ABSTRACT
The interferon-induced transmembrane protein (IFITM) family of proteins inhibit infection of several different enveloped viruses in cell culture by virtue of their ability to restrict entry and fusion from late endosomes. As few studies have evaluated the importance of Ifitm3 in vivo in restricting viral pathogenesis, we investigated its significance as an antiviral gene against West Nile virus (WNV), an encephalitic flavivirus, in cells and mice. Ifitm3−/− mice were more vulnerable to lethal WNV infection, and this was associated with greater virus accumulation in peripheral organs and central nervous system tissues. As no difference in viral burden in the brain or spinal cord was observed after direct intracranial inoculation, Ifitm3 likely functions as an antiviral protein in nonneuronal cells. Consistent with this, Ifitm3−/− fibroblasts but not dendritic cells resulted in higher yields of WNV in multistep growth analyses. Moreover, transcomplementation experiments showed that Ifitm3 inhibited WNV infection independently of Ifitm1, Ifitm2, Ifitm5, and Ifitm6. Beyond a direct effect on viral infection in cells, analysis of the immune response in WNV-infected Ifitm3−/− mice showed decreases in the total number of B cells, CD4+ T cells, and antigen-specific CD8+ T cells. Finally, bone marrow chimera experiments demonstrated that Ifitm3 functioned in both radioresistant and radiosensitive cells, as higher levels of WNV were observed in the brain only when Ifitm3 was absent from both compartments. Our analyses suggest that Ifitm3 restricts WNV pathogenesis likely through multiple mechanisms, including the direct control of infection in subsets of cells.
IMPORTANCE As part of the mammalian host response to viral infections, hundreds of interferon-stimulated genes (ISGs) are induced. The inhibitory activity of individual ISGs varies depending on the specific cell type and viral pathogen. Among ISGs, the genes encoding interferon-induced transmembrane protein (IFITM) have been reported to inhibit multiple families of viruses in cell culture. However, few reports have evaluated the impact of IFITM genes on viral pathogenesis in vivo. In this study, we characterized the antiviral activity of Ifitm3 against West Nile virus (WNV), an encephalitic flavivirus, using mice with a targeted gene deletion of Ifitm3. Based on extensive virological and immunological analyses, we determined that Ifitm3 protects mice from WNV-induced mortality by restricting virus accumulation in peripheral organs and, subsequently, in central nervous system tissues. Our data suggest that Ifitm3 restricts WNV pathogenesis by multiple mechanisms and functions in part by controlling infection in different cell types.
INTRODUCTION
The interferon (IFN)-induced transmembrane protein (IFITM) genes consist of a family of genes encoding related proteins: Ifitm1, 2, 3, 5, 6, 7, and 10 in mice and IFITM1, 2, 3, 5, and 10 in humans (1, 2). The expression of several IFITM genes (e.g., those encoding IFITM1, 2, and 3) can be induced by type I, II, or III IFNs (3, 4). Although initial studies described possible roles of IFITM1, IFITM2, and IFITM3 in development, apoptosis, cell proliferation, and cell signaling (5–13), a subsequent report suggested that ectopic expression of IFITM1 in mouse L cells could restrict infection of vesicular stomatitis virus (VSV) (14). A decade later, gene silencing and ectopic expression studies established that IFITM proteins have antiviral activity in cell culture against members of the Flaviviridae, Orthomyxoviridae, Filoviridae, Rhabdoviridae, Retroviridae, Bunyaviridae, Reoviridae, Togaviridae, and Paramyxoviridae families (15–28). Nonetheless, some enveloped and nonenveloped viruses appear to be resistant to the actions of IFITM proteins, including arenaviruses, papillomaviruses, cytomegaloviruses, and adenoviruses (16, 17, 29).
IFITM proteins are transmembrane proteins. Although their precise membrane topology remains uncertain (5, 6, 15, 30–36), recent studies suggest that they are type II membrane proteins (35–37). Moreover, the cellular sublocalization of the IFITM proteins varies among family members, with IFITM1 expressed primarily at the plasma membrane, and IFITM2 and IFITM3 colocalizing predominantly with late endosomes (20, 32). Based on the cellular localization and effects on specific steps in viral life cycles, IFITM1, IFITM2, and IFITM3 appear to restrict fusion and uncoating of viruses into the cytoplasm (33, 38, 39), with different IFITM proteins inhibiting specific viruses in distinct membrane compartments. Despite the intensive study of the IFITM proteins in cell culture, the precise mechanism of restriction of viral fusion has remained elusive. It has been suggested that IFITM proteins can increase cholesterol accumulation in endosomes, alter membrane fluidity, or make fusion events energetically unfavorable (17, 33, 38, 40). IFITM1, IFITM2, and IFITM3 also can become incorporated into virions and restrict viral infection, as has been demonstrated with HIV (41, 42).
Although IFITM proteins can restrict infection of many viruses in cell culture, their importance in vivo in the context of a complex IFN response with hundreds of other interferon-stimulated genes (ISGs) remains less well characterized. Two publications have reported that Ifitm3−/− mice are more susceptible to influenza A virus (IAV) infection (3, 43). These studies described increased IAV titers in the lung, increased pathology, and decreased CD4+ T cells, CD8+ T cells, and NK cells in Ifitm3−/− compared to wild-type (WT) mice. One of these studies described a human polymorphism in IFITM3 (SNP-rs12252-C, where SNP is single nucleotide polymorphism) that results in an altered splice acceptor site, which truncates the N-terminal 21 amino acids of IFITM3. This truncated IFITM3 protein showed altered cellular localization and reduced antiviral activity against IAV (32, 43, 44). A second study demonstrated that CD8+ resident memory T cells expressed high levels of Ifitm3 in the lung following IAV infection and that Ifitm3 expression was important for memory T cell survival against virus rechallenge (45). Ifitm3 also reportedly has an antiviral role against respiratory syncytial virus in vivo, as Ifitm3−/− mice sustained a higher viral burden in the lungs than did WT mice (46). To date, no studies have described an antiviral role of Ifitm3 in vivo apart from viruses that preferentially infect the lung.
West Nile virus (WNV) is a neurotropic, mosquito-transmitted, positive-stranded, enveloped RNA virus in the Flaviviridae family, which includes several viruses of global concern such as dengue (DENV), Zika (ZIKV), yellow fever (YFV), and Japanese encephalitis (JEV) viruses. Whereas most infections with WNV in humans are asymptomatic, ∼30% develop a febrile illness, which can progress to severe neurological disease, including meningitis, flaccid paralysis, encephalitis, and death (47, 48). Several studies have established that IFN signaling and induction of downstream antiviral effector proteins (e.g., IFIT2, viperin, protein kinase R [PKR], RNase L, and Ifi27l2a) restrict the tropism and dissemination of WNV (49–52). Here, we examined the role of Ifitm3 in vivo in restricting infection of WNV using Ifitm3−/− mice. Extensive virological and immunological analysis revealed that Ifitm3−/− mice were more vulnerable to WNV infection than WT mice, with greater lethality, higher viral burden, and altered immune induction. Our study demonstrates a contribution of Ifitm3 to controlling WNV in peripheral organs prior to dissemination to the brain and infection and injury of target neuron populations.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (Assurance number A3381-01). Dissections and footpad injections were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize suffering.
Virus propagation.
The WNV strain New York 1999 (53, 54) was passaged in Vero cells to generate a mammalian-cell-derived stock. The WNV strain from Madagascar (DakAnMg 798, WNV-MAD) was isolated in 1978 and also passaged in Vero cells (55). Titration of viral stocks was performed using a focus-forming assay as described previously (56).
Mouse experiments and tissue preparation.
Wild-type C57BL/6 (000664) or B6.SJL (002014) mice were purchased from Jackson Laboratory. Ifitm3−/− mice were generated previously (57), backcrossed using speed congenics onto a C57BL/6J background, and bred in a pathogen-free animal facility at the Washington University School of Medicine. Eight- to 10-week-old age- and sex-matched mice were used. Mice were infected via a subcutaneous (102 focus-forming units [FFU] in 50 μl) or intracranial (101 FFU in 10 μl) route with virus diluted in phosphate-buffered saline (PBS). For survival studies, infected mice were monitored for 21 days. For viral-burden studies, at specified time points after infection, serum was collected and animals were perfused with 20 ml of PBS. Subsequently, the spleen, kidney, draining lymph node, brain, and spinal cord were harvested, weighed, homogenized by bead dissociation using a MagNA Lyzer (Roche). The virus titer was determined by plaque assay on Vero cells (58, 59), or levels of viral RNA were measured by reverse transcription-quantitative PCR (qRT-PCR) as described previously (49, 60).
Measurement of WNV-specific antibodies.
WNV-specific IgM and IgG titers were determined by an enzyme-linked immunosorbent assay (ELISA) using purified WNV E protein as described previously (61). Focus reduction neutralization assays were performed on Vero cells after mixing serial dilutions of serum with a fixed amount (102 FFU) of WNV (56).
Analysis of cellular immune responses.
Splenocytes were harvested from WT or Ifitm3−/− mice on day 7 or 8 after WNV infection. Lymphocyte populations were stained using anti-CD3-V500 (560711; BD Bioscience), anti-CD8α- or anti- CD8β-peridinin chlorophyll protein (PerCP) Cy5.5 (100734; Biolegend), anti-CD19-Alexa Fluor 700 (115528; Biolegend), anti-CD4-BV421 (100437; Biolegend), anti-granzyme B (GRB04; Invitrogen), and tetramers specific for a Db-restricted immunodominant peptide in NS4B (62). Brains were harvested from WT or Ifitm3−/− mice on day 8 after WNV infection. Central nervous system (CNS) leukocytes were isolated by Percoll gradient centrifugation as described previously (62, 63) and stained with the antibodies and tetramer described above in addition to anti-CD11b-phycoerythrin (PE)-Cy7 (101216; Biolegend) and anti-CD45-BV605 (103139; Biolegend). Cells were analyzed on a BD LSRII flow cytometer, and data were processed using FlowJo software.
To examine intracellular cytokine production by CD4+ and CD8+ T cells, anti-CD3 or antigen-specific peptide restimulation was performed (62). In brief, splenocytes were isolated and stimulated with anti-CD3 (100207; Biolegend) or the immunodominant NS4B peptide or were subjected to no stimulation. Cells were incubated for 6 h at 37°C in the presence of brefeldin A and then fixed and permeabilized using the FoxP3/transcription factor staining buffer set (00-5523-00; eBioscience). Subsequently, cells were stained with anti-CD4-BV421 (100437; Biolegend), anti-CD8β-PerCP-Cy5.5 (126609; Biolegend), anti-CD19-Alexa Fluor 700 (115528; Biolegend), anti-IFN-γ-APC (51-73-1182; eBioscience), and anti-tumor necrosis factor alpha-phycoerythrin (anti-TNF-α-PE; 506306l; Biolegend) antibodies and analyzed.
Primary cell isolation and infection.
Primary macrophages (Mϕ), dendritic cells (DCs), and mouse embryonic fibroblasts (MEFs) were generated from WT and Ifitm3−/− mice as described previously (64). For single-step and multistep growth curves, cells were infected at a multiplicity of infection (MOI) of 5 and 0.01, respectively. In some experiments, cells were pretreated with IFN-β (12400-1; PBL Assay Science) for 16 h before infection. Viral supernatants were harvested, and their titers were determined using a focus-forming assay.
Transformation of MEFs.
MEFs were transfected with the plasmid SV2, which encodes the large T antigen of simian virus 40 (SV40) polyomavirus (65). Cells were passaged ∼10 times and used for subsequent experiments.
Transcomplementation of MEFs. (i) Production of pFCIV-containing lentivirus.
293T cells were seeded at 5 × 106 cells per well in a 6-well plate. One day later, cells were transfected with 0.4 μg of pMD2.G (Addgene 12259), 0.8 μg of pSPAX2 (Addgene 12260), and 0.8 μg of pFCIV-c-Myc-Ifitm3 or pFCIV-c-Myc-Firefly luciferase using FuGENE (Roche) according to the manufacturer's instructions (66, 67). Two to 3 days after transfection, lentivirus was collected from the supernatant of cells, centrifuged at 1,000 × g for 10 min at 4°C to remove cellular debris, and then stored at −80°C.
(ii) Generation of MEF transfectants.
Transformed MEFs were seeded at 0.5 × 104 cells per well in a 96-well plate. Six hours after plating, 100 μl of lentivirus and 1 μg Polybrene (sc-134220; Santa Cruz Biotech) were added to each well, and cells were spinoculated at 1,000 × g for 30 min at 24°C. Six hours later, lentivirus was removed and replaced with Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). Cells were passaged and expanded into a T-75 tissue culture flask. Transduced cells were sorted for green fluorescent protein (GFP) expression (pFCIV encodes GFP under an internal ribosome entry site [IRES] promoter) on a BD FACsAria II flow cytometer. Cells were passaged five times to confirm stable expression of GFP and c-Myc-tagged proteins using flow cytometry and then used for subsequent experiments.
Western blotting of Ifitm3 expression.
WT or Ifitm3−/− MEFs, or Ifitm3−/− MEFs transcomplemented with c-Myc-Ifitm3 were seeded at 5 × 104 cells per well in a 24-well plate. Cells were stimulated with or without 10 U/ml of IFN-β for 16 h at 37°C. Cells were washed with PBS and then lysed in 200 μl of radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors (9806S; Cell Signaling). Cellular debris was removed from the sample by centrifugation at 10,000 × g for 10 min at 4°C. The clarified supernatant was resuspended in 4× LDS sample buffer (NP0008; NuPAGE), boiled (5 min at 90°C), and then electrophoresed on a 12% bis-Tris gel (NP0343BOX; NuPAGE) at 200 V for 40 min in MES (morpholineethanesulfonic acid) buffer (NP0002; NuPAGE). Protein was transferred to a polyvinylidene difluoride (PVDF) membrane (IB24002; Invitrogen) using an Iblot2 (IB21001; Life Technologies) with a standard 7-min transfer. A 1:1,000 dilution of mouse anti-β-actin (11714-1-AP; Proteintech) and rabbit anti-Ifitm3 (3700; Cell Signaling) was prepared in Tris-buffered saline with Tween 20 (TBST) with 4% milk and used to stain the membrane overnight at room temperature. Subsequently, after washing, a 1:10,000 dilution of donkey anti-mouse IRDye 800 (925-32212; LI-COR) or anti-rabbit IRdye 680 (926-68073; LI-COR) was prepared in TBST with 4% milk and used to stain the membrane for 2 h at room temperature. An Odyssey machine (LI-COR Biosciences) was used to visualize the bands on the membrane.
Infection of MEFs.
WT, gene deletion, and transcomplemented MEFs were seeded at 2 × 104 cells per 96-well plate. Sixteen hours later, cells were infected with WNV at an MOI of 5. Twenty-four hours later, cells were harvested, fixed, and permeabilized using the Foxp3/transcription factor staining buffer set. Subsequently, cells were incubated with rabbit anti-c-Myc (71D10; Cell Signaling) antibody or humanized E16 (68), a WNV E protein-specific monoclonal antibody. After cells were washed, Alexa Fluor 647-conjugated anti-rabbit-IgG (A-21245; Invitrogen) and Alexa Fluor 568-conjugated anti-human IgG (A-21090; Invitrogen) secondary antibodies were added. Cells were analyzed on a BD LSRII flow cytometer array using FlowJo software.
Bone marrow chimera.
Ten million donor bone marrow cells from CD45.1 (B6.SJL) and CD45.2 (Ifitm3−/−) mice were transferred adoptively by intravenous injection into 8-week-old recipient B6.SJL and Ifitm3−/− mice that had been irradiated with 900 cGγ. Mice were infected 8 weeks later with WNV. Tissues were harvested 7 days after infection for viral burden and cellular immune response analysis as described above.
Statistical analysis.
Data were analyzed using Prism Software (GraphPad4, San Diego, CA). Kaplan-Meier survival curves were analyzed by the log rank test. A one-way or two-way analysis of variance (ANOVA) was used to determine statistically significant differences for in vitro viral growth experiments. The Mann-Whitney test was used to analyze differences in viral burden, cell numbers, and antibody titers. A one-way ANOVA with a multiple-comparison correction was used to analyze differences in viral burden, cell numbers, and antibody titers for bone marrow chimera experiments.
RESULTS
Ifitm3 restricts WNV infection and pathogenesis in mice.
Ifitm3 is an ISG that has been reported to restrict infection of several flaviviruses in cell culture, including WNV and DENV (15, 19, 21–23, 69–71). To determine the role of Ifitm3 in restricting pathogenesis of a flavivirus in vivo, we inoculated 8- to 9-week-old WT and congenic Ifitm3−/− C57BL/6 mice via a subcutaneous route with 102 FFU of a pathogenic strain of WNV (New York 1999). Ifitm3−/− mice were more vulnerable to WNV infection than WT mice, as a greater percentage of the former (90% versus 40% lethality, P < 0.001) succumbed to disease than the latter (Fig. 1A). The mean time to death of Ifitm3−/− was shorter than that of WT mice (10.6 versus 11.8 days, P < 0.05), which is consistent with an accelerated disease course. One to 2 days before death, WT and Ifitm3−/− mice that ultimately would die similarly exhibited fur ruffling, hunched posture, and diminished movement. Within 1 day of death, some of these animals developed clinical evidence of hind limb paralysis.
FIG 1.
Survival and viral burden analysis of WT and Ifitm3−/− mice. (A) Eight- to 10-week-old age-matched WT (n = 28) and Ifitm3−/− (n = 23) C57BL/6 mice were infected via the subcutaneous route with 102 FFU of WNV (New York 1999) and monitored for mortality for 21 days. Survival differences were analyzed by the log rank test (P < 0.005). (B to F) WNV tissue burden in WT and Ifitm3−/− mice after subcutaneous infection. WNV levels in serum (B), draining lymph node (C), spleen (D), brain (E), and spinal cord (F) were measured by qRT-PCR (B and C) or plaque assay (D to F). Solid lines represent the median viral titers, and dotted lines denote the limit of detection of the assay. Two or three mice were used per independent experiment,and 5 to 7 independent experiments were performed. Asterisks indicate statistically significant differences by the Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Ifitm3 limits WNV infection in different tissues.
To begin to determine how an absence of Ifitm3 results in enhanced WNV pathogenesis, we measured viral burden in the serum, spleen, kidney, draining lymph nodes, brain, and spinal cord at days 2, 4, 6, and 8 postinfection after subcutaneous infection.
Relatively small, yet statistically significant differences in viral burden were observed in some peripheral organs. For example, viremia was equivalent at day 2 after WNV infection in the two sets of mice but higher at days 4 and 6 in Ifitm3−/− than in WT mice (Fig. 1B, P < 0.01). Increased viral titers also were detected in the draining lymph node (DLN) on days 6 and 8 after infection (Fig. 1C, P < 0.05). However, an increase in WNV infection was not detected in the spleen at any of the time points (Fig. 1D, P > 0.05). Viral yield remained near the limit of detection in the kidney of Ifitm3−/− mice (data not shown); in WT mice, this organ is normally resistant to WNV infection, but it becomes susceptible in the absence of an intact type I IFN signaling response (72, 73).
We next assessed the impact of Ifitm3 on viral replication in the brain and spinal cord, which are targets of WNV infection in mice and humans (74). Higher viral titers were observed in the brains of Ifitm3−/− than in those of WT mice at day 6 after infection (Fig. 1E, P < 0.05) and in the spinal cord of Ifitm3−/− mice on days 6 and 8 after infection (Fig. 1F, P < 0.05). Overall, an absence of Ifitm3 resulted in enhanced viral replication in the CNS.
Ifitm3 does not directly restrict WNV infection in the CNS.
The high WNV titers in the brain and spinal cord of Ifitm3−/− mice could reflect enhanced replication in peripheral organs and viremia, which results in increased seeding of CNS tissues. Alternatively, it could be due to intrinsic antiviral effects of Ifitm3 in cells of the CNS. Although Ifitm3 is expressed in neurons at baseline and after type I IFN induction (74), it remains unknown whether Ifitm3 has an antiviral role in neurons in vivo. To evaluate this question, WT and Ifitm3−/− mice were infected with 101 FFU of WNV directly into the cerebral cortex via an intracranial route, and viral burden in the cerebral cortex, white matter, brain stem, cerebellum, and spinal cord was measured on days 3 and 5 after infection. Notably, no difference in viral burden was detected in CNS tissues on day 3 or day 5 after infection (Fig. 2A to F, P > 0.05). Because the virulent strain of WNV strain New York can antagonize IFN responses (75, 76), we repeated intracranial infection studies with an attenuated strain (Madagascar 1978 [WNV-MAD]) that is more sensitive to type I IFN than strain New York (77). Again, no significant difference in viral burden was observed in CNS tissues of WNV-MAD-infected WT and Ifitm3−/− mice (Fig. 2G to L, P > 0.05). These data suggest that the increased CNS titers in Ifitm3−/− mice after subcutaneous WNV infection were not due to direct antiviral effects in neuronal cells.
FIG 2.
No difference in CNS viral burden via intracranial route of inoculation. Eight- to 10-week-old age-matched WT and Ifitm3−/− C57BL/6 mice were inoculated via an intracranial route with 101 FFU of WNV (New York 1999) (A to F) or WNV-MAD (Madagascar 1978) (G to L). Tissues were harvested at day 3 or day 5 after infection. WNV levels in olfactory bulb (A, G), brain stem (B, H), cerebellum (C, I), cerebral cortex white matter (D, J), cerebral cortex gray matter (E, K), and spinal cord (F, L) were measured by plaque assay. Solid lines represent the median viral titers, and dotted lines denote the limit of detection of the assay. Two or three mice were used per independent experiment, and 3 or 4 independent experiments were performed. None of the comparisons were statistically different as determined by the Mann-Whitney test (A to L, P > 0.05).
T cell responses in Ifitm3−/− mice.
A previous study reported reduced CD8+ T cell numbers in Ifitm3−/− mice during acute IAV infection (43). Another study demonstrated that lung resident memory CD8+ T cells with reduced Ifitm3 expression were vulnerable to IAV infection and were lost selectively during a secondary challenge (45). As depressed antiviral CD8+ T cells also can facilitate enhanced replication of WNV in the CNS (62, 78, 79), we investigated whether an absence of Ifitm3 influenced the development of an adaptive immune response during infection.
At day 7 after WNV infection, splenocytes were harvested and the percentage and number of bulk CD4+ and CD8+ T cells and NS4B tetramer-specific CD8+ T cells were determined. Although differences in T cell numbers were not detected at baseline in naive mice, the number of CD4+ T cells, CD8+ T cells, and NS4B tetramer-specific CD8 T cells was decreased in Ifitm3−/− mice after WNV infection (Fig. 3A to F and M). To assess whether antigen-specific CD8+ T cells showed qualitative defects in the absence of Ifitm3, splenocytes from WNV-infected mice were stimulated ex vivo with an immunodominant WNV NS4B peptide and tested for cytokine production. However, equivalent percentages, numbers, and geometric mean fluorescence intensities of IFN-γ- and TNF-α-producing CD8+ T cells were detected in WT and Ifitm3−/− mice (Fig. 3G to L and N, P > 0.05).
FIG 3.
Ifitm3−/− mice have blunted T cell responses in the spleen after WNV infection. Immune responses were examined in the spleen after subcutaneous inoculation of 102 FFU of WNV (New York 1999). Spleens were harvested on day 7 (A to H) or 8 (I to N) after infection. The percentage of live cells and total cell population of CD3+ CD4+ T cells (A and B), CD3+ CD8+ T cells (C and D), and NS4B-specific CD3+ CD8+ T cells (E and F) were measured. The percentage of CD8+ T cells and total numbers of CD3+ CD8+ T cells that expressed IFN-γ (G and H), TNF-α (I and J), or TNF-α and IFN-γ (K and L) after ex vivo stimulation with NS4B peptide are indicated. Representative flow cytometry plots of NS4B-specific CD3+ CD8+ T cells (M) and CD3+ CD8+ that express TNF-α and IFN-γ after peptide restimulation (N) are shown. Four or five mice were used per independent experiment, and 4 independent experiments were performed. Statistical significance was determined by the Mann-Whitney test (*, P < 0.05; **, P < 0.01).
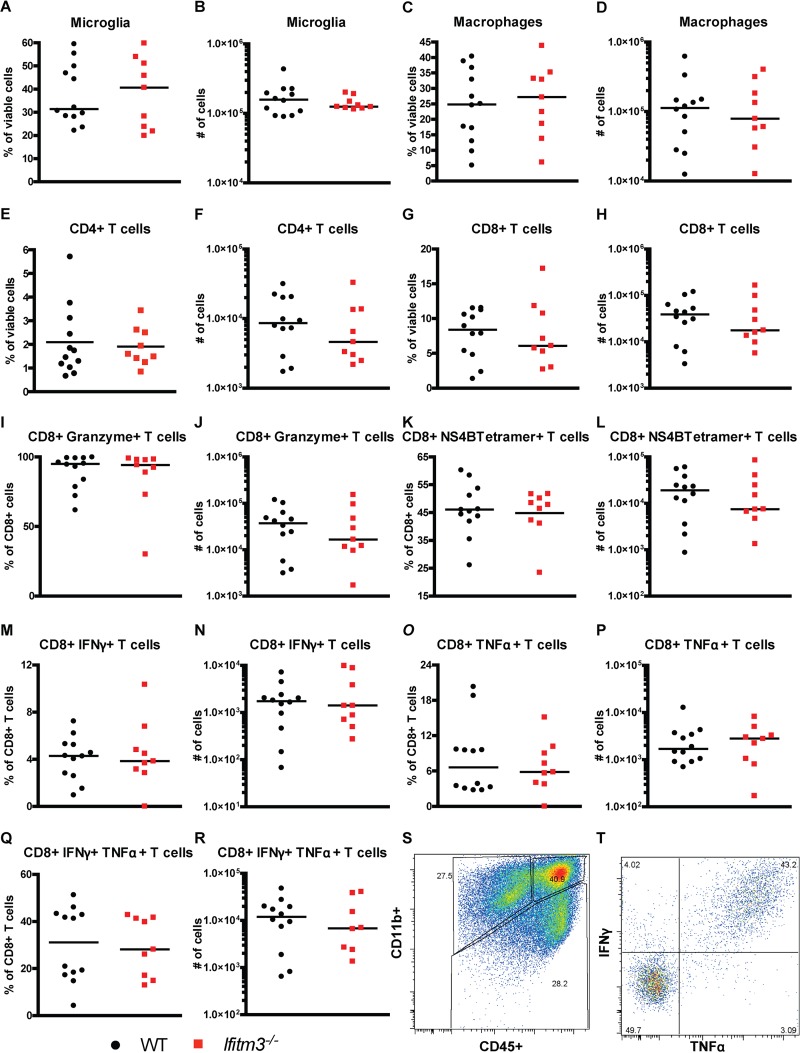
A small decrease in the number of antigen-specific CD8+ T cells in the periphery of Ifitm3−/− mice could impact the accumulation of these cells in the CNS and viral clearance. As such, we compared the numbers of immune cells in the brains of WT and Ifitm3−/− mice at day 8 after infection by flow cytometry. We did not observe differences in the numbers of activated microglia (CD11bhigh CD45low) or macrophages (CD11bhigh CD45high) (Fig. 4A to D and S, P > 0.05) in brains of WNV-infected WT and Ifitm3−/− mice. Moreover, similar percentages and numbers of NS4B tetramer+ and granzyme B+ CD8+ T cells were detected (Fig. 4E to L, P > 0.05). Also, after ex vivo restimulation with the NS4B peptide, equivalent percentages and numbers of IFN-γ- and TNF-α-secreting CD8+ T cells were measured from the brains of WT and Ifitm3−/− mice (Fig. 4M to R and T, P > 0.05). Thus, while a small decrease in CD8+ T cell response in the periphery of WNV-infected Ifitm3−/− mice was observed, it did not appear to impact the clearance of virus in the spleen (Fig. 1C) or the accumulation of antigen-specific immune cells in the brain.
FIG 4.
Ifitm3−/− mice have normal levels of immune cell populations in the brain after WNV infection. CNS immune responses were examined after subcutaneous inoculation of of 102 FFU of WNV (New York 1999). Brains were harvested day 8 postinfection. The percentage and total number of CD11b+ CD45lo microglia (A and B), CD11b+ CD45hi macrophages (C and D), CD4+ T cells (E and F), CD8+ T cells (G and H), CD8+ granzyme B+ T cells (I and J), and NS4B-specific CD8+ T cells (K and L) are shown. The percentage and total number of CD8+ T cells that expressed IFN-γ (M and N), TNF-α (O and P), or both IFN-γ and TNF-α (Q and R) after NS4B peptide restimulation are shown. Representative flow cytometry plots of myeloid cell gating (S) and CD8+ IFN-γ+ and TNF-α+ cells (T) are shown. Three mice were used per independent experiment, and 3 independent experiments were performed. None of the comparisons were statistically different as determined by the Mann-Whitney test (P > 0.05).
B cell responses in Ifitm3−/− mice.
Antiviral antibody responses restrict dissemination of WNV in vivo into the CNS (59, 80). As such, we determined whether Ifitm3 expression modulated antiviral antibody responses. Whereas baseline numbers of CD19+ B cells were similar in naive mice, we observed a decrease in their number in the spleen in Ifitm3−/− mice at day 7 after infection (Fig. 5A and B). To assess the effect on antiviral antibody responses, we analyzed sera from WT and Ifitm3−/− mice on day 8 after infection for binding to WNV E protein. Although anti-WNV IgM titers were equivalent in WT and Ifitm3−/− mice (Fig. 5C, P > 0.05), the anti-WNV IgG response was greater in Ifitm3−/− mice at day 8 after infection (Fig. 5D, P < 0.05), possibly due to the higher levels of virus in peripheral organs. To evaluate the functional quality of anti-WNV antibody from WT and Ifitm3−/− mice, we assessed neutralizing activity. No difference in inhibitory activity was detected in sera from WT and Ifitm3−/− mice at day 8 postinfection (Fig. 5E; P > 0.05). These data suggest that while there might be small effects of Ifitm3 on B cell responses, these do not translate into depressed antiviral antibody responses and thus likely do not contribute to the increased WNV infection observed in different organs in Ifitm3−/− mice.
FIG 5.
Ifitm3−/− mice have reduced B cell numbers but not reduced antiviral antibody titers. (A and B) Spleens were harvested at day 7 after subcutaneous inoculation of WNV (New York 1999). The percentage (A) and total number (B) of B cells were determined by flow cytometry. (C to E) Serum was harvested at day 8 after infection. (C and D) IgM and IgG antibody titers were determined using an ELISA against purified WNV E protein. (E) Serum neutralization activity was evaluated using a focus reduction neutralization test, and data were fitted by regression analysis to obtain 50% effective concentration (EC50) values. Two to 5 mice were used per independent experiment, and 4 independent experiments were performed. Statistical significance was determined by the Mann-Whitney test (*, P < 0.05).
Ifitm3 controls WNV infection in some primary cells.
To gain further insight in which cells require Ifitm3 for optimal restriction of WNV infection, we compared multistep growth kinetics and effects of IFN-β treatment in different primary cells derived from WT and Ifitm3−/− mice, including Mϕ, DCs, and fibroblasts (MEFs). Although we did not observe statistically increased viral yields in unstimulated Ifitm3−/− Mϕ (Fig. 6A), pretreatment of IFITM3−/− Mϕ with low doses of IFN-β to stimulate ISG production did result in marginally higher levels of virus than in similarly treated WT Mϕ (Fig. 6B and C). In comparison, unstimulated or IFN-β-treated Ifitm3−/− DCs did not support statistically increased WNV infection compared to WT cells (Fig. 6D to F). However, multistep growth analysis demonstrated an increased WNV yield in the supernatants of Ifitm3−/− compared to WT MEFs (Fig. 6G, P < 0.05), and this difference was amplified under conditions of IFN-β pretreatment (Fig. 6H, P < 0.05). Single-step growth kinetic analysis failed to demonstrate increased WNV yield of unstimulated IFITM3−/− MEFs (Fig. 6I, P > 0.05), although pretreatment with IFN-β did show an effect (Fig. 6J, P < 0.05). Thus, Ifitm3 restricts WNV infection in some cell types, although its expression must be induced to observe this phenotype.
FIG 6.
WNV infection in Mϕ, DC, and MEFs. Bone marrow-derived Mϕ (A to C) and DCs (D to F) were infected at an MOI of 0.01 with WNV (New York 1999), and MEFs (G to J) were infected at an MOI of 5 or 0.01 with WNV (New York 1999). In some experiments, cells were pretreated for 16 h with 0 0.1, 1, or 10 U/ml of IFN-β. Viral titers were determined by focus-forming assays, and the results reflect at least three independent experiments. (K to M) Transformed WT, Ifitm12356−/−, or Ifitm3−/− MEFs were transduced with lentiviruses expressing a c-Myc-tagged Ifitm3 or firefly luciferase (FLUC). Lentivirus-mediated expression of Ifitm3 was validated by Western blotting and compared to endogenous Ifitm3 expression levels in WT MEFs pretreated for 16 h with 10 U/ml of IFN-β (K). Transduced MEFs were infected with WNV (New York 1999) at an MOI of 0.01 for 24 h. Cells were harvested, and flow cytometry was used to detect expression of c-Myc-tagged proteins and WNV-infected cells. Shown are representative flow cytometry plots (L) and pooled data (M) from at least 3 independent experiments performed in duplicate. Statistical significance was determined by a one-way (M) or two-way (A to J) ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To confirm that the increased permissiveness of Ifitm3−/− MEFs for WNV was due to the loss of Ifitm3, we transcomplemented cells with an N-terminal c-Myc-tagged form of Ifitm3 (c-Myc-Ifitm3) and compared this to a control c-Myc-tagged form of firefly luciferase (c-Myc-FLUC) (Fig. 6K to M). Lentivirus-mediated expression of c-Myc-tagged Ifitm3 first was validated by Western blotting (Fig. 6K). Untransduced and c-Myc-FLUC-transduced Ifitm3−/− and Ifitm12356−/− (locus deletion) MEFs sustained higher WNV infection as judged by viral antigen staining and flow cytometry than did the comparable Ifitm3-sufficient WT MEFs (3- to 4-fold, P < 0.05). However, ectopic expression of c-Myc-Ifitm3 reduced WNV infection in WT, Ifitm3−/− (13-fold, P < 0.01), and Ifitm12356−/− (Ifitm locus deletion) (21-fold, P < 0.001) MEFs compared to expression of FLUC in these cells. These data suggest that the relative susceptibility of Ifitm3−/− and Ifitm12356−/− MEFs to WNV infection is due largely to the loss of expression of Ifitm3 and that Ifitm3 can function as an antiviral molecule in the absence of expression of Ifitm1, Ifitm2, Ifitm5, and Ifitm6.
Ifitm3 expression on radioresistant and radiosensitive cells restricts WNV infection.
Our data in primary cells suggested that Ifitm3 might have antiviral effects in cells of different lineages. To assess the relative importance of Ifitm3 in different cellular compartments in vivo, we generated reciprocal bone marrow chimeras using congenic CD45.1+ (WT) and CD45.2+ (Ifitm3−/−) donor and recipient mice, and viral burden was measured at 7 days after infection (Fig. 7A and B). Due to the process of irradiation and reconstitution, the chimeric mice necessarily were infected at an older age, 16 weeks, than in the other studies (Fig. 1). Bone marrow chimeras with Ifitm3−/− or WT radioresistant nonhematopoietic cells (WT→Ifitm3−/− or Ifitm3−/−→WT) had equivalent levels of virus in serum, draining lymph node, and spinal cord (Fig. 7C to F) at day 7 after infection compared to WT mice (WT→WT), whereas the Ifitm3−/− mice (Ifitm3−/−→Ifitm3−/−) sustained higher titers in the brain (Fig. 7E, P < 0.05), as seen with the direct gene deletions (Fig. 1). Analysis of the adaptive immune response revealed minor differences in the chimeric mice. The percentage of CD19+ B cells was higher in Ifitm3−/−→Ifitm3−/− mice (Fig. 7G), but no difference in the absolute number of CD19+ B cells was observed (Fig. 7H). CD4+ T cell numbers and percentages in the spleen also were similar (Fig. 7I and J). The percentage of CD8+ T cells was reduced in Ifitm3−/−→WT and Ifitm3−/−→Ifitm3−/− mice (Fig. 7K), but this did not affect the total number of CD8+ T cells or the number or percentage of NS4B tetramer+ CD8+ T cells (Fig. 7L to N). Thus, Ifitm3 expression in both radioresistant and radiosensitive cell compartments contributes to the control of WNV infection, and only when there is a lack of Ifitm3 in both compartments is the phenotype of enhanced infection in the CNS revealed.
FIG 7.
Ifitm3 has functions in both the hematopoietic and nonhematopoietic cell compartments to restrict WNV infection. (A) CD45.1+ B6.SJL and CD45.2+ Ifitm3−/− mice were sublethally irradiated and reconstituted with either CD45.1+ B6.SJL or CD45.2+ Ifitm3−/− bone marrow (107 cells). (B) Representative flow cytometry plots of CD45.1 versus CD45.2 for CD19+ B cells and CD3+ T cells after reconstitution. Sixteen-week-old chimeric mice were inoculated via the subcutaneous route with 102 FFU of WNV (New York 1999), and tissues were harvested at day 7 after infection. WNV levels in serum (C), draining lymph node (D), brain (E), and spinal cord (F) were measured by qRT-PCR (C and D) or by infectious plaque assay (E and F). Solid lines represent the median viral titers, and dotted lines denote the limit of detection of the assay. Percentage and total cell numbers in the spleen of CD19+ B cells (G and H), CD3+ CD4+ T cells (I and J), CD3+ CD8+ T cells (K and L), and NS4B-specific CD3+ CD8+ T cells (M and N). Two to 4 animals were used per independent experiment, and 2 independent experiments were performed. Asterisks indicate values that are statistically significant by a nonparametric (C to F) or parametric (H to N) ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
Based primarily on studies in cell culture, the ISG IFITM3 has been suggested to have antiviral activity against a variety of viral pathogens, including flaviviruses (16–27). A demonstrable antiviral role for IFITM3/Ifitm3 in vivo has been established previously only for IAV and RSV (3, 43, 45, 46). We explored the function of Ifitm3 in restricting WNV pathogenesis in mice. Survival analysis revealed that Ifitm3−/− mice were more vulnerable to lethal WNV infection. This enhanced susceptibility was associated with increased viral infection in both peripheral and CNS tissues. Our culture studies with primary cells confirm an antiviral role of IFITM3 against WNV and extend these findings to a physiologically relevant in vivo system.
Several studies have examined the interaction between IFITM proteins and Flaviviridae members in cell culture (15, 19, 23, 69–71). DENV, WNV, YFV, and recently ZIKV were shown to be susceptible to IFITM-mediated antiviral restriction, with IFITM3 being the most potent antiviral IFITM protein in cell culture (15, 71, 81). Mechanism of action studies demonstrated that IFITM3 had no significant effect of WNV RNA replication (19). Rather, flavivirus infection likely is inhibited by IFITM3 at the fusion step in the late endosome, analogous to its antiviral actions on IAV (33, 38, 39). Other work has shown that IFITM1, IFITM2, and IFITM3 all can restrict DENV infection, including under conditions of antibody-dependent enhancement (23). Our transcomplementation experiments in Ifitm12356−/− MEFs extend these findings by showing that Ifitm3 can inhibit infection of WNV independently of expression of Ifitm1, Ifitm2, Ifitm5, and Ifitm6. These data argue strongly against an essential role for heteromeric complexes of different IFITM proteins in the antiviral activity of IFITM3 (70, 82).
Few studies have examined the function of IFITM3 in vivo. Increased lethality of Ifitm3−/− mice was observed after challenge with IAV (43), and this was associated with increased viral titers in the lung, increased lung pathology, and lymphopenia compared to WT mice. An additional study observed that Ifitm3−/− and Ifitm12356−/− mice were equally susceptible to IAV, suggesting that Ifitm3 may be the primary IFITM restriction (3). Of note, we were unable to study WNV infection in Ifitm12356−/− mice, as virtually all of these animals were stillborn when crossed onto a pure C57BL/6J background (M. J. Gorman and M. S. Diamond, unpublished observations), which is consistent with the original description of several IFITM proteins having a role in germ cell development (57, 83–85). Ifitm3−/− mice were also susceptible to another respiratory virus, RSV, with increased viral titers in the lung after infection (46). Ifitm3, however, does not restrict all pathogens, as Ifitm3−/− mice were not more susceptible to Salmonella enterica serovar Typhimurium, Citrobacter rodentium, Mycobacterium tuberculosis, or Plasmodium berghei (46).
Prior studies have demonstrated Ifitm3 expression throughout the lung, spleen, lymph node, liver, and intestine, suggesting that this protein may have broad antiviral functions in many tissues (3, 46). One of these functions may be the protection of immune cells such as CD8+ resident memory T cells or dendritic cells, as increased IAV infection and reduced survival were observed in Ifitm3-deficient T cells and dendritic cells (45, 86). Our studies demonstrate that neurotropic viruses also are restricted by Ifitm3 in vivo, although this phenotype appeared to be largely independent of a cell-intrinsic antiviral effect in the CNS. Our data are more consistent with a dominant antiviral effect of Ifitm3 in cells of peripheral organs, which then limits viremia and seeding of neurons in the brain and spinal cord. Our bone marrow chimera experiments suggest that Ifitm3 expression on both radioresistant and radiosensitive cells contributes to the control of WNV infection and pathogenesis in vivo. A definitive identification of the cell types in vivo that require Ifitm3 for antiviral protection awaits the generation of conditional gene deletion mice.
Similar to previously reported results, we discovered several adaptive immune deficiencies during viral infection. IAV challenge models had demonstrated that Ifitm3−/− mice had reduced CD4+ T cell and CD8+ T cell response in the lung during infection (43, 45). Our data demonstrated a similar phenotype, with CD4+ T cells, CD8+ T cells, and CD19+ B cells being reduced in the spleen during WNV infection. Despite this, antibody responses and CNS infiltration by antigen-specific CD8+ T cells showed little defect in WNV-infected Ifitm3−/− mice. Our bone marrow chimera experiments demonstrated that while the radiosensitive cells of the adaptive immune response may be important, they are not the dominant cell type leading to an increased CNS viral burden.
IFITM3 is one of several ISGs that restrict WNV in vivo in mice. Antiviral functions against WNV have been documented for protein kinase R (PKR), RNase L, Ifit2, Ifit27l2a, and viperin (49–52). Ifit2, Ifi27l2a, and viperin had antiviral functions that were limited largely to neurons in the CNS. Our data suggest that the dominant antiviral function of Ifitm3 likely occurs before WNV reaches the brain. Individual ISGs appear to have compartmentalized functions and restrict WNV infection in a tissue-specific manner. The basis of this remains uncertain but could be determined by relative expression or interaction with partner proteins that are differentially expressed. Moreover, despite a highly intricate IFN-dependent signaling cascade in which hundreds of ISGs are induced in a single cell, the deletion of single effector genes still can impact the control of viral infection and disease outcome.
In summary, our study has demonstrated that Ifitm3 has an important antiviral function in vivo against WNV. As IFITM3 is one of several IFITM family members, which differ in their cellular localization, expression, and range of viral restriction, additional work is needed to determine whether other IFITM proteins have analogous antiviral functions in vivo.
ACKNOWLEDGMENTS
National Institutes of Health grants U19 AI083019, U19 AI106772, R01 AI104972, and R01 AI104002 supported this study.
We thank Tiffany Lucas and Amelia Pinto for experimental advice and help with flow cytometry analysis. Experimental support for the speed congenic backcrossing was provided by the Washington University Facility of the Rheumatic Diseases Core Center. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases, part of the National Institutes of Health, under award number P30AR048335.
REFERENCES
- 1.Hickford DE, Frankenberg SR, Shaw G, Renfree MB. 2012. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics 13:155. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z, Liu J, Li M, Yang H, Zhang C. 2012. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 7:e49265. doi: 10.1371/journal.pone.0049265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey CC, Huang I-C, Kam C, Farzan M. 2012. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog 8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, Guo J-T. 2014. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci U S A 111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans SS, Lee DB, Han T, Tomasi TB, Evans RL. 1990. Monoclonal antibody to the interferon-inducible protein Leu-13 triggers aggregation and inhibits proliferation of leukemic B cells. Blood 76:2583–2593. [PubMed] [Google Scholar]
- 6.Takahashi S, Doss C, Levy S, Levy R. 1990. TAPA-1, the target of an antiproliferative antibody, is associated on the cell surface with the Leu-13 antigen. J Immunol 145:2207–2213. [PubMed] [Google Scholar]
- 7.Bradbury LE, Goldmacher VS, Tedder TF. 1993. The CD19 signal transduction complex of B lymphocytes. Deletion of the CD19 cytoplasmic domain alters signal transduction but not complex formation with TAPA-1 and Leu 13. J Immunol 151:2915–2927. [PubMed] [Google Scholar]
- 8.Frey M, Appenheimer MM, Evans SS. 1997. Tyrosine kinase-dependent regulation of L-selectin expression through the Leu-13 signal transduction molecule: evidence for a protein kinase C-independent mechanism of L-selectin shedding. J Immunol 158:5424–5434. [PubMed] [Google Scholar]
- 9.Tanaka SS, Matsui Y. 2002. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech Dev 119(Suppl):S261–S267. doi: 10.1016/S0925-4773(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 10.Zucchi I, Prinetti A, Scotti M, Valsecchi V, Valaperta R, Mento E, Reinbold R, Vezzoni P, Sonnino S, Albertini A, Dulbecco R. 2004. Association of rat8 with Fyn protein kinase via lipid rafts is required for rat mammary cell differentiation in vitro. Proc Natl Acad Sci U S A 101:1880–1885. doi: 10.1073/pnas.0307292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martensen PM, Justesen J. 2004. Small ISGs coming forward. J Interferon Cytokine Res 24:1–19. doi: 10.1089/107999004772719864. [DOI] [PubMed] [Google Scholar]
- 12.Yang G, Xu Y, Chen X, Hu G. 2007. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene 26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 13.Daniel-Carmi V, Makovitzki-Avraham E, Reuven E-M, Goldstein I, Zilkha N, Rotter V, Tzehoval E, Eisenbach L. 2009. The human 1-8D gene (IFITM2) is a novel p53 independent pro-apoptotic gene. Int J Cancer 125:2810–2819. doi: 10.1002/ijc.24669. [DOI] [PubMed] [Google Scholar]
- 14.Alber D, Staeheli P. 1996. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J Interferon Cytokine Res 16:375–380. doi: 10.1089/jir.1996.16.375. [DOI] [PubMed] [Google Scholar]
- 15.Brass AL, Huang I-C, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey CC, Zhong G, Huang I, Farzan M. 2014. IFITM-family proteins: the cell's first line of antiviral defense. Annu Rev Virol 1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidner JM, Jiang D, Pan X-B, Chang J, Block TM, Guo J-T. 2010. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol 84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang D, Weidner JM, Qing M, Pan X-B, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi P-Y, Block TM, Guo J-T. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol 84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang I-C, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, Elledge SJ, Bavari S, Denison MR, Choe H, Farzan M. 2011. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. 2011. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol 85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkins C, Woodward J, Lau DT-Y, Barnes A, Joyce M, McFarlane N, Tyrrell DL, Gale M. 2013. IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57:461–469. doi: 10.1002/hep.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan YK, Huang I-C, Farzan M. 2012. IFITM proteins restrict antibody-dependent enhancement of dengue virus infection. PLoS One 7:e34508. doi: 10.1371/journal.pone.0034508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mudhasani R, Tran JP, Retterer C, Radoshitzky SR, Kota KP, Altamura LA, Smith JM, Packard BZ, Kuhn JH, Costantino J, Garrison AR, Schmaljohn CS, Huang I-C, Farzan M, Bavari S. 2013. IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J Virol 87:8451–8464. doi: 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SE, Gibson MS, Wash RS, Ferrara F, Wright E, Temperton N, Kellam P, Fife M. 2013. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro. J Virol 87:12957–12966. doi: 10.1128/JVI.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Zhang L, Zan Y, Du N, Yang Y, Tien P. 2015. Human respiratory syncytial virus infection is inhibited by IFN-induced transmembrane proteins. J Gen Virol 96:170–182. doi: 10.1099/vir.0.066555-0. [DOI] [PubMed] [Google Scholar]
- 27.Qian J, Duff Le Y, Wang Y, Pan Q, Ding S, Zheng Y-M, Liu S-L, Liang C. 2015. Primate lentiviruses are differentially inhibited by interferon-induced transmembrane proteins. Virology 474:10–18. doi: 10.1016/j.virol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weston S, Czieso S, White IJ, Smith SE, Wash RS, Diaz-Soria C, Kellam P, Marsh M. 24 May 2016. Alphavirus restriction by IFITM proteins. Traffic doi: 10.1111/tra.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren CJ, Griffin LM, Little AS, Huang I-C, Farzan M, Pyeon D. 2014. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS One 9:e96579. doi: 10.1371/journal.pone.0096579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YX, Welte K, Gebhard DH, Evans RL. 1984. Induction of T cell aggregation by antibody to a 16kd human leukocyte surface antigen. J Immunol 133:2496–2501. [PubMed] [Google Scholar]
- 31.Yount JS, Karssemeijer RA, Hang HC. 2012. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J Biol Chem 287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia R, Pan Q, Ding S, Rong L, Liu S-L, Geng Y, Qiao W, Liang C. 2012. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol 86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li K, Markosyan RM, Zheng Y-M, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu S-L. 2013. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hach JC, McMichael T, Chesarino NM, Yount JS. 2013. Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J Virol 87:9923–9927. doi: 10.1128/JVI.00621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey CC, Kondur HR, Huang I-C, Farzan M. 2013. Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J Biol Chem 288:32184–32193. doi: 10.1074/jbc.M113.514356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weston S, Czieso S, White IJ, Smith SE, Kellam P, Marsh M. 2014. A membrane topology model for human interferon inducible transmembrane protein 1. PLoS One 9:e104341. doi: 10.1371/journal.pone.0104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling S, Zhang C, Wang W, Cai X, Yu L, Wu F, Zhang L, Tian C. 2016. Combined approaches of EPR and NMR illustrate only one transmembrane helix in the human IFITM3. Sci Rep 6:24029. doi: 10.1038/srep24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. 2014. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen L-M, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, Brass AL. 2011. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amini-bavil-olyaee S, Choi YJ, Lee JH, Shi M, Huang I, Farzan M. 2013. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, Liang C, Casartelli N, Schwartz O. 2014. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 16:736–747. doi: 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tartour K, Appourchaux R, Gaillard J, Nguyen X-N, Durand S, Turpin J, Beaumont E, Roch E, Berger G, Mahieux R, Brand D, Roingeard P, Cimarelli A. 2014. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology 11:103. doi: 10.1186/s12977-014-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P. 2012. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng Y-M, Liu S-L, Guo F, Geng Y, Qiao W, Liang C. 2014. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol 16:1080–1093. doi: 10.1111/cmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakim LM, Gupta N, Mintern JD, Villadangos JA. 2013. Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol 14:238–245. doi: 10.1038/ni.2525. [DOI] [PubMed] [Google Scholar]
- 46.Everitt AR, Clare S, McDonald JU, Kane L, Harcourt K, Ahras M, Lall A, Hale C, Rodgers A, Young DB, Haque A, Billker O, Tregoning JS, Dougan G, Kellam P. 2013. Defining the range of pathogens susceptible to ifitm3 restriction using a knockout mouse model. PLoS One 8:e80723. doi: 10.1371/journal.pone.0080723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armah HB, Wang G, Omalu BI, Tesh RB, Gyure KA, Chute DJ, Smith RD, Dulai P, Vinters HV, Kleinschmidt-DeMasters BK, Wiley CA. 2007. Systemic distribution of West Nile virus infection: postmortem immunohistochemical study of six cases. Brain Pathol 17:354–362. doi: 10.1111/j.1750-3639.2007.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omalu BI, Shakir AA, Wang G, Lipkin WI, Wiley CA. 2003. Fatal fulminant pan-meningo-polioencephalitis due to West Nile virus. Brain Pathol 13:465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BRG, Silverman RH, Gale M, Diamond MS. 2006. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol 80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, Diamond MS. 2011. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J Virol 85:11557–11566. doi: 10.1128/JVI.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho H, Shrestha B, Sen GC, Diamond MS. 2013. A role for Ifit2 in restricting West Nile virus infection in the brain. J Virol 87:8363–8371. doi: 10.1128/JVI.01097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lucas TM, Richner JM, Diamond MS. 2016. The interferon-stimulated gene Ifi27l2a restricts West Nile virus infection and pathogenesis in a cell-type- and region-specific manner. J Virol 90:2600–2615. doi: 10.1128/JVI.02463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanciotti RS, Ebel GD, Deubel V, Kerst AJ, Murri S, Meyer R, Bowen M, McKinney N, Morrill WE, Crabtree MB, Kramer LD, Roehrig JT. 2002. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298:96–105. doi: 10.1006/viro.2002.1449. [DOI] [PubMed] [Google Scholar]
- 54.Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. 2004. Genetic and phenotypic variation of West Nile virus in New York, 2000-2003. Am J Trop Med Hyg 71:493–500. [PubMed] [Google Scholar]
- 55.Beasley DWC, Li L, Suderman MT, Barrett ADT. 2002. Mouse neuroinvasive phenotype of West Nile virus strains varies depending upon virus genotype. Virology 296:17–23. doi: 10.1006/viro.2002.1372. [DOI] [PubMed] [Google Scholar]
- 56.Brien JD, Lazear HM, Diamond MS. 2013. Propagation, quantification, detection and storage of West Nile virus. Curr Protoc Microbiol 31:15D3.1–15D3.18. doi: 10.1002/9780471729259.mc15d03s31. [DOI] [PubMed] [Google Scholar]
- 57.Lange UC, Adams DJ, Lee C, Barton S, Schneider R, Bradley A, Surani MA. 2008. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol Cell Biol 28:4688–4696. doi: 10.1128/MCB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szretter KJ, Daffis S, Patel J, Suthar MS, Klein RS, Gale M, Diamond MS. 2010. The innate immune adaptor molecule MyD88 restricts West Nile virus replication and spread in neurons of the central nervous system. J Virol 84:12125–12138. doi: 10.1128/JVI.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diamond MS, Shrestha B, Marri A, Mahan D, Engle M. 2003. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol 77:2578–2586. doi: 10.1128/JVI.77.4.2578-2586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 38:4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehlhop E, Diamond MS. 2006. Protective immune responses against West Nile virus are primed by distinct complement activation pathways. J Exp Med 203:1371–1381. doi: 10.1084/jem.20052388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purtha WE, Myers N, Mitaksov V, Sitati E, Connolly J, Fremont DH, Hansen TH, Diamond MS. 2007. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. Eur J Immunol 37:1845–1854. doi: 10.1002/eji.200737192. [DOI] [PubMed] [Google Scholar]
- 63.Szretter KJ, Samuel MA, Gilfillan S, Fuchs A, Colonna M, Diamond MS. 2009. The immune adaptor molecule SARM modulates tumor necrosis factor alpha production and microglia activation in the brainstem and restricts West Nile virus pathogenesis. J Virol 83:9329–9338. doi: 10.1128/JVI.00836-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lazear HM, Pinto AK, Vogt MR, Gale M, Diamond MS. 2011. Beta interferon controls West Nile virus infection and pathogenesis in mice. J Virol 85:7186–7194. doi: 10.1128/JVI.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dora S, Schwarz C, Baack M, Graessmann A, Knippers R. 1989. Analysis of a large-T-antigen variant expressed in simian virus 40-transformed mouse cell line mKS-A. J Virol 63:2820–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Araki T, Sasaki Y, Milbrandt J. 2004. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 67.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system. J Virol 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med 11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu X, He Z, Yuan J, Wen W, Huang X, Hu Y, Lin C, Pan J, Li R, Deng H, Liao S, Zhou R, Wu J, Li J, Li M. 2015. IFITM3-containing exosome as a novel mediator for anti-viral response in dengue virus infection. Cell Microbiol 17:105–118. doi: 10.1111/cmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.John SP, Chin CR, Perreira J, Feeley EM, Aker A, Savidis G, Smith SE, Elia AEH, Everitt AR, Vora M, Pertel T, Elledge SJ, Kellam P, Brass AL. 2013. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J Virol 87:7837–7852. doi: 10.1128/JVI.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja M-K, Diamond MS, Gale M. 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog 6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daffis S, Samuel MA, Keller BC, Gale M, Diamond MS. 2007. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog 3(7):e106. doi: 10.1371/journal.ppat.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cho H, Proll SC, Szretter KJ, Katze MG, Gale M, Diamond MS. 2013. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat Med 19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, García-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fredericksen BL, Gale M. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol 80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol 80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brien JD, Uhrlaub JL, Nikolich-Zugich J. 2007. Protective capacity and epitope specificity of CD8(+) T cells responding to lethal West Nile virus infection. Eur J Immunol 37:1855–1863. doi: 10.1002/eji.200737196. [DOI] [PubMed] [Google Scholar]
- 79.Shrestha B, Diamond MS. 2004. Role of CD8+ T cells in control of West Nile virus infection. J Virol 78:8312–8321. doi: 10.1128/JVI.78.15.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, Engle M. 2003. A critical role for induced IgM in the protection against West Nile virus infection. J Exp Med 198:1853–1862. doi: 10.1084/jem.20031223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Savidis G, Perreira JM, Portmann JM, Meraner P, Guo Z, Green S, Brass AL. 2016. The IFITMs inhibit Zika virus replication. Cell Rep 15:2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 82.Perreira JM, Chin CR, Feeley EM, Brass AL. 2013. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol 425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H, Tam PPL. 2005. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev Cell 9:745–756. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 84.Lange UC, Saitou M, Western PS, Barton SC, Surani MA. 2003. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol 3:1. doi: 10.1186/1471-213X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saitou M, Barton SC, Surani MA. 2002. A molecular programme for the specification of germ cell fate in mice. Nature 418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 86.Infusini G, Smith JM, Yuan H, Pizzolla A, Ng WC, Londrigan SL, Haque A, Reading PC, Villadangos JA, Wakim LM. 2015. Respiratory DC use IFITM3 to avoid direct viral infection and safeguard virus-specific CD8+ T cell priming. PLoS One 10:e0143539. doi: 10.1371/journal.pone.0143539. [DOI] [PMC free article] [PubMed] [Google Scholar]