiP)b | 100 (20/20) | 100 (20/20) | 80 (16/20) | 80 (16/20) |
| CE(NiP)ΔP2-5b | 100 (20/20) | 100 (20/20) | 35c (7/20) | 30c (6/20) |
A group of 5 mice was inoculated with a diluent.
Groups of 20 mice were inoculated with each virus.
P value of <0.01 compared to the CE(NiP) strain group by Fisher's exact test.
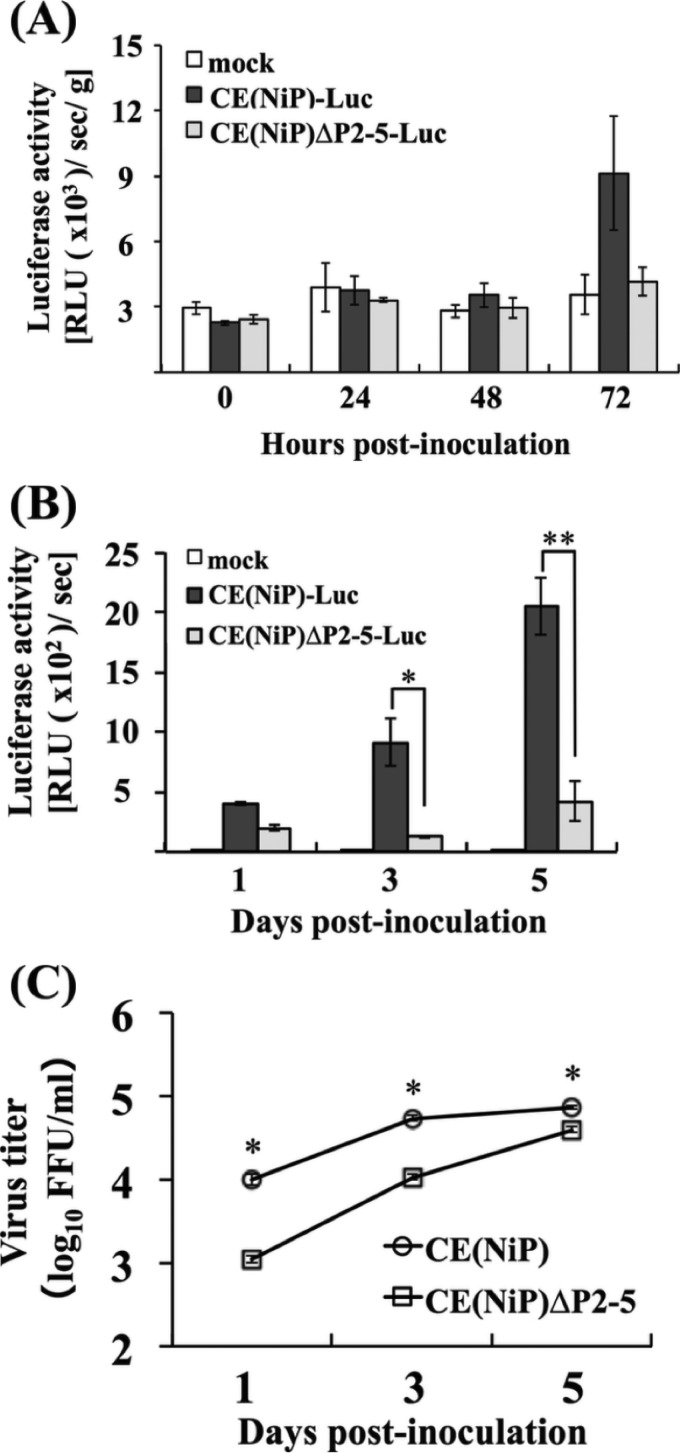
To compare the replication efficiencies of CE(NiP)ΔP2-5 and CE(NiP) in the mouse brain, we inoculated CE(NiP)-Luc and CE(NiP)ΔP2-5-Luc expressing firefly luciferase into mice via the i.c. route and checked the activities of luciferase expressed in the inoculated brains after sequential collection. We found that luciferase activities detected in the mouse brains infected with the respective viruses increased over time (Fig. 5A). We also found that the levels of luciferase activities in the brains infected with CE(NiP)ΔP2-5 tended to be lower than those in the CE(NiP)-infected brains, although there were no statistically significant difference between the activities (P ≧ 0.05). In addition, we compared viral titers in the mouse brains infected with CE(NiP)ΔP2-5 and CE(NiP) at 5 days after i.c. inoculation. The titers of both viruses were comparable to each other, reaching over 106 FFU/g (Fig. 5B). Notably, there was no statistically significant difference between the titers of CE(NiP)ΔP2-5 and CE(NiP) in the brains (P ≧ 0.05). Taken together, these findings demonstrated that tPs do not make a major contribution to neurovirulence as well as viral replication in the brain.
FIG 5.
Comparison of viral replication in brains infected with CE(NiP)- and CE(NiP)ΔP2-5-Luc in vivo. (A) Brains of mice inoculated with 104 FFU of CE(NiP)-Luc and CE(NiP)ΔP2-5-Luc via the i.c. route were collected at 1, 3, and 5 dpi. Luciferase activities in the brains were measured and calculated as relative light units (log10 RLU/s/g brain weight). (B) Propagation of CE(NiP) and CE(NiP)ΔP2-5 in adult mouse brains. Mice were inoculated with 104 FFU of each virus via the i.c. route. Viral titers in brains collected at 5 dpi were determined by focus assays (FFU/g brain weight).
Examination of the pathogenicity of CE(NiP)ΔP2-5 in mice by i.m. inoculation.
To examine the importance of tPs in viral neuroinvasiveness, we next inoculated mice with CE(NiP)ΔP2-5 and CE(NiP) via the i.m. route. We found that, after the i.m. inoculation, CE(NiP)ΔP2-5 caused symptomatic infection in 35% and then killed 30% of the inoculated mice, whereas CE(NiP) caused lethal infection in 80% of the mice after inducing severe neurological symptoms (Fig. 4B and Table 1). There were statistically significant differences in the morbidity and mortality rates between CE(NiP)- and CE(NiP)ΔP2-5-infected mice (P < 0.01) (Table 1). These findings demonstrated that tPs play a critical role in viral neuroinvasiveness.
Biodistribution of CE(NiP)ΔP2-5 in mice.
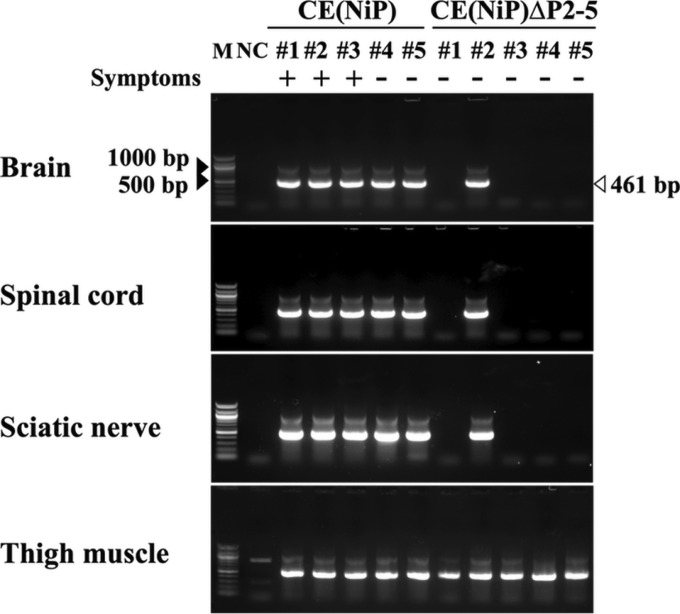
To elucidate the mechanism underlying the difference between CE(NiP)ΔP2-5 and CE(NiP) in neuroinvasiveness, we compared biodistributions of the two viruses in mice after i.m. inoculation. At 5 dpi, when 0% of CE(NiP)ΔP2-5-infected mice (0/5 mice) and 60% of CE(NiP)-infected mice (3/5 mice) developed symptoms, brains, spinal cords, sciatic nerves, and thigh muscles of the inoculated mice were collected for the detection of viral genomic RNA by using a highly sensitive RT-nested PCR method previously reported (22). In all of the mice inoculated with CE(NiP)ΔP2-5 and CE(NiP), viral genomic RNA was detected in thigh muscles, where the respective viruses had been inoculated (Fig. 6). Because of the high sensitivity of this RT-nested PCR, of which the detection limit is equal to 10 FFU of infectious viruses (22), there is the possibility that not only genomic RNAs of replicating viruses but also those of nonreplicating viruses contained in the inoculum were detected in muscles. Notably, the brains, spinal cords, and sciatic nerves were positive for viral genomic RNA in 100% of the mice inoculated with CE(NiP) (5/5 mice) but in only 20% of the mice inoculated with CE(NiP)ΔP2-5 (1/5 mice) (Fig. 6). These results indicated that after i.m. inoculation, CE(NiP)ΔP2-5 infected peripheral nerves of mice less efficiently than did CE(NiP), strongly suggesting the importance of tPs in infection of peripheral nerves.
FIG 6.
Comparison of viral distribution in mice inoculated with CE(NiP) and CE(NiP)ΔP2-5. Tissues of mice infected with 106 FFU of each virus via the i.m. route were collected at 5 dpi and analyzed for the existence of viral genomic RNAs by RT-nested PCR targeting the N gene region (461 bp). M, marker; #1 to #5, identification numbers of mice; NC, negative control (each tissue i.m. inoculated with medium). Symptoms: +, mouse with symptoms such as body weight loss on the day of tissue collection; −, mouse without any symptoms on the day of tissue collection.
Replication of CE(NiP)ΔP2-5 in muscle cells in vivo and in vitro.
Our recent study demonstrated a strong correlation between the abilities of RABV to infect peripheral nerves and to replicate in muscle cells (22). We therefore compared replication efficiency of CE(NiP)ΔP2-5 in muscle cells with that of CE(NiP) by using both in vivo and in vitro models. First, to examine the replication ability of each virus in vivo, we inoculated CE(NiP)ΔP2-5-Luc and CE(NiP)-Luc into thigh muscles of mice and then compared the activities of luciferase expressed in the inoculated thigh muscles after sequential collection. In the CE(NiP)-Luc-infected mice, luciferase activity detected in muscles tended to gradually increase with time, while in the CE(NiP)ΔP2-5-Luc-infected mice, the activity in muscles tended to remain at a low level (Fig. 7A). These results suggested that CE(NiP)ΔP2-5 replicated less efficiently in muscle in vivo than did CE(NiP).
FIG 7.
Comparison of viral replication in muscle cells infected with CE(NiP)- and CE(NiP)ΔP2-5-Luc in vivo and in vitro. (A) Thigh muscles of mice inoculated with 106 FFU of CE(NiP)-Luc and CE(NiP)ΔP2-5-Luc via the i.m. route were collected at 0, 24, 48, and 72 hpi. Lysates of muscles were prepared and used to measure luciferase activities calculated as relative light units (RLU [×103]/s/g muscle weight). (B) Muscle G-8 cells inoculated with CE(NiP)-Luc and CE(NiP)ΔP2-5-Luc at an MOI of 1 were collected at 1, 3, and 5 dpi. Cell lysates were prepared and used to measure luciferase activities, calculated as relative light units (RLU [×102]/s). (C) Growth curves of CE(NiP) and CE(NiP)ΔP2-5 in G-8 cells. Each virus was inoculated into G-8 cells at an MOI of 1. Viral titers in culture supernatants collected at 1, 3, and 5 dpi were determined by focus assays. All assays were carried out in triplicate, and the values in the graph are shown as means ± standard errors of the means. *, Significant difference at a P value of <0.05; **, significant difference at a P value of <0.01.
To check whether the same phenomenon can be observed in vitro, we inoculated CE(NiP)ΔP2-5-Luc and CE(NiP)-Luc to mouse muscle-derived G-8 cells and then compared the luciferase activities in the cells infected with the respective viruses. We found that levels of luciferase activity in CE(NiP)-Luc-infected G-8 cells were higher than those in CE(NiP)ΔP2-5-Luc-infected cells at 1, 3, and 5 dpi (Fig. 7B). There were statistically significant differences between the activities detected in CE(NiP)- and CE(NiP)ΔP2-5-Luc-infected cells at both 3 dpi (P < 0.05) and 5 dpi (P < 0.01). These results indicated that replication efficiency of CE(NiP)ΔP2-5 in muscle cells was lower than that of CE(NiP).
We next compared efficiencies of infectious virus production in G-8 cells infected with CE(NiP)ΔP2-5 and CE(NiP). Although the titers of both viruses in the culture supernatants gradually increased with time, the titers of CE(NiP)ΔP2-5 were always significantly lower than those of CE(NiP) during the observation period (P < 0.05) (Fig. 7C). The most prominent difference between titers of the viruses was observed at 1 dpi: the titer of CE(NiP) reached about 104 FFU/ml, whereas that of CE(NiP)ΔP2-5 was about 103 FFU/ml. These results indicated that CE(NiP)ΔP2-5 produced infectious viruses less efficiently in muscle cells than did CE(NiP).
Taken together, the findings obtained from both in vivo and in vitro models demonstrated that CE(NiP)ΔP2-5 replicated less efficiently in muscle cells than did CE(NiP), indicating the contribution of tPs to efficient viral replication in muscle cells.
IFN induction and response in muscle cells infected with CE(NiP)ΔP2-5.
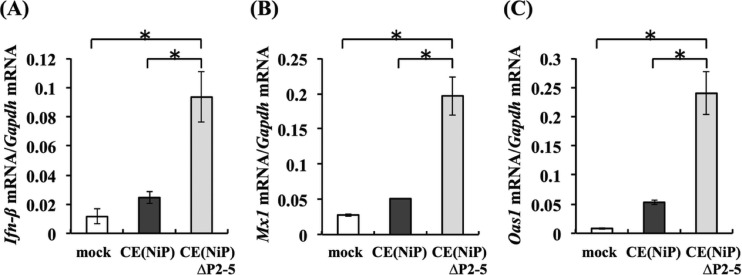
Our previous findings strongly suggested that inhibition of IFN induction is important for efficient replication of RABV in muscle cells (22), leading to the hypothesis that CE(NiP)ΔP2-5 has an impaired ability to antagonize IFN-mediated innate immunity in muscle cells. To prove this hypothesis, we compared expression levels of Ifn-β mRNA in G-8 cells infected with CE(NiP)ΔP2-5 and CE(NiP) at 1 dpi, at which time the difference between viral titers of the two viruses was most prominent (Fig. 7C). Consistent with the finding that CE(NiP)ΔP2-5 replicated less efficiently in G-8 cells than did CE(NiP), infection with CE(NiP)ΔP2-5 induced a significantly higher level of expression of Ifn-β mRNA than did infection with CE(NiP) (P < 0.01) (Fig. 8A). To check whether the induced IFN in CE(NiP)ΔP2-5-infected G-8 cells stimulates expression of ISGs, we next compared the expression levels of Mx1 and Oas1 mRNAs in G-8 cells infected with CE(NiP)ΔP2-5 and CE(NiP) (Fig. 8B and C). Consistent with the results showing strong IFN induction (Fig. 8A), significantly higher levels of Mx1 and Oas1 mRNAs were expressed in CE(NiP)ΔP2-5-infected cells than in CE(NiP)-infected cells (P < 0.01). These results indicated that CE(NiP)ΔP2-5 had a defect in the ability to antagonize IFN induction in muscle cells.
FIG 8.
Comparison of relative expression levels of IFN-related genes in muscle G-8 cells infected with CE(NiP) and CE(NiP)ΔP2-5. G-8 cells inoculated with each virus at an MOI of 1 were lysed at 24 hpi and used for RNA extraction. The expression levels of Ifn-β (A), Mx1 (B), and Oas1 (C) genes in each cell lysate were measured and are indicated as the number of copies of specific mRNA per copy of mouse Gapdh mRNA. All assays were carried out in triplicate, and the values in the graph are shown as means ± standard errors of the means. *, Significant difference at a P value of <0.01.
Functions of P1 of CE(NiP)ΔP2-5 as a viral antagonist to inhibit IFN induction and a cofactor of viral RNA polymerase in muscle cells.
While the above-described findings suggest that tPs contribute to IFN antagonism in muscle cells, the findings raised another possibility that the Met-to-Ile mutations in P1 of CE(NiP)ΔP2-5 impair its function to antagonize IFN induction in the cells. To check this possibility, we conducted IFN-β promoter-reporter assays by using G-8 cells transfected with pEGFP-P1ΔP2-5 or pEGFP-P1. However, even GFP-P1 expressed from pEGFP-P1 failed to inhibit poly(I·C)-induced IFN-β promoter activity (data not shown). Accordingly, instead of pEGFP-P1ΔP2-5 and pEGFP-P1, pCAGGS-P1ΔP2-5 and pCAGGS-P1, respectively, were used for the reporter assay. Compared to the IFN-β promoter activity in control cells transfected with an empty vector, the promoter activity was significantly lower in cells transfected with pCAGGS-P1ΔP2-5 and in cells transfected with pCAGGS-P1 (Fig. 9A, top) (P < 0.01). Importantly, there was no statistically significant difference between the poly(I·C)-induced promoter activities in cells transfected with these plasmids (P ≧ 0.05). Notably, Western blotting of the lysates used for the above-described reporter assay revealed that P1s, but not tPs, were present in the cells transfected with the respective plasmids (Fig. 9A, bottom). These results strongly suggest that the Met-to-Ile mutations in P1 of CE(NiP)ΔP2-5 did not affect its function to antagonize IFN induction in muscle cells.
FIG 9.
Effects of mutations introduced into CE(NiP)ΔP2-5 on the activities of P1 as an IFN antagonist and a cofactor of viral RNA polymerase in muscle cells. (A) IFN-β promoter-reporter assay to compare the activities of P1s of CE(NiP) and CE(NiP)ΔP2-5 to suppress IFN induction. G-8 cells were transfected with an empty plasmid, pCAGGS-P1, or pCAGGS-P1ΔP2-5, together with the reporter plasmid IFNB-pGL3. The cells were transfected with poly(I·C) at 24 h posttransfection and incubated for 6 h. Cell lysates then were prepared and used to measure luciferase activities. (Bottom) P1s and tubulin in the cell lysates were detected by Western blotting. (B) Minigenome assay to compare polymerase cofactor activities of P1s of CE(NiP) and CE(NiP)ΔP2-5 in muscle cells. G-8 cells were transfected with an empty plasmid, pCAGGS-P1, or pCAGGS-P1ΔP2-5, together with plasmids expressing luciferase-based minigenome RNA and viral N and L proteins. At 48 h posttransfection, luciferase activities in cell lysates were measured. All assays were carried out in triplicate, and the values in the graph are shown as means ± standard errors of the means. *, Significant difference at a P value of <0.01; ns, not significant (P ≧ 0.05).
In addition, to check whether the Met-to-Ile mutations in P1 of CE(NiP)ΔP2-5 affect its activity as a cofactor of viral RNA polymerase in muscle cells, we conducted a minigenome assay by using G-8 cells transfected to express P1ΔP2-5 or P1, together with minigenome RNA and N and L proteins. In the cells transfected with pCAGGS-P1ΔP2-5, the level of luciferase expression was statistically indistinguishable from that in the cells transfected with pCAGGS-P1 (Fig. 9B) (P ≧ 0.05).
Taken together, these findings strongly suggest that the impaired abilities of CE(NiP)ΔP2-5 to replicate efficiently (Fig. 7) and to suppress IFN induction (Fig. 8A) in muscle cells were not due to a functional defect of P1 as a viral antagonist of IFN induction or as a cofactor of viral RNA polymerase but rather were due to the impairment of the expression of tPs in muscle cells.
Inhibition of IFN induction by respective tPs in muscle cells.
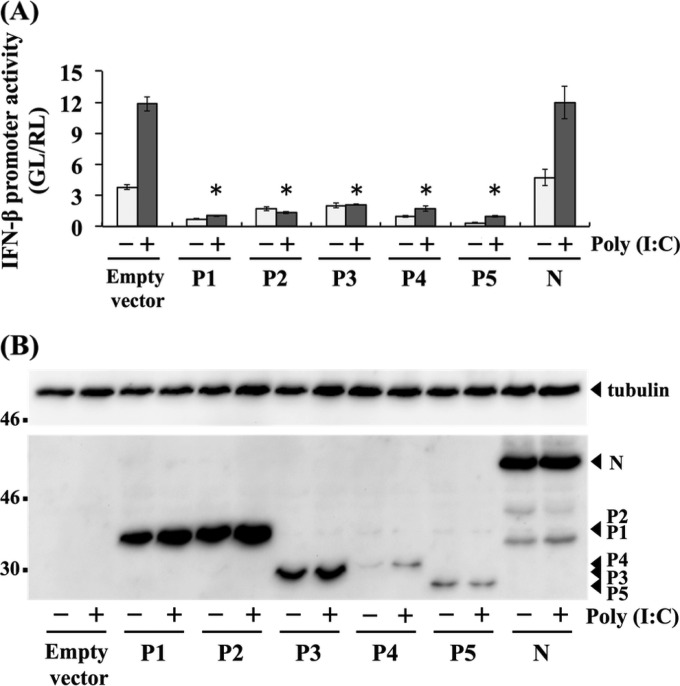
The above-described findings strongly suggested that tPs function to antagonize IFN induction in muscle cells. To confirm this function, we checked whether P1 and the respective tPs had activities to inhibit IFN-β promoter activity in G-8 cells by a luciferase-based reporter assay. In cells transfected with an empty vector or N protein-expressing plasmid, which were used as negative controls, IFN-β promoter activity was clearly increased by transfection with poly(I·C) (Fig. 10A). Notably, compared with the results of negative controls, the promoter activity induced by poly(I·C) stimulation was significantly lower in cells transfected with pCAGGS-P1 to -P5 to express P1 and the respective tPs (P < 0.005) (Fig. 10A). To check expression of the recombinant P1 and the respective tPs in cell lysates prepared for the above-described reporter assay, we conducted Western blot analysis and found that only the largest isoform was present in each lysate sample (Fig. 10B). These results strongly suggested that, in addition to P1, all of the tPs had activities to antagonize IFN induction in muscle cells.
FIG 10.
IFN antagonist activities of respective P protein isoforms to suppress IFN induction in muscle cells. (A) G-8 cells were transfected with a plasmid expressing each P protein isoform (pCAGGS-P1, -P2, -P3, -P4, or -P5) or N protein (pCAGGS-N), or an empty plasmid, together with IFNB-pGL3. The cells were transfected with poly(I·C) at 24 h posttransfection and incubated for 6 h. Cell lysates then were prepared and used to measure luciferase activities. GL, firefly luciferase activity; RL, Renilla luciferase activity. All assays were carried out in triplicate, and the values in the graph are shown as means ± standard errors of the means. *, Significant difference at a P value of <0.005. (B) P protein isoforms and N protein expressed in the above-described transfected cells were analyzed by Western blotting. Tubulin in each sample was also detected as a loading control.
DISCUSSION
To the best of our knowledge, we have shown for first time a substantial role of RABV P protein isoforms (tPs) in pathogenesis in vivo by exploiting a reverse genetics approach. To date, studies on tPs of RABV have mainly focused on functions of P2 and P3 as viral IFN antagonists. The results of those studies have indicated that P2 is involved in IFN resistance (18) and that P3 has functions to inhibit IFN signaling via various mechanisms targeting STATs (11, 17). While these previous findings highlight the roles of tPs in IFN antagonism, the actual contribution of tPs to viral pathogenesis in an in vivo setting has remained to be elucidated. Therefore, the present study provides novel insights into the molecular basis of RABV-host interactions and pathogenesis.
In this study, to examine the importance of tPs in the pathogenesis of RABV, we utilized chimeric CE(NiP), which has the P gene of highly neurovirulent and neuroinvasive Ni in the genome of the attenuated derivative Ni-CE. The fact that CE(NiP) is more neurovirulent and neuroinvasive than Ni-CE (21, 22) indicated the importance of P protein in viral pathogenesis. Since CE(NiP) was characterized in detail in our previous studies, we believe that CE(NiP) provides a good model to evaluate the roles of P protein, including tPs, in pathogenesis.
Western blot analysis of cultured cells transfected to express each Ni P protein isoform revealed that mobilities of these isoforms did not correspond to their actual molecular sizes (Fig. 1C). This phenomenon was not previously observed in the P protein isoforms of other RABV strains, CVS and SAD L16 (13, 18). Notably, similar anomalous SDS-PAGE migration of P protein was found in our previous study: Ni-CE P1 migrated slower than did Ni P1, although both P1s consist of 297 amino acids (7), suggesting a unique structural property of Ni and Ni-CE P proteins. The mechanism underlying the molecular size-independent migration of P protein isoforms has remained unclear, but phosphorylation of these isoforms may be involved in this phenomenon. Alternatively, the difference among these isoforms in SDS-binding capacity that is influenced by the protein structure might cause the anomalous migration. Notably, Rath et al. (26) reported that anomalous SDS-PAGE migration of various membrane proteins can be explained by their difference in SDS-binding capacity. The same mechanism might act on the non-membrane-associated Ni P protein isoforms. In any case, biochemical analysis of the P protein isoforms will be required to elucidate the mechanism.
In this study, we demonstrated that P1, P2, and P3, but not P4 and P5, were expressed at detectable levels in the CE(NiP)-infected cells (Fig. 1C). The same phenomenon was observed in a previous study with another RABV strain, CVS (13). Notably, in that study, P4 and P5 were detected in purified virus particles, which probably contain higher concentrations of those isoforms than do infected cells, indicating that P4 and P5 were indeed expressed, but at quite low levels, in CVS-infected cells. According to those previous findings, we designed experiments in this study based on the assumption that P4 and P5 were expressed at low levels in CE(NiP)-infected cells.
Interestingly, while P3 was clearly expressed in CE(NiP)-infected NA cells (Fig. 1C, lane 8), only the largest isoform was detected in cells transfected with the respective plasmid expressing P1 or one of the tPs (lanes 2 to 6). This fact leads to the hypothesis that a stimulus from viral infection is required for efficient expression of tPs by a ribosomal leaky scanning mechanism. This hypothesis is supported by a previous finding that infection with a vaccinia virus strain expressing T7 RNA polymerase induced expression of tPs in cells transfected with a plasmid expressing CVS P protein under the control of the T7 promoter (13). Notably, Shabman et al. recently showed by functional analysis of the L gene mRNA of Ebola virus, which contains two overlapped open reading frames (ORFs), that cell stress facilitates its translation from the 2nd start codon, resulting in efficient expression of L protein encoded by the 2nd ORF (27). Therefore, cell stress caused by RABV infection might be involved in the enhanced expression of tPs in infected cells.
To establish CE(NiP)ΔP2-5 by gene manipulation of CE(NiP), we replaced all of the start codons (AUG, in positive sense) for tPs with AUA, encoding Ile. We chose this Ile for the following reasons: (i) the Ile is naturally found in minor RABV strains such as V123.DG (GenBank accession no. AF369318) and V124.DG (AF369319) (16), which do not have a start codon for P3, and (ii) replacement of Met with Ile is not expected to drastically affect the molecular structure and function of P1, since both amino acid residues are classified to the same hydrophobic amino acid. Notably, it was previously reported that a mutant of RABV strain SAD L16, of which the ability to express P2, P3, and P4 was impaired, was established by introduction of the same Met-to-Ile mutations and also did not show significant attenuation in its replication ability in vitro (5). In this study, we demonstrated that CE(NiP)ΔP2-5 had a defect in the ability to express tPs in infected cells (Fig. 1C) without attenuation of its ability to replicate in neuroblastoma NA cells (Fig. 2A). In addition to similar growth abilities of the two viruses, our results indicated that both activities of CE(NiP)ΔP2-5 P1 as a cofactor of viral RNA polymerase and a viral IFN antagonist were intact (Fig. 2C and 3). These findings indicate that CE(NiP) and CE(NiP)ΔP2-5 are useful for examination of the roles of tPs in the pathogenesis of RABV.
Comparison of the pathogenicities of CE(NiP) and CE(NiP)ΔP2-5 in mice with that of CE(NiP) demonstrated that CE(NiP)ΔP2-5 was less pathogenic than CE(NiP) in infection via the i.m. route (Fig. 4), indicating the importance of tPs in neuroinvasiveness. Further examinations by comparing biological characteristics of the two viruses demonstrated that CE(NiP)ΔP2-5 had a defect in the ability to spread to peripheral nerves, which was concomitant with the impaired abilities of this virus to efficiently replicate and to suppress IFN induction in muscle cells (Fig. 6 to 8). These findings strongly suggest that tPs function to support viral replication in muscle cells by their IFN antagonist activities and thereby to facilitate infection of peripheral nerves. Consistent with our previous results obtained from comparative analysis of CE(NiP) and Ni-CE (22), these findings highlight the contribution of IFN antagonism to viral replication in muscle cells and subsequently to efficient infection of peripheral nerves.
Our data obtained from an IFN-β promoter reporter assay strongly suggest that all of the tPs, as well as P1, have activity to antagonize IFN induction in muscle cells (Fig. 10). While P2 and P3 were strongly suggested by results of previous studies to inhibit IFN responses (11, 17, 18), to our knowledge, we have provided here the first evidence demonstrating the function of tPs to suppress IFN induction. Notably, despite the fact that expression levels of recombinant P4 and P5 in the lysates were obviously lower than the levels of P1, P2, and P3 (Fig. 10B), the expression of P4 and P5 inhibited the IFN-β promoter activities stimulated by poly(I·C) treatment with efficiencies similar to those of other isoforms (Fig. 10A). These results imply that P4 and P5 have strong activities to antagonize IFN induction in muscle cells. Although their expression levels appear to be quite low in infected cells, these isoforms might play an important role in IFN antagonism in muscle cells.
The findings obtained in this study indicated the importance of tPs in neuroinvasiveness but, on the other hand, not in neurovirulence: after i.c. inoculation, both CE(NiP)ΔP2-5 and CE(NiP) caused lethal infection in 100% of the mice (Fig. 4A). This observation is consistent with the findings that the two strains replicated with similar efficiencies in mouse brains (Fig. 5) and neuroblastoma cell lines, NA cells (Fig. 2A), and SYM-I cells (data not shown). These data indicate the possibility that the IFN antagonist function of tPs is cell type specific. However, our preliminary data indicate that P1 and all of the tPs retain activity to inhibit IFN induction in SYM-I cells (data not shown), implying that the mechanism underlying the cell type-dependent function of tPs is complex. Further investigations to elucidate the mechanism are in progress.
In conclusion, this study has demonstrated that tPs play an important role in the pathogenesis of RABV, especially its neuroinvasiveness. Furthermore, our results strongly suggest that tPs support stable viral replication in muscle cells via their activities to antagonize IFN induction and consequently enhance infection of peripheral nerves. We believe that these findings are useful to establish the molecular basis for development of a novel prophylaxis approach and also an effective therapeutic approach for rabies.
ACKNOWLEDGMENTS
We thank Akihiko Kawai (Research Institute for Production and Development, Japan) for providing SYM-I cells and anti-P protein rabbit polyclonal antibody. We also thank Yoshihiro Kawaoka (University of Tokyo, Tokyo, Japan) and Rongtuan Lin (McGill University, Montreal, Canada) for providing plasmids.
This study was partially supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (no. 23380179, 24580424, and 15K07720), and by a grant from the Ministry of Education, Culture, Sports, Science and Technology, Japan, for the Joint Research Program of the Research Center for Zoonosis Control, Hokkaido University. S.Y. and H.E. are supported by the Division of Intramural Research, NIAID, NIH.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Jackson AC, Fu ZF. 2013. Pathogenesis, p 299–349. In Jackson AC. (ed), Rabies, 3rd ed Academic Press, London, United Kingdom. [Google Scholar]
- 2.Hampson K, Coudeville L, Lembo T, Sambo M, Kieffer A, Attlan M, Barrat J, Blanton JD, Briggs DJ, Cleaveland S, Costa P, Freuling CM, Hiby E, Knopf L, Leanes F, Meslin FX, Metlin A, Miranda ME, Muller T, Nel LH, Recuenco S, Rupprecht CE, Schumacher C, Taylor L, Vigilato MA, Zinsstag J, Dushoff J, Global Alliance for Rabies Control Partners for Rabies Prevention . 2015. Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis 9:e0003709. doi: 10.1371/journal.pntd.0003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wunner WH, Conzelmann KK. 2013. Rabies virus, p 17–60. In Jackson AC. (ed), Rabies, 3rd ed Academic Press, London, United Kingdom. [Google Scholar]
- 4.Blondel D, Regad T, Poisson N, Pavie B, Harper F, Pandolfi PP, De The H, Chelbi-Alix MK. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957–7970. doi: 10.1038/sj.onc.1205931. [DOI] [PubMed] [Google Scholar]
- 5.Brzozka K, Finke S, Conzelmann KK. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J Virol 79:7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brzozka K, Finke S, Conzelmann KK. 2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J Virol 80:2675–2683. doi: 10.1128/JVI.80.6.2675-2683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito N, Moseley GW, Blondel D, Shimizu K, Rowe CL, Ito Y, Masatani T, Nakagawa K, Jans DA, Sugiyama M. 2010. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J Virol 84:6699–6710. doi: 10.1128/JVI.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieder M, Brzozka K, Pfaller CK, Cox JH, Stitz L, Conzelmann KK. 2011. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J Virol 85:842–852. doi: 10.1128/JVI.01427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu K, Ito N, Sugiyama M, Minamoto N. 2006. Sensitivity of rabies virus to type I interferon is determined by the phosphoprotein gene. Microbiol Immunol 50:975–978. doi: 10.1111/j.1348-0421.2006.tb03875.x. [DOI] [PubMed] [Google Scholar]
- 10.Vidy A, Chelbi-Alix M, Blondel D. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J Virol 79:14411–14420. doi: 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidy A, El Bougrini J, Chelbi-Alix MK, Blondel D. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J Virol 81:4255–4263. doi: 10.1128/JVI.01930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiltzer L, Okada K, Yamaoka S, Larrous F, Kuusisto HV, Sugiyama M, Blondel D, Bourhy H, Jans DA, Ito N, Moseley GW. 2014. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J Infect Dis 209:1744–1753. doi: 10.1093/infdis/jit829. [DOI] [PubMed] [Google Scholar]
- 13.Chenik M, Chebli K, Blondel D. 1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J Virol 69:707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chenik M, Schnell M, Conzelmann KK, Blondel D. 1998. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J Virol 72:1925–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi Y, Okuda H, Nakamura K, Sato G, Itou T, Carvalho A, Silva M, Mota C, Ito F, Sakai T. 2007. Genetic analysis of phosphoprotein and matrix protein of rabies viruses isolated in Brazil. J Vet Med Sci 69:1145–1154. doi: 10.1292/jvms.69.1145. [DOI] [PubMed] [Google Scholar]
- 16.Nadin-Davis SA, Abdel-Malik M, Armstrong J, Wandeler AI. 2002. Lyssavirus P gene characterisation provides insights into the phylogeny of the genus and identifies structural similarities and diversity within the encoded phosphoprotein. Virology 298:286–305. doi: 10.1006/viro.2002.1492. [DOI] [PubMed] [Google Scholar]
- 17.Moseley GW, Lahaye X, Roth DM, Oksayan S, Filmer RP, Rowe CL, Blondel D, Jans DA. 2009. Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J Cell Sci 122:3652–3662. doi: 10.1242/jcs.045542. [DOI] [PubMed] [Google Scholar]
- 18.Marschalek A, Drechsel L, Conzelmann KK. 2012. The importance of being short: the role of rabies virus phosphoprotein isoforms assessed by differential IRES translation initiation. Eur J Cell Biol 91:17–23. doi: 10.1016/j.ejcb.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Chelbi-Alix M, Quignon F, Pelicano L, Koken M, de Thé H. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol 72:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regad T, Chelbi-Alix M. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu K, Ito N, Mita T, Yamada K, Hosokawa-Muto J, Sugiyama M, Minamoto N. 2007. Involvement of nucleoprotein, phosphoprotein, and matrix protein genes of rabies virus in virulence for adult mice. Virus Res 123:154–160. doi: 10.1016/j.virusres.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Yamaoka S, Ito N, Ohka S, Kaneda S, Nakamura H, Agari T, Masatani T, Nakagawa K, Okada K, Okadera K, Mitake H, Fujii T, Sugiyama M. 2013. Involvement of the rabies virus phosphoprotein gene in neuroinvasiveness. J Virol 87:12327–12338. doi: 10.1128/JVI.02132-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol Immunol 47:613–617. doi: 10.1111/j.1348-0421.2003.tb03424.x. [DOI] [PubMed] [Google Scholar]
- 24.Minamoto N, Tanaka H, Hishida M, Goto H, Ito H, Naruse S, Yamamoto K, Sugiyama M, Kinjo T, Mannen K, Mifune K. 1994. Linear and conformation-dependent antigenic sites on the nucleoprotein of rabies virus. Microbiol Immunol 38:449–455. doi: 10.1111/j.1348-0421.1994.tb01806.x. [DOI] [PubMed] [Google Scholar]
- 25.Masatani T, Ito N, Shimizu K, Ito Y, Nakagawa K, Sawaki Y, Koyama H, Sugiyama M. 2010. Rabies virus nucleoprotein functions to evade activation of the RIG-I-mediated antiviral response. J Virol 84:4002–4012. doi: 10.1128/JVI.02220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rath A, Glibowicka M, Nadeau V, Chen G, Deber C. 2009. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci U S A 106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shabman RS, Hoenen T, Groseth A, Jabado O, Binning JM, Amarasinghe GK, Feldmann H, Basler CF. 2013. An upstream open reading frame modulates Ebola virus polymerase translation and virus replication. PLoS Pathog 9:e1003147. doi: 10.1371/journal.ppat.1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]