ABSTRACT
The Gag protein is the main retroviral structural protein, and its expression alone is usually sufficient for production of virus-like particles (VLPs). In this study, we sought to investigate—in parallel comparative analyses—Gag cellular distribution, VLP size, and basic morphological features using Gag expression constructs (Gag or Gag-YFP, where YFP is yellow fluorescent protein) created from all representative retroviral genera: Alpharetrovirus, Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus, Lentivirus, and Spumavirus. We analyzed Gag cellular distribution by confocal microscopy, VLP budding by thin-section transmission electron microscopy (TEM), and general morphological features of the VLPs by cryogenic transmission electron microscopy (cryo-TEM). Punctate Gag was observed near the plasma membrane for all Gag constructs tested except for the representative Beta- and Epsilonretrovirus Gag proteins. This is the first report of Epsilonretrovirus Gag localizing to the nucleus of HeLa cells. While VLPs were not produced by the representative Beta- and Epsilonretrovirus Gag proteins, the other Gag proteins produced VLPs as confirmed by TEM, and morphological differences were observed by cryo-TEM. In particular, we observed Deltaretrovirus-like particles with flat regions of electron density that did not follow viral membrane curvature, Lentivirus-like particles with a narrow range and consistent electron density, suggesting a tightly packed Gag lattice, and Spumavirus-like particles with large envelope protein spikes and no visible electron density associated with a Gag lattice. Taken together, these parallel comparative analyses demonstrate for the first time the distinct morphological features that exist among retrovirus-like particles. Investigation of these differences will provide greater insights into the retroviral assembly pathway.
IMPORTANCE Comparative analysis among retroviruses has been critically important in enhancing our understanding of retroviral replication and pathogenesis, including that of important human pathogens such as human T-cell leukemia virus type 1 (HTLV-1) and HIV-1. In this study, parallel comparative analyses have been used to study Gag expression and virus-like particle morphology among representative retroviruses in the known retroviral genera. Distinct differences were observed, which enhances current knowledge of the retroviral assembly pathway.
INTRODUCTION
The Gag polyprotein is the primary retroviral structural protein responsible for orchestrating retrovirus assembly. HIV-1 Gag has arguably been the most extensively studied Gag polyprotein to date (1–7); however, distinct differences in the biology of retroviral Gag proteins have emphasized the importance of comparative analyses in order to gain further insights (8).
The Gag polyprotein is functionally conserved across the two retroviral subfamilies (i.e., Orthoretrovirinae and Spumaretrovirinae). While the six genera in the Orthoretrovirinae subfamily (Alpha-, Beta-, Delta-, Epsilon-, Gamma-, and Lentivirus) and the single genus in the Spumaretrovirinae subfamily (Spumavirus) all encode Gag and have similar genomic organizations, distinct differences among the Gag proteins impact their replication and assembly processes. Specifically, while the Gag polyprotein serves the same function for all seven genera, analysis of Gag tertiary structures reveals differences between the two subfamilies. Analysis of the Gag N-terminal domain (NTD) of human foamy virus (HFV; Spumavirus) has revealed a structure that is unrelated to orthoretroviral Gag proteins, containing many β-sheet structures whereas orthoretroviral Gags contain α-helical structures (9).
The Gag polyprotein of the Orthoretrovirinae consists of three structurally distinct but functionally overlapping structural domains. The N-terminal domain of Gag (matrix [MA]) facilitates both Gag-membrane and Gag-RNA interactions (10). The capsid (CA) domain is primarily responsible for Gag-Gag interactions, and the C-terminal region encodes the nucleocapsid (NC) domain, which is particularly crucial for retroviral RNA packaging and stabilizing CA-CA interactions (11, 12). Other domains are encoded by some orthoretroviral Gag proteins. For example, lentiviruses such as HIV-1 encode the spacer peptide 1 (SP1) and spacer peptide 2 (SP2), which flank the NC domain, and the p6 domain, which is located at the C terminus of HIV-1 Gag. During or at completion of virus budding, the viral protease is activated, cleaving the Gag polyprotein into mature viral proteins. MA remains associated with the viral membrane, and CA forms a capsid core structure that encapsulates the NC protein bound to the viral RNA along with reverse transcriptase and integrase. Unlike the orthoretroviral Gag proteins, spumaviral Gag proteins do not undergo extensive proteolytic processing and typically remain in a polyprotein form during the virus assembly process (13).
In this study, we sought to devise a parallel comparative analysis of Gag proteins and the resultant Gag-based VLPs from representatives of the various retroviral genera. The goal of this comparative analysis was to determine whether there were differences in the general subcellular distribution of the various Gag proteins and whether variations existed regarding the general morphology of VLPs produced in a parallel manner. Differences were observed among both the Orthoretrovirinae and the representative member of the Spumaretrovirinae. Key observations include the nuclear localization of Epsilonretrovirus Gag, Deltaretrovirus-like particles with flat regions of electron density representative of the immature Gag lattice, and HIV-2 Lentivirus-like particles with a narrow range of particle size and consistent electron density below the viral membrane, suggesting a tightly packed Gag lattice. These results support the argument for how comparative analyses can be critically important for gaining new insights into the retroviral assembly pathway.
MATERIALS AND METHODS
Plasmids and cell lines.
Codon-optimized gag genes of HIV-1 (Lentiretrovirus), HIV-2 (Lentiretrovirus), murine leukemia virus (MLV; Gammaretrovirus), mouse mammary tumor virus (MMTV; Betaretrovirus), human foamy virus (HFV; Spumavirus), and walleye dermal sarcoma virus (WDSV; Epsilonretrovirus) were designed using the UpGene program (14) and synthesized by Genscript Co. (Piscataway, NJ). Gag genes were based on the following sequences: HIV-1 (bases 1 to 500 from GenBank accession number NP_057850.1), HIV-2 (bases 1103 to 2668 from GenBank accession number M30502.1), MMTV (bases 313 to 2088 from GenBank accession number AF033807.1), MLV (bases 357 to 1973 from GenBank accession number AF033811.1), HFV (bases 1869 to 2815 from GenBank accession number Y07725.1), and WDSV (bases 800 to 2548 in GenBenk accession number AF033822.1). Each gag gene expression cassette was created to contain a 5′ Kozak sequence (GCACCATG, start codon in bold) (15, 16). HindIII and BamHI restriction sites were engineered at the 5′ and 3′ ends of the genes, respectively, for subcloning. Each Gag expression plasmid was created by cloning the gag gene into the peYFP-N3 vector, creating a carboxy-terminal Gag-eYFP fusion (where eYFP is enhanced yellow fluorescent protein), or by cloning into the pN3 vector, where the eYFP tag was removed by deletion of the BamHI-NotI restriction fragment. The human T-cell leukemia virus type 1 (HTLV-1; Deltaretrovirus) codon-optimized Gag-eYFP plasmid has been previously described (17). A codon-optimized Rous sarcoma virus (RSV; Alpharetrovirus) gagpro was created (bases 381 to 2485 from GenBank accession number AF033808.1) using the same general strategy as that of the gag expression constructs in order to create peYFP-N3-RSV gagpro and pN3-RSV gagpro. The RSV pro sequence was removed from pN3-RSV gagpro by engineering a stop codon at position 2110 using primers 5′-CCTCCGGCCGTGTCCTAAGCGATGACCATGG-3′ and 5′-CCATGGTCATCGCTTAGGACACGGCCGGAGG-3′ (stop codon in bold). To create peYFP-N3-RSV gag, the following dinucleotide sequence was synthesized as a gBlock (Integrated DNA Technologies, Coralville, IA) containing a 5′ flanking BglII site and a 3′ flanking BamHI site (sequence, 5′-CGGGATGGGGCATAACGCTAAGCAGTGCCGAAAGCGAGACGGGAACCAGGGACAGCGCCCTGGCAGGGGGCTGTCCTCCGGCCCATGGCCGGGTCCCGAGCCTCCGGCCGTGTCCGGATCCATCGCCACCATGGTGAGC-3′). The restriction sites were used to clone the sequence into the N3-eYFP vector, removing the pro sequence.
A codon-optimized WDSV env gene (bases 5974 to 9651 in GenBank accession number AF033822.1) was constructed as described above for the gag gene expression constructs. The HFV env plasmid, pCiES (18), was a kind gift from Maxine Linial. The MMTV env expression plasmid, Q61 (19), was a kind gift from Jackie Dudley and Susan Ross. The RSV env expression plasmid was kindly provided by Marc Johnson (20). The HTLV-1 env expression construct, CMV-ENV, was kindly provided by Kathryn Jones and Marie-Christine Dokhelar (21). The MLV env expression construct, SV-A-MLV-env, has been previously described (22). HeLa cells and HEK293T cells were purchased from ATCC (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FetalClone III (FC3; GE Healthcare Lifesciences, Logan, UT).
Confocal microscopy.
HeLa cells were grown in six-well plates with 1.5-mm glass slides coated with poly-l-lysine. The cells were transiently transfected with peYFP-N3-gag plasmids and pN3-gag plasmids at a 1:4 weight ratio using Genjet according to the manufacturer's instructions (SignaGen, Gaithersburg, MD). The homologous env expression construct for each retroviral Gag (e.g., HIV-2 Env and HIV-2 Gag) was cotransfected into the HeLa cells at a plasmid weight ratio of 1:10 for the Env and Gag expression plasmids for RSV, HTLV-1, MLV, and HIV-2. The MMTV and WDSV Env and Gag expression plasmids were cotransfected at a weight ratio of 1:1, respectively. HFV was cotransfected at an Env-to-Gag expression plasmid weight ratio of 1:16. Sixteen to 24 h posttransfection, cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 30 min. Cells were then washed with PBS containing 0.05% Triton X-100 (Sigma-Aldrich, St. Louis, MN). The actin proteins were stained using ActinRed 555 ReadyProbes Reagent (Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. Coverslips were mounted on slides and preserved with ProLong Diamond Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies). Subcellular localization of Gag-eYFP was determined using a Zeiss LSM 700 confocal laser scanning microscope with a Plan-Apochromat 63×/1.40 numerical aperture (NA) oil objective at 1.2 zoom. A range of WDSV Env and Gag expression plasmid cotransfection ratios were tested, and at least 50 cells were imaged for each transfection ratio. The percentage of cells containing YFP Gag outside the nucleus were determined by qualitative evaluation of the images. Each replicate was normalized to the percentage of cells containing cytoplasmic Gag in the transfection of no Env, and the fold increases were calculated for 1:5 and 1:10 Env:Gag transfections.
Transmission electron microscopy of 293T cells producing virus-like particles.
Transfection of 293T cells with pN3-gag plasmids was done at a plasmid weight ratio of 10:1 for the Gag and Env expression plasmids, respectively, unless otherwise noted previously. Forty-eight hours posttransfection, cells were harvested and washed two times with 0.1 M sodium cacodylate. Cells were pelleted, fixed with 2.5% glutaraldehyde for 40 min, and then washed three times with 0.1 M sodium cacodylate. Samples were then postfixed with 1% OsO4 for 30 min. Cells were washed three times before they were dehydrated in a grade series of ethanol. Samples were then embedded in EMbed 812 resin. Ultrathin sections (65 nm) were stained with uranyl acetate and lead citrate. Samples were examined with a JEOL 1200EX II transmission electron microscope.
Cryo-TEM of virus-like particles.
Cotransfection of 293T cells with the pN3-gag expression constructs and their homologous Env expression plasmids was done using GenJet at a weight ratio of 10:1, respectively, unless otherwise noted previously. Forty-eight hours posttransfection, the cell culture supernatant was harvested and centrifuged at 3,000 × g for 5 min to remove large cellular debris. The supernatant was then passed through a 0.2-μm filter to remove any other debris or vesicles. VLPs were concentrated by ultracentrifugation in a 50.2 Ti rotor (Beckman) at 35,000 × g for 90 min through an 8% OptiPrep (Sigma-Aldrich) cushion. The VLP pellets were resuspended in 200 μl 1× STE (10 mM Tris-Cl [pH 7.4], 100 mM NaCl, 1 mM EDTA). The concentrated VLPs were then centrifuged through a 10 to 30% OptiPrep gradient in an SW55 Ti rotor (Beckman) at 45,000 × g for 3 h. The visible VLP band was extracted and pelleted in 1× STE at 35,000 × g for 1 h using a SW55 Ti rotor. The pellet was resuspended in 10 μl 1× STE and frozen at −80°C. The sample was thawed on ice prior to analysis by cryogenic transmission electron microscopy (cryo-TEM).
VLP samples were prepared for cryo-TEM as previously described (17). Briefly, 3 μl of concentrated VLP sample was applied to a glow-discharged c-flat holey carbon grid (Ted Pella, Redding, CA) and then blotted with filter papers to remove the sample excess. The grid was then plunged frozen into liquid ethane (23) with an FEI MarkIII Vitrobot system. The frozen grids were then transferred to an FEI TF30 field emission gun transmission electron microscope at liquid nitrogen temperature (FEI Company, Hillsboro, OR). Images were then recorded at a nominal magnification of ×39,000 and ×59,000 under low-dose (∼30 electrons/Å2) and 1- to 5-μm underfocus conditions using a Gatan 4k by 4k charge-coupled-device (CCD) camera (Gatan Inc., Pleasanton, CA).
Image analysis of virus-like particles.
Cryo-TEM images of VLPs were used for all analyses. VLP diameter was measured using ImageJ software (NIH, Bethesda, MD). For each VLP, two perpendicular diameters were measured and used to calculate an average diameter. The histogram was generated using GraphPad Prism 5 software (GraphPad, La Jolla, CA). Radial density profiles were calculated using the Radial Profile Extended applet in ImageJ. The ImageJ circle tool was used to encompass a VLP of average size for each genus tested, with the center of the circle lying in the center of the particle. The average image intensity for each radial point was obtained from running the applet. This was repeated for three VLPs of the same size and shape. The background was subtracted, and both the average and standard deviation were normalized to 1. The radial density profiles for all three VLPs measured were aligned based on the electron-dense peak for the lipid bilayer. The averaged radial density profiles were plotted using GraphPad Prism 5 software.
RESULTS
Gag subcellular localization in cells.
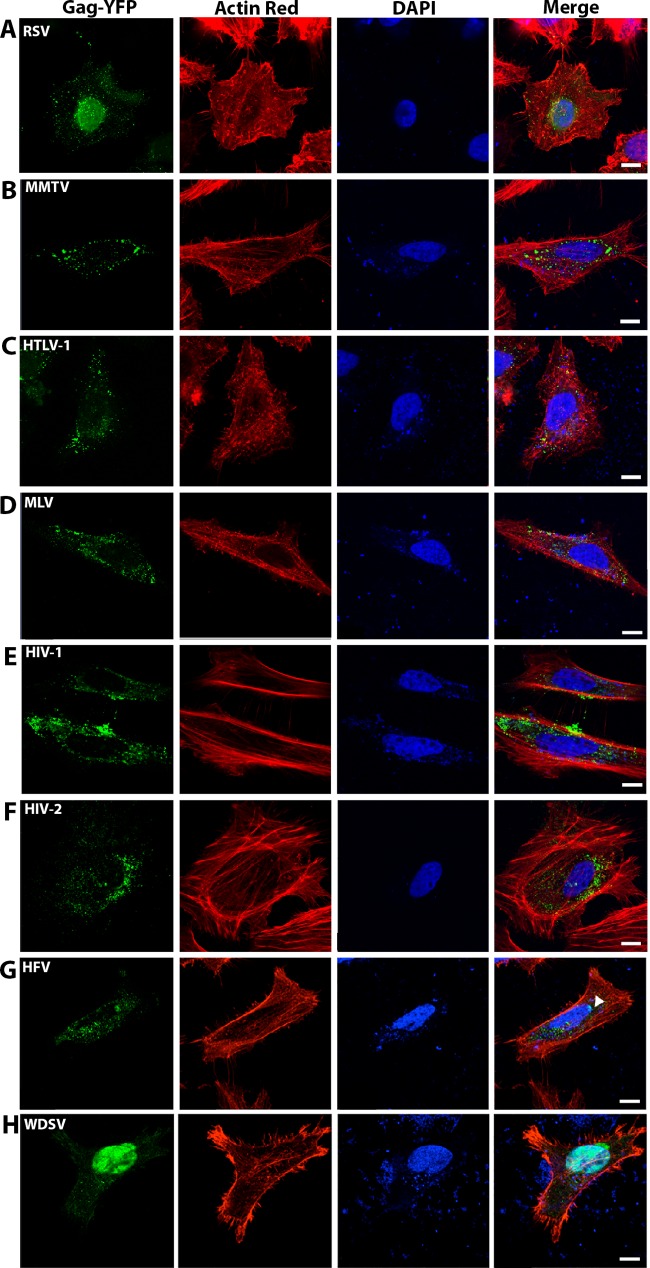
In order to investigate the subcellular distribution of the eight different retroviral Gag proteins, peYFP-N3-Gag expression constructs were engineered to express a Gag-YFP fusion for each of the retroviral genera. The tagged Gags were cotransfected at a 1:4 ratio with untagged Gags to maintain untagged Gag distribution phenotypes. Transiently transfected HeLa cells were analyzed using confocal microscopy 16 to 24 h posttransfection. The cell perimeter was visualized using actin staining. All images shown are optical sections of a representative cell at the widest section of the cell. More than 50 cells were qualitatively analyzed before representative cells were selected for imaging. While some variability existed based on hours posttransfection and Gag expression level, Gag distribution within the cells remained fairly consistent within genera. RSV had puncta that appeared in the cytoplasm and around the plasma membrane in addition to a low level of diffuse fluorescence found in the nucleus (Fig. 1A). HTLV-1, MLV, HIV-1, and HIV-2 all have puncta appearing both near the cell membrane and throughout the cytoplasm (Fig. 1C to F). Unlike the other retroviral Gag proteins analyzed, both MMTV and HFV were primarily found in the cytoplasm (Fig. 1B and G), with very few puncta appearing around the membrane. This was expected for MMTV and HFV, as these Gag proteins are known to traffic to intracellular membranes to initiate particle assembly (3, 24). HFV was also found on the apical side of the nucleus (Fig. 1G, arrow).
FIG 1.
Cellular localization of transient Gag-YFP expression in HeLa cells. HeLa cells were transiently cotransfected with 4:1 weight ratios of untagged Gag to YFP-tagged Gag in addition to homologous envelope plasmids as described in Materials and Methods. (A) Rous sarcoma virus (RSV); (B) mouse mammary tumor virus (MMTV); (C) human T-cell leukemia virus type 1 (HTLV-1); (D) murine leukemia virus (MLV); (E) HIV-1; (F) HIV-2; (G) human foamy virus (HFV); (H) walleye dermal sarcoma virus (WDSV). Optical sections close to the bottom of the cells were standardly used for collecting representative images of Gag-YFP localization. Nuclei were identified with DAPI stain (blue), and actin filaments were identified with the ActinRed 555 ReadyProbes reagent (red). Gag-YFP expression was identified by green fluorescence. Scale bar, 10 μm.
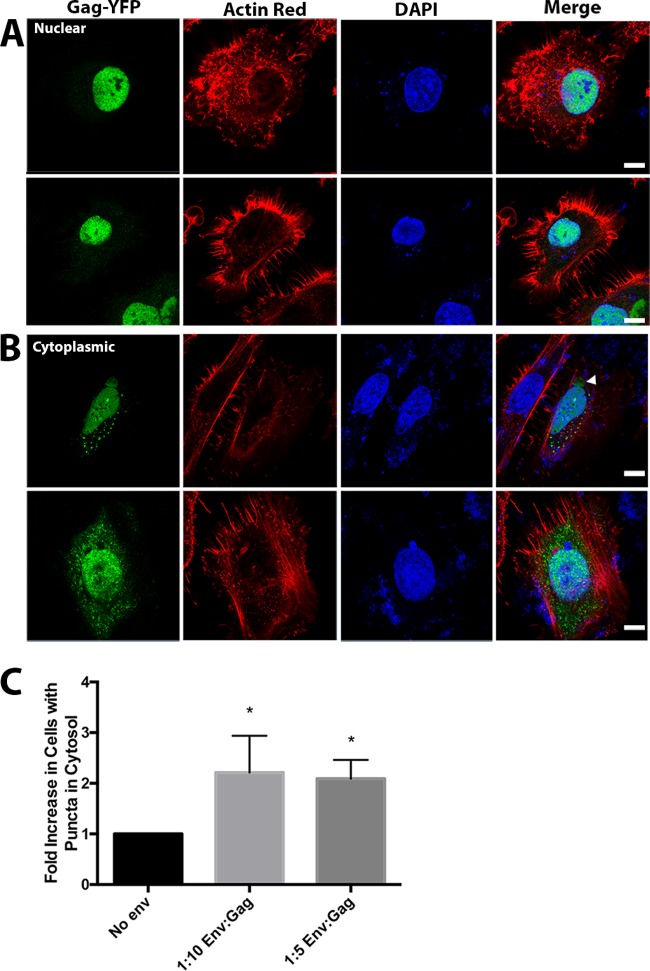
Cells transiently transfected with peYFP-N3-WDSV Gag primarily had fluorescence localized in or around the nucleus (Fig. 1F). The subcellular localization of WDSV Gag has not been previously described, and the primarily nuclear localization appears to be novel among the other retroviral genera tested here. Diffuse eYFP-WDSV Gag was also observed to be closely associated with the nucleus but not overlapping DAPI-stained regions (Fig. 1G; Fig. 2B, arrow). In the absence of WDSV Env expression, fewer than 15% of transfected cells were observed to have eYFP-WDSV Gag puncta in the cytoplasm. Coexpression of WDSV Gag and Env at 1:10 and 1:5 ratios, respectively, increased the proportion of cells expressing cytoplasmic Gag puncta (Fig. 2B and C); however, the overall percentage of cells with Gag in the cytoplasm was still relatively low compared to what was seen in other retroviral genera. This indicates that the WDSV Env influenced Gag localization in cells.
FIG 2.
Differential walleye dermal sarcoma virus (WDSV) Gag-YFP localization by WDSV Env expression. HeLa cells were transiently transfected as described in Materials and Methods with 1:4 weight ratios of WDSV Gag-YFP and untagged WDSV Gag in addition to WDSV Env expression plasmids at ratios of either 1:10 or 1:5. Cells were fixed, DAPI stained, and stained with ActinRed 16 h posttransfection. Images collected by confocal microscopy were used to evaluate cells for subcellular localization of WDSV Gag-YFP. (A) Representative examples of HeLa cells with no WDSV Gag-YPF puncta observed in the cytosol from transfections with no Env. (B) Representative examples of HeLa cells with WDSV Gag puncta observed in the cytosol from 1:10 or 1:5 cotransfections. Scale bar, 10 μm. (C) Fold increase in percentage of cells with Gag in the cytoplasm in different Env cotransfection ratios based on analysis of 50 cells (n = 3).
Thin-section transmission electron microscopy.
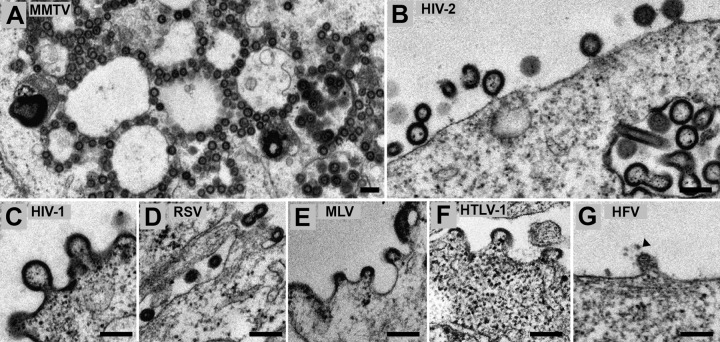
Thin-section TEM of 293T cells was performed to visualize VLPs produced by cells transiently transfected with a Gag and corresponding Env expression constructs. Forty-eight hours posttransfection, cells were fixed and stained in preparation for thin-section TEM. Electron densities, consistent with the immature Gag lattice, were observed in cells transfected with RSV, MLV, MMTV, HTLV-1, HIV-1, and HIV-2 Gags (Fig. 3A to F). No evidence of particle assembly was observed in or around cells transfected with WDSV Gag, presumably due to the nuclear localization of this Gag protein.
FIG 3.
Transmission electron microscopy of 293T cells transiently transfected with retroviral Gag expression constructs. 293T cells were transiently transfected as described in Materials and Methods with a Gag expression construct along with the homologous Env expression construct. Cells were prepared and analyzed by transmission electron microscopy as described in Materials and Methods. Shown are virus-like particle productions from 293T cells of mouse mammary tumor virus (MMTV) Gag (A), HIV-2 Gag (B), HIV-1 Gag (C), Rous sarcoma virus (RSV) Gag (D), murine leukemia virus Gag (E), human T-cell leukemia virus type 1 (HTLV-1) Gag (F), human foamy virus (HFV) Gag (G). Scale bar, 200 nm.
As expected, electron-dense MMTV-like particles were observed primarily intracellularly and were associated with large, electron-lucent vesicles (Fig. 3A). This has been previously observed for viruses found in mouse mammary tumor cells and more recently demonstrated for Mason-Pfizer monkey viruses produced from a full-length MPMV genome (25, 26). The electron densities consistent with the Gag lattice were approximately 90 nm in diameter and were abundant in transfected cells. Few electron densities resembling the MMTV Gag lattice were observed near the plasma membrane. Transfection with HIV-1, HIV-2, RSV, MLV, and HTLV-1 Gag resulted in VLP production at the plasma membrane (Fig. 3B to F). HIV-2 VLPs were found budding at both the cell membrane and into intracellular vesicles, which has been observed before in 293T cells transfected with HIV-1 Gag (27). Observations of cells transfected with HIV-1, HTLV-1, MLV, and RSV Gags correlated with previous observations for these viral proteins (28–30).
Unlike what was seen in the orthoretroviruses, VLPs produced by HFV Gag were not observed to be associated with a distinct electron density (Fig. 3G), but particles with diffuse electron density that were approximately 80 to 90 nm in diameter were observed. In addition, these VLPs contained small electron-dense regions around them that were consistent with HFV Env protein spikes (Fig. 3G, arrow).
Cryo-TEM of Gag-based VLPs.
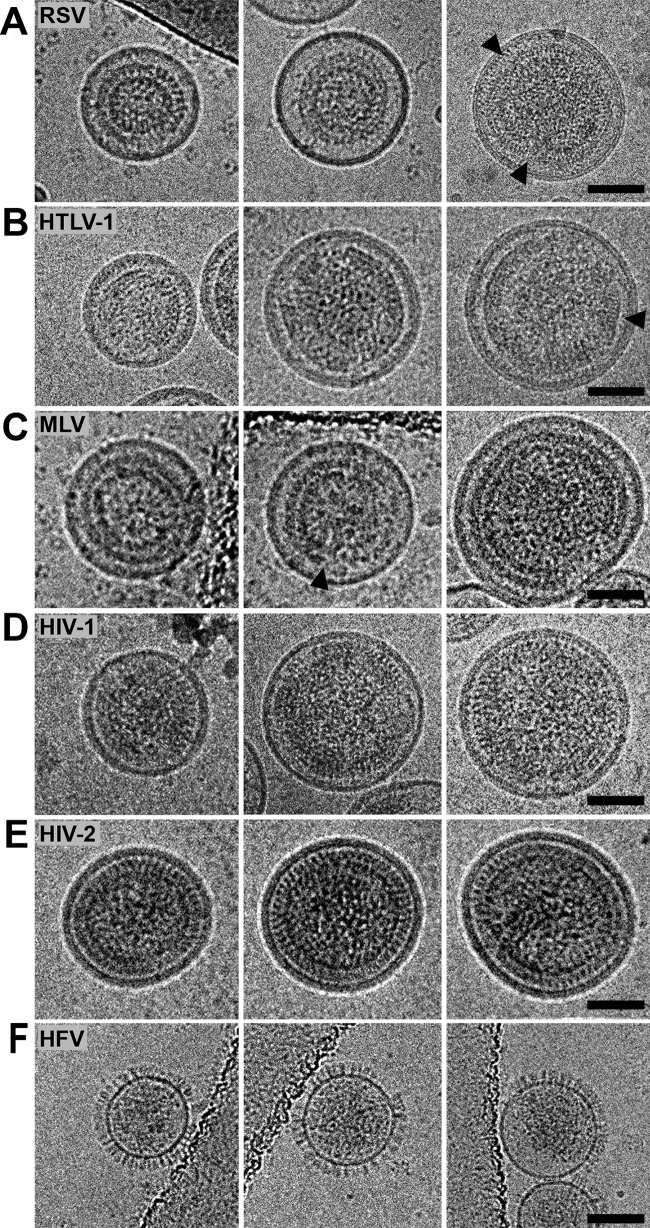
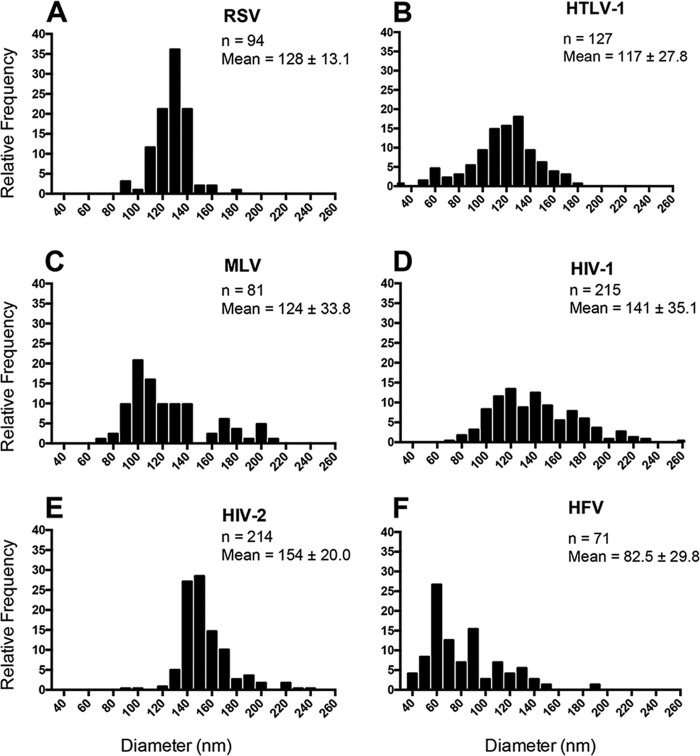
Cryo-TEM was done to analyze VLPs produced from 293T cells that had been transiently transfected with Gag and Env expression constructs for each of the representative retroviruses under study. Forty-eight hours posttransfection, the VLPs were purified on an Opti-Prep gradient and visualized using cryo-TEM. VLPs were not readily produced by transfection of 293T cells with MMTV Gag or WDSV Gag, so no particle analysis was performed. RSV, HTLV-1, MLV, HIV-1, HIV-2, and HFV VLPs were produced at sufficient levels to allow for morphological analyses (Fig. 4A to F). With the exception of VLPs produced by HFV Gag (Fig. 4E), all observed VLPs were spherical with electron density below the VLP membrane. In addition, VLPs produced from all Gag proteins resulted in a distribution of particle sizes, with a general range from 81 to 200 nm in diameter (Fig. 5A to F).
FIG 4.
Cryo-TEM of retrovirus-like particles produced by transient expression of Gag in 293T cells. 293T cells were transiently transfected with selected retroviral Gag expression plasmids and homologous Env expression constructs (ratio of 10:1 for RSV, HTLV-1, MLV, HIV-1, and HIV-2; ratio of 16:1 for HFV) as described in Materials and Methods. Virus-like particles were collected and purified from the cell culture supernatant. (A) Rous sarcoma virus (RSV); (B) human T-cell leukemia virus type 1 (HTLV-1); (C) murine leukemia virus (MLV); (D) HIV-1; (E) HIV-2; (F) human foamy virus (HFV). Scale bar, 50 nm.
FIG 5.
Analysis of the distribution of retrovirus-like particle diameters. Using images collected by cryo-TEM, retrovirus-like particle diameters were measured in two perpendicular directions and averaged. A minimum of two independently prepared particle preparations were used for the particle diameter analyses. (A) Rous sarcoma virus (RSV); (B) human T-cell leukemia virus type 1 (HTLV-1); (C) murine leukemia virus; (D) HIV-1; (E) HIV-2; (F) human foamy virus (HFV).
VLPs produced from RSV Gag were found to be 128 ± 13 nm in diameter (Fig. 5A), which is similar to previously reported values (5). The majority of RSV VLPs had electron density consistent with an immature Gag lattice that followed the curvature of the inner viral membrane, but there were some instances of particles where there were discontinuities in electron density or where the density did not strictly follow the curvature of the inner viral membrane (Fig. 4A, arrow). Similar to RSV-like particles, the VLPs produced by HTLV-1 Gag expression also had regions where there were discontinuities in electron density below the viral membrane (Fig. 4B). MLV-like particles also contained discontinuities in electron density beneath the viral membrane (Fig. 4C, arrow), with an average particle diameter of 124 nm. Intriguingly, there appeared to be two distinct particle subpopulations among the MLV-like particles. The majority (∼80%) of these particles were observed in the range of 70 to 140 nm. A minority (∼20%) of the particles were observed in the range of 160 to 210 nm. The particle morphologies for the two subpopulations were comparable (Fig. 4C). Of the five orthoretrovirus VLPs observed, VLPs produced by HTLV-1 Gag were the smallest, with an average size of 116 ± 28 nm in diameter (Fig. 5B). HTLV-1-like particles had regions of electron density resembling the Gag lattice that were flat and did not follow the curvature of the lipid bilayer (Fig. 4B, arrow) (31).
HIV-2-like particles were found to have an average particle diameter of 154 ± 20 nm in diameter (Fig. 5E), which was significantly larger than the observed diameter of HIV-1-like particles (141 ± 35.1 nm) (Table 1). The diameter of HIV-1-like particles falls within the range of previous reports of the diameter of HIV-1 or HIV-1-like particles (32, 46). Compared to the other retrovirus-like particles in this study, the HIV-2-like particles had a narrow size distribution and a uniform particle morphology. In particular, these VLPs were observed to have a very organized electron density beneath the viral membrane. This organized density is suggestive of a tightly packed immature Gag lattice that closely follows the inner viral membrane.
TABLE 1.
Summary of the dimensionsa of virus-like particles and Gag lattice structures based upon cryo-TEM
| Dimension | RSV | MLV | HTLV-1 | HIV-1 | HIV-2 | HFV |
|---|---|---|---|---|---|---|
| Avg particle diam | 128 (±13.1) | 125 (±33.8) | 116.6 (±27.8) | 141 (±35.1)b | 154.5 (±20.0)b | 82.5 (±29.8) |
| Radius to inner CA subdomain | ∼26 | ∼41 | ∼39 | ∼49 | ∼53 | NDc |
| Radius to outer CA subdomain | ∼39 | ∼51 | ∼44 | ∼55 | ∼60 | ND |
| Distance from lipid bilayer to outer CA subdomain | ∼18 | ∼12 | ∼10 | ∼10 | ∼9 | ND |
| Length of Env protrusions | ND | ND | ND | ND | ND | 16 (±2.1) |
All values are averages (± standard deviations) in nanometers.
HIV-2-like particles are significantly larger than HIV-1-like particles (t test, P = 0.0002).
ND, not determined.
HFV-like particles had a unique particle morphology that was clearly distinguishable from the other retrovirus-like particles analyzed in this parallel comparative study. First, there was no distinguishable electron density found within the particles that would be consistent with an immature Gag lattice. The vast majority of particles observed were studded with electron-dense projections (16 ± 2 nm) that were morphologically consistent with HFV Env proteins. In addition, more than 50% of the visible particle surface was decorated with these electron-dense projections. It is interesting that the production of HFV-like particles was from a 1:16 cotransfection of the HFV Env expression plasmid (pCiEs) to that of the HFV Gag expression plasmid (pN3-HFV-gag), respectively, which is a lower level of Env plasmid than for the other Gags. The HFV-like particles were also distinct in their relatively small size, with an average diameter of 82.5 ± 30 nm, excluding the length of the Env projections (Fig. 5F).
Radial density profile analysis.
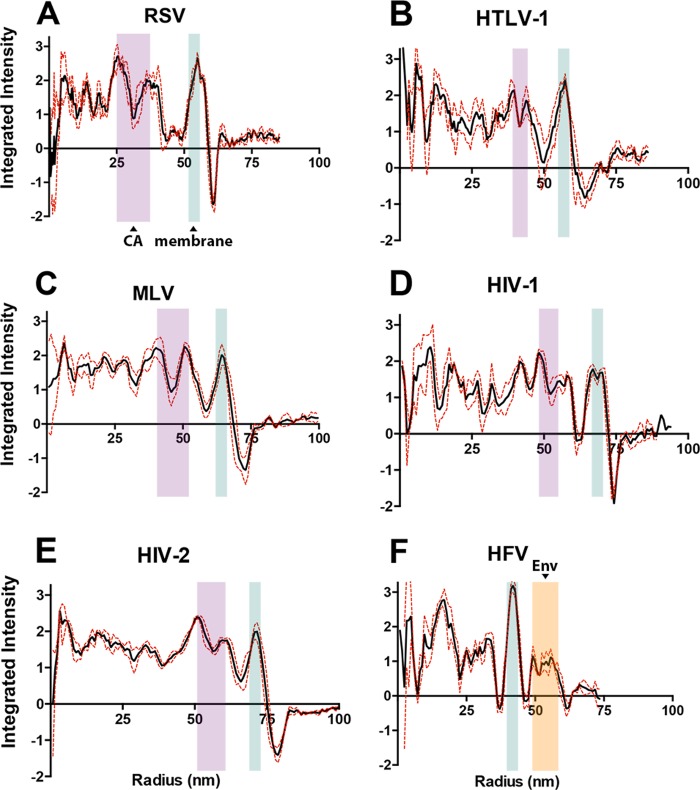
Particles characterized by cryo-TEM were further analyzed to determine their radial density profiles. For each retrovirus-like particle, 3 representative particles that were within 2 nm of each other in particle diameter were analyzed using the Radial Profile Extended applet in ImageJ. The integrated intensity units were corrected for background, and the average and standard deviation were normalized to a value of 1. The integrated intensity units for each of the three VLPs were then averaged and plotted with the standard error of the mean (Fig. 6A to E).
FIG 6.
Radial density profile analysis of retrovirus-like particles. The solid black line indicates the average retrovirus-like particle density measured. The dashed red line indicates the standard error of the mean. The radial density profiles shown were averaged from three particles of median size. (A) Rous sarcoma virus (RSV); (B) human T-cell leukemia virus type 1 (HTLV-1); (C) murine leukemia virus (MLV); (D) HIV-1; (E) HIV-2; (F) human foamy virus (HFV). As points of reference, the blue horizontal box indicates the location of the lipid bilayer density, the purple box indicates the location of CA densities, and the orange box indicates the Env density.
A distinct electron-dense peak, which is representative of the lipid bilayer, was seen in all radial density profiles (indicated by the blue box) (Fig. 6A to F). It is generally assumed that the MA domain for orthoretroviral Gags remains associated with the viral membrane and therefore has no distinct peak. Distinct electron density peaks that are consistent with the CA NTD and C-terminal domain (CTD) for the VLPs of RSV, HTLV-1, MLV, HIV-1, and HIV-2 were observed. In contrast, no electron density peaks consistent with the CA NTD or CTD were observed for HFV-like particles. Analysis of MLV- and RSV-like particles revealed a large space in electron density between the viral membrane and the presumed CA subunit, 12 and 18 nm, respectively. This space in electron density could be due to a long flexible linker connecting the MA domain and the CA NTD (p12 in MLV and p10 in RSV) (Table 1) (33, 34). In addition, the MLV- and RSV-like particles also have a relatively large space between the two electron density peaks representing the inner and outer CA subdomains. While there were no distinct peaks visible that would be consistent for a CA NTD or CTD for HFV-like particles, which may be due to the lack of a typical orthoretroviral Gag conformation, there was a distinct density from 49 nm to 52 nm that would be consistent with the HFV Env protein on the surface of particles.
DISCUSSION
To our knowledge, this is the first comprehensive comparative study of retrovirus-like particle morphology performed in parallel using a Gag expression construct along with the corresponding retroviral Env expression plasmid (e.g., HIV-2 Gag expression plasmid and HIV-2 Env expression plasmid). In particular, we selected at least one representative virus from each of the retroviral genera to analyze the differences in Gag subcellular localization as well as in general particle morphology and size. A practical advantage of this experimental system is that it helps to limit variables related to biological differences in virus replication among the different retroviruses. While authentic viral RNA for each virus was not available for packaging into particles, the system is standardized in that the same cellular RNA is available for all RNA packaging, limiting cell type variability as well. Through the use of this system, we observed both distinct differences and similarities in the characteristics of the retrovirus-like particles produced and the distribution of Gag in cells.
In this study, WDSV (Epsilonretrovirus) Gag appeared to localize in the nucleus, and perhaps near the nucleus, which is a new observation. There have been no previous analyses of WDSV Gag localization or protein structure, but based on sequence homology, WDSV Gag has similarities to other orthoretrovirus Gag proteins; however, the distinct subcellular localization distinguishes this Gag. We also observed that the WDSV Env appears to influence WDSV Gag localization, as coexpression of both Gag and Env resulted in the cytoplasmic appearance of WDSV Gag, though the majority remained in the nucleus regardless of Env expression. When investigated by thin-section TEM, there was no evidence of intracellular assembly of particles. The WDSV accessory protein, open reading frame (Orf) A (rv-cyclin), has also been observed to localize in the nucleus and may interact with the Gag protein there (35). Further studies are needed to better understand the nature of the nuclear localization of WDSV Gag, whether WDSV Gag nuclear localization may facilitate interaction with RNA as it does for RSV (36), and the general requirements for virus particle release from cells.
Many previous studies have evaluated the immature particle morphology of MLV (Gammaretrovirus), HIV-1 (Lentiretrovirus), RSV (Alpharetrovirus), and MPMV (Betaretrovirus) (37, 38) (5) (4, 39). In contrast, limited information is available regarding MMTV (Betaretrovirus), HTLV-1 (Deltaretrovirus), HIV-2 (Lentiretrovirus), HFV (Spumavirus), and WDSV (Epsilonretrovirus) immature or mature particle morphology. For example, MMTV particles were previously purified from murine cells and visualized by thin-section microscopy (40), and authentic HTLV-1, HFV, and MLV have been visualized by cryo-TEM or cryo-electron tomography (41–43). Our study provides, to our knowledge, the first information regarding HIV-2-like particle morphology, and no reports are available on WDSV particle morphology.
Based on the structural similarities among most Gags, HIV-1 Gag trafficking and localization are often used as a reference model for many different retroviruses. Our findings here indicate that HIV-1 Gag packaging may not be a good model for many retroviral Gag studies, as we found a wide range of particle sizes and morphologies in our analysis. This is highlighted by our analysis of HIV-2 Gag-based VLPs, which is a lentivirus closely related to HIV-1. HIV-2 Gag has a subcellular distribution similar to what is seen for HIV-1 based on confocal analysis, but it produces particles that are distinct in size and morphology, indicating differences between particle assembly and/or the nature of the immature Gag lattice. Intriguingly, HIV-1 and HIV-2 Gags can assemble together into the same VLPs, indicating that the Gags still share many similarities (44).
Our observations indicate that orthoretrovirus VLPs share some morphological features, such as a spherical shape, a distribution of diameter sizes, and distinct inner and outer CA ring densities. This is not particularly surprising given that retroviral CA structures are markedly similar even with little amino acid sequence homology. Despite these similarities, all of the orthoretrovirus VLPs observed here have distinguishing morphological features. These differences may be in part due to distinguishing features of the Gags outside the CA domain, such as the linker domains that connect the RSV and MLV MA and CA, which can lead to a large distance from the lipid bilayer to the outer CA ring. We observed that this may be different for HIV-2, however, as the electron density remained closely associated with the viral membrane, possibly due to a smaller linker between HIV-2 MA and CA.
HFV VLPs were markedly different from the Orthoretrovirus VLPs. This could be due to the nature of HFV particle assembly, which occurs intracellularly at the endoplasmic reticulum and requires expression of the homologous HFV Env protein (3, 45). The requirement of Env protein expression for HFV-like particle assembly perhaps explains the high occurrence of spike-like protrusions on the surface of HFV-like particles. Furthermore, the lack of electron density that would correspond to an immature Gag lattice may be due to an unusual structure and folding of HFV Gag, which helps to distinguish spumaviral Gag proteins from orthoretroviral Gag proteins.
Taken together, these parallel comparative analyses have identified distinct features that exist among retrovirus-like particles. In particular, differences were observed with WDSV, HTLV-1, and HIV-2. These studies emphasize the differences that exist among retroviruses in regard to particle assembly/morphology, and the importance of their study in order to gain deeper insights into retroviral assembly.
ACKNOWLEDGMENTS
We thank Jonathan Rawson for vectors and reagents and Michelle Christian for technical assistance in the construction of the HIV-2 reagents. We thank Fang Zhou for performing thin-section TEM experiments. The cryo-TEM images were recorded using a Tecnai TF30 TEM maintained by the Characterization Facility, College of Science and Engineering, University of Minnesota.
This work is supported by NIH grant R01 GM098550. J.L.M. has been supported by NIH grant T32 DA007097. J.O.M. has been supported from NIH grants T32 AI083196 (Institute for Molecular Virology Training Program) and F30 DE022286.
REFERENCES
- 1.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 2.Wilk T, Gross I, Gowen BE, Rutten T, de Haas F, Welker R, Krausslich HG, Boulanger P, Fuller SD. 2001. Organization of immature human immunodeficiency virus type 1. J Virol 75:759–771. doi: 10.1128/JVI.75.2.759-771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietschmann T, Heinkelein M, Heldmann M, Zentgraf H, Rethwilm A, Lindemann D. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J Virol 73:2613–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bharat TA, Davey NE, Ulbrich P, Riches JD, de Marco A, Rumlova M, Sachse C, Ruml T, Briggs JA. 2012. Structure of the immature retroviral capsid at 8 A resolution by cryo-electron microscopy. Nature 487:385–389. doi: 10.1038/nature11169. [DOI] [PubMed] [Google Scholar]
- 5.Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM. 2006. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J Mol Biol 355:157–168. doi: 10.1016/j.jmb.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Na Struct Mol Biol 11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 7.Martin-Serrano J, Zang T, Bieniasz PD. 2003. Role of ESCRT-I in retroviral budding. J Virol 77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Cao S, Martin JL, Mueller JD, Mansky LM. 2015. Morphology and ultrastructure of retrovirus particles. AIMS Biophys 2:343–369. doi: 10.3934/biophy.2015.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstone DC, Flower TG, Ball NJ, Sanz-Ramos M, Yap MW, Ogrodowicz RW, Stanke N, Reh J, Lindemann D, Stoye JP, Taylor IA. 2013. A unique spumavirus Gag N-terminal domain with functional properties of orthoretroviral matrix and capsid. PLoS Pathog 9:e1003376. doi: 10.1371/journal.ppat.1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M, Grigsby IF, Gorelick RJ, Mansky LM, Musier-Forsyth K. 2014. Retrovirus-specific differences in matrix and nucleocapsid protein-nucleic acid interactions: implications for genomic RNA packaging. J Virol 88:1271–1280. doi: 10.1128/JVI.02151-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, Rein A, Rouzina I, Williams MC. 2006. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res 34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J Virol 72:1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flugel RM, Pfrepper KI. 2003. Proteolytic processing of foamy virus Gag and Pol proteins. Curr Top Microbiol Immunol 277:63–88. [DOI] [PubMed] [Google Scholar]
- 14.Gao W, Rzewski A, Sun H, Robbins PD, Gambotto A. 2004. UpGene: application of a web-based DNA codon optimization algorithm. Biotechnol Prog 20:443–448. [DOI] [PubMed] [Google Scholar]
- 15.Kozak M. 1984. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res 12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigsby IF, Zhang W, Johnson JL, Fogarty KH, Chen Y, Rawson JM, Crosby AJ, Mueller JD, Mansky LM. 2010. Biophysical analysis of HTLV-1 particles reveals novel insights into particle morphology and Gag stoichiometry. Retrovirology 7:75. doi: 10.1186/1742-4690-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trobridge G, Josephson N, Vassilopoulos G, Mac J, Russell DW. 2002. Improved foamy virus vectors with minimal viral sequences. Mol Ther 6:321–328. doi: 10.1006/mthe.2002.0672. [DOI] [PubMed] [Google Scholar]
- 19.Golovkina TV, Dudley JP, Ross SR. 1998. B and T cells are required for mouse mammary tumor virus spread within the mammary gland. J Immunol 161:2375–2382. [PubMed] [Google Scholar]
- 20.Jorgenson RL, Vogt VM, Johnson MC. 2009. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol 83:4060–4067. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delamarre L, Rosenberg AR, Pique C, Pham D, Dokhelar MC. 1997. A novel human T-leukemia virus type 1 cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J Virol 71:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landau NR, Littman DR. 1992. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol 66:5110–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker TS, Olson NH, Fuller SD. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev 63:862–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabransky A, Hadravova R, Stokrova J, Sakalian M, Pichova I. 2009. Premature processing of mouse mammary tumor virus Gag polyprotein impairs intracellular capsid assembly. Virology 384:33–37. doi: 10.1016/j.virol.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 25.Smith GH, Wivel NA. 1973. Intracytoplasmic A particles: mouse mammary tumor virus nucleoprotein cores? J Virol 11:575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stansell E, Apkarian R, Haubova S, Diehl WE, Tytler EM, Hunter E. 2007. Basic residues in the Mason-Pfizer monkey virus gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J Virol 81:8977–8988. doi: 10.1128/JVI.00657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigorov B, Arcanger F, Roingeard P, Darlix JL, Muriaux D. 2006. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J Mol Biol 359:848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Johnson MC, Scobie HM, Vogt VM. 2001. PR domain of Rous sarcoma virus Gag causes an assembly/budding defect in insect cells. J Virol 75:4407–4412. doi: 10.1128/JVI.75.9.4407-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortuza GB, Dodding MP, Goldstone DC, Haire LF, Stoye JP, Taylor IA. 2008. Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation. J Mol Biol 376:1493–1508. doi: 10.1016/j.jmb.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Machesky NJ, Mansky LM. 2004. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J Virol 78:1503–1512. doi: 10.1128/JVI.78.3.1503-1512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maldonado JO, Cao S, Zhang W, Mansky LM. 2016. Distinct morphology of human T-cell leukemia virus type 1-like particles. Viruses 8(5):pii:E132. doi: 10.3390/v8050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Wu B, Musier-Forsyth K, Mansky LM, Mueller JD. 2009. Fluorescence fluctuation spectroscopy on viral-like particles reveals variable gag stoichiometry. Biophys J 96:1961–1969. doi: 10.1016/j.bpj.2008.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyere SK, Joseph PR, Summers MF. 2008. The p12 domain is unstructured in a murine leukemia virus p12-CA(N) Gag construct. PLoS One 3:e1902. doi: 10.1371/journal.pone.0001902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nandhagopal N, Simpson AA, Johnson MC, Francisco AB, Schatz GW, Rossmann MG, Vogt VM. 2004. Dimeric Rous sarcoma virus capsid protein structure relevant to immature Gag assembly. J Mol Biol 335:275–282. doi: 10.1016/j.jmb.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 35.Rovnak J, Casey JW, Quackenbush SL. 2001. Intracellular targeting of walleye dermal sarcoma virus Orf A (rv-cyclin). Virology 280:31–40. doi: 10.1006/viro.2000.0731. [DOI] [PubMed] [Google Scholar]
- 36.Garbitt-Hirst R, Kenney SP, Parent LJ. 2009. Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging. J Virol 83:6790–6797. doi: 10.1128/JVI.00101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JA. 2015. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 38.Schur FK, Dick RA, Hagen WJ, Vogt VM, Briggs JA. 2015. The structure of immature virus-like Rous sarcoma virus Gag particles reveals a structural role for the p10 domain in assembly. J Virol 89:10294–10302. doi: 10.1128/JVI.01502-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schur FK, Hagen WJ, de Marco A, Briggs JA. 2013. Determination of protein structure at 8.5A resolution using cryo-electron tomography and sub-tomogram averaging. J Struct Biol 184:394–400. doi: 10.1016/j.jsb.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 40.Menendez-Arias L, Risco C, Pinto da Silva P, Oroszlan S. 1992. Purification of immature cores of mouse mammary tumor virus and immunolocalization of protein domains. J Virol 66:5615–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao S, Maldonado JO, Grigsby IF, Mansky LM, Zhang W. 2015. Analysis of human T-cell leukemia virus type 1 particles by using cryo-electron tomography. J Virol 89:2430–2435. doi: 10.1128/JVI.02358-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamann MV, Mullers E, Reh J, Stanke N, Effantin G, Weissenhorn W, Lindemann D. 2014. The cooperative function of arginine residues in the prototype foamy virus Gag C-terminus mediates viral and cellular RNA encapsidation. Retrovirology 11:87. doi: 10.1186/s12977-014-0087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeager M, Wilson-Kubalek EM, Weiner SG, Brown PO, Rein A. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci U S A 95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyko V, Leavitt M, Gorelick R, Fu W, Nikolaitchik O, Pathak VK, Nagashima K, Hu WS. 2006. Coassembly and complementation of Gag proteins from HIV-1 and HIV-2, two distinct human pathogens. Mol Cell 23:281–287. doi: 10.1016/j.molcel.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 45.Goepfert PA, Shaw K, Wang G, Bansal A, Edwards BH, Mulligan MJ. 1999. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J Virol 73:7210–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]