ABSTRACT
The high incidence of AIDS cases and the dominance of HIV-1 subtype C infections are two features that distinguish the HIV-1 epidemic in the two southernmost Brazilian states (Rio Grande do Sul [RS] and Santa Catarina [SC]) from the epidemic in other parts of the country. Nevertheless, previous studies on HIV molecular epidemiology were conducted mainly in capital cities, and a more comprehensive understanding of factors driving this unique epidemic in Brazil is necessary. Blood samples were collected from individuals in 13 municipalities in the Brazilian southern region. HIV-1 env and pol genes were submitted to phylogenetic analyses for assignment of subtype, and viral population phylodynamics were reconstructed by applying Skygrid and logistic coalescent models in a Bayesian analysis. A high prevalence of subtype C was observed in all sampled locations; however, an increased frequency of recombinant strains was found in RS, with evidence for new circulating forms (CRFs). In the SC state, subtype B and C epidemics were associated with distinct exposure groups. Although logistic models estimated similar growth rates for HIV-1 subtype C (HIV-1C) and HIV-1B, a Skygrid plot reveals that the former epidemic has been expanding for a longer time. Our results highlight a consistent expansion of HIV-1C in south Brazil, and we also discuss how heterosexual and men who have sex with men (MSM) transmission chains might have impacted the current prevalence of HIV-1 subtypes in this region.
IMPORTANCE The AIDS epidemic in south Brazil is expanding rapidly, but the circumstances driving this condition are not well known. A high prevalence of HIV-1 subtype C was reported in the capital cities of this region, in contrast to the subtype B dominance in the rest of the country. This study sought to comparatively investigate the HIV-1 subtype B and C epidemics by sampling individuals from several cities in the two states with the highest AIDS incidences in Brazil. Our analyses showed distinct epidemic growth curves for the two epidemics, and we also found evidence suggesting that separate transmission chains may be impacting the viral phylodynamics and the emergence of new recombinant forms.
INTRODUCTION
High-level molecular diversity is one of the main features of HIV-1. Due to the very high replication rate and the lack of a proofreading activity of the viral reverse transcriptase (RT) enzyme, HIV-1 group M, the pandemic strain, has evolved into a diverse number of variants (1). Among 9 HIV-1 group M subtypes, HIV-1 subtype C (HIV-1C) is responsible for >50% of infections worldwide, occurring mostly in southern and east African countries and India. HIV-1B, in turn, is widely dispersed throughout the world, being the dominant strain in Europe and in the Americas, but with a worldwide prevalence of only 10% (1, 2). The high level of variability of HIV-1 is also a consequence of the genetic recombination process, which can occur in unique recombinant forms (URFs), when fewer than three individuals are infected, or in circulating forms (CRFs), when three individuals not epidemiologically related are infected with the same mosaic strain (3). Currently, more than 70 CRFs have been described, encompassing around 20% of HIV-1 infections around the world (see http://www.hiv.lanl.gov/ [accessed June 2016]).
The HIV-1 molecular epidemic in Brazil is driven mainly by subtype B, with a minor circulation of subtype F1, BF1 recombinant forms, and subtype C (4). However, in the two southernmost states, Rio Grande do Sul (RS) and Santa Catarina (SC), a high prevalence of HIV-1C is observed (40 to 80%), followed by BC mosaic forms (20 to 30%, including CRF31_BC) and subtype B (15 to 30%) (5). The HIV-1C prevalence decreases in the northward direction, representing up to 30% of infections in Paraná state and between 5 and 20% of infections in other nearby states (5). Besides the high prevalence of HIV-1C, RS and SC states registered the highest AIDS incidence rates among the Brazilian states in the last 10 years, notifying around 35 new cases per 100,000 habitants per year (6). The AIDS incidence was also observed to have increased more than two times in countryside cities in RS and SC over 10 years (7). The same process has also been observed for other Brazilian states but at a much lower rate.
A decade of epidemiological studies described the HIV-1C epidemic in south Brazil, investigating its origin and highlighting its dispersion throughout the country and continent (8–13). However, those studies included samples mainly from individuals in the capital cities, and very little molecular information is available for countryside locations in the southern region. In the face of this interesting epidemiological scenario, where a high AIDS incidence is observed, along with unique molecular features, a better characterization of the HIV-1 epidemic in a broad sampling area is necessary. The present study aims to describe the HIV-1 molecular epidemiology in countryside cities of RS and SC states. To do so, we obtained samples from individuals in several cities, including border towns, in an effort to construct a comprehensive data bank with viral sequences and patient information. In addition, we analyzed the phylodynamics of HIV-1C and -B using demographic reconstruction of multiple genetic loci to describe the historical epidemic growth trends of these two dominant strains in the region.
MATERIALS AND METHODS
Patients and samples.
HIV-1-seropositive patients were invited to enroll in this study throughout 13 cities of the Brazilian states of SC and RS (Fig. 1). The inclusion criteria were diagnosis of HIV infection and no previous exposure to antiretroviral (ARV) treatment. The Universidade Federal de Santa Catarina (UFSC) and the Fundação Estadual de Pesquisa e Produção em Saúde (FEPPS-RS) ethics committees assessed and approved this study. Informed consent was obtained from all enrolled volunteers.
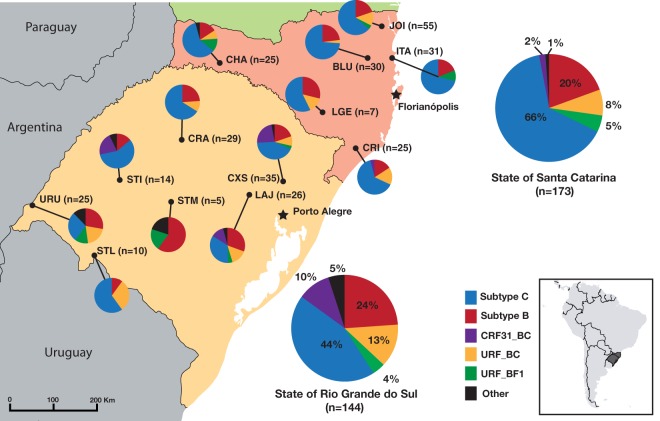
FIG 1.
HIV-1 molecular diversity in countryside cities from the states of Santa Catarina and Rio Grande do Sul, Brazil. The locations of sampled cities are shown by dots on the map, and the locations of capital cities are marked with black stars. Pie chart colors are defined in the key, and an inset map shows the location of the sampled region in South America. Abbreviations for cities: BLU, Blumenau; CHA, Chapecó; CRA, Cruz Alta; CRI, Criciúma; CXS, Caxias do Sul; ITA, Itajaí; JOI, Joinville; LAJ, Lajeado; LGE, Lages; STI, Santiago; STL, Santana do Livramento; STM, Santa Maria; URU, Uruguaiana. (Based on maps from the Natural Earth public domain data set and drawn using QGIS [https://creativecommons.org/licenses/by-sa/3.0/legalcode].)
Blood samples were collected onto FTA cards (Whatman Inc.) and dried at room temperature. Three to five 0.5-mm dried blood spot (DBS) punches were washed and dried according to the manufacturer's instructions. Provirus DNA of HIV-1 was amplified by nested PCR directly from the washed punches, and primers and reaction conditions were as follows (14). The HIV-1 envelope (env) (HXB2 bp 6846 to 7360) and polymerase (pol) (HXB2 bp 2274 to 3545) genes were the genome target regions. Purified PCR products were sequenced by using a BigDye Terminator v3.1 cycle sequencing system (Applied Biosystems) and read by using an ABI 3100 genetic analyzer (Applied Biosystems). The generated chromatograms were then assembled into contigs and visually inspected by using SeqMan software in the Lasergene package (DNASTAR).
Subtype classification.
Sequences generated here were submitted to the REGA, RIP, and SCUEAL online subtyping tools (15–17). The subtype assigned by these automated tools was confirmed by phylogenetic tree reconstruction. Subtype reference sequences were downloaded from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/) and aligned with the new sequences by using the Muscle algorithm (18). Maximum likelihood (ML) tree reconstruction was performed by using PhyML with the GTR+I+Γ4 (general time reversible [GTR] substitution model assuming an estimated proportion of invariant sites [I] and four gamma-distributed rate categories [Γ4]) nucleotide substitution model, as implemented in the SeaView program (19). Aiming to identify the recombination breakpoints, env and pol sequences identified as recombinant sequences by using the online tools and in the ML trees were submitted to bootscanning analysis using Simplot 3.5.1 software (20).
Bayesian phylodynamics analyses.
In order to better understand the expansion of the HIV-1 epidemic in south Brazil, time-scaled phylogenetic trees were reconstructed for subtype B and C sequences by using BEAST/BEAGLE software (21). To improve the accuracy of demographic history reconstruction, only samples classified as belonging to subtype B or C for both the env and pol genes were analyzed. Additional sequences from SC and RS states, which met the same criteria as those for the sequences generated here, were downloaded from the Los Alamos HIV Database and aligned with the countryside sequence data set, compounding a data set sampled from 2008 to 2014. Trees were reconstructed by using an uncorrelated log-normal relaxed molecular clock and the GTR+I+Γ4 nucleotide substitution model. Due to the lack of temporal signal in the data sets analyzed here, as determined by using Path-O-Gen software (http://tree.bio.ed.ac.uk/software/pathogen/), normal distributed priors were applied to the tree root height for the Brazilian HIV-1C (mean of 41 and standard deviation of 4.1 for Pol and mean of 43 and standard deviation of 4.1 for Env) and HIV-1B (mean of 47 and standard deviation of 8.7 for Pol and mean of 50 and standard deviation of 4.1 for Env) clades, as previously estimated (11, 22).
The nonparametric Bayesian Skygrid coalescent model was used to estimate HIV effective population size (Ne) trajectories (23). This model bases estimations on data from multiple genetic loci to improve the accuracy of the analyses. To estimate the epidemic growth rates (r) of HIV-1B and HIV-1C, trees were also reconstructed under different parametric demographic models. Marginal likelihood estimation (MLE) was applied to test for the best parametric model in a Bayesian framework (24, 25). As distinct genetic loci from an organism are supposed to share the same demographic dynamics without following the same evolutionary history, HIV-1 env and pol sequences were analyzed in BEAST as separated partitions with unlinked substitution and clock models but sharing the same coalescent prior.
Monte Carlo Markov chains (MCMCs) were run sufficiently long enough to ensure that the effective sample sizes (ESSs) for all the parameters were above 200. For the HIV-1C data set, five independent runs of 100 million steps were performed and later combined by using LogCombiner (http://beast.bio.ed.ac.uk/LogCombiner). For HIV-1B, a single run of 200 million steps was performed. Tracer software (http://tree.bio.ed.ac.uk/software/tracer/) was used to diagnose MCMCs, adjust the initial burn-in, and perform demographic reconstructions.
Transmission dynamics among exposure groups.
The dynamic of viral transmissions among the main HIV-1 exposure groups was also investigated by using the same data set as the one used in the phylodynamics analysis. To do so, ancestral trait reconstruction was performed by using a discrete model of transition with the Bayesian stochastic search variable selection (BSSVS) procedure and asymmetric transition rates in an analysis where traits were the exposure group of each taxon (26). This analysis was complemented with Markov jump estimation of the number of trait transitions throughout evolutionary history (27). RStudio (http://www.rstudio.org/) was used to summarize the posterior densities of the highly supported transitions from the BEAST log files.
Statistical analysis.
Patients were invited to answer a questionnaire at the time of enrollment. Information about gender, age, exposure group, ethnicity, education level, and marital status were included in the questionnaire, while clinical data were retrieved from patient medical records. A multivariable logistic regression model was applied to determine associations between variables and HIV-1 subtype. To deal with the perfect separation of some variables, the logistic regression was conducted in a Bayesian framework (28). All analyses were executed with RStudio, and statistical significance was achieved when P values were ≤0.05. Data relative to new cases of AIDS annually reported in Brazil were retrieved from the DATASUS system, a data bank hosted by the Brazilian Ministry of Health (http://datasus.saude.gov.br/).
Accession numbers.
The sequences generated in the present study were deposited in GenBank under accession numbers KR065788 to KR066336 and KP224476 to KP224501.
RESULTS
A total of 359 samples were collected between October 2009 and February 2014: 163 (45%) samples were from RS state, and 196 (55%) samples were from SC state. HIV-1 sequences from the pol region were obtained for 253 (71%) samples and from the env region for 284 (79%) samples, totaling 317 (88%) patients with molecular data for pol, env, or both. Among 317 patients with subtype information, 45% were women, and 55% were men. The mean date of diagnosis was May 2010, and the mean patient age was 40 years. The most frequently related HIV-1 exposure group was heterosexual individuals (HET) (71%), followed by men who have sex with men (MSM) (20%). Seventy-two percent of the enrolled patients were self-defined white, while 17% defined themselves as African descendants or Amerindians.
Phylogenetic analysis revealed that 56% of patients had HIV-1 subtype C infections, 22% had subtype B, 10% had URF_BC, 5% had CRF31_BC, 4% had URF_BF1, and 3% had other viral forms (subtypes D, F1, and URF_CF1 and complex unique recombinants). In SC, we observed a higher prevalence of HIV-1C (66%, versus 44% in RS), while BC recombinant forms (CRF31_BC and URF_BC) were more frequently found in RS state (23%, versus 10% in SC) (Fig. 1). HIV-1B has roughly the same prevalence in the two states.
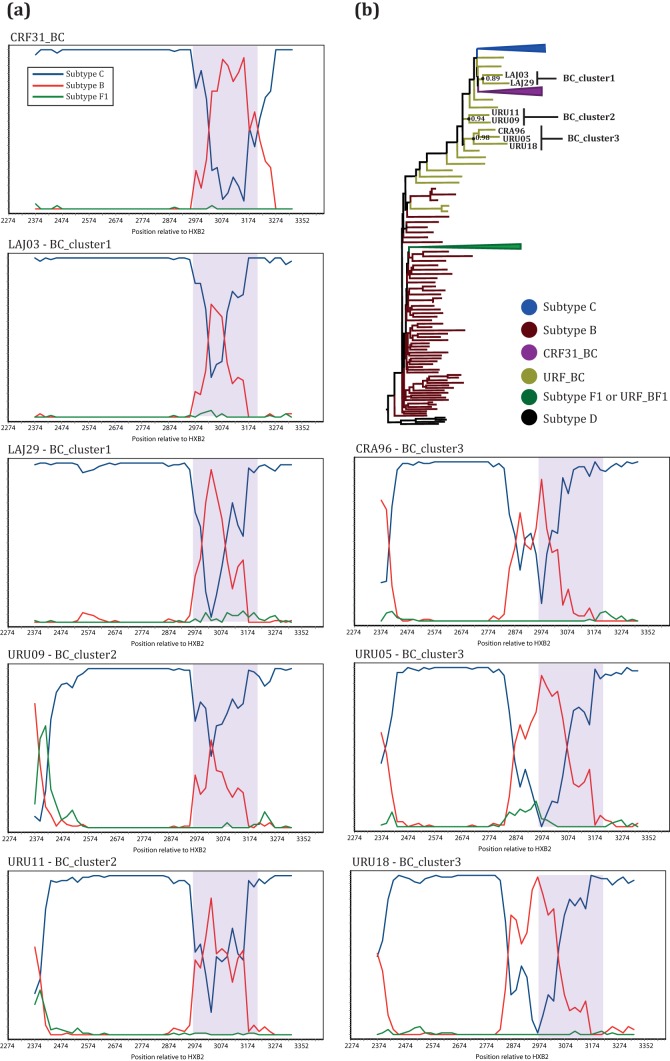
Recombinant strains were observed in 21% (66/317) of the patients studied here, as 77% of them (51/66) were infected by unique forms (URFs). Among the URF_BC samples (n = 33), 30% were intergenic recombinants whose recombination breakpoints were not able to be observed in the pol and env fragments sequenced here. For URF_BF1 (n = 14), intergenic mosaic strains represented 57% of the infections. Some of the URFs identified here were closely related in phylogenetic trees and also showed similar recombination breakpoints in the bootscanning analysis. Two sequences from Chapecó city were intergenic URF_BF1 sequences. They were very closely related in the trees, but direct transmission is unlikely, as the samples were obtained from two women reporting heterosexual exposure to HIV. Among the intragenic recombinant mosaic forms, all identified in pol sequences, three distinct groups of URF_BC strains were identified with similar breakpoints and clustered together in trees (Fig. 2). These groups comprised the LAJ03 and LAJ29 sequences from Lajeado city, named BC_cluster1; URU09 and URU11 from Uruguaiana city, named BC_cluster2; and CRA96, URU05, and URU18 from Cruz Alta and Uruguaiana cities, named BC_cluster3. Heterosexual men and women composed each of these clusters.
FIG 2.
Related BC mosaic strains identified by bootscanning and phylogenetic analyses. (a) Bootscanning plots of the sequences in BC_cluster1 to -3 and a CRF31_BC reference sequence (top left). Line colors for the reference sequences are defined in the key at the top left. The y axis shows the percentage of permuted trees for one of the reference clusters with the query. The x axis represents the positions (relative to HXB2) of the fragment being analyzed. Purple shading in plots indicates the CRF31_BC recombination region. (b) Maximum likelihood tree of the pol gene indicating the positions of the sequences in BC_cluster1 to -3 and their respective Shimodaira-Hasegawa approximate likelihood ratio test (SH-aLRT) node supports. Branches are colored according to the key, and some of the subtype clades are condensed.
A CRF31_BC-related recombination pattern was observed for BC_cluster1 and BC_cluster2 sequences (Fig. 2). The sequences in these clusters share the first recombination breakpoint, between positions 2965 and 2984 relative to the HXB2 strain, with CRF31_BC. However, they have a smaller subtype B insertion in a pol subtype C backbone than in CRF31_BC. In BC_cluster2, it was still possible to detect another recombination breakpoint, although caution is necessary when interpreting data from bootscanning analysis for the extremities of the sequences. BC_cluster3, in turn, shows a distinct mosaic display with a subtype B insertion in positions close to positions 2850 and 3060 (HXB2). It is also possible to observe an additional recombination breakpoint close to the beginning of the analyzed fragment.
A statistically significant association was observed among HIV-1 subtypes (constrained to comparisons between HIV-1B and HIV-1C) and epidemiological data (Table 1). In SC, the multivariable analysis showed that HIV-1C infections were associated with the heterosexual exposure group and negatively associated with the white ethnic group (Table 1). The odds of being infected with HIV-1B, in turn, were higher among MSM and lower for the African descendant/Amerindian ethnic group. Exposure group and ethnicity were not related to each other in this location (data not shown). In RS state, the odds of being infected with HIV-1C were significantly lower for individuals with a higher level of education (>12 years) and individuals living with a partner but not legally married. Also, note the borderline significance (P = 0.07) of the negative association between subtype C and male gender in RS state.
TABLE 1.
Demographic and epidemiological data for the population infected with HIV-1B and HIV-1C in RS and SC states, Brazil, and their association with HIV-1C infectiona
| Predictor | RS |
SC |
||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of patients infected with HIV-1 subtype: |
OR (95% CI) | P | No. (%) of patients infected with HIV-1 subtype: |
OR (95% CI) | P | |||
| B | C | B | C | |||||
| Gender | ||||||||
| Female | 15 (43) | 35 (55) | 1.0 (Ref) | 11 (32) | 46 (40) | 1.0 (Ref) | ||
| Male | 20 (57) | 29 (45) | 0.4 (0.1–1.1) | 0.07 | 23 (68) | 68 (60) | 1.1 (0.4–3.2) | 0.85 |
| Transmission route | ||||||||
| MSM | 5 (14) | 7 (11) | 1.0 (Ref) | 14 (41) | 27 (24) | 1.0 (Ref) | ||
| Heterosexual | 30 (86) | 46 (72) | 0.3 (0.1–2.1) | 0.24 | 16 (47) | 79 (69) | 3.3 (1.1–10.2) | 0.04 |
| IDU | 0 (0) | 4 (6) | 1.9 (0.1–65.6) | 0.73 | 0 (0) | 2 (2) | 3.1 (0.1–85.8) | 0.50 |
| Other | 0 (0) | 2 (3) | 2.1 (0.1–67.7) | 0.67 | 2 (6) | 1 (1) | 0.5 (0.1–3.9) | 0.47 |
| NA | 0 (0) | 5 (8) | 2 (6) | 5 (4) | ||||
| Ethnicity | ||||||||
| African descendants/Amerindians/other | 7 (20) | 13 (20) | 1.0 (Ref) | 0 (0) | 21 (18) | 1.0 (Ref) | ||
| White | 21 (60) | 41 (64) | 1.0 (0.3–3.0) | 0.95 | 33 (97) | 91 (80) | 0.0 (0.0–0.8) | 0.04 |
| NA | 7 (20) | 10 (16) | 1 (3) | 2 (2) | ||||
| No. of yr of education | ||||||||
| ≤8 | 19 (54) | 39 (61) | 1.0 (Ref) | 12 (35) | 49 (43) | 1.0 (Ref) | ||
| 9–11 | 5 (14) | 19 (30) | 1.5 (0.4–5.2) | 0.55 | 14 (41) | 40 (35) | 1.1 (0.4–3.0) | 0.91 |
| ≥12 | 4 (11) | 1 (2) | 0.1 (0.0–0.6) | 0.02 | 8 (24) | 22 (19) | 0.9 (0.3–2.8) | 0.79 |
| NA | 7 (20) | 5 (8) | 0 (0) | 3 (3) | ||||
| Marital status | ||||||||
| Single | 13 (37) | 31 (48) | 1.0 (Ref) | 18 (53) | 52 (46) | 1.0 (Ref) | ||
| Married | 7 (20) | 19 (30) | 0.8 (0.2–2.6) | 0.69 | 7 (21) | 30 (26) | 0.7 (0.2–2.1) | 0.52 |
| Not legally married but living together | 12 (34) | 6 (9) | 0.2 (0.0–0.8) | 0.02 | 7 (21) | 26 (23) | 0.9 (0.3–2.7) | 0.92 |
| Other | 3 (9) | 3 (5) | 0.4 (0.1–2.7) | 0.34 | 1 (3) | 5 (4) | 1.3 (0.2–9.4) | 0.78 |
| NA | 0 (0) | 5 (8) | 1 (3) | 1 (1) | ||||
| Age (yr) | ||||||||
| ≤24 | 6 (17) | 10 (16) | 1.0 (Ref) | 4 (12) | 17 (15) | 1.0 (Ref) | ||
| >24 | 29 (83) | 54 (84) | 1.0 (0.3–3.9) | 0.99 | 30 (88) | 97 (85) | 0.6 (0.2–1.9) | 0.36 |
OR, odds ratio; CI, confidence interval; NA, data not available; MSM, men who have sex with men; IDU, injection drug users; Ref, reference category. Values in parentheses are relative percentages of the total.
Differences between HIV-1B and HIV-1C regarding the epidemic expansion rate were further investigated. Skygrid coalescent analysis reconstructed very similar growth curves for both subtypes, from their independent introduction until the early1990s (Fig. 3a). From this point forward, the Ne value for HIV-1B decreased until around the mid-2000s, when it started to increase again. By its turn, the Ne value for HIV-1C continued to increase in until the mid- to late 2000s, when the epidemic growth rate seemed to have stabilized. Considering that AIDS symptoms arise typically 6 to 10 years after infection (29), the numbers of new cases (incidences) of AIDS annually reported by the Brazilian Ministry of Health in RS and SC, stratified by MSM and HET, follow similar epidemic growth trends for HIV-1B and -C, respectively (Fig. 3b). The incidence of AIDS among MSM annually increased until the mid-1990s, matching the period when the growth rate of the HIV-1B epidemic decreased. The more recent increase of HIV-1B Ne also seems to be reflected in the MSM epidemic after the mid-2000s. The incidence of AIDS among HET, on the other hand, seems to be correlated with the HIV-1C epidemic; however, after the stabilization of new cases in HET from the 2000s, the Ne value for HIV-1C continued to increase, possibly reflecting expansion throughout the MSM group, whose incidence did not follow the shrinkage of HIV-1B Ne since the 1990s. To investigate this hypothesis, an analysis to count the number of transitions among HIV-1 exposure groups was performed. While the number of transitions between HET and MSM groups has remained constant through time for HIV-1B (data not shown), an increase in HET-to-MSM transitions was observed for HIV-1C, reaching the densest period in the late 1990s (Fig. 3a). These findings may illustrate the period of the most effective introduction of HIV-1C in the MSM group, which might have allowed the HIV-1C epidemic to keep expanding in the following years.
FIG 3.
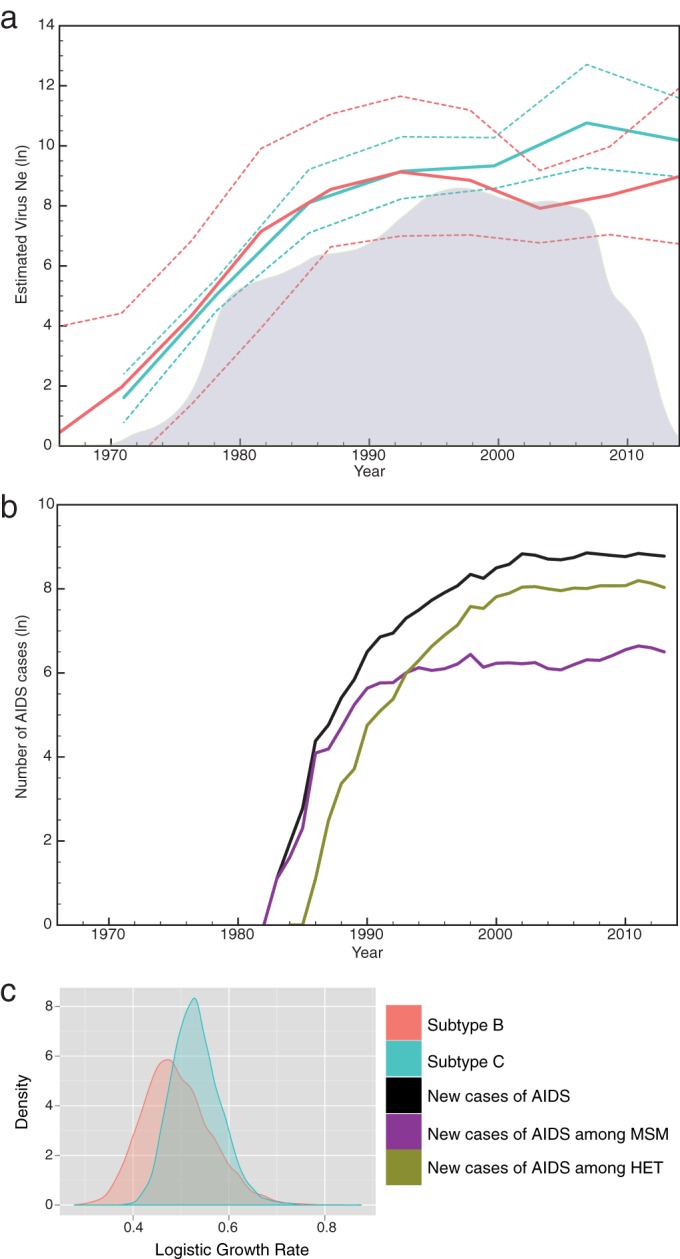
Phylodynamics analyses of env and pol genes from HIV-1 subtype C and B sequences isolated in Rio Grande do Sul (RS) and Santa Catarina (SC) states, Brazil. (a) Bayesian Skygrid plots representing changes in virus effective population sizes (Ne) through time and density of subtype C viral transitions from heterosexual individuals to MSM (gray plot in the background). Solid lines represent the mean estimated Ne, and dashed lines represent the 95% HPD. (b) Numbers of new cases of AIDS reported yearly in SC and RS states by exposure category. (c) Density plots of the estimated logistic growth rates for HIV-1C and -B. Colors are defined in the key.
MLE indicated the logistic growth model as the best coalescent parametric model. This analysis estimated very similar logistic growth rates for HIV-1C (0.53 year−1; 95% high posterior density [HPD], 0.43 to 0.63) and HIV-1B (0.49 year−1; 95% HPD, 0.36 to 0.63), with large overlapping HPD intervals (Fig. 3c).
DISCUSSION
The study presented here describes HIV-1 molecular epidemiology in a large area, comprising 13 municipalities from the two southernmost Brazilian states. In terms of molecular diversity, our results corroborate previous studies that also reported a large dominance of subtype C (∼65%) in SC state (30–33) and a more diverse epidemic in RS state, mainly because of the high prevalence of BC mosaic forms in the latter (∼25%) (34–36). In both states, a low prevalence of subtype F1 and BF1 recombinants (∼3%) was observed, while this frequency can reach up to 20% in northern regions of Brazil (37, 38) and 40% in Argentina (39). It is interesting to note that the locations with higher subtype F1 or BF1 prevalences were cities close to the border with Argentina (i.e., Uruguaiana and Chapecó) and the port city of Itajaí, where a high international population flux may foster contact with new HIV-1 strains (Fig. 1).
The CRF31_BC strain was almost exclusively found in RS state, more precisely in cities near Porto Alegre (i.e., Lajeado and Caxias do Sul) (Fig. 1), where the CRF31_BC frequency is around 22% and may reach 35% in the metropolitan region (34, 40). Despite the more pronounced occurrence in Porto Alegre and nearby cities, CRF31_BC is not limited to this region, as it was also observed in more distant locations in the present study. Among the BC mosaic forms, some displayed a CRF31_BC-related recombination pattern, which likely originated from a second recombination event within the CRF31_BC pol fragment, as previously reported for other samples from Porto Alegre (41). In our sampling, this pattern was observed in two clusters of sequences (Fig. 2) and in some other isolated sequences classified here as URF_BC (data not shown). Although very similar, only a thorough analysis of the recombination region can confirm if these sequences are indeed second-generation recombinants (SGRs) derived from CRF31_BC.
Three possible new CRFs were identified here: BC_cluster1 and BC_cluster2, which might be SGRs from CRF31_BC, and BC_cluster3, with a different recombination pattern. Further studies aiming to sequence the full-length genomes of these strains are needed to confirm this hypothesis, but our findings point to a greater diversity of circulating BC mosaic strains than previously reported (41). Eight distinct BC CRFs have been described worldwide so far, with six of them in the Chinese province of Yunnan, a region near India, where subtypes C and B cocirculate (42). The high prevalence of AIDS in injection drug users (IDU) makes Yunnan province a hot spot for HIV recombination. The very low prevalence of AIDS cases among IDU in Brazil (6) could explain the low level of diversity of BC CRFs observed in the southern region. Nevertheless, due to the coprevalence of HIV-1C and -B, it is likely that CRFs other than CRF31_BC are circulating in the states of SC and RS.
Indeed, separate transmission chains for subtypes B and C might be acting on the compartmentalization of the epidemic in south Brazil. In SC, we observed an association between the heterosexual group and HIV-1C, and the same association was previously reported in the state capital city of Florianópolis (30). The lower prevalence of BC mosaic strains in SC might be a consequence of the segregation of HIV-1C and -B in separate transmission chains, a scenario that was not observed in our sampling from RS state, where a higher number of mosaic strains was observed. Moreover, it was reported that IDU could have had an initial role in the dispersion of subtype C in RS, promoting admixture with the subtype B epidemic in the early stages of the epidemic (43). A second demographic factor that may be shaping viral transmissions in south Brazil is ethnicity. In SC state, all patients who self-declared as African descendants or Amerindians were infected with HIV-1C. Although this demographic feature does not represent an HIV exposure group, factors related to ethnicity or socioeconomic group might be segregating HIV-1 subtypes in separate epidemics. Future studies better designed to capture representative samples from different socioeconomic groups may be more effective in clarifying this issue.
The phylodynamic analysis performed here reconstructed distinct Ne trajectories for HIV-1C and HIV-1B since the early 1990s (Fig. 3a). Before then, the rates of both epidemics increased very similarly, as also captured by the logistic parametric model (Fig. 3b). However, from the early 1990s, HIV-1C experienced a continued expansion for >15 years, with a decrease of the growth rate only in the mid-2000s. The similar curve of AIDS incidence in HET and the association of HIV-1C infections with this group indicate that the HIV-1C epidemic in Brazil might have been propelled by heterosexual transmissions. Although our sampling data did not find a segregated epidemic in RS, other studies with samples from the early 2000s did (35, 36, 44), which supports that in RS, HIV-1C also disseminated mainly among HET in the past. In fact, in Porto Alegre (capital city of RS), a study reported that the prevalence of HIV-1C significantly increased in HET but also subtly increased in MSM in the period from 1998 to 2008, while at the same time, the prevalence of HIV-1B significantly decreased in both groups (43). Our phylodynamic analysis also captured a shrinkage in HIV-1B Ne, which could reflect competition with HIV-1C spreading throughout the MSM group after a period of intense transitions from HET to MSM (likely fostered by bisexual activity) in the late 1990s (Fig. 3a). In our sampling, representative mostly of the early 2010s, 64% of MSM were infected with HIV-1C, reinforcing that this subtype was also effective in dispersing in this group.
Although our findings highlight the success of subtype C in the HIV-1 epidemic in south Brazil, this does not mean that HIV-1C is spreading faster than subtype B. The logistic growth rates estimated here for HIV-1C (0.53 year−1; 95% HPD, 0.43 to 0.63) and HIV-1B (0.49 year−1; 95% HPD, 0.36 to 0.63) were similar, corroborating data reported previously by Bello et al., who also did not find a significant difference between rates of epidemic expansion of HIV-1C and -B (22, 45). Rather, it is conceivable that an efficient introduction of HIV-1C in the HET group allowed progressive spread through a large and expanding network of host individuals, more recently also effectively spreading among MSM. This scenario might have fostered HIV-1C to continuously disseminate in south Brazil and, more recently, also to other parts of the country (5). As recently reported, RS was the likely point of onset of the HIV-1C epidemic in Brazil and has spread northward from that state (46). Because progressive intermixing of partially separated epidemics seems to be a natural trend (47), the segregation of HIV-1C in the HET group can no longer be observed in RS. In time, it should also weaken the association among subtypes and exposure categories in other locations.
It is also interesting to note that HET transmission might be driving HIV-1C epidemic expansion in other parts of the country. In the southern states, where HIV-1C prevails, numbers of cases of AIDS in HET are 4 to 6 times higher than in MSM (6). In turn, in southeastern states, where the frequency of HIV-1C is <10% (5), the HET/MSM ratio is 1.5 to 3.5. Finally, in the central western region, where recent studies reported a frequency of HIV-1C of up to 20%, the HET/MSM ratio of AIDS cases is 4 to 8 (5, 6). These epidemiological data, taken together with our results, support HET transmission as a factor impacting HIV-1C diffusion in Brazil, responsible for successful introduction and establishment in the southern region, the recent spread to the central western states, and imposing constraints to viral dispersion toward the southeast.
In summary, our study depicts the molecular epidemic in several cities in SC and RS states. This is the first time that such a broad sampling area was investigated in south Brazil. Although the sample size from some of the cities is small (e.g., Santa Maria and Lages), we were able to capture the diversity of HIV-1 in the region. Here, we describe the epidemic growth trends of HIV-1C and -B, showing how MSM and HET transmission chains might have impacted the phylodynamics of the viral populations. Our analyses, distinct from previous studies, provided more precise estimations, mainly because our demographic reconstruction was informed by two loci analyzed as separate partitions but linked by the coalescent prior. This approach highlights the importance of sequencing more than one gene when it comes to phylodynamic analysis applied to HIV. Due to the small sample size, estimations for HIV-1B showed larger HPD intervals than those for HIV-1C (Fig. 3a). Because of this, many of the Ne trajectories are overlapping, even though a distinct trend has been observed since the 1990s. In addition, we also described potential new circulating BC mosaic strains whose recombination breakpoints can be derived from CRF31_BC. The region studied here is the region most heavily affected by AIDS in Brazil, and our findings can be valuable for the development of new public health policies that aim to slow HIV dissemination in south Brazil and beyond.
ACKNOWLEDGMENTS
We thank all collaborating municipal health centers from the states of Santa Catarina and Rio Grande do Sul, Brazil. We also thank Philippe Lemey for his helpful suggestions and assistance.
We have no conflicts of interest.
Funding Statement
Tiago Gräf received funding in 2016 from the VIROGENESIS Project as part of the European Union's Horizon 2020 Research and Innovation Programme under grant agreement 634650 and from a Flagship grant from the Medical Research Council (MRC) of the Republic of South Africa (MRC-RFA-FSP-01-2013/UKZN HIVEPI).
REFERENCES
- 1.Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 2.Hemelaar J. 2013. Implications of HIV diversity for the HIV-1 pandemic. J Infect 66:391–400. doi: 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Vuilleumier S, Bonhoeffer S. 2015. Contribution of recombination to the evolutionary history of HIV. Curr Opin HIV AIDS 10:84–89. doi: 10.1097/COH.0000000000000137. [DOI] [PubMed] [Google Scholar]
- 4.Alencar CS, Sabino EC, Carvalho SMF, Leao SC, Carneiro-Proietti AB, Capuani L, Oliveira CL, Carrick D, Birch RJ, Gonçalez TT, Keating S, Swanson PA, Hackett J, Busch MP, NHLBI Retrovirus Epidemiology Donor Study-II, International Component. 2013. HIV genotypes and primary drug resistance among HIV-seropositive blood donors in Brazil: role of infected blood donors as sentinel populations for molecular surveillance of HIV. J Acquir Immune Defic Syndr 63:387–392. doi: 10.1097/QAI.0b013e31828ff979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gräf T, Pinto AR. 2013. The increasing prevalence of HIV-1 subtype C in Southern Brazil and its dispersion through the continent. Virology 435:170–178. doi: 10.1016/j.virol.2012.08.048. [DOI] [PubMed] [Google Scholar]
- 6.Brazilian Ministry of Health. 2015. AIDS epidemiological bulletin July 2014-June 2015. Brazilian Ministry of Health, Brasília, DF, Brazil. [Google Scholar]
- 7.Brazilian Ministry of Health. 2009. AIDS epidemiological bulletin July 2008-June 2009. Brazilian Ministry of Health, Brasília, DF, Brazil. [Google Scholar]
- 8.de Oliveira T, Pillay D, Gifford RJ. 2010. The HIV-1 subtype C epidemic in South America is linked to the United Kingdom. PLoS One 5:e9311. doi: 10.1371/journal.pone.0009311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veras NMC, Gray RR, de Macedo Brigido LF, Rodrigues R, Salemi M. 2011. High-resolution phylogenetics and phylogeography of human immunodeficiency virus type 1 subtype C epidemic in South America. J Gen Virol 92:1698–1709. doi: 10.1099/vir.0.028951-0. [DOI] [PubMed] [Google Scholar]
- 10.Bello G, Zanotto PMA, Iamarino A, Gräf T, Pinto AR, Couto-Fernandez JC, Morgado MG. 2012. Phylogeographic analysis of HIV-1 subtype C dissemination in Southern Brazil. PLoS One 7:e35649. doi: 10.1371/journal.pone.0035649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delatorre E, Couto-Fernandez JC, Guimarães ML, Vaz Cardoso LP, de Alcantara KC, Stefani MMDA, Romero H, Freire CCM, Iamarino A, de A Zanotto PM, Morgado MG, Bello G. 2013. Tracing the origin and northward dissemination dynamics of HIV-1 subtype C in Brazil. PLoS One 8:e74072. doi: 10.1371/journal.pone.0074072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones LR, Dilernia DA, Manrique JM, Moretti F, Salomón H, Gomez-Carrillo M. 2009. In-depth analysis of the origins of HIV type 1 subtype C in South America. AIDS Res Hum Retroviruses 25:951–959. doi: 10.1089/aid.2008.0293. [DOI] [PubMed] [Google Scholar]
- 13.Carrion G, Aguayo N, Avila MM, Ruchansky D, Pando MA, Vinoles J, Perez J, Barboza A, Chauca G, Romero A, Galeano A, Blair PJ, Weissenbacher M, Birx DL, Sanchez JL, Olson JG, Carr JK. 2004. Documentation of subtype C HIV type 1 strains in Argentina, Paraguay, and Uruguay. AIDS Res Hum Retroviruses 20:1022–1025. doi: 10.1089/aid.2004.20.1022. [DOI] [PubMed] [Google Scholar]
- 14.Bello G, Passaes CP, Guimarães ML, Lorete RS, de Matos Almeida SE, Medeiros RM, Alencastro PR, Morgado MG. 2008. Origin and evolutionary history of HIV-1 subtype C in Brazil. AIDS 22:1993–2000. doi: 10.1097/QAD.0b013e328315e0aa. [DOI] [PubMed] [Google Scholar]
- 15.de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, Seebregts C, Snoeck J, van Rensburg EJ, Wensing AMJ, van de Vijver DA, Boucher CA, Camacho R, Vandamme AM. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 16.Pond SLK, Posada D, Stawiski E, Chappey C, Poon AFY, Hughes G, Fearnhill E, Gravenor MB, Brown AJL, Frost SDW. 2009. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol 5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siepel AC, Halpern AL, Macken C, Korber BT. 1995. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res Hum Retroviruses 11:1413–1416. doi: 10.1089/aid.1995.11.1413. [DOI] [PubMed] [Google Scholar]
- 18.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 20.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bello G, Eyer-Silva WA, Couto-Fernandez JC, Guimarães ML, Chequer-Fernandez SL, Teixeira SLM, Morgado MG. 2007. Demographic history of HIV-1 subtypes B and F in Brazil. Infect Genet Evol 7:263–270. doi: 10.1016/j.meegid.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol 30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. 2013. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol 30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien JD, Minin VN, Suchard MA. 2009. Learning to count: robust estimates for labeled distances between molecular sequences. Mol Biol Evol 26:801–814. doi: 10.1093/molbev/msp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelman A, Jakulin A, Pittau MG, Su YS. 2008. A weakly informative default prior distribution for logistic and other regression models. Ann Appl Stat 2:1360–1383. doi: 10.1214/08-AOAS191. [DOI] [Google Scholar]
- 29.Langford SE, Ananworanich J, Cooper DA. 2007. Predictors of disease progression in HIV infection: a review. AIDS Res Ther 4:11. doi: 10.1186/1742-6405-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gräf T, Passaes CPB, Ferreira LGE, Grisard EC, Morgado MG, Bello G, Pinto AR. 2011. HIV-1 genetic diversity and drug resistance among treatment naïve patients from Southern Brazil: an association of HIV-1 subtypes with exposure categories. J Clin Virol 51:186–191. doi: 10.1016/j.jcv.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Locateli D, Stoco PH, De Queiroz ATL, Alcântara LCJ, Ferreira LGE, Zanetti CR, Rodrigues R, Grisard EC, Pinto AR. 2007. Molecular epidemiology of HIV-1 in Santa Catarina State confirms increases of subtype C in southern Brazil. J Med Virol 79:1455–1463. doi: 10.1002/jmv.20955. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigues R, Manenti S, Romao PRT, de Paula Ferreira JL, Batista JPG, Siqueira AFAC, de Macedo Brigido LF. 2010. Young pregnant women living with HIV/AIDS in Criciuma, Southern Brazil, are infected almost exclusively with HIV type 1 clade C. AIDS Res Hum Retroviruses 26:351–357. doi: 10.1089/aid.2009.0214. [DOI] [PubMed] [Google Scholar]
- 33.Brigido LFM, Nunes CC, Oliveira CM, Knoll RK, Ferreira JLP, Freitas CA, Alves MA, Dias C. 2007. HIV type 1 subtype C and CB Pol recombinants prevail at the cities with the highest AIDS prevalence rate in Brazil. AIDS Res Hum Retroviruses 23:1579–1585. doi: 10.1089/aid.2007.0102. [DOI] [PubMed] [Google Scholar]
- 34.de Medeiros RM, Junqueira DM, Matte MCC, Barcellos NT, Chies JAB, de Matos Almeida SE. 2011. Co-circulation HIV-1 subtypes B, C, and CRF31-BC in a drug-naïve population from southernmost Brazil: analysis of primary resistance mutations. J Med Virol 83:1682–1688. doi: 10.1002/jmv.22188. [DOI] [PubMed] [Google Scholar]
- 35.Silveira J, Santos AF, Martínez AMB, Góes LR, Mendoza-Sassi R, Muniz CP, Tupinambás U, Soares MA, Greco DB. 2012. Heterosexual transmission of human immunodeficiency virus type 1 subtype C in southern Brazil. J Clin Virol 54:36–41. doi: 10.1016/j.jcv.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 36.Dias CF, Nunes CC, Freitas IO, Lamego IS, De Oliveira IMR, Gilli S, Rodrigues R, Brigido LF. 2009. High prevalence and association of HIV-1 non-B subtype with specific sexual transmission risk among antiretroviral naïve patients in Porto Alegre, RS, Brazil. Rev Inst Med Trop Sao Paulo 51:191–196. [DOI] [PubMed] [Google Scholar]
- 37.Brindeiro RM, Diaz RS, Sabino EC, Morgado MG, Pires IL, Brigido L, Dantas MC, Barreira D, Teixeira PR, Tanuri A. 2003. Brazilian Network for HIV Drug Resistance Surveillance (HIV-BResNet): a survey of chronically infected individuals. AIDS 17:1063–1069. doi: 10.1097/00002030-200305020-00016. [DOI] [PubMed] [Google Scholar]
- 38.Bongertz V, Bou-Habib DC, Brígido LF, Caseiro M, Chequer PJ, Couto-Fernandez JC, Ferreira PC, Galvão-Castro B, Greco D, Guimarães ML, Linhares de Carvalho MI, Morgado MG, Oliveira CA, Osmanov S, Ramos CA, Rossini M, Sabino E, Tanuri A, Ueda M. 2000. HIV-1 diversity in Brazil: genetic, biologic, and immunologic characterization of HIV-1 strains in three potential HIV vaccine evaluation sites. Brazilian Network for HIV Isolation and Characterization. J Acquir Immune Defic Syndr 23:184–193. doi: 10.1097/00042560-200002010-00011. [DOI] [PubMed] [Google Scholar]
- 39.Pando MA, Gómez-Carrillo M, Vignoles M, Rubio AE, dos Ramos Farias MS, Vila M, Rossi D, Ralón G, Marone R, Reynaga E, Sosa J, Torres O, Maestri M, Avila MM, Salomón H. 2011. Incidence of HIV type 1 infection, antiretroviral drug resistance, and molecular characterization in newly diagnosed individuals in Argentina: a Global Fund Project. AIDS Res Hum Retroviruses 27:17–23. doi: 10.1089/aid.2010.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon D, Béria JU, Tietzmann DC, de Carli R, Stein AT, Lunge VR. 2010. Prevalência de subtipos do HIV-1 em amostra de pacientes de um centro urbano no sul do Brasil. Rev Saude Publica 44:1094–1101. doi: 10.1590/S0034-89102010005000039. [DOI] [PubMed] [Google Scholar]
- 41.Passaes CPB, Bello G, Lorete RS, de Matos Almeida SE, Junqueira DM, Veloso VG, Morgado MG, Guimarães ML. 2009. Genetic characterization of HIV-1 BC recombinants and evolutionary history of the CRF31_BC in Southern Brazil. Infect Genet Evol 9:474–482. doi: 10.1016/j.meegid.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Chen M, Yang L, Ma Y, Su Y, Yang C, Luo H, Chen H, Chen L, Yan W, Shi Y, Jia M, Lu L. 2013. Emerging variability in HIV-1 genetics among recently infected individuals in Yunnan, China. PLoS One 8:e60101. doi: 10.1371/journal.pone.0060101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almeida SE, de Medeiros RM, Junqueira DM, Gräf T, Passaes CPB, Bello G, Morgado MG, Guimarães ML. 2012. Temporal dynamics of HIV-1 circulating subtypes in distinct exposure categories in southern Brazil. Virol J 9:306. doi: 10.1186/1743-422X-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soares MA, De Oliveira T, Brindeiro RM, Diaz RS, Sabino EC, Brigido L, Pires IL, Morgado MG, Dantas MC, Barreira D, Teixeira PR, Cassol S, Tanuri A, Brazilian Network for Drug Resistance Surveillance . 2003. A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS 17:11–21. doi: 10.1097/00002030-200301030-00004. [DOI] [PubMed] [Google Scholar]
- 45.Bello G, Guimarães ML, Passaes CPB, de Matos Almeida SE, Veloso VG, Morgado MG. 2009. Evidences of recent decline in the expansion rate of the HIV type 1 subtype C and CRF31_BC epidemics in southern Brazil. AIDS Res Hum Retroviruses 25:1065–1069. doi: 10.1089/aid.2009.0106. [DOI] [PubMed] [Google Scholar]
- 46.Gräf T, Vrancken B, Maletich Junqueira D, de Medeiros RM, Suchard MA, Lemey P, Esteves de Matos Almeida S, Pinto AR. 2015. Contribution of epidemiological predictors in unraveling the phylogeographic history of HIV-1 subtype C in Brazil. J Virol 89:12341–12348. doi: 10.1128/JVI.01681-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tovanabutra S, Beyrer C, Sakkhachornphop S, Razak MH, Ramos GL, Vongchak T, Rungruengthanakit K, Saokhieo P, Tejafong K, Kim B, De Souza M, Robb ML, Birx DL, Jittiwutikarn J, Suriyanon V, Celentano DD, McCutchan FE. 2004. The changing molecular epidemiology of HIV type 1 among northern Thai drug users, 1999 to 2002. AIDS Res Hum Retroviruses 20:465–475. doi: 10.1089/088922204323087705. [DOI] [PubMed] [Google Scholar]