ABSTRACT
Since influenza C virus was first isolated in 1947, the virus has been only occasionally isolated by cell culture; there are only four strains for which complete genome sequences are registered. Here, we analyzed a total of 106 complete genomes, ranging from the first isolate from 1947 to recent isolates from 2014, to determine the genetic lineages of influenza C virus, the reassortment events, and the rates of nucleotide substitution. The results showed that there are six lineages, named C/Taylor, C/Mississippi, C/Aichi, C/Yamagata, C/Kanagawa, and C/Sao Paulo. They contain both antigenic and genetic lineages of the hemagglutinin-esterase (HE) gene, and the internal genes PB2, PB1, P3, NP, M, and NS are divided into two major lineages, a C/Mississippi/80-related lineage and a C/Yamagata/81-related lineage. Reassortment events were found over the entire period of 68 years. Several outbreaks of influenza C virus between 1990 and 2014 in Japan consisted of reassortant viruses, suggesting that the genomic constellation is related to influenza C virus epidemics. The nucleotide sequences were highly homologous to each other. The minimum percent identity between viruses ranged from 91.1% for the HE gene to 96.1% for the M gene, and the rate of nucleotide substitution for the HE gene was the highest, at 5.20 × 10−4 substitutions/site/year. These results indicate that reassortment is an important factor that increases the genetic diversity of influenza C virus, resulting in its ability to prevail in humans.
IMPORTANCE Influenza C virus is a pathogen that causes acute respiratory illness in children and results in hospitalization of infants. We previously demonstrated (Y. Matsuzaki et al., J Clin Virol 61:87–93, 2014, http://dx.doi.org/10.1016/j.jcv.2014.06.017) that periodic epidemics of this virus occurred in Japan between 1996 and 2014 and that replacement of the dominant antigenic group occurred every several years as a result of selection by herd immunity. However, the antigenicity of the HE glycoprotein is highly stable, and antigenic drift has not occurred for at least 30 years. Here, we analyzed a total of 106 complete genomes spanning 68 years for the first time, and we found that influenza C viruses are circulating worldwide while undergoing reassortment as well as selection by herd immunity, resulting in an increased ability to prevail in humans. The results presented in this study contribute to the understanding of the evolution, including reassortment events, underlying influenza C virus epidemics.
INTRODUCTION
Influenza C virus was first isolated in 1947 from a human with upper respiratory symptoms in the United States (1). This virus has been classified as a member of the Orthomyxoviridae family of enveloped and segmented negative-sense RNA viruses together with influenza A virus and influenza B virus. The genome of influenza C virus consists of seven RNA segments, which encode three polymerase proteins (PB2, PB1, and P3), a hemagglutinin-esterase (HE) glycoprotein, a nucleoprotein (NP), a matrix (M) protein, and a nonstructural (NS) protein. HE glycoprotein is the counterpart of both hemagglutinin (HA) and neuraminidase (NA) in influenza A and B viruses.
Whereas influenza A virus infects a variety of hosts, influenza C viruses are predominantly found in humans and, rarely, in pigs (2). Influenza C virus usually causes a mild upper respiratory tract illness in children but can also cause lower respiratory tract illness, such as bronchitis and pneumonia, particularly in children <2 years old (3–7). Despite the ubiquitous distribution of influenza C virus, which is demonstrated by its high rate of seroprevalence (8–16), the virus has been isolated by cell culture only occasionally, and there are only four strains for which complete genome sequences are registered in GenBank at present: C/Ann Arbor/1/50, C/Johannesburg/1/66, C/Eastern India/1202/2011, and C/Victoria/2/2012.
Since 1988, we have monitored influenza C virus infections in Japan using a cell culture method, and we succeeded in repeatedly detecting outbreaks by 2014 (17–25). This long-term surveillance revealed that there are several antigenic groups among influenza C viruses, and this antigenic difference is cross-reactive but distinguishable by analysis with human antibodies and anti-HE monoclonal antibodies (17–22, 24–26). Influenza C viruses belonging to the different antigenic groups cocirculated in the community, and replacement of the dominant antigenic group occurred every few years as a result of selection by herd immunity (24). However, antigenic drift between viruses due to immune pressure has not occurred for the influenza C viruses for at least 30 years (24). These findings clearly differ from those for the influenza A viruses. Influenza C virus is antigenically stable compared with influenza A virus.
Reassortment, the exchange of genome segments between two different strains, is one way for influenza viruses to increase their genetic diversity (27–30). We demonstrated that frequent reassortment events occurred among influenza C viruses from 1990 to 2000 in Japan (21, 22). Our observation that influenza C outbreaks have consisted of reassortant viruses suggests that the genomic composition of influenza C viruses influences their ability to spread in humans. In our previous works, we performed phylogenetic analysis of internal gene segments, such as the PB2, PB1, P3, and NP genes, using partial sequences of the coding region (469 nucleotides for the PB2 gene, 376 nucleotides for the PB1 gene, 372 nucleotides for the P3 gene, and 600 nucleotides for the NP gene) (18–22, 31, 32). Here, we extended this analysis to the complete genome sequence of each coding region (2,325 nucleotides for the PB2 gene, 2,265 nucleotides for the PB1 gene, 2,130 nucleotides for the P3 gene, and 1,698 nucleotides for the NP gene). In addition, influenza C viruses ranging from the first isolate in 1947 to recent isolates in 2014 were included in this analysis.
In this study, we analyzed a total of 106 complete genomes spanning 68 years to determine the genetic lineages of influenza C virus, the reassortment events, and the rates of nucleotide substitutions. The results will further the understanding of the evolution, including reassortment events, of influenza C virus.
MATERIALS AND METHODS
Viruses.
A total of 102 influenza C viruses isolated between 1947 and 2014 were subjected to complete genome sequencing. Twenty-five strains isolated before 1988 were used that had been used in our previous studies (17, 19, 26, 31, 33) and kept in our laboratory. The remaining strains were isolated by our surveillance in Japan as described previously (17–25, 32). The viruses were all propagated in the amniotic cavity of 8- or 9-day-old embryonated hen eggs, and they were stored at −80°C until use for sequencing. The HE, NP, M, and/or NS gene sequences of some viruses had been determined in our previous studies (17–25, 31–37). Nucleotide sequences determined in our laboratory and registered in DDBJ/GenBank were used for C/Ann Arbor/1/50 (38, 39).
Nucleotide sequencing and phylogenetic analysis.
Viral RNA was extracted from 100 μl of amniotic fluid using the RNeasy minikit (Qiagen). The viral RNA was then transcribed into cDNA using a universal primer (5′-AGCAGAAGCAGG-3′) that is complementary to positions 1 to 12 at the 3′ end of all influenza C virus RNA segments. Using the synthesized cDNA as a template, the complete coding regions of the individual RNA segments were amplified using gene-specific primers. The PCR products were purified using a QIAquick PCR purification kit (Qiagen) and were sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Life Technologies) and an ABI Prism 3130 sequencer (Applied Biosystems). The primer sequences used for amplification and sequencing are presented in Table 1.
TABLE 1.
Primers used for amplifications and sequencing of influenza C viruses in this study
| Target gene | Primer | Sequence (5′ to 3′) |
|---|---|---|
| PB2 | FluCPB2F1 | AGCAGAAGCAGAGGATTGGAAAT |
| FluCPB2R1130 | CTCCAGTATTCAAATTGTAC | |
| FluCPB2F604 | GAAATGAGATCAAAGTTTGC | |
| FluCPB2R2365 | AGCAGTAGCAAGAGGATTTTTA | |
| FluCPB2R492 | TACAGGCTGAGTGTCAAC | |
| FluCPB2F292 | AAAAGGGATCATGTTCTTGC | |
| FluCPB2F909 | TATGAGAATTGGAGAAACAG | |
| FluCPB2F1505 | CTGTCACAATACAATCAGG | |
| FluCPB2F2103 | TTTAGTTGTTGGAGATGAAC | |
| PB1 | FluCPB1F1 | AGCAGAAGCAGAGGATTATGG |
| FluCPB1R1402 | CAATGTATATTTGACCAGTTTG | |
| FluCPB1F1228 | TGCCTGGAGGAATGCTTATG | |
| FluCPB1R2363 | AGCAGTAGCAAGAGGATTTTTTCA | |
| FluCPB1R469 | TCAACTGTCAGTTGCAAAGC | |
| FluCPB1R939 | TGTCTCCAGTTATATTAACTGC | |
| FluCPB1F784 | TTGTTGAAACTGTAGCACAG | |
| FluCPB1F1228 | TGCCTGGAGGAATGCTTATG | |
| FluCPB1F1801 | GTGGGAAACTGATGAACAAC | |
| P3 | FluCP3F1 | AGCAGAAGCAGGGGATCCGAA |
| FluCP3R1170 | ATTGAACCATTTAGGGAAAG | |
| FluCP3F989 | CAATAGCCGATAGAACCATG | |
| FluCP3R2183 | AGCAGTAGCAAGGGGATTTTTTC | |
| FluCP3R515 | CTAAGAAAATCAGCAGCAG | |
| FluCP3R1170 | ATTGAACCATTTAGGGAAAG | |
| FluCP3F989 | CAATAGCCGATAGAACCATG | |
| FluCP3F1604 | GACACACAACAGTTAGAATG | |
| HE | FluCHEKN8 | ATAATGTTTTTCTCATTACT |
| FluCHER2025 | TAAAACTGTACAAAATATTG | |
| FluCHER345 | GCCAAACATACTCAACATCA | |
| FluCHEF213 | ATGGATTGGCTTTGGAGATT | |
| FluCHEF810 | TGGAAAAGTTGTTGGAGGGC | |
| FluCHEF1262 | CCTTGGCTGCAAAGGAAGAA | |
| NP | FluCNPF6 | AAGCAGGAGATTTGGTTTTC |
| FluCNPR1088 | ATAAGCAAGTCCAAAGCATG | |
| FluCNPF546 | GGGGCTGGAATCGAAACTAG | |
| FluCNPR1809 | AGCAGTAGCAAGGAGATTTTTGAA | |
| FluCNPR496 | CTTCCTCTTTGATAAATAATTG | |
| FluCNPF321 | GGCACCAATATAACCTACCA | |
| FluCNPF934 | CATATGCAGGAAGAAGAGCC | |
| FluCNPF1503 | ATAAATTTCAGGTCTGGAGC | |
| M | FluCMF4 | AGAAGCAGGGGATTTCAAAA |
| FluCMR1170 | GGGGATTTTTTCAAGGTAATTA | |
| FluCMR548 | GCTGTGCTGGCTTTTCTTAC | |
| FluCMF313 | GCAAATGAAAGCAGCTGGAG | |
| FluCMF676 | AATGAGACCCCACTTGGAAA | |
| NS | FluCNSF1 | AGCAGAAGCAGGGGTACTTTTCC |
| FluCNSR925 | AAGGGGATTTTTAACTTTGG | |
| FluCNSR586 | CCTGTTTCAATTCCGGCCAC | |
| FluCNSF360 | CCATGTATTCCTAACTTTGATGG |
The sequence data were analyzed with MEGA6 software (40). Based on the best-fit models of nucleotide substitution, maximum-likelihood phylogenetic trees of the individual genes were constructed with 1,000 bootstrapped replicates using the same software.
Evolutionary rate.
The evolutionary rates were estimated using the Bayesian Markov chain Monte Carlo methods (MCMC) framework in BEAST version 1.8.2 (41). A maximum-likelihood phylogenetic tree was constructed using the general time-reversible model. A discrete gamma distribution was used to model the evolutionary rate differences among sites, and the rate variation model allowed for some sites to be evolutionarily invariable. A constant population size model with a strict clock, which assumes a uniform evolutionary rate among the branches of the tree, or a relaxed uncorrelated lognormal clock, which allows the evolutionary rates to change among the branches of the tree, were utilized. In the MCMC analysis, 20 million iterations were performed, and samples were obtained every 1,000 steps.
Nucleotide sequence accession numbers.
Nucleotide sequences determined in this study have been submitted to the DDBJ/GenBank database and assigned the accession numbers LC123283 to LC123384 for the PB2 gene, LC123385 to LC123486 for the PB1 gene, LC123487 to LC123588 for the P3 gene, LC122632 to LC122678 for the HE gene, LC123722 to LC123814 for the NP gene, LC123815 to LC123882 for the M gene, and LC124977 to LC125046 for the NS gene. The accession numbers for nucleotide sequences presented in this study are shown in Fig. 1 and 3.
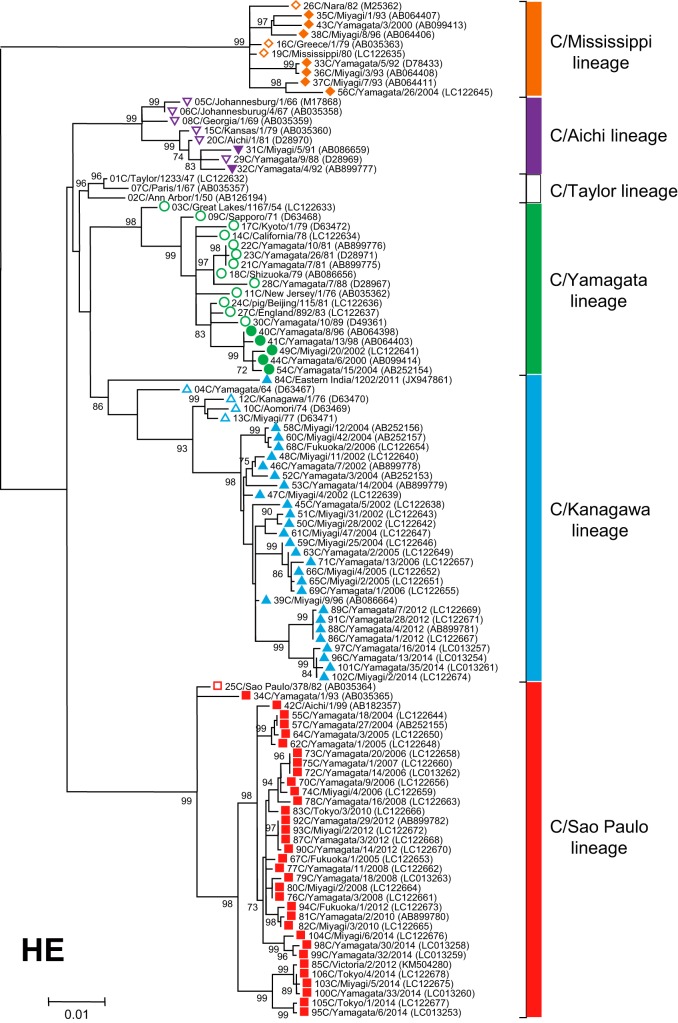
FIG 1.
Phylogenetic tree of the influenza C virus hemagglutinin-esterase (HE) gene. The coding region without a signal peptide in RNA segment 4 was used for the analysis (corresponding to nucleotide positions 64 to 1989 of the HE gene). Numbers below or above the branches indicate the confidence level of the bootstrap analysis with 1,000 replicates as a percentage, and values greater than 70% are shown. Strains belonging to each lineage are marked with the corresponding symbol on the head of the strain name: the C/Mississippi lineage is represented as an orange diamond, the C/Aichi lineage is represented as a purple inverse triangle, the C/Yamagata lineage is represented as a green circle, the C/Kanagawa lineage is represented as a blue triangle, and the C/Sao Paulo lineage is represented as a red square. Strains isolated before 1990 are indicated by open symbols, and strains isolated after 1990 are marked with filled symbols. Numerals prior to the strain name indicate the order of the isolation year. GenBank/DDBJ accession numbers are presented in parentheses.
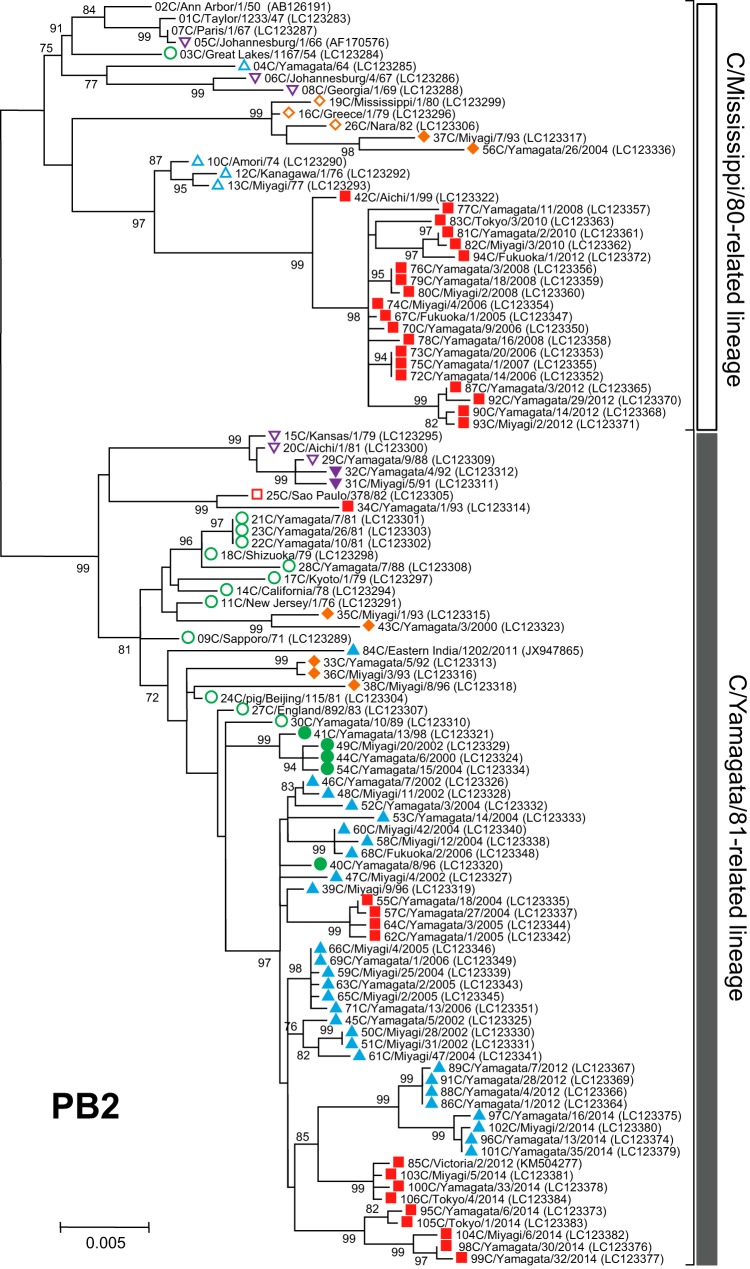
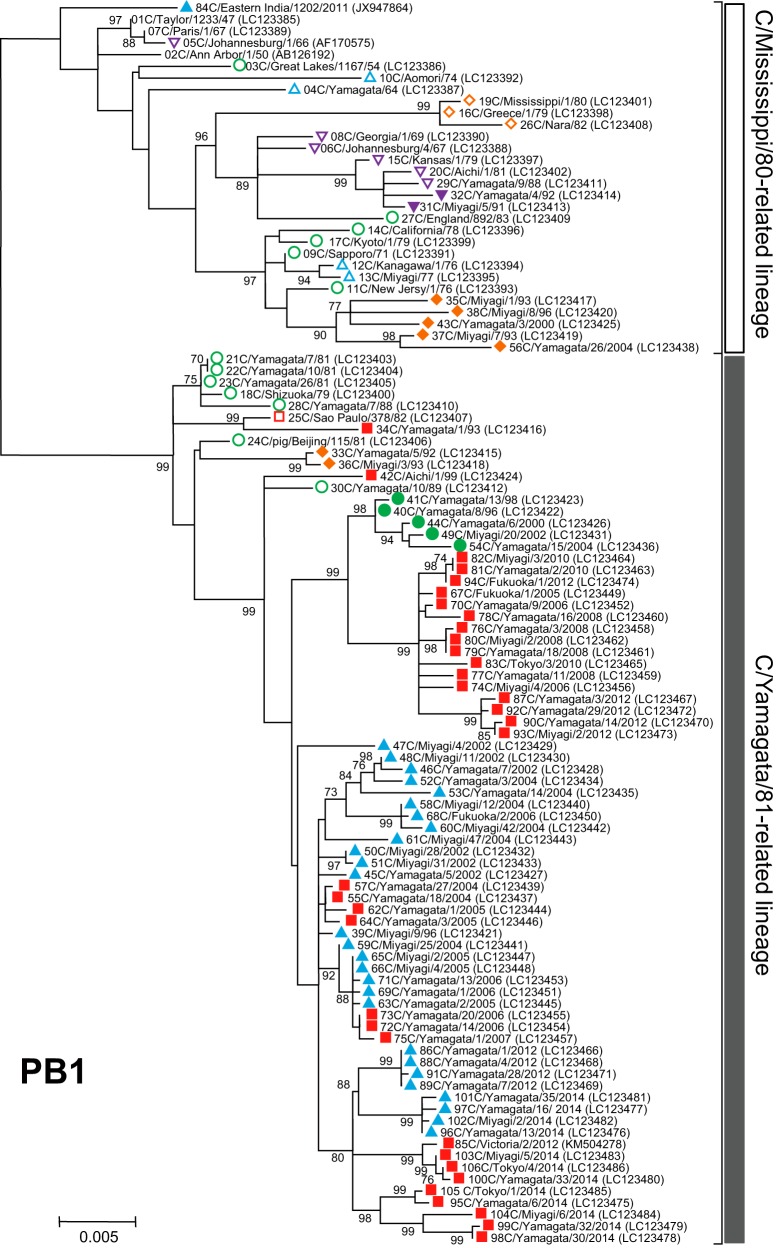
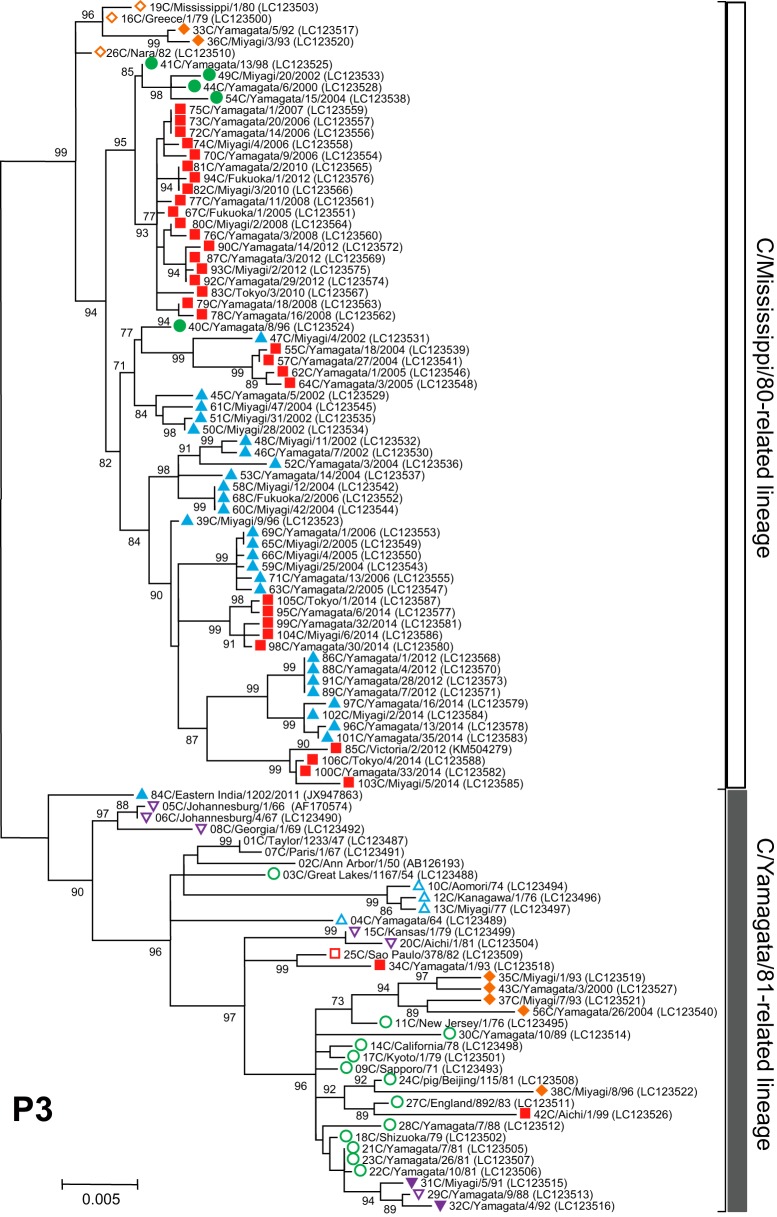
FIG 3.
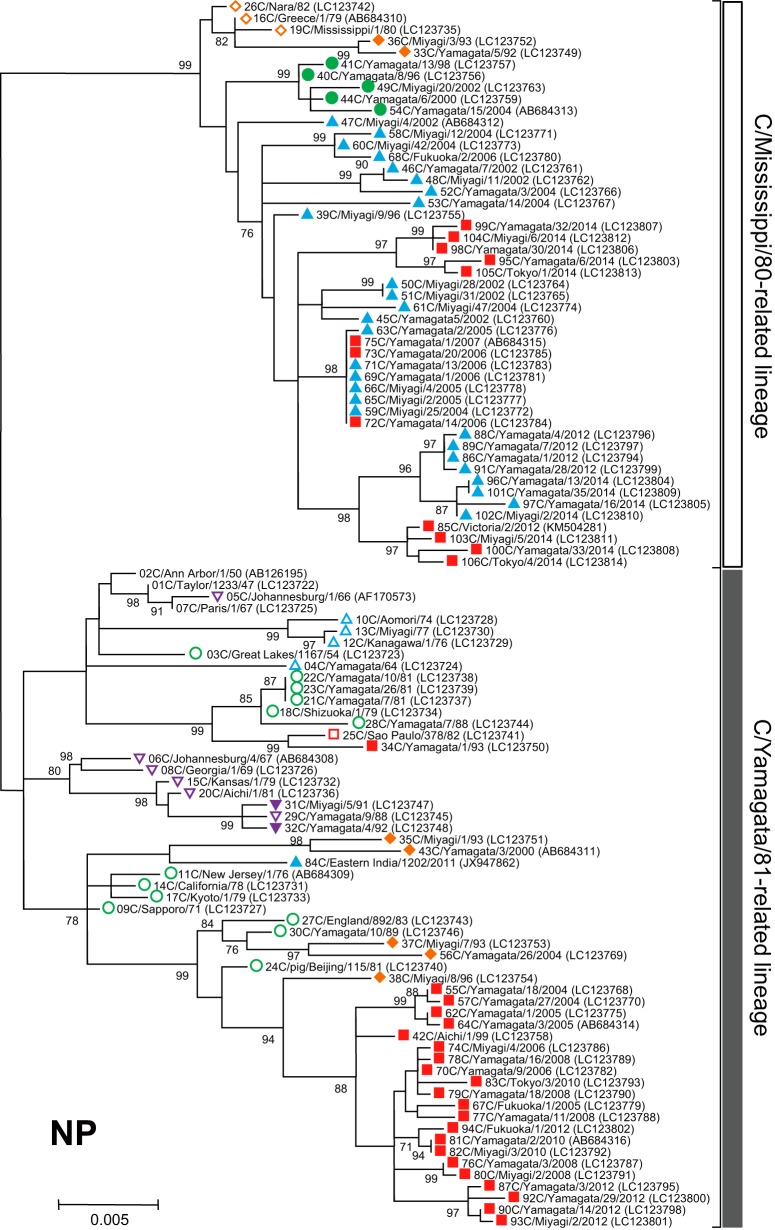
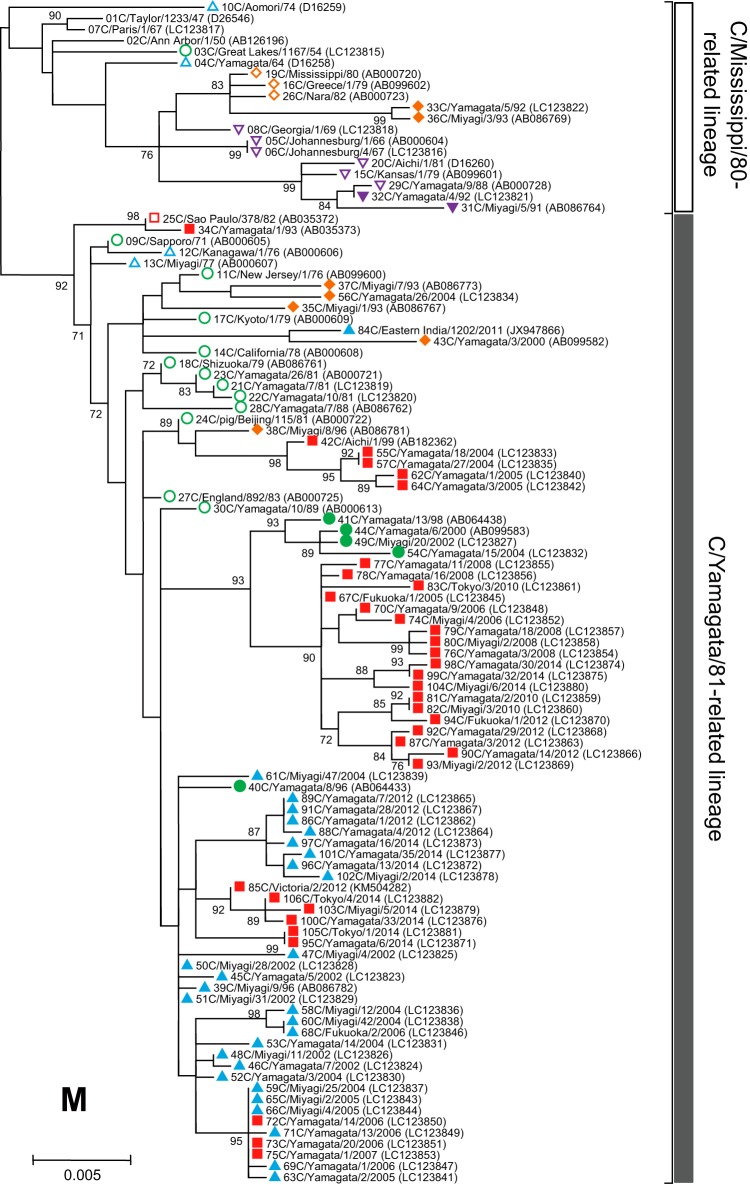
Phylogenetic trees of internal genes of influenza C viruses. The nucleotide sequence of the whole coding region was used for the analysis: nucleotide positions 22 to 2343 for the PB2 gene, 18 to 2279 for the PB1 gene, 22 to 2148 for the P3 gene, 30 to 1724 for the NP gene, 26 to 1147 for the M gene, and 28 to 886 for the NS gene. Numbers below or above the branches indicate the confidence levels of bootstrap analysis with 1,000 replicates as a percentage, and values greater than 70% are shown. Each lineage of the HE gene is shown with the corresponding symbol on the head of the strain name. The C/Mississippi lineage is represented as an orange diamond, the C/Aichi lineage is represented as a purple inverse triangle, the C/Yamagata lineage is represented as a green circle, the C/Kanagawa lineage is represented as a blue triangle, and the C/Sao Paulo lineage is represented as a red square. Strains isolated before 1990 are indicated by open symbols, and strains isolated after 1990 are marked with filled symbols. Numerals prior to the strain name indicate the order of the isolation year. GenBank/DDBJ accession numbers are presented in parentheses.
RESULTS
Six discrete lineages of the HE gene.
To study the lineage of the HE gene in relation to antigenicity, a phylogenetic tree was constructed using 106 HE gene sequences (nucleotides 64 to 1989, corresponding to the complete coding region excluding the signal peptide) from viruses isolated between 1947 and 2014. The sequences of three strains (C/Johannesburg/1/66, C/Eastern India/1202/2011, and C/Victoria/2/2012) were obtained from GenBank. The sequences of the other 103 strains were determined in our laboratory. As shown in Fig. 1, the HE genes were divided into six discrete lineages, C/Taylor/1233/47, C/Mississippi/80, C/Aichi/1/81, C/Yamagata/26/81, C/Kanagawa/1/76, and C/Sao Paulo/378/82. For the influenza C virus, the phylogenetic classification based on the HE gene corresponds to the antigenic classification (19, 21, 24). Therefore, in this study, we named the six HE lineages, the C/Taylor lineage, C/Mississippi lineage, C/Aichi lineage, C/Yamagata lineage, C/Kanagawa lineage, and C/Sao Paulo lineage, as both antigenic and genetic lineages of the HE gene.
The prototype strain, C/Taylor/1233/47, the first isolate from a human, who lived in New York, formed the C/Taylor lineage together with C/Ann Arbor/1/50 and C/Paris/1/67, which were isolated in the United States in 1950 and in France in 1967, respectively. However, there was no virus belonging to the C/Taylor lineage after 1967, suggesting that this lineage died out. Among the 30 strains isolated before 1990, four strains isolated in South Africa (C/Johannesburg/1/66 and C/Johannesburg/4/67) and the United States (C/Georgia/1/69 and C/Kansas/1/79) belong to the C/Aichi lineage together with two Japanese strains (C/Aichi/1/81 and C/Yamagata/9/88); however, this lineage of strains disappeared from Japan and from the world after 1992 (18). Of the other 21 strains isolated before 1990, three strains from Greece (C/Greece/1/79), the United States (C/Mississippi/80), and Japan (C/Nara/82) belong to the C/Mississippi lineage, and four strains from Japan (C/Yamagata/64, C/Aomori/74, C/Kanagawa/1/76, and C/Miyagi/77) and one strain from Brazil (C/Sao Paulo/378/82) form the C/Kanagawa lineage and the C/Sao Paulo lineage, respectively. The remaining 13 strains belong to the C/Yamagata lineage: three strains (C/Great Lakes/1167/54, C/New Jersey/1/76, and C/California/78) from the United States, one swine strain (C/pig/Beijing/115/81) from China, one strain (C/England/892/83) from England, and eight strains (C/Sapporo/71, C/Kyoto/1/79, C/Shizuoka/79, C/Yamagata/7/81, C/Yamagata/10/81, C/Yamagata/26/81, C/Yamagata/7/88, and C/Yamagata/10/89) from Japan.
There were almost no reports of influenza C virus isolation from the middle of the 1980s until the late 2000s, when molecular techniques were used. As shown in Fig. 2, most of the Japanese strains isolated from 1988 to 2000 by our surveillance system belong to the C/Aichi, C/Mississippi, or C/Yamagata lineage (17–22). Viruses isolated in Tokyo, Fukuoka, Miyagi, and Yamagata, Japan, from 2002 to 2014 mainly belong to the C/Kanagawa or C/Sao Paulo lineage (23–25). A strain from Australia deposited in 2012 (C/Victoria/2/2012) belongs to the C/Sao Paulo lineage and forms one cluster together with the Japanese strains (C/Tokyo/4/2014, C/Miyagi/5/2014, and C/Yamagata/33/2014). Another recently reported virus from India in 2011 (C/Eastern India/1202/2011) (42) was classified in the C/Kanagawa lineage but forms a different branch apart from the other strains belonging to the C/Kanagawa lineage.
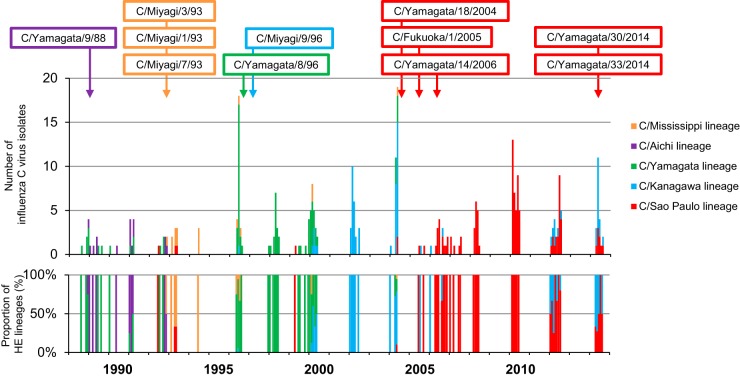
FIG 2.
Monthly distribution of influenza C viruses in Japan between 1988 and 2014. The number of influenza C virus isolates was obtained from our previous reports: Matsuzaki et al. (17, 19, 20, 21, 22, 24, 32), Kimura et al. (18), and Tanaka et al. (25). The colored open squares indicate the representative reassortant viruses that emerged during influenza C virus epidemics.
Two major lineages of internal genes.
We performed phylogenetic analysis of individual RNA segments of the internal genes PB2, PB1, P3, NP, M, and NS. Although partial sequences of the PB2, PB1, P3, and NP genes were used in our previous works (18–22, 31, 32), to extend these studies to the analysis of complete sequences, whole sequences of the coding region for six individual RNA segments were determined for the 102 strains. The sequences of four strains (C/Ann Arbor/1/50, C/Johannesburg/1/66, C/Eastern India/1202/2011, and C/Victoria/2/2012) were obtained from GenBank; however, the M and NS gene sequences of C/Johannesburg/1/66 were determined in our laboratory (35, 36).
As shown in Fig. 3, two major lineages, C/Mississippi/80 and C/Yamagata/26/81, were identified in each of six internal gene trees. Although our previous studies showed some sublineages in the phylogenetic trees (21, 22), more than 80% bootstrap probability at the branches could not be obtained by complete sequence analysis. Therefore, we divided the genetic lineages of the internal genes into two major lineages, the C/Mississippi/80-related lineage and the C/Yamagata/81-related lineage.
Our previous studies demonstrated that the NS gene sequences obtained from 34 strains isolated between 1947 and 1992 were split into two groups, A and B, which correspond to the C/Mississippi/80-related lineage and the C/Yamagata/81-related lineage, respectively. Most influenza C viruses had acquired group B NS genes until the early 1980s irrespective of their HE gene lineage (36). This study confirmed the previous results, although two strains (C/Miyagi/7/93 and C/Yamagata/26/2004) that had HE genes belonging to the C/Mississippi lineage were classified into the C/Mississippi/80-related lineage (correspond to group A) on the NS gene tree. On the other five phylogenetic trees based on the PB2, PB1, P3, NP, and M gene sequences, the sequences seemed to be split into two lineages independently of the year of isolation.
Reassortment of influenza C viruses between 1947 and 2014.
To study the reassortment of influenza C viruses circulating from 1947 to 2014, their internal genome compositions, which were determined by dividing them into C/Mississippi/80-related and C/Yamagata/81-related lineages by phylogenetic analysis, were investigated for the 106 strains according to HE lineages. As shown in Fig. 4A, all strains isolated by the 1960s, whose HE genes belonged to the C/Taylor, C/Aichi, C/Yamagata, and C/Kanagawa lineages, had the same composition of internal genes. After 1970, reassortment events occurred among the viruses from the C/Aichi, C/Yamagata, and C/Kanagawa HE lineages. However, among the viruses belonging to the same HE lineage, viruses isolated in various areas of the world had the same composition during the same period. For example, in the C/Mississippi lineage, C/Mississippi/80 isolated in the United States in 1980 had the same genome composition as C/Greece/1/79 isolated in Greece in 1979 and C/Nara/82 isolated in Japan in 1982. In other lineages, C/Aichi/1/81 isolated in Japan in 1981 had the same genome composition as C/Kansas/1/79 isolated in the United States in 1979, and C/Yamagata/26/81 isolated in Japan in 1981 had the same genome composition as C/pig/Beijing/115/81 isolated from pigs in China in 1981.
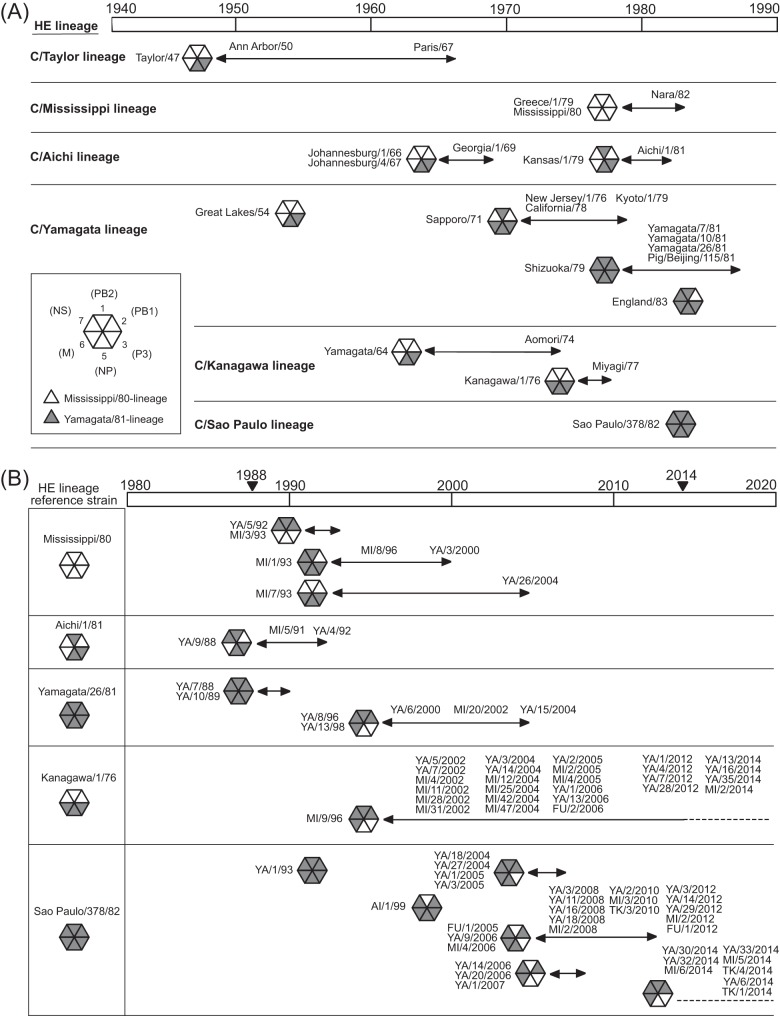
FIG 4.
Internal genome constellations of the influenza C viruses for which the full genome was analyzed in this study. (A) Influenza C viruses circulating between 1947 and 1983 around the world are shown. (B) Influenza C viruses isolated in Japan from 1988 to 2014 are shown. Virus particles are indicated by hexagons containing triangles for the six internal gene segments (clockwise: PB2, PB1, P3, NP, M, and NS) and colored according to their lineage: white, Mississippi/80-related lineage; black, Yamagata/81-related lineage. Abbreviations in panel B: YA, Yamagata; MI, Miyagi; AI, Aichi; FU, Fukuoka; TK, Tokyo.
Since 1988, we have carried out long-term surveillance of influenza C virus in Japan (Fig. 2). In previous studies based on partial sequences of the PB2, PB1, P3, and NP genes, we reported two reassortment events. One hypothesis is that multiple reassortment occurred between 1992 and 1996 among viruses with the HE gene of the C/Mississippi lineage (21), and reassortant viruses of the C/Yamagata lineage, which obtained the P3 and NP genes from a C/Mississippi/80-like virus, caused two outbreaks in Yamagata, Japan, in 1996 and 1998 (20). As shown in Fig. 4B, these findings are consistent with the present results obtained based on complete sequences of internal genes.
In this study, we analyzed the phylogenetic position of each strain circulating between 2000 and 2014 in Japan. On the HE gene tree (Fig. 1), strains of the C/Yamagata lineage isolated between 1996 and 2004, strains of the C/Kanagawa lineage isolated between 1996 and 2014, and strains of the C/Sao Paulo lineage isolated between 1999 and 2014 formed one cluster within the respective lineage. As shown in Fig. 4B, the genomic composition of C/Kanagawa lineage strains such as C/Miyagi/9/96, C/Yamagata/35/2014, and C/Miyagi/2/2014 did not change from 1996 to 2014, and it is the same as that of the C/Yamagata lineage strains circulating between 1996 and 2004. Conversely, some different reassortant viruses of the C/Sao Paulo lineage were found during the period from 1999 to 2014. However, interestingly, the internal genome composition of all strains of the C/Sao Paulo lineage isolated in 2014, such as C/Yamagata/6/2014, C/Miyagi/6/2014, and C/Tokyo/4/2014, is the same as that of the C/Yamagata lineage and C/Kanagawa lineage strains isolated between 1996 and 2014. One C/Sao Paulo lineage strain reported in Australia in 2012, C/Victoria/2/2012, was found to be highly homologous to the Japanese C/Sao Paulo lineage strains isolated in 2014, such as C/Yamagata/33/2014, C/Miyagi/5/2014, and C/Tokyo/4/2014, in the phylogenetic trees constructed with the HE, PB2, PB1, P3, NP, M, and NS genes. One strain of the C/Kanagawa lineage reported in India in 2011, C/Eastern India/1202/2011, had unique phylogenetic positions. In the trees constructed with the HE, PB1, and P3 genes, it was especially closely related to the strains isolated by the 1960s, such as C/Yamagata/64, C/Ann Arbor/1/50, and C/Johannesburg/1/66, respectively. The internal genome composition of C/Eastern India/1202/2011 was found to be similar to that of C/England/892/83.
Nucleotide and amino acid sequence identities and evolutionary rates.
The identities of the nucleotides and deduced amino acid sequences among 106 strains are summarized in Table 2. The nucleotide sequences of influenza C virus isolates were highly homologous to each other; mean percent identities were greater than 95% for all segments. The sequences in the HE gene had lower homology, at 95.3%, and the minimum identity of 91.1% was between C/Yamagata/26/2004 (C/Mississippi lineage) and C/Yamagata/16/2014 (C/Kanagawa lineage). The PB2, PB1, P3, HE, and NP genes each encode single proteins, and the M and NS genes each encode two proteins, M1 and CM2 as well as NS1 and NS2, respectively. The HE protein is composed of two subunits that are cleaved by a host protease: HE1 is the globular region of the HE protein, and HE2 is the stalk region. The amino acid sequences were also highly homologous for all proteins. The minimum amino acid sequence identity was 93.3% in the HE1 region of the HE protein found between C/Yamagata/14/2004 (C/Kanagawa lineage) and C/Victoria/2/2012 (C/Sao Paulo lineage).
TABLE 2.
Identities and nucleotide substitution rates for all segments of influenza C viruses circulating between 1947 and 2014
| Segment | Coding region | Nucleotide length | Amino acid length | Mean % identity (range) |
Nucleotide substitution rate (10−4 substitutions/site/year), 95% HPDb |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Strict clock |
Lognormal relaxed clock |
||||||||
| Mean | Lower | Upper | Mean | Lower | Upper | ||||||
| 1 | PB2 | 2,322 | 774 | 97.8 (95.7–100) | 99.6 (98.7–100) | 3.87 | 3.31 | 4.44 | 3.96 | 3.37 | 4.55 |
| 2 | PB1 | 2,262 | 754 | 97.6 (94.4–100) | 99.5 (98.3–100) | 4.11 | 3.56 | 4.71 | 4.38 | 3.73 | 5.04 |
| 3 | P3 | 2,127 | 709 | 97.8 (95.1–100) | 99.3 (97.9–100) | 3.76 | 3.25 | 4.27 | 3.87 | 3.29 | 4.44 |
| 4 | HE | 1,923 | 641a | 95.3 (91.1–100) | 97.1 (94.7–100) | 5.20 | 4.51 | 5.88 | 5.23 | 4.51 | 5.98 |
| HE1 | 1,296 | 432 | 94.9 (89.9–100) | 96.2 (93.3–100) | |||||||
| HE2 | 627 | 209 | 96.1 (92.7–100) | 98.9 (95.7–100) | |||||||
| 5 | NP | 1,695 | 565 | 97.7 (95.7–100) | 99.4 (98.4–100) | 4.03 | 3.44 | 4.66 | 4.04 | 3.40 | 4.69 |
| 6 | M | 1,122 | 98.3 (96.1–100) | 4.04 | 3.28 | 4.83 | 4.05 | 3.27 | 4.81 | ||
| M1 | 726 | 242 | 98.3 (95.1–100) | 99.7 (98.3–100) | |||||||
| CM2 | 345 | 115a | 98.7 (96.1–100) | 99.4 (95.7–100) | |||||||
| 7 | NS | 859 | 98.1 (95.6–100) | 4.31 | 3.36 | 5.30 | 4.48 | 3.49 | 5.56 | ||
| NS1 | 738 | 246 | 98.4 (96.4–100) | 98.6 (95.5–100) | |||||||
| NS2 | 546 | 182 | 98.0 (95.2–100) | 97.8 (94.0–100) | |||||||
Signal peptide is excluded from length.
HPD, highest posterior density.
The nucleotide substitution rates for all of the segments were estimated using BEAST (Table 2). The rates of nucleotide substitution of individual segments ranged from 3.76 × 10−4 to 5.20 × 10−4 (strict clock) and from 3.87 × 10−4 to 5.23 × 10−4 (relaxed clock) substitutions/site/year, with the lowest and highest rates found for the P3 and HE genes, respectively.
DISCUSSION
In this study, we determined the genetic lineages of influenza C virus by analyzing complete genome sequences of isolates, ranging from the first, C/Taylor/1233/47, to recent isolates of 2014. Moreover, the relationship between internal genome composition and antigenic lineage (HE lineage) was studied. The results suggested that there are some advantageous genome constellations and that they may be related to influenza C virus epidemics.
We previously suggested that the C/Kanagawa lineage died out, similar to the C/Taylor lineage, because no strain of the C/Kanagawa lineage was isolated since 1977 (34). However, a C/Kanagawa lineage virus reemerged in Sendai City, Japan, in 1996 (C/Miyagi/9/96) that had acquired its internal genome composition from epidemic strains of the C/Yamagata lineage, such as C/Yamagata/8/96, and spread to various areas of Japan during 2000 (21, 22). The present study revealed that such C/Kanagawa lineage viruses continue to circulate as a predominant virus over the C/Yamagata lineage viruses (Fig. 2 and 4B).
In 1992 and 1993, three strains belonging to the C/Sao Paulo lineage, which have internal genome sequences phylogenetically similar to that of C/Sao Paulo/378/82, were isolated in Japan for the first time; one of the three strains is C/Yamagata/1/93 (19). In 1999, one strain belonging to the C/Sao Paulo lineage (C/Aichi/1/99) was isolated in Japan from a traveler who had visited Malaysia (32). Therefore, C/Aichi/1/99 was circulating in Malaysia and had a genome composition distinct from that of C/Yamagata/1/93 (32). The increase in isolates belonging to the C/Sao Paulo lineage in Japan started in 2004 (Fig. 2) (24). This study revealed that multiple HE gene reassortants of the C/Sao Paulo lineage coexisted in Japan during the short period between 2004 and 2012, and these strains are phylogenetically more similar to C/Aichi/1/99 than C/Yamagata/1/93. Among the internal gene sequences of 23 strains belonging to the C/Sao Paulo lineage isolated between 2004 and 2012, the NP and M gene sequences of 4 strains isolated in 2004 and 2005 (C/Yamagata/18/2004, C/Yamagata/27/2004, C/Yamagata/1/2005, and C/Yamagata/3/2005) were very similar to those of C/Aichi/1/99, and the PB2 gene sequences of the remaining 19 strains isolated in Tokyo, Fukuoka, Miyagi, and Yamagata, Japan, form one cluster together with C/Aichi/1/99 in the C/Mississippi/80-related lineage. This result suggests that a newly introduced influenza C virus spread widely throughout Japan while undergoing reassortment events.
All C/Sao Paulo lineage strains isolated in 2014 had the same genome constellation, which is similar to that of the C/Kanagawa lineage isolated in 2014. However, their HE gene sequences could be divided into two distinct clusters within the C/Sao Paulo lineage. One cluster is represented by C/Aichi/1/99, and the other cluster is represented by C/Victoria/2/2012 (Fig. 1). On the HE gene tree, three strains (C/Yamagata/30/2014, C/Yamagata/32/2014, and C/Miyagi/6/2014) were classified into a cluster together with C/Aichi/1/99 and all strains of the C/Sao Paulo lineage isolated between 2004 and 2012. However, their PB2, PB1, P3, and NP gene sequences are closely related to those of the C/Kanagawa lineage circulating between 2012 and 2014 in Japan. Therefore, these strains seem to have emerged by acquiring the gene segments that encode the polymerase complex and the nucleoprotein from viruses of the C/Kanagawa lineage circulating in Japan. Five strains (C/Yamagata/6/2014, C/Yamagata/33/2014, C/Miyagi/5/2014, C/Tokyo/1/2014, and C/Tokyo/4/2014) form a clearly distinct cluster together with C/Victoria/2/2012 on the HE gene tree. Of these strains, C/Yamagata/33/2014, C/Miyagi/5/2014, and C/Tokyo/4/2014 have genes that are highly homologous to those of C/Victoria/2/2012 on the internal gene trees, suggesting that they were derived from a strain that was introduced into Japan from a foreign country. Interestingly, the internal genome composition of C/Victoria/2/2012 is the same as that of strains having HE genes belonging to the C/Kanagawa lineage, such as C/Miyagi/9/96. Odagiri et al. previously reported that 10 strains isolated in the Philippines in 2011 and 2013 had HE genes belonging to the C/Sao Paulo lineage and had the same internal genome composition as C/Miyagi/9/96 (43). Therefore, the viruses isolated in the Philippines are closely related to C/Victoria/2/2012, suggesting that such viruses of the C/Sao Paulo lineage were circulating in Australia and the Philippines in 2011 to 2013 before they spread to Japan. Although antigenic differences between the two distinct clusters within the C/Sao Paulo lineage cannot be detected using polyclonal immune sera, these clusters could be distinguished from each other by their reactivity with an anti-HE monoclonal antibody in hemagglutinin inhibition testing (25). Observation over a longer period is required to reveal whether either or both of the two clusters survive in a community.
In Japan in 2014, influenza C viruses belonging to the C/Kanagawa and C/Sao Paulo lineages were cocirculating (Fig. 2) (25), and these lineages had the same internal genome constellation as the C/Yamagata lineage, which was the dominant lineage in Japan between 1996 and 2000 (Fig. 2 and 4B). Therefore, this genome composition certainly seems to provide some advantage for viral spread in humans. Interestingly, as shown in Fig. 4A, this genome composition is the reverse of that of each virus isolated before 1970. Although various genome constellations appeared after 1970, the P3 and NP gene segments always belong to the same lineage (C/Mississippi/80-related lineage or C/Yamagata/81-related lineage) except for a small number of exceptions, such as viruses of the C/Sao Paulo lineage isolated between 2004 and 2012. This observation suggests that the genome constellation of influenza C viruses restricts viral fitness. Reassortment events might play a role in the acquisition of an advantageous internal gene composition under this restriction. Further analyses using a larger number of influenza C viruses, including foreign isolates, and also functional analysis, including the generation of reassortant viruses, are required to confirm this suggestion.
This study provides the first examination of the rates of nucleotide substitution for influenza C viruses spanning 68 years from complete coding region sequences. The highest rate was that of the HE gene. The comparison performed using similar methodology yielded a rate of 5.20 × 10−4 substitutions/site/year, which is approximately 8- to 11-fold lower than the estimated rates of 3.99 × 10−3 substitutions/site/year (30) or 5.72 × 10−3 substitutions/site/year (28) obtained for the HA gene from influenza A (H3N2) viruses and approximately 4-fold lower than the estimated rate of 2.15 × 10−3 substitutions/site/year obtained for the HA gene from influenza B viruses (29). The amino acid sequence identity of more than 93% obtained for the HE1 protein suggests that the antigenicity of influenza C virus is highly stable. PB2, PB1, P3, and NP proteins, which construct a ribonucleoprotein complex together with vRNA, have a high identity of more than 98%. Although both M1 and CM2 protein sequences are conserved fairly well, the amino acid sequence of the M1 protein, which gives rigidity to the virion (39), has higher identity than that of the CM2 protein, which plays a role in the genome packaging and uncoating processes (44). The divergence of the amino acid sequence of the NS2 protein, which possesses nuclear export activity (45), is higher than that of the NS1 protein and comparable to that of the HE protein. The precise role of each protein of the influenza C virus has yet to be determined. Further analysis of the relationship between conserved regions in amino acid sequences and biological activity might help to clarify the functions of the respective proteins.
ACKNOWLEDGMENTS
We thank Yoko Kadowaki for her assistance with sequence analysis.
This work was supported by JSPS KAKENHI grant 26461563.
We have no potential conflict of interest to declare related to this article.
REFERENCES
- 1.Taylor RM. 1949. Studies on survival of influenza virus between epidemics and antigenic variants of the virus. Am J Public Health Nations Health 39:171–178. doi: 10.2105/AJPH.39.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo YJ, Jin FG, Wang P, Wang M, Zhu JM. 1983. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J Gen Virol 64:177–182. doi: 10.1099/0022-1317-64-1-177. [DOI] [PubMed] [Google Scholar]
- 3.Katagiri S, Ohizumi A, Homma M. 1983. An outbreak of type C influenza in a children's home. J Infect Dis 148:51–56. doi: 10.1093/infdis/148.1.51. [DOI] [PubMed] [Google Scholar]
- 4.Moriuchi H, Katsushima N, Nishimura H, Nakamura K, Numazaki Y. 1991. Community-acquired influenza C virus infection in children. J Pediatr 118:235–238. doi: 10.1016/S0022-3476(05)80489-5. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzaki Y, Katsushima N, Nagai Y, Shoji M, Itagaki T, Sakamoto M, Kitaoka S, Mizuta K, Nishimura H. 2006. Clinical features of influenza C virus infection in children. J Infect Dis 193:1229–1235. doi: 10.1086/502973. [DOI] [PubMed] [Google Scholar]
- 6.Calvo C, García-García ML, Borrell B, Pozo F, Casas I. 2013. Prospective study of influenza C in hospitalized children. Pediatr Infect Dis J 32:916–919. doi: 10.1097/INF.0b013e31828fca10. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu Y, Abiko C, Ikeda T, Mizuta K, Matsuzaki Y. 2015. Influenza C virus and human metapneumovirus infections in hospitalized children with lower respiratory tract illness. Pediatr Infect Dis J 34:1273–1275. doi: 10.1097/INF.0000000000000863. [DOI] [PubMed] [Google Scholar]
- 8.Dykes AC, Cherry JD, Nolan CE. 1980. A clinical, epidemiologic, serologic, and virologic study of influenza C virus infection. Arch Intern Med 140:1295–1298. [PubMed] [Google Scholar]
- 9.O'Callaghan RJ, Gohd RS, Labat DD. 1980. Human antibody to influenza C virus: its age-related distribution and distinction from receptor analogs. Infect Immun 30:500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homma M, Ohyama S, Katagiri S. 1982. Age distribution of the antibody to type C influenza virus. Microbiol Immunol 26:639–642. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura H, Sugawara K, Kitame F, Nakamura K, Sasaki H. 1987. Prevalence of the antibody to influenza C virus in a northern Luzon Highland Village, Philippines. Microbiol Immunol 31:1137–1143. doi: 10.1111/j.1348-0421.1987.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 12.Manuguerra JC, Hannoun C, Aymard M. 1992. Influenza C virus infection in France. J Infect 24:91–99. doi: 10.1016/0163-4453(92)91150-A. [DOI] [PubMed] [Google Scholar]
- 13.Manuguerra JC, Hannoun C, Sáenz Mdel C, Villar E, Cabezas JA. 1994. Sero-epidemiological survey of influenza C virus infection in Spain. Eur J Epidemiol 10:91–94. doi: 10.1007/BF01717459. [DOI] [PubMed] [Google Scholar]
- 14.Motta FC, Luiz MO, Couceiro JN. 2000. Serological analysis reveals circulation of influenza C viruses, Brazil. Rev Saude Publica 34:204–205. doi: 10.1590/S0034-89102000000200017. [DOI] [PubMed] [Google Scholar]
- 15.Yano T, Maeda C, Akachi S, Matsuno Y, Yamadera M, Kobayashi T, Nagai Y, Iwade Y, Kusuhara H, Katayama M, Fukuta M, Nakagawa Y, Naraya S, Takahashi H, Hiraoka M, Yamauchi A, Nishinaka T, Amano H, Yamaguchi T, Ochiai H, Ihara T, Matsuzaki Y. 2014. Phylogenetic analysis and seroprevalence of influenza C virus in Mie Prefecture, Japan in 2012. Jpn J Infect Dis 67:127–131. doi: 10.7883/yoken.67.127. [DOI] [PubMed] [Google Scholar]
- 16.Salez N, Mélade J, Pascalis H, Aherfi S, Dellagi K, Charrel RN, Carrat F, de Lamballerie X. 2014. Influenza C virus high seroprevalence rates observed in 3 different population groups. J Infect 69:182–189. doi: 10.1016/j.jinf.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki Y, Muraki Y, Sugawara K, Hongo S, Nishimura H, Kitame F, Katsushima N, Numazaki Y, Nakamura K. 1994. Cocirculation of two distinct groups of influenza C virus in Yamagata City, Japan. Virology 202:796–802. doi: 10.1006/viro.1994.1401. [DOI] [PubMed] [Google Scholar]
- 18.Kimura H, Abiko C, Peng G, Muraki Y, Sugawara K, Hongo S, Kitame F, Mizuta K, Numazaki Y, Suzuki H, Nakamura K. 1997. Interspecies transmission of influenza C virus between humans and pigs. Virus Res 48:71–79. doi: 10.1016/S0168-1702(96)01427-X. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzaki Y, Mizuta K, Kimura H, Sugawara K, Tsuchiya E, Suzuki H, Hongo S, Nakamura K. 2000. Characterization of antigenically unique influenza C virus strains isolated in Yamagata and Sendai cities, Japan, during 1992-1993. J Gen Virol 81:1447–1452. doi: 10.1099/0022-1317-81-6-1447. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki Y, Sugawara K, Mizuta K, Tsuchiya E, Muraki Y, Hongo S, Suzuki H, Nakamura K. 2002. Antigenic and genetic characterization of influenza C viruses which caused two outbreaks in Yamagata City, Japan, in 1996 and 1998. J Clin Microbiol 40:422–429. doi: 10.1128/JCM.40.2.422-429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuzaki Y, Mizuta K, Sugawara K, Tsuchiya E, Muraki Y, Hongo S, Suzuki H, Nishimura H. 2003. Frequent reassortment among influenza C viruses. J Virol 77:871–881. doi: 10.1128/JVI.77.2.871-881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuzaki Y, Takao S, Shimada S, Mizuta K, Sugawara K, Takashita E, Muraki Y, Hongo S, Nishimura H. 2004. Characterization of antigenically and genetically similar influenza C viruses isolated in Japan during the 1999-2000 season. Epidemiol Infect 132:709–720. doi: 10.1017/S0950268804002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuzaki Y, Abiko C, Mizuta K, Sugawara K, Takashita E, Muraki Y, Suzuki H, Mikawa M, Shimada S, Sato K, Kuzuya M, Takao S, Wakatsuki K, Itagaki T, Hongo S, Nishimura H. 2007. A nationwide epidemic of influenza C virus infection in Japan in 2004. J Clin Microbiol 45:783–788. doi: 10.1128/JCM.01555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzaki Y, Sugawara K, Abiko C, Ikeda T, Aoki Y, Mizuta K, Katsushima N, Katsushima F, Katsushima Y, Itagaki T, Shimotai Y, Hongo S, Muraki Y, Nishimura H. 2014. Epidemiological information regarding the periodic epidemics of influenza C virus in Japan (1996-2013) and the seroprevalence of antibodies to different antigenic groups. J Clin Virol 61:87–93. doi: 10.1016/j.jcv.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka S, Aoki Y, Matoba Y, Yahagi K, Mizuta K, Itagaki T, Katsushima F, Katsushima Y, Matsuzaki Y. 2015. The dominant antigenic group of influenza C infections changed from C/Sao Paulo/378/82-lineage to C/Kanagawa/1/76-lineage in Yamagata, Japan, in 2014. Jpn J Infect Dis 68:166–168. doi: 10.7883/yoken.JJID.2014.520. [DOI] [PubMed] [Google Scholar]
- 26.Sugawara K, Nishimura H, Kitame F, Nakamura K. 1986. Antigenic variation among human strains of influenza C virus detected with monoclonal antibodies to gp88 glycoprotein. Virus Res 6:27–32. doi: 10.1016/0168-1702(86)90054-7. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki Y, Sugawara K, Takashita E, Muraki Y, Hongo S, Katsushima N, Mizuta K, Nishimura H. 2004. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J Med Virol 74:132–140. doi: 10.1002/jmv.20156. [DOI] [PubMed] [Google Scholar]
- 28.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R, Holmes EC. 2008. The evolutionary dynamics of human influenza B virus. J Mol Evol 66:655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westgeest KB, Russell CA, Lin X, Spronken MI, Bestebroer TM, Bahl J, van Beek R, Skepner E, Halpin RA, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Smith DJ, Wentworth DE, Fouchier RA, de Graaf M. 2014. Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J Virol 88:2844–2857. doi: 10.1128/JVI.02163-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng G, Hongo S, Kimura H, Muraki Y, Sugawara K, Kitame F, Numazaki Y, Suzuki H, Nakamura K. 1996. Frequent occurrence of genetic reassortment between influenza C virus strains in nature. J Gen Virol 77:1489–1492. doi: 10.1099/0022-1317-77-7-1489. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzaki Y, Sato K, Sugawara K, Takashita E, Muraki Y, Morishita T, Kumagai N, Suzuki S, Hongo S. 2005. Isolation of an influenza C virus introduced into Japan by a traveler from Malaysia. J Clin Microbiol 43:993–995. doi: 10.1128/JCM.43.2.993-995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adachi K, Kitame F, Sugawara K, Nishimura H, Nakamura K. 1989. Antigenic and genetic characterization of three influenza C strains isolated in the Kinki district of Japan in 1982-1983. Virology 172:125–133. doi: 10.1016/0042-6822(89)90114-1. [DOI] [PubMed] [Google Scholar]
- 34.Muraki Y, Hongo S, Sugawara K, Kitame F, Nakamura K. 1996. Evolution of the haemagglutinin-esterase gene of influenza C virus. J Gen Virol 77:673–679. doi: 10.1099/0022-1317-77-4-673. [DOI] [PubMed] [Google Scholar]
- 35.Tada Y, Hongo S, Muraki Y, Sugawara K, Kitame F, Nakamura K. 1997. Evolutionary analysis of influenza C virus M genes. Virus Genes 15:53–59. doi: 10.1023/A:1007915215958. [DOI] [PubMed] [Google Scholar]
- 36.Alamgir AS, Matsuzaki Y, Hongo S, Tsuchiya E, Sugawara K, Muraki Y, Nakamura K. 2000. Phylogenetic analysis of influenza C virus nonstructural (NS) protein genes and identification of the NS2 protein. J Gen Virol 81:1933–1940. doi: 10.1099/0022-1317-81-8-1933. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzaki Y, Ikeda T, Abiko C, Aoki Y, Mizuta K, Shimotai Y, Sugawara K, Hongo S. 2012. Detection and quantification of influenza C virus in pediatric respiratory specimens by real-time PCR and comparison with infectious viral counts. J Clin Virol 54:130–134. doi: 10.1016/j.jcv.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Muraki Y, Washioka H, Sugawara K, Matsuzaki Y, Takashita E, Hongo S. 2004. Identification of an amino acid residue on influenza C virus M1 protein responsible for formation of the cord-like structures of the virus. J Gen Virol 85:1885–1893. doi: 10.1099/vir.0.79937-0. [DOI] [PubMed] [Google Scholar]
- 39.Muraki Y, Murata T, Takashita E, Matsuzaki Y, Sugawara K, Hongo S. 2007. A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein. J Virol 81:8766–8773. doi: 10.1128/JVI.00075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee TR, Mukherjee A, Mullick S, Chawla-Sarkar M. 2013. Full genome analysis and characterization of influenza C virus identified in eastern India. Infect Genet Evol 16:419–425. doi: 10.1016/j.meegid.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 43.Odagiri T, Matsuzaki Y, Okamoto M, Suzuki A, Saito M, Tamaki R, Lupisan SP, Sombrero LT, Hongo S, Oshitani H. 2015. Isolation and characterization of influenza C viruses in the Philippines and Japan. J Clin Microbiol 53:847–858. doi: 10.1128/JCM.02628-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furukawa T, Muraki Y, Noda T, Takashita E, Sho R, Sugawara K, Matsuzaki Y, Shimotai Y, Hongo S. 2011. Role of the CM2 protein in the influenza C virus replication cycle. J Virol 85:1322–1329. doi: 10.1128/JVI.01367-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paragas J, Talon J, O'Neill RE, Anderson DK, García-Sastre A, Palese P. 2001. Influenza B and C virus NEP (NS2) proteins possess nuclear export activities. J Virol 75:7375–7383. doi: 10.1128/JVI.75.16.7375-7383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]