ABSTRACT
Equine herpesvirus 1 (EHV-1) is a major pathogen affecting equines worldwide. The virus causes respiratory disease, abortion, and, in some cases, neurological disease. EHV-1 strain KyA is attenuated in the mouse and equine, whereas wild-type strain RacL11 induces severe inflammation of the lung, causing infected mice to succumb at 4 to 6 days postinfection. Our previous results showed that KyA immunization protected CBA mice from pathogenic RacL11 challenge at 2 and 4 weeks postimmunization and that KyA infection elicited protective humoral and cell-mediated immune responses. To investigate the protective mechanisms of innate immune responses to KyA, KyA-immunized mice were challenged with RacL11 at various times postvaccination. KyA immunization protected mice from RacL11 challenge at 1 to 7 days postimmunization. Immunized mice lost less than 10% of their body weight and rapidly regained weight. Virus titers in the lungs of KyA-immunized mice were 1,000-fold lower at 2 days post-RacL11 challenge than virus titers in the lungs of nonimmunized mice, indicating accelerated virus clearance. Affymetrix microarray analysis revealed that gamma interferon (IFN-γ) and 16 antiviral interferon-stimulated genes (ISGs) were upregulated 3.1- to 48.2-fold at 8 h postchallenge in the lungs of RacL11-challenged mice that had been immunized with KyA. Murine IFN-γ inhibited EHV-1 infection of murine alveolar macrophages and protected mice against lethal EHV-1 challenge, suggesting that IFN-γ expression is important in mediating the protection elicited by KyA immunization. These results suggest that EHV-1 KyA may be used as a live attenuated EHV-1 vaccine as well as a prophylactic agent in horses.
IMPORTANCE Viral infection of cells initiates a signal cascade of events that ultimately attempts to limit viral replication and prevent infection through the expression of host antiviral proteins. In this study, we show that EHV-1 KyA immunization effectively protected CBA mice from pathogenic RacL11 challenge at 1 to 7 days postvaccination and increased the expression of IFN-γ and 16 antiviral interferon-stimulated genes (ISGs). The administration of IFN-γ blocked EHV-1 replication in murine alveolar macrophages and mouse lungs and protected mice from lethal challenge. To our knowledge, this is the first report of an attenuated EHV-1 vaccine that protects the animal at 1 to 7 days postimmunization by innate immune responses. Our findings suggested that IFN-γ serves as a novel prophylactic agent and may offer new strategies for the development of anti-EHV-1 agents in the equine.
INTRODUCTION
Equine herpesvirus 1 (EHV-1), a member of the family Herpesviridae and the subfamily Alphaherpesvirinae, is the causative agent of a number of equine pathological states, including severe disease of the central nervous system, respiratory infections, and abortion storms (1–4). Respiratory transmission of this highly contagious virus has resulted in disastrous outbreaks of disease in domestic horse populations and has had significant economic impact on the equine industry. EHV-1 infection of the horse generates a short-lived humoral response but does not confer long-term protection, since disease often occurs following natural infection (5, 6).
A model that closely mimics EHV-1 infection in the natural host has been established with various strains of mice (7). Common features include replication in the respiratory mucosae, the development of pneumonitis, cell-associated viremia, and abortion (7, 8). Various EHV-1 glycoproteins, including gB, gC, gD, and gH, were capable of inducing neutralizing antibodies (9–14). EHV-1-specific CD8+ class I major histocompatibility complex (MHC)-restricted cytotoxic T lymphocytes (CTL) were identified in the peripheral blood mononuclear cells of infected horses, reaching maximal levels 2 to 3 weeks postinfection (15, 16). Intranasal inoculation of mice with an attenuated EHV-1 strain, KyA, resulted in the production of a primary virus-specific CTL response in the draining mediastinal lymph nodes (17). In this model, EHV-1-specific CTL activity was mediated by class I MHC-restricted CD8+ T cells (17).
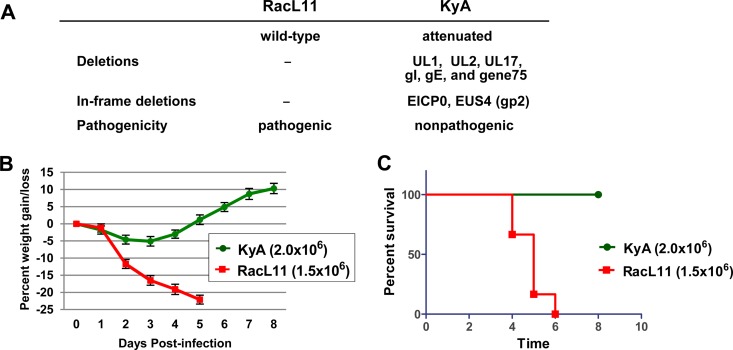
The nonpathogenic EHV-1 strain Kentucky A (KyA) is attenuated in mice and horses, whereas the wild-type pathogenic strain RacL11 induces severe inflammatory cell infiltration in the lung, such that infected mice succumb at 4 to 6 days postinfection (dpi) (Fig. 1) (18–23). RacL11 was isolated from an aborted fetus (24). KyA has a long history of passage outside the natural host and was generated by passage through Syrian hamsters and L-M mouse fibroblasts in cell culture (22, 25–27). Our individual DNA sequencing analyses of KyA (GenBank accession numbers M87497.2, U81154.1, and AY762536.1) revealed several genomic deletions (UL1, UL2, UL17, gI, gE, and gene 75) and in-frame deletions (EICP0 and EUS4 [gp2]) relative to the sequence of the wild-type Ab4 strain of EHV-1 (Fig. 1A) (28–34). Our previous results showed that EHV-1 glycoproteins gI, gE, and full-length gp2 contribute to the pathogenesis of the RacL11 strain (19, 20, 22). Likewise, in the case of pseudorabies virus (PRV), the mutant JS-2012-ΔgE/gI was avirulent in suckling piglets but was able to provide full protection for young piglets against challenge with virulent PRV (35).
FIG 1.
Challenge of CBA mice with the attenuated EHV-1 strain KyA or the pathogenic strain RacL11. (A) The Kentucky A (KyA) strain of EHV-1 lacks the genes encoding gI and gE and is attenuated in both equines and mice. —, no deletion. (B and C) Average body weights (B) and percentages of survival (C) of CBA mice infected with KyA or RacL11. Groups of CBA mice (n = 8) were infected intranasally with 2 × 106 PFU/mouse of KyA or 1.5 × 106 PFU/mouse of RacL11, which is the standard protocol used in our previous publications (14, 17, 19). Mice were monitored daily for clinical signs of infection, weight gain or loss, and morbidity.
There are three types of interferons (IFNs), types I, II, and III, each of which displays distinct expression patterns and has myriad roles in innate and adaptive immunity. Type I IFNs are a family of cytokines with a critical role in controlling the innate and adaptive immune responses to infection, cancer, and other inflammatory stimuli. Many viruses trigger the type I IFN system, leading to the transcription of hundreds of interferon-stimulated genes (ISGs) (36). The products of these ISGs perform numerous antiviral effector functions, many of which have not been fully described yet. ISGs can target almost any step in a virus life cycle. Interferon-inducible protein 16 (IFI16; its murine homolog is IFI204) functions as a DNA sensor and detects herpesvirus genomes replicating in the nucleus, such as those of Kaposi's sarcoma-associated herpesvirus (KSHV), Epstein-Barr virus (EBV), and herpes simplex virus 1 (HSV-1), leading to inflammasome formation (37–40). Interferon gamma (IFN-γ) is a type II interferon that is pivotal in the regulation of the host immune response to viral and intracellular bacterial pathogens. High levels of IFN-γ are secreted by type 1 T helper cells (Th1 cells), CD8+ CTL, and NK cells during active infection (41). IFN-γ has a major effect on the regulation of antigen presentation by macrophages and dendritic cells, as well as in the induction of class switching of B cells (42, 43). As a proinflammatory cytokine, IFN-γ directly activates phagocytic cells and stimulates the oxidative burst and the release of degradative enzymes, thereby supporting host defense responses to intracellular pathogens (44). IFN-γ also induces the production of proinflammatory cytokines and chemokines in endothelial cells, epithelial cells, and fibroblasts.
Despite regular and widespread vaccination, outbreaks of EHV-1 continue to occur, frequently with fatal outcomes due to the development of neurological disease, and inflict substantial suffering on the animals. Several EHV-1 vaccines, which are based mainly on preparations of inactivated viruses, are available for protection against the respiratory and abortogenic forms of EHV-1; however, they do not elicit long-term immunity or protection for the horse (45–47). Outbreaks of infection occur within the horse population despite active vaccination with currently available EHV-1 vaccines. In order to develop a safe and effective EHV-1 vaccine, an attenuated strain, KyA, was investigated for the ability to elicit protection by innate immune responses. KyA immunization effectively protected CBA mice from challenge with the highly pathogenic EHV-1 strain RacL11 at 1 to 7 days postimmunization. Affymetrix microarray analyses revealed that IFN-γ and 16 antiviral ISGs were significantly upregulated at 8 h postchallenge in the lungs of RacL11-challenged mice that had been immunized with KyA. IFN-γ inhibited EHV-1 replication in murine alveolar macrophages and protected CBA mice from lethal EHV-1 challenge. These results suggest that IFN-γ and the upregulated antiviral proteins function to inhibit EHV-1 replication within mouse lungs and protect the mice from RacL11 infection.
MATERIALS AND METHODS
Viruses and cell culture.
The KyA strain of EHV-1 was propagated in suspension cultures of L-M mouse fibroblasts as described previously (48, 49). Pathogenic strains RacL11 and Ab4 were propagated in equine NBL6 cells. L-M and NBL6 cells were maintained at 37°C in complete Eagle's minimum essential medium (EMEM) supplemented with 100 U/ml of penicillin, 100 μg/ml of streptomycin, nonessential amino acids, and 5% fetal bovine serum. The murine alveolar macrophage cell line MH-S (CRL-2019; American Type Culture Collection) was maintained at 37°C in RPMI 1640 medium containing 100 U/ml of penicillin, 100 μg/ml of streptomycin, 0.02 mM 2-mercaptoethanol, and 5% fetal bovine serum.
Ethics statement.
The guidelines of the Louisiana State University Health Sciences Center at Shreveport (LSUHSC-Shreveport) Institutional Animal Care and Use Committee (IACUC) were followed for all animal experiments. These guidelines are in strict adherence to the Public Health Service Policy on Humane Care and Use of Laboratory Animals, and the LSUHSC-Shreveport OLAW assurance number is A3095-01. All animal studies were approved by the LSUHSC-Shreveport IACUC under protocols P-13-024 and P-15-038.
Mice.
Female CBA mice, 3 to 4 weeks old, obtained from Harlan Sprague Dawley (Indianapolis, IN), were maintained in filter-topped cages within the Animal Resource Facility of the LSUHSC-Shreveport. All mice rested for a period of 5 to 7 days prior to use. Experimental groups consisted of eight (for body weight changes and percentages of survival) or six (for virus titers in lungs) mice, and all experiments were repeated a minimum of three times.
Infection and assessment of respiratory disease.
CBA mice were anesthetized by isoflurane (Sigma Chemical Co., St. Louis, MO) inhalation and were inoculated intranasally either with 2 × 106 PFU/mouse of KyA or with 1.5 × 106 PFU/mouse of wild-type RacL11 in a total inoculum of 40 μl. Following infection, mice were observed daily for clinical signs of respiratory disease, including labored breathing, ruffled fur, and huddling. For weight loss experiments, mice were weighed individually immediately prior to infection and daily thereafter at the same time each day. Animals exhibiting profound wasting or an inability to move or function were humanely sacrificed to prevent undue suffering.
Isolation of mRNA.
At various times postinfection, mice were sacrificed by prolonged isoflurane inhalation, and the lungs were removed. Total lung RNA was obtained by grinding the lungs in glass tissue grinders in the presence of TRIzol (Life Technologies, Grand Island, NY). Poly(A)+ RNA was isolated from total RNA using the FastTrack 2.0 kit (Invitrogen, San Diego, CA) according to the manufacturer's instructions.
Histopathology.
Lungs from CBA mice infected with 1.5 × 106 PFU/mouse of KyA or RacL11 were harvested and were processed for histopathological analysis as described previously (22). Six-micrometer-thick sections were cut using a Leica CM1850 cryostat (Leica, Bannockburn, IL), placed on precleaned Superfrost Plus slides (VWR, West Chester, NY), air dried overnight, and stored at −20°C until use. For hematoxylin and eosin (H&E) staining, slides were fixed for 10 min in 10% buffered formalin and were stained using a standard H&E protocol. Slides were viewed and photographed using a Nikon Eclipse TE300 inverted microscope equipped with a Nikon DXM 1200F digital camera (Nikon, Tokyo, Japan).
Microarray analysis.
CBA mice were immunized with 1.5 × 106 PFU KyA and were challenged with 1.5 × 106 PFU of RacL11. The quality and quantity of RNA were determined, and RNA was processed with the Mouse Affymetrix Genome 430 2.0 arrays (Affymetrix, Santa Clara, CA) at the Microarray Core Facility at the University of Texas Southwestern Medical Center. Three independent experimental arrays and three control arrays were analyzed with GeneChip operating software (GCOS; Affymetrix), GeneSpring, version 7.0 (Silicon Genetics), significance analysis of microarrays (SAM; Stanford University, Palo Alto, CA, USA), and Spotfire Decision Site, version 8.2 (Spotfire, Somerville, MA). Biotinylated cRNA was prepared using the Illumina RNA amplification kit (Ambion, Inc., Austin, TX) according to the manufacturer's directions, starting with ∼200 ng total RNA. Hybridization to Affymetrix Mouse Genome 430 2.0 arrays, washing, and scanning were performed according to the Illumina BeadStation 500× manual (revision C). Arrays were scanned with an Illumina BeadArray Reader confocal scanner, and the data were analyzed using Illumina's BeadStudio software (version 3). Analysis of variance was performed with Spotfire. The data were normalized by mean values (for Spotfire pairwise comparisons and SAM two-class comparisons) or percentile values (for GeneSpring analyses). Gene expression changes were considered significant if the P value was less than 0.05, the fold difference between infected and mock-infected lungs was at least 2.0, and changes in gene expression were reproducible in all replicate comparisons. Genes expressed at different levels in untreated controls were excluded from analysis.
qRT-PCR assays.
Quantitative real-time reverse transcription-PCR (qRT-PCR) assays were performed as described previously (50). Briefly, total RNA was extracted from lung tissue by using the RNeasy minikit (Qiagen, CA). qRT-PCR amplification was carried out with the CFX96 Real-Time PCR detection system by using the iTaq Universal SYBR green one-step kit according to the manufacturer's instructions (Bio-Rad Laboratories, Hercules, CA). Reverse transcription was carried out at 10 min at 50°C and 1 min at 95°C, followed by 40 cycles of PCR for 10 s at 95°C for denaturation and 30 s at 60°C for annealing/extension. For the melting curve analysis, after 40 reaction cycles, the temperature ramp was programmed from 65 to 95°C in increments of 0.5°C, with a 5-s hold before each acquisition. The primers used in qRT-PCR are shown in Table 1. Mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Each sample was assayed in triplicate.
TABLE 1.
List of primers for real-time RT-PCR analysis
| Gene name | Primer orientation | Primer sequence | Location |
|---|---|---|---|
| RSAD2 | Forward | TGCTGGCTGAGAATAGCATTAGG | 97–119 |
| Reverse | GCTGAGTGCTGTTCCCATCT | 208–189 | |
| MX1 | Forward | GACCATAGGGGTCTTGACCAA | 621–141 |
| Reverse | AGACTTGCTCTTTCTGAAAAGCC | 802–780 | |
| IRF7 | Forward | GAGACTGGCTATTGGGGGAG | 38–57 |
| Reverse | GACCGAAATGCTTCCAGGG | 139–121 | |
| IFI44L | Forward | GGGTCTGACGAAGCAGTATC | 700–721 |
| Reverse | CCCCATTGAAGAATCACACAGCAT | 801–781 | |
| ISG15 | Forward | GGTGTCCGTGACTAACTCCAT | 48–68 |
| Reverse | CTGTACCACTAGCATCACTGTG | 222–201 | |
| OAS1a | Forward | GCCTGATCCCAGAATCTATGC | 495–515 |
| Reverse | GAGCAACTCTAGGGCGTACTG | 711–691 | |
| IFN-γ | Forward | ATGAACGCTACACACTGCATC | 1–21 |
| Reverse | CCATCCTTTTGCCAGTTCCTC | 182–162 | |
| IFI204 | Forward | GACAACCAAGAGCAATACACCA | 112–133 |
| Reverse | ATCAGTTTGCCCAATCCAGAAT | 197–176 | |
| GAPDH-m | Forward | ATCACTGCCACTCAGAAGACTGT | 541–563 |
| Reverse | ACCAGTGGATGCAGGGATGATGTT | 636–613 |
Microarray data accession number.
The microarray data obtained in this study were deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE81219 (http://www.ncbi.nlm.nih.gov/geo).
RESULTS
Immunization with attenuated EHV-1 KyA protects CBA mice from pathogenic RacL11 challenge at 1 to 7 days postimmunization.
EHV-1 strain KyA is attenuated in mice and horses, whereas wild-type strain RacL11 induces severe inflammatory infiltration of the lung (18–22). To compare the pathogenicities of EHV-1 strains KyA and RacL11, groups of CBA mice (n = 8) were infected intranasally with 2 × 106 PFU/mouse of KyA or 1.5 × 106 PFU/mouse of RacL11 and were monitored for weight gain or loss and morbidity. RacL11-infected mice lost more than 20% of their preinfection body weight by day 5 postinfection and succumbed to RacL11 infection by 4 to 6 days postinfection (dpi) (Fig. 1B and C). In contrast, KyA-infected mice lost less than 5% of their preinfection body weight and rapidly regained weight, and none of the mice succumbed to KyA infection (Fig. 1B and C). These observations are consistent with our previous results (14, 18, 22).
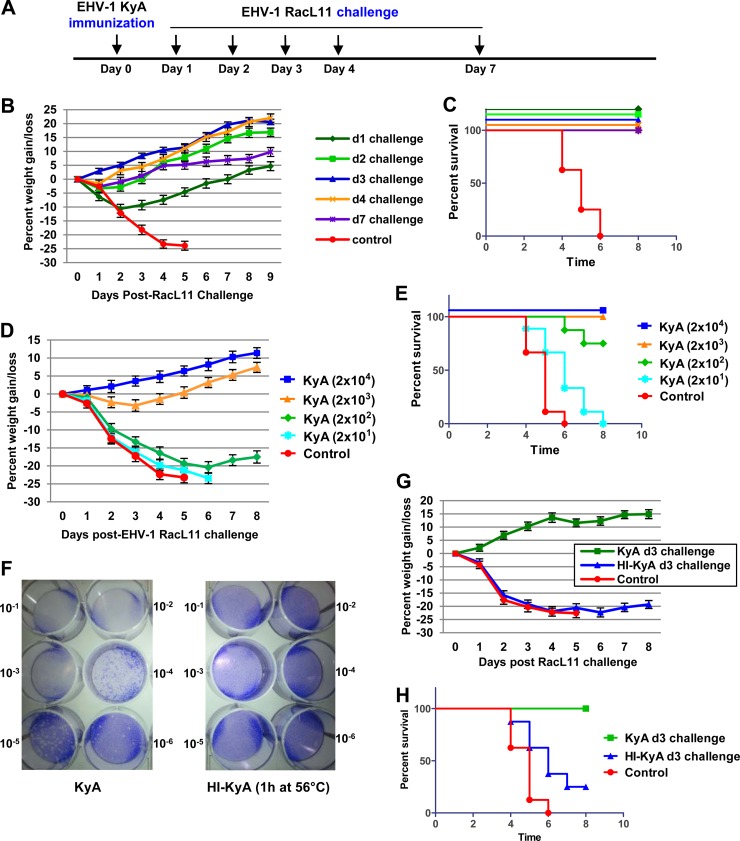
EHV-1 KyA immunization protected CBA mice from pathogenic RacL11 challenge at 4 weeks postimmunization via adaptive immune responses (14, 18, 22). To investigate the protective mechanisms of innate immune responses elicited by KyA, the immunized CBA mice were infected with EHV-1 RacL11 at times earlier than 2 weeks postimmunization (Fig. 2A). CBA mice (n = 8) were first immunized intranasally either with a control medium or with 2.0 × 106 PFU/mouse of KyA and then challenged with 1.5 × 106 PFU/mouse of RacL11 at 1, 2, 3, 4, and 7 days postimmunization. All of the KyA-immunized mice lost less than 10% of their preinfection body weight and rapidly regained weight (Fig. 2B). None of the immunized mice died upon challenge with RacL11 (Fig. 2C). However, the control nonimmunized mice lost more than 20% of their preinfection body weight by day 4 post-RacL11 challenge (Fig. 2B), and all succumbed to RacL11 infection by days 4 to 6 postchallenge (Fig. 2C). These results indicated that KyA immunization effectively protected CBA mice from RacL11 challenge at 1 to 7 days postimmunization. To determine the minimal dose of KyA required for protection, mice were immunized with 2 × 101, 2 × 102, 2 × 103, or 2 × 104 PFU/mouse and were challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 days postimmunization. Immunization with as little as 2,000 PFU of KyA protected CBA mice from RacL11 challenge (Fig. 2D and E).
FIG 2.
Live EHV-1 KyA, but not heat-inactivated KyA, protected CBA mice from challenge with pathogenic EHV-1 RacL11 at 1 to 7 days postimmunization. (A) Schematic representation of immunization and EHV-1 challenge infection of CBA mice. (B and C) Average body weights (B) and percentages of survival (C) of CBA mice challenged with RacL11 at 1 to 7 days postimmunization. (D and E) Low-dose KyA immunization protects CBA mice from challenge with 1.5 × 106 PFU/mouse of RacL11. Average body weights (D) and percentages of survival (E) are shown. CBA mice (n = 8) were immunized with 2 × 101, 2 × 102, 2 × 103, or 2 × 104 PFU/mouse of KyA and were challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 days postimmunization. (F to H) Heat-inactivated KyA (HI-KyA) could not protect CBA mice from RacL11 challenge. (F) EHV-1 KyA was inactivated by incubation at 56°C for 1 h (14). The amount of infectious EHV-1 was determined in equine NBL6 cells by standard plaque titration (50). Crystal violet stains the cells, allowing visualization of the clear plaques. The viral concentrations in the stained plates decrease from the top left (10−1) to the bottom right (10−6). (G and H) Average body weights (G) and percentages of survival (H) of CBA mice challenged with RacL11 at 3 days postimmunization. CBA mice (n = 8) were first immunized with medium (control), KyA (2 × 106 PFU/mouse), or HI-KyA (equivalent to 2 × 106 PFU prior to inactivation/mouse) and then challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 and 14 days postimmunization.
Since most commercially available EHV-1 vaccines are inactivated forms of the virus, we tested immunization with heat-inactivated KyA (HI-KyA) for the ability to protect mice from RacL11 challenge. EHV-1 KyA was inactivated by incubation at 56°C for 1 h (51), and the heat-treated virus was not able to infect equine NBL6 cells (Fig. 2F). CBA mice were immunized intranasally with KyA or HI-KyA and were challenged with RacL11 at 3 days postimmunization. As expected, mice immunized with KyA lost less than 5% of their body weight (Fig. 2G), and none of them died (Fig. 2H). In contrast, mice immunized with HI-KyA lost more than 20% of their body weight by day 3 post-challenge infection (Fig. 2G), and more than 75% of these mice succumbed to RacL11 infection by 7 dpi (Fig. 2H).
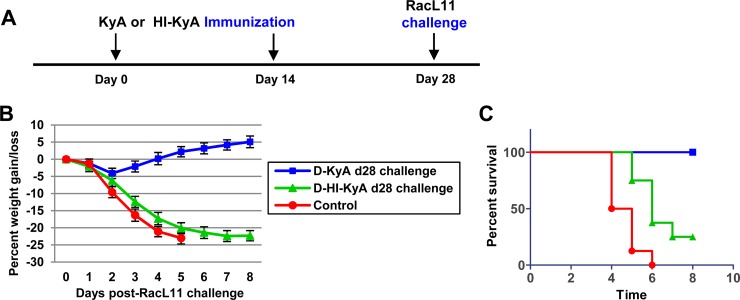
CBA mice immunized twice with KyA or HI-KyA (at day 0 and at day 14) were challenged with RacL11 at 28 days after the first immunization. The mice immunized twice with HI-KyA lost more than 20% of their body weight by day 5 post-challenge infection (Fig. 3B), and 75% of them succumbed to RacL11 infection (Fig. 3C). These results indicated that immunization with heat-inactivated KyA does not effectively protect CBA mice from pathogenic RacL11 infection.
FIG 3.
Heat-inactivated KyA (HI-KyA) does not protect CBA mice from RacL11 challenge. (A) Schematic representation of HI-KyA immunization and RacL11 challenge infection of CBA mice. CBA mice (n = 8) were immunized with medium (control), KyA (2 × 106 PFU/mouse), or HI-KyA (equivalent to 2 × 106 PFU prior to inactivation/mouse) at day 0 and day 14 and were challenged with RacL11 (1.5 × 106 PFU/mouse) at 28 days after the first immunization. (B and C) Average body weights (B) and percentages of survival (C) of CBA mice challenged with RacL11 at 28 days postimmunization. D, twice immunized.
Rapid virus clearance from the lungs of mice immunized with KyA.
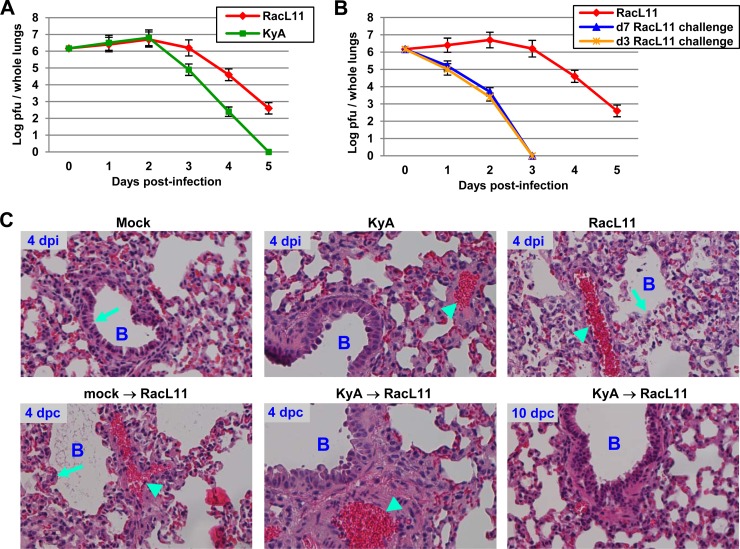
CBA mice were infected intranasally with KyA (2 × 106 PFU/mouse) or RacL11 (1.5 × 106 PFU/mouse), and the amount of infectious EHV-1 in the lungs was determined. Virus titers in the lungs of KyA-infected mice were 20-fold lower at 3 dpi and 158-fold lower at 4 dpi than virus titers in the lungs of RacL11-infected mice (Fig. 4A). When the KyA-immunized mice were challenged with RacL11 at 3 or 7 days postimmunization, the virus titers in their lungs were 3 log units lower than those for RacL11-infected mice at 2 days post-challenge infection (Fig. 4B), indicating accelerated virus clearance from the lungs.
FIG 4.
Immunization with KyA is protective against challenge with pathogenic RacL11. (A and B) Rapid virus clearance from the lungs of CBA mice immunized with KyA. Groups of CBA mice (n = 6) were infected intranasally with KyA (2 × 106 PFU/mouse) and were challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 or 7 days postimmunization. On days 1 to 5 postchallenge, the lungs were removed and homogenized, and the amount of infectious EHV-1 was determined by standard plaque titration as described previously (50). KyA, KyA-infected mice; RacL11, RacL11-infected mice. (C) Histological sections of infected mouse lungs. CBA mice were intranasally mock infected or infected with KyA and were challenged with RacL11 at 3 days postimmunization (mock → RacL11 or KyA → RacL11, respectively). On days 4 and 10 p.i., the mice were sacrificed, and the lungs were removed. The lung tissue was mounted and processed for histological evaluation as described in Materials and Methods. The resulting slides were photographed at ×20 magnification with a Nikon TE300 Eclipse microscope. dpi, days postinfection; dpc, days post-RacL11 challenge; B, bronchus. Bronchial epithelial (arrows) and inflammatory infiltration (arrowheads) regions are indicated.
Mice were either mock infected or infected with KyA and were challenged with RacL11 at 3 days postimmunization. Lungs were collected at 4 and 10 days post-RacL11 challenge and were assessed for cytopathology. While the lungs of mice infected with KyA exhibited almost undetectable inflammatory infiltration at day 4 postinfection (p.i.), the lungs of mice infected with RacL11 exhibited extensive peribronchial cuffing and interstitial inflammatory infiltration (Fig. 4C). RacL11 infection also disrupted the bronchial epithelium (Fig. 4C). When the KyA-immunized mice were challenged with RacL11 at 3 days postimmunization, the lungs exhibited almost undetectable inflammatory infiltration at 4 days post-challenge infection and showed normal lung architecture at 10 days post-challenge infection.
Innate immunity-related genes and antiviral interferon-stimulated genes are upregulated in the lungs of KyA-immunized mice.
To investigate the mechanism by which KyA immunization protects CBA mice from subsequent challenge with pathogenic RacL11 at 1 to 7 days postimmunization, Affymetrix microarray analyses were performed with RNA from infected mouse lungs. RNA from mock-infected lungs was used as the control. The lungs of mice infected with 1.5 × 106 PFU/mouse of EHV-1 KyA were harvested at 6, 8, and 12 h postinfection (hpi) and were analyzed using the Affymetrix mouse genome 430 2.0 array, which offers complete coverage of the expression of >39,000 transcripts (Microarray Core, University of Texas Southwestern Medical Center, Dallas, TX). As shown in Table 2, genes associated with immune and inflammatory responses were significantly upregulated in the lungs of KyA-infected mice relative to expression in the lungs of control mice. SLFN3 and SLFN4, which are involved in both T cell and macrophage activation, were upregulated 10.7-fold and 6.2-fold at 8 hpi, respectively. Both CC chemokines (MCP-1, MCP-3, MCP-5, and MIP-3α) and CXC chemokines (CXCL5, CXCL10, MIP-2α, and MIP-2β) were also significantly upregulated. The cytokine gene IL-6 (interleukin 6) was significantly upregulated 45-fold at 6 hpi, 33.2-fold at 8 hpi, and 46.4-fold at 12 hpi (Table 2). Tumor necrosis factor (TNF) was upregulated 14.5-fold at 6 hpi and was reduced to 2.2-fold at 12 hpi.
TABLE 2.
Genes upregulated by EHV-1 KyA in the lungs of CBA mice
| Gene function and name | Fold changea in expression at: |
Description | ||
|---|---|---|---|---|
| 6 hpi | 8 hpi | 12 hpi | ||
| Immune and inflammatory responses | ||||
| SLFN4 | 5.5 | 10.7 | 42.4 | Schlafen 4: macrophage activation |
| SLFN3 | 4.1 | 6.2 | 24.9 | Schlafen 3: T cell activation |
| TNFRSF9 | 7.2 | 4.9 | 8.8 | TNF receptor superfamily, member 9: T cell development |
| PTX3 | 9.4 | 8.4 | 9.8 | Pentraxin 3: innate immune response |
| IRG1 | 133.1 | 7.8 | 17.7 | Immunoresponsive gene 1: inhibition of the inflammatory response |
| CD177 | 5.7 | 6.0 | — | CD177 antigen: neutrophil activation |
| S100A9 | 2.2 | 7.4 | 13.1 | S100 calcium binding protein A9: neutrophil chemotaxis and adhesion |
| S100A8 | 2.9 | 7.1 | 12.7 | S100 calcium binding protein A8: neutrophil chemotaxis and adhesion |
| CLEC4D | 5.1 | 5.3 | 7.8 | C-type lectin domain family 4, member d: inflammation and immune response |
| CLEC4E | 10.0 | 4.1 | 7.8 | C-type lectin domain family 4, member e: cell surface receptor for ligands |
| SOCS3 | 3.6 | 4.2 | 5.9 | Suppressor of cytokine signaling 3 |
| LCN2 | 5.8 | 3.6 | 6.5 | Lipocalin 2: macrophage deactivation |
| Orm2 | 31.8 | 5.8 | 7.9 | Orosomucoid 1: immunosuppression |
| CC chemokines | ||||
| CCL2 (MCP-1) | 16.0 | 11.2 | 20.0 | Chemokine (C-C motif) ligand 2 |
| CCL7 (MCP-3) | 14.3 | 8.8 | 19.8 | Chemokine (C-C motif) ligand 7 |
| CCL20 (MIP-3α) | 53.4 | 8.3 | 3.7 | Chemokine (C-C motif) ligand 20 |
| CCL12 (MCP-5) | 7.0 | 4.0 | 8.6 | Chemokine (C-C motif) ligand 12 |
| CXC chemokines | ||||
| CXCL5 | 141.3 | 31.2 | 25.0 | Chemokine (C-X-C motif) ligand 5 |
| CXCL1 | 12.6 | 5.5 | 15.6 | Chemokine (C-X-C motif) ligand 1 |
| CXCL10 | 6.2 | 20.3 | 95.0 | Chemokine (C-X-C motif) ligand 10 |
| CXCL3 (MIP-2β) | 106.9 | 10.2 | 25.5 | Chemokine (C-X-C motif) ligand 3 |
| CXCL2 (MIP-2α) | 50.8 | 11.0 | 12.4 | Chemokine (C-X-C motif) ligand 2 |
| Cytokines | ||||
| IL-6 | 45.0 | 33.2 | 46.4 | Interleukin 6 |
| IL-1β | 11.2 | 8.3 | — | Interleukin 1β |
| IL-1RN | 3.7 | 2.7 | — | Interleukin 1 receptor antagonist |
| IL-10 | 5.5 | — | — | Interleukin 10 |
| IL-11 | 7.2 | — | — | Interleukin 11 |
| TNF | 14.5 | — | 2.2 | Tumor necrosis factor |
| TLR-associated factorsb | ||||
| TLR2 | 3.4 | 2.1 | 3.1 | Toll-like receptor 2 |
| CD14 | 13.2 | 5.9 | 6.0 | CD14 antigen |
| MyD88 | 3.1 | 2.2 | 2.6 | Myeloid differentiation primary response gene 88 |
| Cell cycle and apoptosis | ||||
| Gadd45g | 5.0 | — | 2.9 | Growth arrest and DNA damage-inducible 45 gamma |
| Casp4 | 4.8 | 2.8 | 3.2 | Caspase 4: inhibition of cell growth and induction of apoptosis |
| Antibacterial factors | ||||
| Reg3g | 74.8 | — | — | Regenerating islet-derived 3 gamma |
| Adm | 10.6 | 4.3 | 4.8 | Adrenomedulin |
Mean fold change from three replicate experiments at 8 or 12 hpi and from two replicate experiments at 6 hpi. —, fold difference of <±2.0 between infected and mock-infected lungs.
TRL, Toll-like receptor.
Sixteen antiviral interferon-stimulated genes (ISGs)—RSAD2 (viperin), MX1, MX2, IRF7, IFI44L, IFIT1, IFIT2, IFIT3, ISG15, ISG20, OAS1a, OAS2, OASL2, GBP2, SLC15A3, and IFI204—were upregulated in the lungs of mice infected with EHV-1 KyA (Table 3 and data not shown). RSAD2 (viperin), which is known to be antiviral against dengue virus, human cytomegalovirus, and hepatitis C virus, was upregulated 6.2-fold at 8 hpi and 12.8-fold at 12 hpi. Influenza virus resistance genes MX1 and MX2 were upregulated 24.6- and 6.1-fold at 12 hpi, respectively (data not shown).
TABLE 3.
EHV-1 KyA immunization increases the expression of antiviral genes in the lungs of CBA mice challenged with RacL11
| Gene name | KyA-infected mice |
RacL11-infected mice |
KyA → RacL11-infected micea |
HI-KyA → RacL11-infected miceb |
||||
|---|---|---|---|---|---|---|---|---|
| Fold changec | P valued | Fold changec | P valued | Fold changec | P valued | Fold changec | P valued | |
| RSAD2 | 6.2 | 0.024032 | 4.7 | 0.004344 | 21.6 | 0.000066 | 5.0 | 0.029294 |
| MX1 | 4.5 | 0.014696 | 4.7 | 0.015646 | 29.4 | 0.000057 | 5.1 | 0.022906 |
| MX2 | 2.0 | 0.046565 | 2.4 | 0.010745 | 5.5 | 0.000022 | - | |
| IRF7 | 2.3 | 0.040911 | 2.7 | 0.002847 | 27.8 | 0.000002 | 2.2 | 0.049218 |
| IFI44L | 2.3 | 0.039495 | 2.4 | 0.005339 | 48.2 | 0.000001 | 2.8 | 0.043189 |
| IFIT1 | 4.9 | 0.022448 | 5.2 | 0.002613 | 12.9 | 0.000130 | 4.7 | 0.011233 |
| IFIT2 | 3.4 | 0.039152 | 2.7 | 0.009578 | 7.9 | 0.000065 | 2.4 | 0.019820 |
| IFIT3 | 2.7 | 0.034187 | 3.1 | 0.000898 | 8.5 | 0.000001 | 2.8 | 0.011494 |
| ISG15 | 3.8 | 0.027831 | 4.4 | 0.004029 | 19.7 | 0.000102 | 2.6 | 0.028449 |
| ISG20 | — | — | 3.9 | 0.000062 | — | |||
| OAS1a | — | 2.0 | 0.001532 | 11.1 | 0.000009 | — | ||
| OAS2 | — | — | 0.006082 | 9.8 | 0.000087 | — | ||
| OASL2 | 2.1 | 0.025561 | 2.5 | 0.000725 | 8.4 | 0.000008 | — | |
| GBP2 | 2.0 | 0.036321 | — | 4.4 | 0.000159 | — | ||
| SLC15A3 | — | — | 3.1 | 0.000657 | — | |||
| IFI204 (IFI16) | 3.0 | 0.053942 | 2.0 | 0.062900 | 29.7 | 0.000098 | 2.5 | 0.038217 |
| IFN-γ | — | — | 8.5 | 0.000413 | — | |||
CBA mice immunized with EHV-1 KyA were challenged with pathogenic RacL11 at 3 days postimmunization.
CBA mice immunized with heat-inactivated EHV-1 KyA were challenged with pathogenic RacL11 at 3 days postimmunization.
Mean fold change in expression from three replicate experiments at 8 h postinfection (for KyA- or RacL11-infected mice) or 8 h post-RacL11 challenge (for KyA → RacL11- or HI-KyA → RacL11-infected mice). —, fold difference of <±2.0 between infected and mock-infected lungs.
Statistics for the three replicates of each gene were calculated independently. Mean signals were compared by using a t test analysis assuming equal variances and 6 degrees of freedom.
KyA immunization increases the expression of 16 antiviral ISGs and the IFN-γ gene upon RacL11 challenge.
Additional microarray analyses of RNA isolated from the lungs of mice challenged with RacL11 at 3 days postimmunization were performed. Levels of antiviral ISG expression in control nonimmunized RacL11-infected mice were very similar to those of KyA-infected mice (Table 3). Interferon gamma (IFN-γ) and 16 antiviral ISGs were upregulated 3.1- to 48.2-fold at 8 h postchallenge in the lungs of RacL11-challenged mice that had been immunized with KyA (Table 3). In contrast, IFN-γ and 16 antiviral ISGs were upregulated only 1- to 5.2-fold in the lungs of nonimmunized mice upon RacL11 challenge (Table 3). Of these genes, the IFN-γ, IRF7, IFI44L, ISG15, and IFI204 genes were upregulated 8.5, 27.8-, 48.2-, 19.7-, and 29.7-fold, respectively, after KyA immunization and RacL11 challenge (Table 3). However, immunization with heat-inactivated KyA was not able to increase the expression of the 16 antiviral ISGs and the IFN-γ gene upon RacL11 challenge over the levels observed in the lungs of KyA-infected mice (Table 3). The lungs of mice immunized with KyA and then challenged with KyA at 3 days postimmunization showed levels of IFN-γ and six ISGs (RSAD, MX1, IRF7, IFI44L, ISG15, and IFI204) that were 2- to 3-fold greater than those in the lungs of RacL11-challenged mice that had been immunized with KyA (qRT-PCR data not shown). These results demonstrated that a second immunization with KyA results in the potent expression of IFN-γ and multiple ISGs.
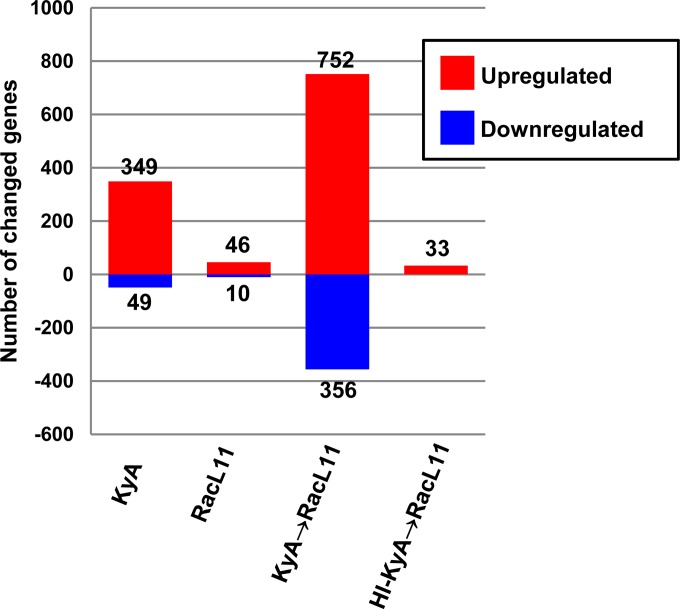
Genome-wide gene expression profiles were obtained for KyA-infected mice, RacL11-infected mice, and RacL11-challenged mice. A total of 349 genes were upregulated, and 49 genes were downregulated, in the lungs of KyA-infected mice (Fig. 5). However, only 46 and 10 genes were upregulated and downregulated in RacL11-infected mice, respectively (Fig. 5). Interestingly, much greater numbers of genes were upregulated (752 genes) and downregulated (356 genes) in the lungs of RacL11-challenged mice that had been immunized with live KyA (Fig. 5). As shown in Fig. 5, only 33 genes were upregulated in the lungs of RacL11-challenged mice that had been immunized with heat-inactivated KyA. These results demonstrated that immunization with infectious KyA results in significant changes in the expression of many cellular genes in the mouse lung, including the upregulation of 16 antiviral ISGs.
FIG 5.
Immunization with EHV-1 KyA increases the number of genes with changed expression in the lungs of CBA mice upon RacL11 challenge. The numbers of genes from the total microarray data sets that showed a >2.0-fold increase (red) or a >2.0-fold reduction (blue) in expression (P ≤ 0.05) are given. Data for KyA and RacL11 were obtained at 8 h postinfection. Data for KyA- or heat-inactivated KyA-immunized mice challenged with RacL11 (KyA → RacL11 or HI-KyA → RacL11, respectively) were obtained at 8 h post-RacL11 challenge.
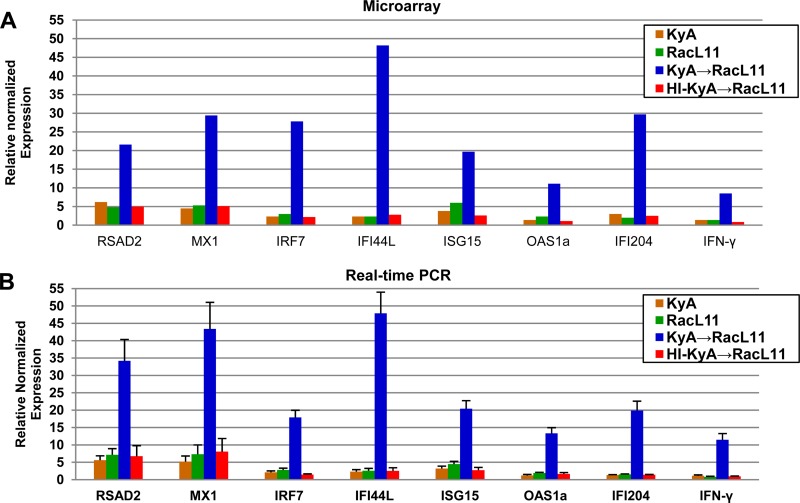
Validation of Affymetrix microarray data by real-time PCR analysis.
To confirm the significance of the results of the microarray data, qRT-PCR analyses were carried out for the relevant genes from the original samples used in the microarray study (Table 3). Seven significantly upregulated ISGs (RSAD2, MX1, IRF7, IFI44L, ISG15, OAS1a, and IFI204) and the IFN-γ gene were selected for qRT-PCR analysis. The data presented in Fig. 6 and Table 4 compare the results obtained from the microarray analysis with those of qRT-PCR and indicate that the changes in expression levels for these eight selected genes as detected by qRT-PCR were consistent with the changes predicted by microarray analysis. However, the fold changes in RSAD2, MX1, IRF7, and IFI204 expression differed between the microarray and qRT-PCR. This discrepancy may be attributed to the technical differences in the methods of analysis and normalization. Despite the difference in the levels of gene expression, both methods showed that the expression of these antiviral genes was increased after RacL11 challenge.
FIG 6.
Validation of Affymetrix microarray data by real-time PCR analysis. The relative mRNA expression of seven of the most highly upregulated ISGs and the IFN-γ gene, shown in Table 3, was quantified using qRT-PCR. Data were normalized to GAPDH expression. The genes selected are seven antiviral ISGs (RSAD2, MX1, IRF7, IFI44L, ISG15, OAS1a, and IFI204) and the IFN-γ gene. Data for KyA and RacL11 were obtained at 8 h postinfection. Data for KyA- or heat-inactivated KyA-immunized mice challenged with RacL11 (KyA → RacL11 or HI-KyA → RacL11, respectively) were obtained at 8 h post-RacL11 challenge. P values for KyA → RacL11 and HI-KyA → RacL11 mice are shown in Table 4. Results are expressed as the fold change from expression in the mock-infected control.
TABLE 4.
Validation of microarray results by qRT-PCR
| Gene name | KyA → RacL11-infected micea |
HI-KyA → RacL11-infected miceb |
||||
|---|---|---|---|---|---|---|
| Fold change by: |
P value for qRT-PCR resulte | Fold change by: |
P value for qRT-PCR resulte | |||
| Microarrayc | qRT-PCRd | Microarrayc | qRT-PCRd | |||
| RSAD2 | 21.6 | 34.2 | 0.0050 | 5.0 | 6.7 | 0.0900 |
| MX1 | 29.4 | 43.2 | 0.0040 | 5.1 | 7.9 | 0.0800 |
| IRF7 | 27.8 | 17.9 | 0.0004 | 2.2 | 1.4 | 0.0610 |
| IFI44L | 48.2 | 45.2 | 0.0003 | 2.8 | 2.3 | 0.1200 |
| ISG15 | 19.7 | 20.4 | 0.0006 | 2.6 | 2.5 | 0.0920 |
| OAS1a | 11.1 | 12.9 | 0.0010 | 4.7 | 1.5 | 0.2200 |
| IFI204 | 29.7 | 19.9 | 0.0069 | 2.5 | 1.4 | 0.0348 |
| IFN-γ | 8.5 | 10.6 | 0.0020 | — | 0.9 | 0.3710 |
CBA mice immunized with EHV-1 KyA were infected with pathogenic RacL11 at 3 days postimmunization.
CBA mice immunized with HI-KyA were infected with pathogenic RacL11 at 3 days postimmunization.
Mean fold change from three replicate experiments at 8 h post-RacL11 challenge. —, fold difference of < ±2.0 between infected and mock-infected lungs.
The relative mRNA expression of seven antiviral interferon-stimulated genes (RSAD2, MX1, IRF7, IFI44L, ISG15, OAS1a, and IFI204) and the IFN-γ gene, shown in Table 3, was quantified using qRT-PCR. RT-PCR data were normalized to GAPDH expression.
Statistics for the three replicates of each gene were calculated independently. Mean signals were compared by using a t test analysis assuming equal variances and 6 degrees of freedom.
IFN-γ inhibits EHV-1 replication in a murine alveolar macrophage cell line.
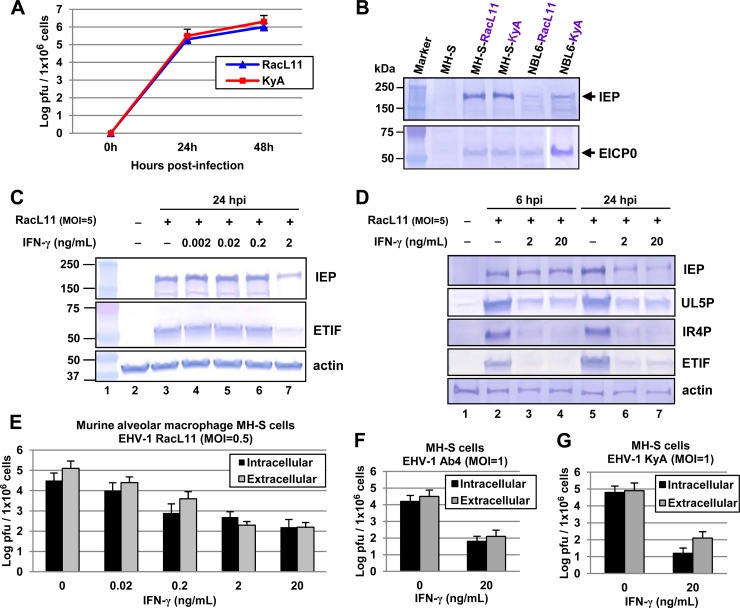
Macrophages in the alveolar lumen provide an innate defense mechanism, and IFN-γ is the prototypical macrophage-activating cytokine. The primary immune response in the lungs of mice infected with wild-type EHV-1 was an alveolar and interstitial inflammation, dominated by the sequential appearance of neutrophils and macrophages, while the numbers of T and B lymphocytes remained unaltered (52). Lung sections taken from EHV-1 RacL11-infected mice revealed massive cellular consolidation of the lung, consisting primarily of lymphocytes, macrophages, and neutrophils (23). Severe murine lung immunopathology elicited by RacL11 correlated with early production of macrophage inflammatory proteins 1α, 1β, and 2 and tumor necrosis factor alpha (23). In a cell culture model of Ebola virus infection of mouse macrophages, murine IFN-γ provided >95% protection with a dose of only 2 ng/ml (53). EHV-1 KyA and RacL11 were able to replicate in MH-S murine alveolar macrophages, and the EHV-1 regulatory proteins IEP (immediate early protein) and EICP0 were expressed at 6 hpi (Fig. 7A and B). To investigate the effects of IFN-γ on EHV-1 infection in this cell type, increasing concentrations of IFN-γ were evaluated for their effects on viral gene expression. Treatment with 2 ng/ml of IFN-γ reduced the expression levels of viral regulatory proteins IEP and ETIF (a late protein) by 70% and 85% (as determined by PhosphorImager analysis) at 24 hpi, respectively, from those of nontreated cells (Fig. 7C). Also, the levels of two viral early proteins (UL5P and IR4P) and one late protein, ETIF, were greatly reduced in these cells at both 6 and 24 hpi (Fig. 7D). Interestingly, IFN-γ was not able to reduce the levels of IEP expression at 6 hpi (Fig. 7D), indicating that IFN-γ may inhibit viral gene expression after the immediate early stage of infection.
FIG 7.
IFN-γ-treated MH-S murine alveolar macrophages are resistant to EHV-1 infection. (A) EHV-1 strains KyA and RacL11 replicate in MH-S murine alveolar macrophages (CRL-2019; American Type Culture Collection). Virus titers were determined at 0, 24, and 48 hpi. MH-S cells (1 × 106) were infected with KyA or RacL11 (multiplicity of infection [MOI], 1). Intracellular virus was released by three freeze-thaw cycles at 0, 24, or 48 hpi, and titers were determined by plaque assays, which were performed as described previously (50). Each sample was assayed in triplicate. Error bars indicate standard deviations. (B) MH-S cells were infected with KyA or RacL11 at an MOI of 5, harvested at 6 hpi, and used for Western blot analyses with the anti-IEP polyclonal antibody OC33 (77) and an anti-EICP0 polyclonal antibody (28). The sizes of molecular mass standards (Bio-Rad Laboratories, Hercules, CA) are given on the left. (C and D) IFN-γ reduced EHV-1 gene expression. (C) MH-S cells were treated with 0.002, 0.02, 0.2, or 2 ng/ml of murine IFN-γ (485-MI; Cell Sciences, Canton, MA) and were infected with EHV-1 RacL11 (MOI, 5) at 24 h posttreatment. The infected cells were harvested at 24 hpi and were used for Western blot analyses with the anti-IEP polyclonal antibody OC33 (77), anti-ETIF (78), and anti-actin (SC-1615-R; Santa Cruz Biotechnology, Santa Cruz, CA). The sizes of molecular mass standards (in kilodaltons) (Bio-Rad Laboratories, Hercules, CA) are given on the left. (D) MH-S cells were treated with 2 or 20 ng/ml of murine IFN-γ and were infected with EHV-1 RacL11 (MOI, 5) at 24 h posttreatment. The infected cells were harvested at 6 or 24 hpi for Western blot analyses using antibodies to the early proteins UL5 (79) and IR4 (80). (E to G) Virus titers were determined by plaque assays on NBL6 cells. (E) MH-S cells (1 × 106) were treated with 0.02, 0.2, 2, or 20 ng/ml of IFN-γ, infected with EHV-1 RacL11 (MOI, 1) at 24 h posttreatment, and washed four times with growth medium without serum. Intracellular virus was released by three freeze-thaw cycles at 24 hpi. (F and G) MH-S cells (1 × 106) were treated with 20 ng/ml of IFN-γ, infected with wild-type EHV-1 Ab4 (MOI, 1) (F) or EHV-1 KyA (MOI, 1) (G) at 24 h posttreatment, and washed four times with growth medium without serum. Each sample was assayed in triplicate. Error bars indicate standard deviations.
To investigate whether murine IFN-γ inhibits EHV-1 replication, increasing concentrations of IFN-γ were evaluated for the inhibition of RacL11 by plaque assays. IFN-γ reduced intracellular and extracellular virus yields at 24 hpi in a dose-dependent manner (Fig. 7E). When MH-S cells were treated with 2 or 20 ng/ml of murine IFN-γ, extracellular virus yields at 24 hpi (Fig. 7E) were very similar to those at 0 hpi (data not shown), which was indicative of no virus release. MH-S cells treated with 20 ng/ml of murine IFN-γ exhibited an intracellular virus yield 199-fold lower than that of untreated cells (Fig. 7E). Treatment with IFN-γ also reduced yields of intracellular wild-type EHV-1 Ab4 (Fig. 7F) and KyA (Fig. 7G) by 251- and 398-fold, respectively, from those of untreated cells. However, murine IFN-γ was not able to reduce virus yields in mouse fibroblast L-M cells (data not shown). These results indicated that IFN-γ blocks EHV-1 replication in MH-S murine alveolar macrophages.
IFN-γ protects CBA mice against lethal EHV-1 challenge.
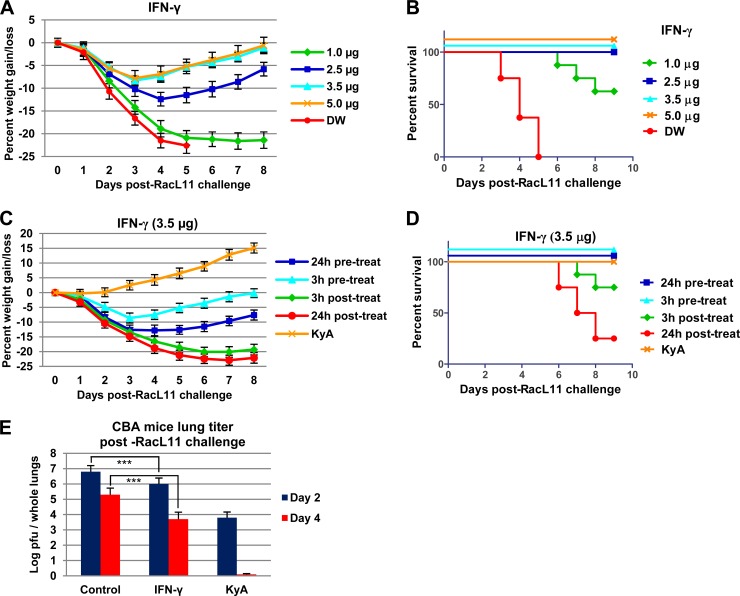
EHV-1 KyA immunization significantly increased IFN-γ and antiviral ISG mRNA levels upon pathogenic RacL11 challenge (Table 3 and Fig. 6). Since IFN-γ inhibited EHV-1 replication in murine alveolar macrophages (Fig. 7), and the alveolar macrophage is an important cell target for EHV-1 infection in the equine (54), the ability of IFN-γ to protect mice from pathogenic RacL11 challenge was investigated. CBA mice were treated intranasally with 1, 2.5, 3.5, or 5 μg of IFN-γ and were challenged with 1.5 × 106 PFU/mouse of EHV-1 RacL11 at 3 h posttreatment. Mice treated with 2.5, 3.5, or 5 μg of IFN-γ lost less than 12% of their preinfection body weight and rapidly regained their body weight (Fig. 8A); all these infected mice survived (Fig. 8B). However, mice treated with 1 μg of IFN-γ lost more than 20% of their preinfection body weight by day 5 p.i. (Fig. 8A), and 3 of the 8 mice in this group succumbed to RacL11 challenge infection by day 8 p.i. (Fig. 8B). Initial IFN-γ dose-response studies indicated that 3.5 μg and 5 μg of IFN-γ similarly protected mice against lethal EHV-1 challenge. Therefore, 3.5 μg of IFN-γ was administered 24 h prior to, 3 h prior to, 3 h after, or 24 h after a lethal dose of EHV-1 RacL11. Mice administered IFN-γ at 3 h or 24 h after a lethal dose of EHV-1 RacL11 had less morbidity and mortality than mice treated with distilled water, but 25% and 75% of these mice succumbed to RacL11 challenge, respectively (Fig. 8C and D). All mice given IFN-γ at 3 h or 24 h prior to a lethal dose of EHV-1 RacL11 survived (Fig. 8C and D), indicating that IFN-γ administration may prove more efficacious before exposure than after exposure. CBA mice were treated intranasally with IFN-γ and were challenged with 1.5 × 106 PFU/mouse of RacL11 at 3 h posttreatment, and the amount of infectious EHV-1 in the lungs was determined. Virus titers in the lungs of IFN-γ-treated mice were 7-fold lower at 2 dpi and 40-fold lower at 4 dpi than those in the lungs of mock-treated mice (Fig. 8E). These results indicated that murine IFN-γ effectively protected CBA mice against lethal EHV-1 challenge.
FIG 8.
IFN-γ protects mice from lethal EHV-1 RacL11 challenge. (A and B) Average body weights (A) and percentages of survival (B) of CBA mice treated with different dosages of IFN-γ. CBA mice (n = 8) were treated intranasally with 1, 2.5, 3.5, or 5 μg of murine IFN-γ per mouse (Cell Sciences, Canton, MA) and were challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 h posttreatment. (C and D) Average body weights (C) and percentages of survival (D) of CBA mice treated with IFN-γ. Mice (n = 8) were treated with 3.5 μg of murine IFN-γ 24 h prior to, 3 h prior to, 3 h following, or 24 h following EHV-1 RacL11 (1.5 × 106 PFU/mouse) infection. Mice immunized with KyA (2 × 106 PFU/mouse) were challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 dpi. (E) Virus titers from the lungs of CBA mice treated with IFN-γ. Mice (n = 6) were treated with 3.5 μg/mouse of murine IFN-γ and were challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 hpi. Mice immunized with KyA (2 × 106 PFU/mouse) were challenged with RacL11 (1.5 × 106 PFU/mouse) at 3 dpi. On days 2 to 4 postchallenge, virus titers in the lungs were determined as described in the legend to Fig. 4. Triple asterisks indicate statistical significance (P < 0.01).
DISCUSSION
We report here that intranasal EHV-1 KyA immunization significantly increased the expression of IFN-γ and 16 antiviral interferon-stimulated genes (ISGs) upon pathogenic RacL11 challenge, accelerated the clearance of virus from the lungs of infected CBA mice, and protected mice at 1 to 7 days postimmunization. Murine IFN-γ treatment effectively protected mice against lethal EHV-1 challenge. These results suggest that IFN-γ expression may be important in mediating the protection elicited by KyA immunization.
The innate immune response provides the first line of defense against viral pathogens. Incoming viruses are sensed by pattern recognition receptors, leading to the activation of interferon regulatory factors (IRFs) and the subsequent transcriptional induction of interferons. IFNs bind their cognate receptors, signal through the JAK-STAT pathway, and transcriptionally induce hundreds of ISGs, many of which function to block virus replication (36, 55, 56). Our study showed that 16 antiviral ISGs were upregulated 3.1- to 48.2-fold at 8 h post-RacL11 challenge (Table 3). Of these genes, RSAD (viperin), MX1, IRF7, IFI44L, ISG15, and IFI204 were upregulated 19.7- to 48.2-fold. The ability of KyA to activate innate immune responses was clearly shown in experiments in which KyA was administered at day 0 and again at day 3. In the lungs of these animals, the six ISGs and IFN-γ were upregulated at 8 h after the second KyA infection to levels 2- to 3-fold greater than those in the lungs of RacL11-challenged mice that had been immunized once with KyA. Interferon regulatory factor 7 (IRF7) can activate ISG expression in the absence of IFN signaling (57) and plays a critical role in the innate immune response against both DNA and RNA viruses (58–60). The ubiquitin-like molecule ISG15 plays a central role in the host's antiviral response to approximately 18 viruses, including herpes simplex virus 1 (HSV-1) and murine gammaherpesvirus 68 (61–64). In agreement with these published findings on the importance of ISG15 in innate immune responses to herpesviruses, the level of ISG15 expression was increased 19.7-fold in KyA-immunized mice following RacL11 challenge (Table 3), and our ongoing experiments reveal that intranasal administration of a recombinant adenovirus expressing murine ISG15 protects CBA mice from lethal challenge with EHV-1 RacL11 (data not shown). These data suggest that ISG15 functions as an antiviral molecule against EHV-1, but its mechanism of action, as well as the identity of the EHV-1 molecules that induce ISG15, remains to be determined. In this regard, comparison of alphaherpesvirus genomes showed that varicella-zoster virus, pseudorabies virus, and HSV-1 are similar in structure to EHV-1 and that each of these viruses encodes a protein that shares extensive homology with the EHV-1 immediate early protein (65, 66). Infection with EHV-1 wild-type strain Ab4 reduced IFN-γ production in primary equine respiratory epithelial cells (67). Our microarray data showed that infection with EHV-1 KyA or RacL11 was not able to induce IFN-γ gene expression (<2-fold increase) in the lungs of CBA mice at 8 h postinfection. Interestingly, the IFN-γ expression level was increased 8.5-fold at 8 h postchallenge in the lungs of RacL11-challenged mice that had been immunized with KyA (Table 3). Since IFN-γ, IFN-α, and lipopolysaccharide (LPS) induce IFI204 expression (68), it may be significant that IFN-γ upregulated IFI204 29.7-fold in the lungs of RacL11-challenged mice that had been immunized with KyA 3 days prior to challenge. IFI16, a human homolog of IFI204, has been shown to interact with STING (stimulator of interferon genes), leading to the phosphorylation and nuclear translocation of IRF3 via the IFI16-STING-TBK signaling axis (69, 70).
Alveolar macrophages, one of the prominent types of cells of the immune system found in the airways, constitute a unique subset of macrophages that serve as a first line of defense against foreign invaders in the lung. IFN-γ treatment inhibited EHV-1 replication in murine alveolar macrophages (Fig. 7) and protected mice from lethal challenge (Fig. 8). A single treatment with IFN-γ protected mice against EHV-1 challenge, but only when given prior to infection (Fig. 8C and D). However, in the case of Ebola virus, postchallenge treatment was more efficacious (53), and other studies have shown that multiple treatments with IFN-γ protected mice from a lethal challenge with vaccinia virus via the respiratory route (71). EHV-1 infection did not cause a decrease in the viability of IFN-γ-treated MH-S alveolar macrophages (data not shown). Also, no virus release was detected (Fig. 7), suggesting that murine IFN-γ inhibits EHV-1 replication in murine alveolar macrophages as well as the spread of the virus to the mouse lung.
Infection with attenuated EHV-1 KyA elicits humoral and cell-mediated immune responses capable of protecting mice from challenge infection (14, 17, 72). A West Nile virus recombinant protein vaccine, which coactivates both innate and adaptive immunity, induced a protective immune response when mice were challenged 10 weeks after two or three immunizations (73). Our data showed that a single immunization with EHV-1 KyA effectively protected CBA mice from pathogenic RacL11 challenge at 1 to 7 days postimmunization. These results suggest that upregulated IFN-γ and the 16 upregulated antiviral proteins function to block EHV-1 replication within the mouse lung. Other factors, yet to be identified, may also be involved in the inhibition of the EHV-1 life cycle. Taken together, our previously published findings and these results indicate that EHV-1 KyA protects CBA mice by both the innate and adaptive immune responses.
EHV-1 infection can be subclinical or can result in economically important outcomes, such as abortion, neonatal death, or neurological disease. EHV-1 neurological disease has been predominant both in the United States and in Europe over the past decade (74), and it has been speculated that this phenomenon might be associated with excessive vaccination using inactivated products that promote a bona fide immunopathological condition. The economic losses from EHV-1 infection have led to increased efforts to understand EHV-1 pathogenesis, with the ultimate goal of developing a safe and effective vaccine. At present, there is no equine vaccine that has a label claim for protection against the neurological strain of the virus. Several inactivated vaccines are available for protection against the respiratory and abortogenic forms of EHV-1; however, they do not elicit long-term immunity or protection in the horse (45–47). Our findings demonstrated that EHV-1 KyA immunization greatly increased the number of genes with changed expression, as well as the levels of expression of the 16 antiviral ISGs and IFN-γ in the lungs of CBA mice. Even low-dose KyA immunization was able to protect mice from RacL11 challenge (Fig. 2D and E). The fact that KyA is able to grow rapidly in a wide variety of cell types, including mouse fibroblast L-M, rabbit kidney RK13, equine NBL6, monkey kidney Vero, and human HeLa cells (17, 27, 48, 50, 75), and replicates to high titers of released virus makes it an attractive vaccine candidate.
Our previous results showed that recombinant EHV-1 glycoprotein D (gD), expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli, elicited both high titers of neutralizing antibody and CD4 T cell proliferative responses following subcutaneous or intranasal immunization of CBA mice but elicited only a weak antibody response after intraperitoneal immunization (14). Intranasal administration of purified recombinant gD induced partial or complete protection against three different EHV-1 strains (AR11, AR52, and HH1) in BALB/c mice (76). Levels of the IFN-γ gene and 16 antiviral ISGs were significantly upregulated upon pathogenic RacL11 challenge (Table 3), suggesting that immunization with attenuated EHV-1 KyA induces innate immune responses upon pathogenic EHV-1 challenge within 2 weeks postimmunization that protect mice. EHV-1 KyA immunization protected CBA mice from pathogenic RacL11 challenge at 4 weeks postimmunization, and the KyA infection elicited protective humoral and cell-mediated immune responses (14, 17, 71), indicating that immunization with attenuated EHV-1 KyA protects mice from pathogenic RacL11 challenge through adaptive immune responses. It is possible that immunization with attenuated EHV-1 KyA protects horses from pathogenic EHV-1 challenge by both the innate and adaptive immune responses. These results suggest that EHV-1 KyA may be used as a live attenuated EHV-1 vaccine as well as a prophylactic agent in horses. These are important hypotheses for our future work.
Elucidation of the mechanisms by which IFN-γ and antiviral ISGs may control EHV-1 replication will provide a basis for more-effective treatment of equine infections and will contribute to the understanding of ISGs as antiherpesvirus proteins. Our initial efforts to generate and characterize recombinant adenoviruses expressing antiviral ISGs in the CBA mouse model have generated encouraging results, and this approach may provide more insight into the importance of specific ISGs in the prevention of EHV-1 infection. Our future studies of KyA immunization and IFN-γ administration in the natural host may offer a new strategy to help control this major equine pathogen.
ACKNOWLEDGMENTS
We thank Matthew D. Woolard and Michelle M. Arnold for their critical readings of the manuscript and comments.
REFERENCES
- 1.Allen GP, Bryans JT. 1986. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog Vet Microbiol Immunol 2:78–144. [PubMed] [Google Scholar]
- 2.Carroll CL, Westbury HA. 1985. Isolation of equine herpesvirus 1 from the brain of a horse affected with paresis. Aust Vet J 62:345–346. doi: 10.1111/j.1751-0813.1985.tb07660.x. [DOI] [PubMed] [Google Scholar]
- 3.Jackson T, Kendrick JW. 1971. Paralysis of horses associated with equine herpesvirus 1 infection. J Am Vet Med Assoc 158:1351–1357. [PubMed] [Google Scholar]
- 4.O'Callaghan DJ, Osterrieder N. 1999. Equine herpesviruses, p 508–515. In Webster RG, Granoff A (ed), Encyclopedia of virology, 2nd ed Academic Press Ltd, London, United Kingdom. [Google Scholar]
- 5.Bryans JT, Allen GP. 1986. Equine viral rhinopneumonitis. Rev Sci Tech 5:837–847. [DOI] [PubMed] [Google Scholar]
- 6.Crandell R, Mock R, Lock T. 1980. Vaccination of pregnant ponies against equine rhinopneumonitis. Am J Vet Res 41:994–996. [PubMed] [Google Scholar]
- 7.Awan AR, Chong Y-C, Field HJ. 1990. The pathogenesis of equine herpesvirus type 1 in the mouse: a new model for studying host responses to the infection. J Gen Virol 71:1131–1140. doi: 10.1099/0022-1317-71-5-1131. [DOI] [PubMed] [Google Scholar]
- 8.Awan A, Gibson J, Field H. 1991. A murine model for studying EHV-1-induced abortion. Res Vet Sci 51:94–99. doi: 10.1016/0034-5288(91)90038-P. [DOI] [PubMed] [Google Scholar]
- 9.Bell CW, Boyle DB, Whalley JM. 1990. Transcript analysis of the equine herpesvirus 1 glycoprotein B gene homologue and its expression by a recombinant vaccinia virus. J Gen Virol 71:1119–1129. doi: 10.1099/0022-1317-71-5-1119. [DOI] [PubMed] [Google Scholar]
- 10.Guo P, Goebel S, Perkus M, Taylor J, Norton E, Allen G, Languet B, Desmettre P, Paoletti E. 1990. Coexpression by vaccinia virus recombinants of equine herpesvirus 1 glycoproteins gp13 and gp14 results in potentiated immunity. J Virol 64:2399–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osterrieder N, Wagner R, Pfeffer M, Kaaden O-R. 1994. Expression of equine herpesvirus type 1 glycoprotein gp14 in Escherichia coli and in insect cells: a comparative study on protein processing and humoral immune responses. J Gen Virol 75:2041–2046. doi: 10.1099/0022-1317-75-8-2041. [DOI] [PubMed] [Google Scholar]
- 12.Stokes A, Alber D, Cameron R, Marshall R, Allen G, Killington R. 1996. The production of a truncated form of baculovirus expressed EHV-1 glycoprotein C and its role in protection of C3H (H-2Kk) mice against virus challenge. Virus Res 44:97–109. doi: 10.1016/0168-1702(96)01339-1. [DOI] [PubMed] [Google Scholar]
- 13.Tewari D, Whalley J, Love D, Field H. 1994. Characterization of immune responses to baculovirus-expressed equine herpesvirus type 1 glycoproteins D and H in a murine model. J Gen Virol 75:1735–1741. doi: 10.1099/0022-1317-75-7-1735. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Smith PM, Tarbet EB, Osterrieder N, Jennings SR, O'Callaghan DJ. 1998. Protective immunity against equine herpesvirus type-1 (EHV-1) infection in mice induced by recombinant EHV-1 gD. Virus Res 56:11–24. doi: 10.1016/S0168-1702(98)00054-9. [DOI] [PubMed] [Google Scholar]
- 15.Allen G, Yeargan M, Costa L, Cross R. 1995. Major histocompatibility complex class I-restricted cytotoxic T-lymphocyte responses in horses infected with equine herpesvirus 1. J Virol 69:606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis JA, Steeves E, Wright AK, Bogdan JR, Davis WC, Kanara EW, Haines DM. 1997. Cell-mediated cytolysis of equine herpesvirus-infected cells by leukocytes from young vaccinated horses. Vet Immunol Immunopathol 57:201–214. doi: 10.1016/S0165-2427(96)05749-2. [DOI] [PubMed] [Google Scholar]
- 17.Smith PM, Zhang Y, Jennings SR, O'Callaghan DJ. 1998. Characterization of the cytolytic T-lymphocyte response to a candidate vaccine strain of equine herpesvirus 1 in CBA mice. J Virol 72:5366–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colle CF, Tarbet EB, Grafton WD, Jennings SR, O'Callaghan DJ. 1996. Equine herpesvirus-1 strain KyA, a candidate vaccine strain, reduces viral titers in mice challenged with a pathogenic strain, RacL. Virus Res 43:111–124. doi: 10.1016/0168-1702(96)01324-X. [DOI] [PubMed] [Google Scholar]
- 19.Frampton AR, Smith PM, Zhang Y, Matsumura T, Osterrieder N, O'Callaghan DJ. 2002. Contribution of gene products encoded within the unique short segment of equine herpesvirus 1 to virulence in a murine model. Virus Res 90:287–301. doi: 10.1016/S0168-1702(02)00245-9. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura T, Kondo T, Sugita S, Damiani AM, O'Callaghan DJ, Imagawa H. 1998. An equine herpesvirus type 1 recombinant with a deletion in the gE and gI genes is avirulent in young horses. Virology 242:68–79. doi: 10.1006/viro.1997.8984. [DOI] [PubMed] [Google Scholar]
- 21.Matsumura T, O'Callaghan D, Kondo T, Kamada M. 1996. Lack of virulence of the murine fibroblast adapted strain, Kentucky A (KyA), of equine herpesvirus type 1 (EHV-1) in young horses. Vet Microbiol 48:353–365. doi: 10.1016/0378-1135(09)59999-3. [DOI] [PubMed] [Google Scholar]
- 22.Smith PM, Kahan SM, Rorex CB, von Einem J, Osterrieder N, O'Callaghan DJ. 2005. Expression of the full-length form of gp2 of equine herpesvirus 1 (EHV-1) completely restores respiratory virulence to the attenuated EHV-1 strain KyA in CBA mice. J Virol 79:5105–5115. doi: 10.1128/JVI.79.8.5105-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith PM, Zhang Y, Grafton WD, Jennings SR, O'Callaghan DJ. 2000. Severe murine lung immunopathology elicited by the pathogenic equine herpesvirus 1 strain RacL11 correlates with early production of macrophage inflammatory proteins 1α, 1β, and 2 and tumor necrosis factor alpha. J Virol 74:10034–10040. doi: 10.1128/JVI.74.21.10034-10040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reczko E, Mayr A. 1963. On the fine structure of a virus of the herpes group isolated from horses. Arch Gesamte Virusforsch 13:591–593. (In German.) doi: 10.1007/BF01267802. [DOI] [PubMed] [Google Scholar]
- 25.Allen GP, Yeargan MR, Bryans JT. 1983. Alterations in the equine herpesvirus 1 genome after in vitro and in vivo virus passage. Infect Immun 40:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Callaghan D, Allen G, Randall C. 1978. Structure and replication of the equine herpesviruses, p 1–32. In Bryans JT, Gerber H (ed), Proceedings, 4th International Conference on Equine Infectious Diseases. Veterinary Publications Inc, Princeton, NJ. [Google Scholar]
- 27.Randall CC, Lawson LA. 1962. Adaptation of equine abortion virus to Earle's L cells in serum-free medium with plaque formation. Exp Biol Med 110:487–489. doi: 10.3181/00379727-110-27558. [DOI] [PubMed] [Google Scholar]
- 28.Bowles DE, Holden VR, Zhao Y, O'Callaghan DJ. 1997. The ICP0 protein of equine herpesvirus 1 is an early protein that independently transactivates expression of all classes of viral promoters. J Virol 71:4904–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colle CF, Flowers CC, O'Callaghan DJ. 1992. Open reading frames encoding a protein kinase, homolog of glycoprotein gX of pseudorabies virus, and a novel glycoprotein map within the unique short segment of equine herpesvirus type 1. Virology 188:545–557. doi: 10.1016/0042-6822(92)90509-N. [DOI] [PubMed] [Google Scholar]
- 30.Flowers CC, O'Callaghan DJ. 1992. The equine herpesvirus type 1 (EHV-1) homolog of herpes simplex virus type 1 US9 and the nature of a major deletion within the unique short segment of the EHV-1 KyA strain genome. Virology 190:307–315. doi: 10.1016/0042-6822(92)91217-I. [DOI] [PubMed] [Google Scholar]
- 31.Matsumura T, Smith RH, O'Callaghan DJ. 1993. DNA sequence and transcriptional analyses of the region of the equine herpesvirus type 1 Kentucky A strain genome encoding glycoprotein C. Virology 193:910–923. doi: 10.1006/viro.1993.1200. [DOI] [PubMed] [Google Scholar]
- 32.Telford EA, Watson MS, McBride K, Davison AJ. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304–316. doi: 10.1016/0042-6822(92)90706-U. [DOI] [PubMed] [Google Scholar]
- 33.Yalamanchili RR, O'Callaghan DJ. 1990. Sequence and organization of the genomic termini of equine herpesvirus type 1. Virus Res 15:149–161. doi: 10.1016/0168-1702(90)90005-V. [DOI] [PubMed] [Google Scholar]
- 34.Yalamanchili RR, Raengsakulrach B, O'Callaghan DJ. 1991. Equine herpesvirus 1 sequence near the left terminus codes for two open reading frames. Virus Res 18:109–116. doi: 10.1016/0168-1702(91)90012-K. [DOI] [PubMed] [Google Scholar]
- 35.Tong W, Li G, Liang C, Liu F, Tian Q, Cao Y, Li L, Zheng X, Zheng H, Tong G. 2016. A live, attenuated pseudorabies virus strain JS-2012 deleted for gE/gI protects against both classical and emerging strains. Antiviral Res 130:110–117. doi: 10.1016/j.antiviral.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh VV, Kerur N, Bottero V, Dutta S, Chakraborty S, Ansari MA, Paudel N, Chikoti L, Chandran B. 2013. Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. J Virol 87:4417–4431. doi: 10.1128/JVI.03282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ansari MA, Singh VV, Dutta S, Veettil MV, Dutta D, Chikoti L, Lu J, Everly D, Chandran B. 2013. Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J Virol 87:8606–8623. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson KE, Chikoti L, Chandran B. 2013. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol 87:5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spellberg B, Edwards JE. 2001. Type 1/type 2 immunity in infectious diseases. Clin Infect Dis 32:76–102. doi: 10.1086/317537. [DOI] [PubMed] [Google Scholar]
- 42.Frucht DM, Fukao T, Bogdan C, Schindler H, O'Shea JJ, Koyasu S. 2001. IFN-γ production by antigen-presenting cells: mechanisms emerge. Trends Immunol 22:556–560. doi: 10.1016/S1471-4906(01)02005-1. [DOI] [PubMed] [Google Scholar]
- 43.Harris DP, Goodrich S, Gerth AJ, Peng SL, Lund FE. 2005. Regulation of IFN-γ production by B effector 1 cells: essential roles for T-bet and the IFN-γ receptor. J Immunol 174:6781–6790. doi: 10.4049/jimmunol.174.11.6781. [DOI] [PubMed] [Google Scholar]
- 44.Koirala J, Adamski A, Koch L, Stueber D, El-Azizi M, Khardori NM, Ghassemi M, Novak RM. 2008. Interferon-gamma receptors in HIV-1 infection. AIDS Res Hum Retroviruses 24:1097–1102. doi: 10.1089/aid.2007.0261. [DOI] [PubMed] [Google Scholar]
- 45.Bürki F, Rossmanith W, Nowotny N, Pallan C, Möstl K, Lussy H. 1990. Viraemia and abortions are not prevented by two commercial equine herpesvirus-1 vaccines after experimental challenge of horses. Vet Q 12:80–86. doi: 10.1080/01652176.1990.9694249. [DOI] [PubMed] [Google Scholar]
- 46.Burrows R, Goodridge D, Denyer M. 1984. Trials of an inactivated equid herpesvirus 1 vaccine: challenge with a subtype 1 virus. Vet Rec 114:369–374. doi: 10.1136/vr.114.15.369. [DOI] [PubMed] [Google Scholar]
- 47.Hannant D, Jessett D, O'Neill T, Dolby C, Cook R, Mumford J. 1993. Responses of ponies to equid herpesvirus-1 ISCOM vaccination and challenge with virus of the homologous strain. Res Vet Sci 54:299–305. doi: 10.1016/0034-5288(93)90126-Z. [DOI] [PubMed] [Google Scholar]
- 48.O'Callaghan DJ, Cheevers WP, Gentry GA, Randall CC. 1968. Kinetics of cellular and viral DNA synthesis in equine abortion (herpes) virus infection of LM cells. Virology 36:104–114. doi: 10.1016/0042-6822(68)90120-7. [DOI] [PubMed] [Google Scholar]
- 49.Perdue ML, Kemp MC, Randall CC, O'Callaghan DJ. 1974. Studies of the molecular anatomy of the LM cell strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology 59:201–216. doi: 10.1016/0042-6822(74)90216-5. [DOI] [PubMed] [Google Scholar]
- 50.Kim SK, Kim S, Dai G, Zhang Y, Ahn BC, O'Callaghan DJ. 2011. Identification of functional domains of the IR2 protein of equine herpesvirus 1 required for inhibition of viral gene expression and replication. Virology 417:430–442. doi: 10.1016/j.virol.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. 2011. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res Hum Retroviruses 27:391–398. doi: 10.1089/aid.2010.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartels T, Steinbach F, Hahn G, Ludwig H, Borchers K. 1998. In situ study on the pathogenesis and immune reaction of equine herpesvirus type 1 (EHV-1) infections in mice. Immunology 93:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhein BA, Powers LS, Rogers K, Anantpadma M, Singh BK, Sakurai Y, Bair T, Miller-Hunt C, Sinn P, Davey RA. 2015. Interferon-γ inhibits Ebola virus infection. PLoS Pathog 11:e1005263. doi: 10.1371/journal.ppat.1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori E, Mori C, Della Libera A, Lara M, Fernandes W. 2003. Evaluation of alveolar macrophage function after experimental infection with equine herpesvirus-1 in horses. Arq Bras Med Vet Zootec 55:271–278. doi: 10.1590/S0102-09352003000300005. [DOI] [Google Scholar]
- 55.Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci U S A 95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sen GC, Peters GA. 2007. Viral stress-inducible genes. Adv Virus Res 70:233–263. doi: 10.1016/S0065-3527(07)70006-4. [DOI] [PubMed] [Google Scholar]
- 57.Schmid S, Mordstein M, Kochs G, García-Sastre A. 2010. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem 285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daffis S, Samuel MA, Suthar MS, Keller BC, Gale M, Diamond MS. 2008. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J Virol 82:8465–8475. doi: 10.1128/JVI.00918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 60.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lenschow DJ. 2010. Antiviral properties of ISG15. Viruses 2:2154–2168. doi: 10.3390/v2102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, García-Sastre A. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A 104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang D, Zhang D-E. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res 31:119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grundy FJ, Baumann RP, O'Callaghan DJ. 1989. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology 172:223–236. doi: 10.1016/0042-6822(89)90124-4. [DOI] [PubMed] [Google Scholar]
- 66.Kim SK, Shakya AK, Kim S, O'Callaghan DJ. 2016. Functional characterization of the serine-rich tract of varicella-zoster virus IE62. J Virol 90:959–971. doi: 10.1128/JVI.02096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soboll Hussey G, Ashton LV, Quintana AM, Lunn DP, Goehring LS, Annis K, Landolt G. 2014. Innate immune responses of airway epithelial cells to infection with equine herpesvirus-1. Vet Microbiol 170:28–38. doi: 10.1016/j.vetmic.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 68.Gariglio M, De Andrea M, Lembo M, Ravotto M, Zappador C, Valente G, Landolfo S. 1998. The murine homolog of the HIN 200 family, Ifi 204, is constitutively expressed in myeloid cells and selectively induced in the monocyte/macrophage lineage. J Leukoc Biol 64:608–614. [DOI] [PubMed] [Google Scholar]
- 69.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu G, Zhai Q, Schaffner DJ, Wu A, Yohannes A, Robinson TM, Maland M, Wells J, Voss TG, Bailey C. 2004. Prevention of lethal respiratory vaccinia infections in mice with interferon-α and interferon-γ. FEMS Immunol Med Microbiol 40:201–206. doi: 10.1016/S0928-8244(03)00358-4. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, Smith PM, Jennings SR, O'Callaghan DJ. 2000. Quantitation of virus-specific classes of antibodies following immunization of mice with attenuated equine herpesvirus 1 and viral glycoprotein D. Virology 268:482–492. doi: 10.1006/viro.2000.0197. [DOI] [PubMed] [Google Scholar]
- 73.McDonald WF, Huleatt JW, Foellmer HG, Hewitt D, Tang J, Desai P, Price A, Jacobs A, Takahashi VN, Huang Y, Nakaar V, Alexopoulou L, Fikrig E, Powell TJ. 2007. A West Nile virus recombinant protein vaccine that coactivates innate and adaptive immunity. J Infect Dis 195:1607–1617. doi: 10.1086/517613. [DOI] [PubMed] [Google Scholar]
- 74.Dunowska M. 2014. A review of equid herpesvirus 1 for the veterinary practitioner. Part B: pathogenesis and epidemiology. N Z Vet J 62:179–188. doi: 10.1080/00480169.2014.899946. [DOI] [PubMed] [Google Scholar]
- 75.Kim SK, Ahn BC, Albrecht RA, O'Callaghan DJ. 2006. The unique IR2 protein of equine herpesvirus 1 negatively regulates viral gene expression. J Virol 80:5041–5049. doi: 10.1128/JVI.80.10.5041-5049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fuentealba N, Zanuzzi C, Scrochi M, Sguazza G, Bravi M, de la Paz VC, Corva S, Portiansky E, Gimeno E, Barbeito C. 2014. Protective effects of intranasal immunization with recombinant glycoprotein D in pregnant BALB/c mice challenged with different strains of equine herpesvirus 1. J Comp Pathol 151:384–393. doi: 10.1016/j.jcpa.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 77.Harty RN, O'Callaghan DJ. 1991. An early gene maps within and is 3′ coterminal with the immediate-early gene of equine herpesvirus 1. J Virol 65:3829–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis JB, Thompson YG, Feng X, Holden VR, O'Callaghan D, Caughman GB. 1997. Structural and antigenic identification of the ORF12 protein (αTIF) of equine herpesvirus 1. Virology 230:369–375. doi: 10.1006/viro.1997.8477. [DOI] [PubMed] [Google Scholar]
- 79.Albrecht RA, Kim SK, Zhang Y, Zhao Y, O'Callaghan DJ. 2004. The equine herpesvirus 1 EICP27 protein enhances gene expression via an interaction with TATA box-binding protein. Virology 324:311–326. doi: 10.1016/j.virol.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 80.Holden VR, Caughman GB, Zhao Y, Harty RN, O'Callaghan DJ. 1994. Identification and characterization of the ICP22 protein of equine herpesvirus 1. J Virol 68:4329–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]