Summary
In a combined analysis of six published GWASs of 12160 lung cancer cases and 16838 controls, we found that a novel genetic variant in GTF2H4 was associated with lung cancer risk, providing a new clue for prevention in lung cancer.
Abstract
DNA repair pathways maintain genomic integrity and stability, and dysfunction of DNA repair leads to cancer. We hypothesize that functional genetic variants in DNA repair genes are associated with risk of lung cancer. We performed a large-scale meta-analysis of 123,371 single nucleotide polymorphisms (SNPs) in 169 DNA repair genes obtained from six previously published genome-wide association studies (GWASs) of 12160 lung cancer cases and 16838 controls. We calculated odds ratios (ORs) with 95% confidence intervals (CIs) using the logistic regression model and used the false discovery rate (FDR) method for correction of multiple testing. As a result, 14 SNPs had a significant odds ratio (OR) for lung cancer risk with P FDR < 0.05, of which rs3115672 in MSH5 (OR = 1.20, 95% CI = 1.14–1.27) and rs114596632 in GTF2H4 (OR = 1.19, 95% CI = 1.12–1.25) at 6q21.33 were the most statistically significant (P combined = 3.99×10−11 and P combined = 5.40×10−10, respectively). The MSH5 rs3115672, but not GTF2H4 rs114596632, was strongly correlated with MSH5 rs3131379 in that region (r 2 = 1.000 and r 2 = 0.539, respectively) as reported in a previous GWAS. Importantly, however, the GTF2H4 rs114596632 T, but not MSH5 rs3115672 T, allele was significantly associated with both decreased DNA repair capacity phenotype and decreased mRNA expression levels. These provided evidence that functional genetic variants of GTF2H4 confer susceptibility to lung cancer.
Introduction
Lung cancer remains a major cause of cancer morbidity and mortality worldwide (1). Although most lung cancer is attributed to tobacco smoking, accumulative evidence suggests that inherited genetic factors also play a pivotal role in lung cancer development (2,3). Notably, individuals with a family history of lung cancer have an increased risk, compared with those without a family history (4). Previous genome-wide association studies (GWASs) in populations of European descent have consistently identified common single nucleotide polymorphisms (SNPs) that confer risk of lung cancer at three independent loci at 5p15.33, 15q25.1 and 6p21.33 (5–8). A more recent GWAS analysis conducted by the Transdisciplinary Research In Cancer of the Lung (TRICL) consortium identified two additional rare variants in BRCA2 and CHEK2 with large effects on susceptibility to lung squamous cell carcinomas (9). These findings have advanced our knowledge of the genetic basis of lung cancer and provided new evidence of the involvement of additional biological pathways and relevance of DNA repair in the etiology of lung cancer. However, these reported loci only account for a fraction of the familial relative risk of lung cancer in Europeans, suggesting that the majority of missing heritability remain to be determined (10).
Because the reported loci by GWAS require a stringent genome-wide significance threshold and they are likely to reflect only the tip of the iceberg in the genetic etiology of lung cancer, many complementary approaches have been applied to the post-GWAS analysis for identifying the missing heritability of the disease, such as pathway-based association analysis (11–13). The pathway-based hypothesis tests for associations of genetic variants in biological pathway genes with the disease risk, in which the genes are involved in complex molecular networks, cellular pathways and cross-talks. Investigating SNPs in a biological pathway rather than individual genes may provide a better chance to identify the genes and mechanisms underlying disease pathogenesis (11).
Many of the biological pathways identified to date have been proposed as important candidate pathways for lung carcinogenesis, including DNA repair pathways, a critical defense mechanism against human carcinogenesis (14). DNA damage is caused by both endogenous oxygen free radicals from metabolic processes and exogenous (both chemical and physical) mutagens. The DNA repair process is an important mechanism to maintain genomic stability and integrity, and unrepaired or incorrectly repaired DNA may result in mutation fixation, thus leading to cancer development (15). In humans, a total of 15 DNA repair pathways have been characterized according to their unique repair processes, such as base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), double-strand break repair (DSBR), direct reversion repair (DRR), and DNA polymerases (15,16).
Although a few previous studies, including ours, have investigated associations between SNPs in DNA repair pathway genes and lung cancer risk, only a limited number of SNPs and candidate genes in the pathways with some small effects on cancer risk have been reported (17–23), due to the limited sample sizes and number of SNPs queried in these prior studies. Here, we examined the associations between genetic variants in 169 DNA repair genes and lung cancer risk comprehensively by using a large-scale meta-analysis, including 12160 cases and 16838 controls derived from six previously published lung cancer GWASs.
Methods
Study populations
We conducted a pooled analysis of datasets from six previously published GWASs of lung cancer in 12160 lung cancer cases and 16838 controls of European ancestry. The present study was part of the TRICL consortium established in 2008 and associated with the International Lung Cancer Consortium (ILCCO). The six studies included in this analysis were: the Institute of Cancer Research (ICR) GWAS; the MD Anderson Cancer Center (MDACC) GWAS; the International Agency for Research on Cancer (IARC) GWAS; the National Cancer Institute (NCI) GWAS; the Samuel Lunenfeld Research Institute study (SLRI) GWAS; and the Germany Lung Cancer study (GLC), which have been published previously (9,24). The detailed recruitments of cases and controls recruitments and their characteristics are summarized in Supplementary Table 1, available at Carcinogenesis Online. Lung cancer diagnosis in these studies was primarily pathologically confirmed with a small proportion of patients diagnosed by clinical history and imaging. Written informed consent was obtained from each participant, and this study was approved by the institutional review boards for each of the participating institutions.
Selection of genes and SNPs from the DNA repair pathways
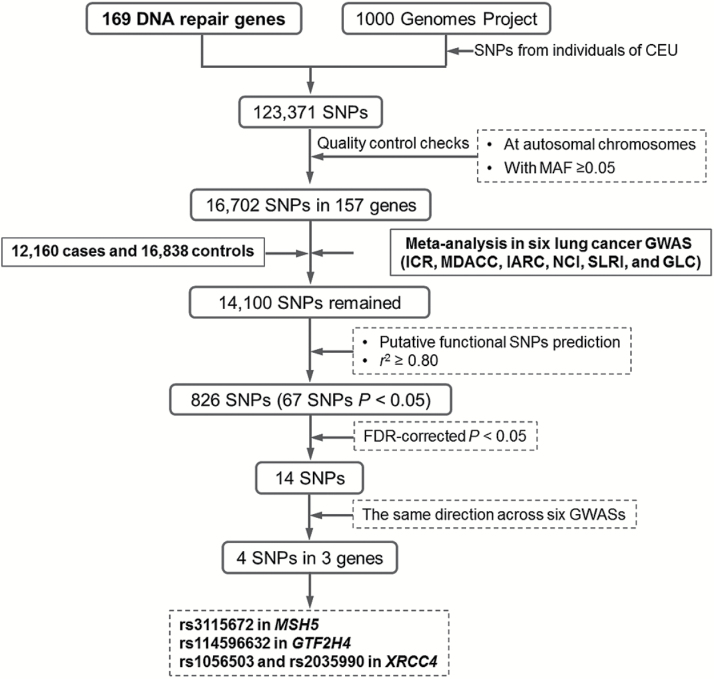
The selection process of the genes and SNPs is shown in Figure 1. We selected 169 candidate genes from 15 main DNA repair pathways from the publically available database MSigDB (Supplementary Table 2 is available at Carcinogenesis Online), which has compiled gene sets from a variety of resources, such as Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and others (25). Additional genes involved in DNA repair were obtained from previously published literature (15,16,26). A total of 123,371 SNPs within these selected genes from 2kb upstream to 2kb downstream were extracted based on CEU data from 1000 Genomes Project (March 2012). Quality control for SNPs fit the following two criteria: (i) SNPs located on autosomal chromosomes; (ii) minor allelic frequency (MAF) ≥ 5% in the CEU populations. As a result, 16702 SNPs in 157 genes were included after quality control processing. Some additional SNPs were excluded due to low quality of imputation in the individual GWASs, leaving a set of 14100 SNPs from the pooled lung cancer GWASs.
Figure 1.
Schematic flow for selecting putative functional genetic variants in DNA repair genes.
Putative functional SNPs were predicted by a web-based tool, SNPinfo, which has incorporated functional predictions of protein structure, gene regulation, splicing and microRNA (miRNA) binding, with consideration of whether the alternative alleles of a SNP were likely to have differential effects on gene function (27). Most functional SNPs could be further validated by an independent software package FunciSNP (28). Only one SNP was selected when multiple SNPs showed strong pair-wise linkage disequilibrium (LD) r 2 ≥ 0.8. After this filtering, a total of 826 putative functional SNPs in 12160 cases and 16838 were retained in the final analysis.
DNA repair capacity analysis
The analysis of DNA repair capacity (DRC) was performed using the host-cell reactivation assay for 869 control subjects from the MDACC study, as described previously (29). The host-cell reactivation assay measures the activity of the CAT gene, a bacterial drug resistance gene, in cells that have been transfected with BPDE-treated plasmids. Before the plasmid transfection, cultured T lymphocytes were isolated from whole peripheral blood samples stimulated by phytohemagglutinin. The activity of the repaired CAT gene was quantified using a scintillation counter to determine the formation of [3H]monoacetylated and [3H]diacetylated chloramphenicols by adding the chloramphenicol and [3H]acetyl-CoA in the cell extracts. DRC was defined as the ratio of the CAT activity of cells transfected with BPDE-treated plasmids (treated) to that of cells transfected with untreated plasmids (untreated): DRC = treated/untreated × 100% (29–31).
Expression analysis
Expression quantitative trait locus (eQTL) analysis was performed to determine the correlations between genotypes of the identified SNPs and expression levels of the nearby genes using several publically available datasets. The data used for eQTLs were obtained from lymphoblostoid cell lines derived from 270 individuals from four ethnic populations (CEU: 90 Utah residents from northern and western Europe; CHB: 45 unrelated Han Chinese in Beijing; JPT: 45 unrelated Japanese in Tokyo; YRI: 90 Yoruba in Ibadan, Nigeria), and the DNA samples from these cell lines were also used for genotyping (32).
We further performed an RNA expression analysis by using lung cancer data from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/) (RNASeqV2.Level_3.1.8.0). In the TCGA database, 489 subjects had lung adenocarcinoma and 489 had lung squamous cell carcinoma, of which 57 and 50 had matching adjacent normal samples, respectively. Differential gene expression was measured only in paired tumor and normal tissues. All individuals included in the TCGA data analysis were of European descents.
Statistical analysis
In each of six lung cancer GWASs, SNP genotyping assays were completed using Illumina HumanHap 300 BeadChips, HumanHap 550 or 610 Quad arrays. We imputed unmeasured genotypes using data from the 1000 Genomes Project (phase I integrated release 3, March 2012) as the reference using IMPUTE2, MACH or minimac software as previously reported (9). The SLRI and GLC studies followed the same protocol for imputation as has previously been followed by MDACC (9). A series of quality control steps were performed before meta-analysis of the results from imputation for each study. Specifically, only imputed SNPs with an information measure ≥0.40 with IMPUTE2 or an RSQR ≥ 0.30 with MACH were included for further analysis. The association between each SNP and lung cancer risk was estimated using an additive genetic model in the logistic regression. The pooled odds ratio (OR) and 95% confidence interval (CI) were calculated by the Mantel-Haenszel procedure assumed a fixed-effects model. A random-effects model was used if there was significant heterogeneity (P < 0.05). The between-study heterogeneity was calculated based on Cochran’s Q statistics and I 2. The false discovery rate (FDR) method was used to correct for multiple comparisons. Recombination rates (cM/Mb) across the 6p21.33 region were estimated from HapMap (Build 35 coordinates). Haploview 4.2 was applied to infer the LD structure of the genome derived from 1000 Genomes Project CEU individuals. Human leukocyte antigen (HLA) genotypes of HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1 genes were sequenced by the 1000 Genomes Project (33) and the results were used to evaluate LD between SNPs and HLA alleles. The relationship between the SNPs and corresponding gene expression was examined using a linear regression model. We used Gene Relationships Across Implicated Loci (GRAIL) to identify subsets of highly related genes in DNA pathways from 37 genes associated with lung cancer risk (34). PLINK1.07 was used for primary statistical analysis of the GWAS datasets.
Results
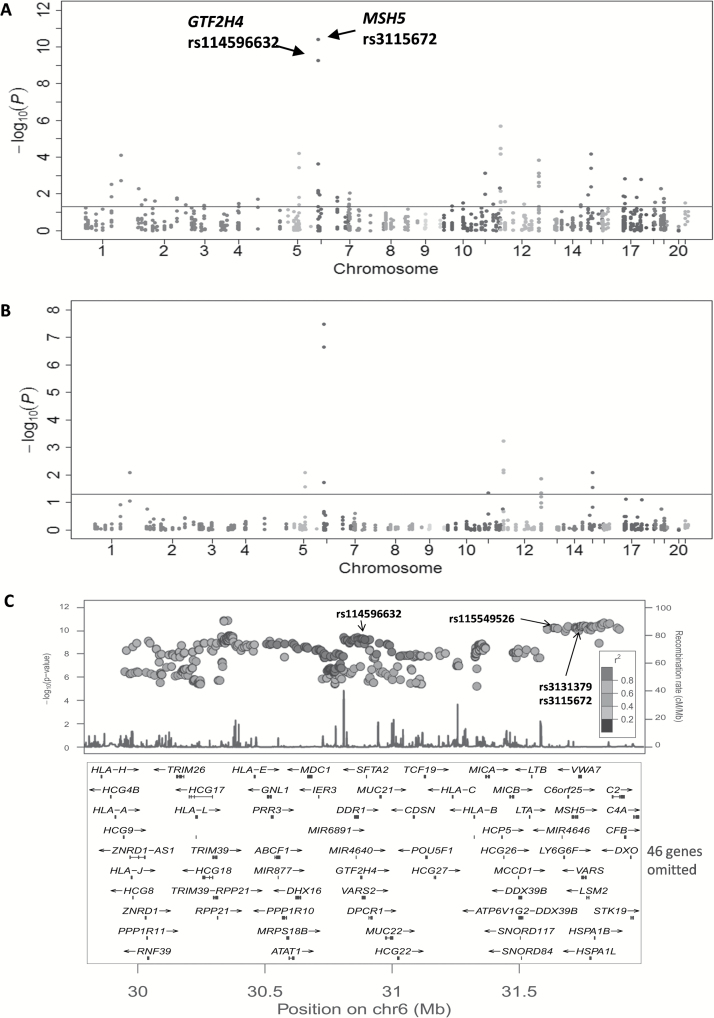
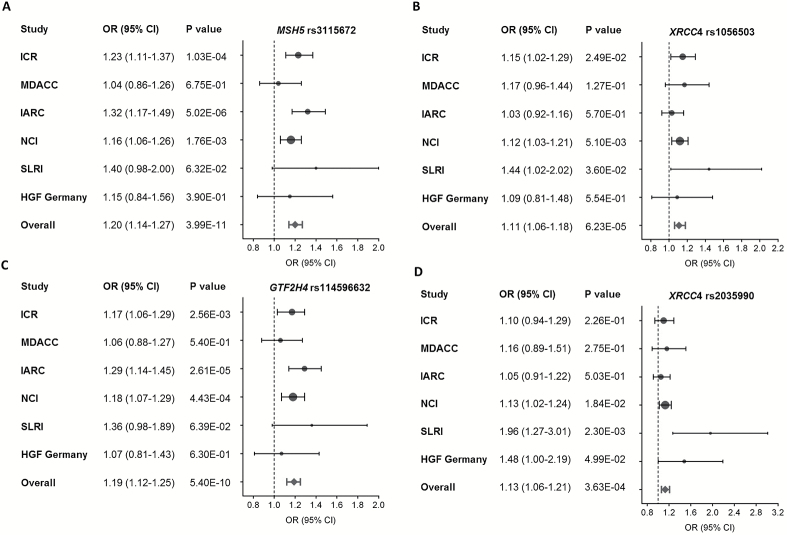
As shown in the Manhattan plot of Figure 2 derived from the additive genetic model of the meta-analysis of the 826 SNPs for 12160 cases and 16838 controls, suggestive evidence for an association was found for many regions harboring DNA repair genes throughout the genome. Across all the genetic variants, multiple signals were nominally associated with lung cancer risk at P < 1.0×10−7 on chromosome 6. Overall, 67 SNPs achieved a significant association of P < 0.05 (Supplementary Table 3 is available at Carcinogenesis Online). However, only 14 SNPs remained statistically significant after the FDR correction (P FDR < 0.05) (Table 1). Notably, four SNPs in three genes had the same direction of effects across all six lung cancer GWASs (rs3115672 in MSH5, rs114596632 in GTF2H4, and rs1056503 and rs2035990 in XRCC4) (Table 2 and Figure 3). There was no evidence of between-study heterogeneity for these four SNPs.
Figure 2.
Association results of SNPs in DNA repair genes and lung cancer risk. (A) Manhattan plot of association results of 826 putative functional SNPs (r 2 < 0.8 with each SNP) in DNA repair genes. Scatter plot of P values in the −log10 scale includes original P values and (B) FDR-corrected P values obtained from the meta-analysis of six lung cancer GWASs. Horizontal line represents the threshold of P FDR = 0.05. (C) Region association of all SNPs (functional and non-functional) in 6p21.33, which showed a moderate LD between GTF2H4 SNP rs114596632 and MSH5 rs3115672, as well as two other reported SNPs (rs3131379 in MSH5 and rs115549526 in BAG6).
Table 1.
Associations between 14 SNPs and lung cancer risk with P < 0.05 after FDR-correction
| SNP | Chr | Positiona | Gene | Alleleb | EAFc | OR (95% CI)d | P d | P FDR e | Effectsf | P hete g | I 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3115672 | 6 | 31727897 | MSH5 | C/T | 0.106 | 1.20 (1.14–1.27) | 3.99E-11 | 3.30E-08 | + + + + + + | 0.266 | 22.30 |
| rs114596632 | 6 | 30879987 | GTF2H4 | C/T | 0.114 | 1.19 (1.12–1.25) | 5.40E-10 | 2.23E-07 | + + + + + + | 0.462 | 0.00 |
| rs3748522 | 12 | 1058688 | RAD52 | C/A | 0.481 | 0.92 (0.89–0.95) | 2.11E-06 | 5.81E-04 | − − − − + + | 0.108 | 44.61 |
| rs11571475 | 12 | 1022352 | RAD52 | A/G | 0.134 | 0.90 (0.85–0.95) | 3.32E-05 | 6.86E-03 | − − − − + − | 0.641 | 0.00 |
| rs1056503 | 5 | 82648977 | XRCC4 | T/G | 0.120 | 1.11 (1.06–1.18) | 6.23E-05 | 8.02E-03 | + + + + + + | 0.515 | 0.00 |
| rs506120 | 15 | 43802024 | TP53BP1 | C/T | 0.279 | 0.92 (0.89–0.96) | 6.53E-05 | 8.02E-03 | − − − − + − | 0.420 | 0.00 |
| rs11571376 | 12 | 1059556 | RAD52 | C/G | 0.290 | 0.92 (0.89–0.96) | 6.96E-05 | 8.02E-03 | − + − − − − | 0.113 | 43.82 |
| rs12563994 | 1 | 155244092 | CLK2 | C/T | 0.243 | 1.09 (1.04–1.13) | 7.77E-05 | 8.02E-03 | + − + + − + | 0.200 | 31.39 |
| rs7334543 | 13 | 32973276 | BRCA2 | A/G | 0.256 | 0.93 (0.89–0.96) | 1.49E-04 | 1.37E-02 | − − − − + − | 0.208 | 30.32 |
| rs707937 | 6 | 31731014 | MSH5 | C/G | 0.180 | 0.91 (0.87–0.96) | 2.30E-04 | 1.90E-02 | − − + − − − | 0.234 | 26.79 |
| rs2035990 | 5 | 82649467 | XRCC4 | T/C | 0.069 | 1.13 (1.06–1.21) | 3.63E-04 | 2.73E-02 | + + + + + + | 0.106 | 44.97 |
| rs28628574 | 15 | 43802038 | TP53BP1 | A/C | 0.103 | 0.90 (0.85–0.95) | 4.05E-04 | 2.79E-02 | − − + − − − | 0.960 | 0.00 |
| rs9534160 | 13 | 32888021 | BRCA2 | G/A | 0.049 | 1.15 (1.06–1.25) | 7.65E-04 | 4.56E-02 | + + + + + − | 0.433 | 0.00 |
| rs4246215 | 11 | 61564299 | FEN1 | G/T | 0.358 | 0.94 (0.91–0.97) | 7.73E-04 | 4.56E-02 | − − − − − + | 0.120 | 42.76 |
SNP, single nucleotide polymorphisms; FDR, false discovery rate; Chr, chromosome.
aBased on NCBI build 37 of the human genome.
bReference allele/effect allele.
cEffect allele frequency.
dMeta-analysis additive model P-value based on six lung cancer GWASs.
eFalse discovery rate (FDR) correction.
fEffects by study: ICR, MDACC, IARC, NCI, SLRI and GLC, respectively. + represents OR > 1.00, and − represents OR < 1.00.
g P value for heterogeneity.
Table 2.
Association results of four SNPs that had consistent effects in the six lung cancer GWASs
| Study population | Sample size | rs3115672 (T) | rs114596632 (T) | rs1056503 (G) | rs2035990 (C) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | EAFa | OR (95% CI) | P additive | EAFa | OR (95% CI) | P additive | EAFa | OR (95% CI) | P additive | EAFa | OR (95% CI) | P additive | |
| ICR | 1952 | 5200 | 0.139 | 1.23 (1.11−1.37) | 1.03E-04 | 0.158 | 1.17 (1.06−1.29) | 2.56E-03 | 0.105 | 1.15 (1.02−1.29) | 2.49E-02 | 0.058 | 1.10 (0.94−1.29) | 2.26E-01 |
| MDACC | 1150 | 1134 | 0.114 | 1.04 (0.86−1.26) | 6.75E-01 | 0.124 | 1.06 (0.88−1.27) | 5.40E-01 | 0.111 | 1.17 (0.96−1.44) | 1.27E-01 | 0.062 | 1.16 (0.89−1.51) | 2.75E-01 |
| IARC | 2533 | 3791 | 0.093 | 1.32 (1.17−1.49) | 5.02E-06 | 0.097 | 1.29 (1.14−1.45) | 2.61E-05 | 0.117 | 1.03 (0.92−1.16) | 5.70E-01 | 0.070 | 1.05 (0.91−1.22) | 5.03E-01 |
| NCI | 5713 | 5736 | 0.094 | 1.16 (1.06−1.26) | 1.76E-03 | 0.094 | 1.18 (1.07−1.29) | 4.43E-04 | 0.132 | 1.12 (1.03−1.21) | 5.10E-03 | 0.077 | 1.13 (1.02−1.24) | 1.84E-02 |
| SLRI | 331 | 499 | 0.098 | 1.40 (0.98−2.00) | 6.32E-02 | 0.112 | 1.36 (0.98−1.89) | 6.39E-02 | 0.120 | 1.44 (1.02−2.02) | 3.60E-02 | 0.070 | 1.96 (1.27−3.01) | 2.30E-03 |
| GLC | 481 | 478 | 0.096 | 1.15 (0.84−1.56) | 3.90E-01 | 0.110 | 1.07 (0.81−1.43) | 6.30E-01 | 0.122 | 1.09 (0.81−1.48) | 5.54E-01 | 0.070 | 1.48 (1.00−2.19) | 4.99E-02 |
| All combinedb | 12160 | 16838 | 0.106 | 1.20 (1.14−1.27) | 3.99E-11 | 0.114 | 1.19 (1.12−1.25) | 5.40E-10 | 0.120 | 1.11 (1.06−1.18) | 6.23E-05 | 0.069 | 1.13 (1.06−1.21) | 3.63E-04 |
aEffect allele frequency.
bThe combined OR and P value were estimated using a fixed-effects model.
Figure 3.
Forest plot of the association between four SNPs and lung cancer risk. (A) rs3115672 in MSH5, (B) rs1056503 in XRCC4, (C) rs114596632 in GTF2H4 and (D) rs2035990 in XRCC4. The circles and horizontal lines show additive OR and 95% CI for each study. The diamond represents the summary OR and 95% CI.
SNP rs3115672 is located in intron 3 of MSH5 at 6q21.33, near the SNP rs3131379, which has been reported in a previous GWAS (5). We also observed strong support for an association between rs3131379 and lung cancer risk across studies (P = 5.36×10−11) (Figure 2C). In the 1000 Genomes Project CEU individuals, rs3115672 is in perfect LD with rs3131379 (r 2 = 1.00), suggesting that they reflect the same signal in this region (Supplementary Table 4 is available at Carcinogenesis Online).
Additionally, one SNP, rs114596632 within in an intron of GTF2H4, reached the genome-wide significance (P = 5.40×10−10 and P FDR = 2.23×10−7). Although rs114596632 is located in the same 6q21.33 region as the reported MSH5 SNP rs3131379 and its completed LD SNP rs115549526 in BAG6, it is located over 700-kb upstream from them. A LD analysis revealed that rs114596632 is only moderately correlated with rs3131379 (r 2 = 0.539) (Figure 2C and Supplementary Table 4 is available at Carcinogenesis Online). The recombination rates across the region including rs114596632 and rs3131379 showed that rs114596632 is an independent susceptibility locus (Supplementary Figure 1 is available at Carcinogenesis Online). As the significant SNP rs114596632 appeared to be located at 6p21.3, in the HLA region, we further performed a LD analysis for the association between rs114596632 and HLA alleles that have been analyzed as a part of the 1000 Genomes Project CEU individuals. We found that rs114596632 is in moderate LD with HLA-B*0801 (r 2 = 0.592) and HLA-C*0701 (r 2 = 0.675), suggesting that some HLA alleles may be partially tagged by this newly identified SNP rs114596632 (Supplementary Table 5 is available at Carcinogenesis Online). These HLA alleles form the ‘8.1 haplotype’, which extends for 4.7 million base pairs and is among the longest known haplotypes in humans (35). However, the HLA-B*0801 and HLA-C*0701 had only weak LD with the reported SNP rs3131379 in MSH5 (data not shown).
We also evaluated mRNA expressions levels of GTF2H4 in lung cancer cases from publicly available TCGA datasets. As shown in Supplementary Figure 2, available at Carcinogenesis Online, the expression levels of GTF2H4 was significantly higher in tumor tissues than in adjacent normal tissues among lung adenocarcinoma (P = 9.27×10−13) and lung squamous cell carcinoma (P = 4.64×10−14) from the TCGA database.
In addition to the two most significant SNPs at 6p, two other SNPs in XRCC4 also showed a suggestive association with lung cancer risk (P = 6.23×10−5 for rs1056503, and P = 3.63×10−4 for rs2035990) (Table 1). SNPs rs1056503 and rs2035990 were moderately correlated with each other (r 2 = 0.390). We previously reported that another SNP rs2075685 in XRCC4 was associated with lung cancer risk (19), but rs1056503 and rs2035990 were not correlated with that SNP (r 2 = 0).
We also had the data on the DNA (NER) repair capacity (DRC) assay with cultured peripheral blood T-lymphocytes from 869 controls (29) whose DNA were used for the GWAS analysis, with which we further analyzed the genotype–phenotype correlation in the present study. As shown in Table 3, there was a significant association between the genotypes of the GTF2H4 (a NER gene) rs114596632 and the DRC phenotype in these 869 control subjects, with the T allele carriers having a lower DRC than the C allele carriers (P = 0.032). Consistently, the T allele was also correlated with decreased mRNA expression of GTF2H4 in the 270 lymphoblastoid cell lines from HapMap. In contrast, the SNP rs3115672 in MSH5 were not significantly associated with both this DRC phenotype, nor with the mRNA expression of MSH5 (Table 3).
Table 3.
Association of two SNPs in GTF2H4 and MSH5 with DNA repair capacity (DRC) among 869 controls with mRNA expression levels of corresponding gene in 270 lymphoblastoid cell lines from HapMap
| rs114596632 in GTF2H4 | P a | P adj b | rs3115672 in MSH5 | P a | P adj b | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | CC | CT | TT | CC | CT | TT | ||||
| DRC | ||||||||||
| N (%) | 676 (77.8) | 181 (20.8) | 12 (1.4) | 688 (79.2) | 172 (19.8) | 9 (1.0) | ||||
| Mean ± SD | 9.16±3.33 | 8.67±2.85 | 7.80±2.30 | 0.027 | 0.032 | 9.07±3.28 | 8.97±3.09 | 7.40±1.88 | 0.34 | 0.277 |
| mRNA expression | ||||||||||
| N (%) | 230 (87.1) | 33 (12.5) | 1 (0.4) | 102 (38.9) | 116 (44.3) | 44 (16.8) | ||||
| Mean ± SD | 8.12±0.24 | 8.01±0.22 | 7.98 | 0.017 | 0.031 | 6.96±0.17 | 6.92±0.19 | 6.92±0.22 | 0.148 | 0.417 |
a P value was estimated from a generalized linear model.
bAdjustment for age, sex, and pack-years of smoking in the DRC correlation analysis; adjustment for populations (CEU, YRI, Asian) in the expression correlation analysis.
Significant correlation was also observed between the XRCC4 SNP rs1056503 and the gene expression levels, with higher expression in individuals with the TT genotype [P = 1.65×10−8 in all populations (n = 270), and P = 0.006 in the CEU population (n = 90)] (Supplementary Figure 3 is available at Carcinogenesis Online).
We also used the GRAIL method for the literature-based pathway analysis to explore the connections between 37 significant DNA repair genes. Overall, 16 regions had significant GRAIL P < 1.35×10−3 (0.05/37) (Supplementary Table 6 is available at Carcinogenesis Online). Pairwise associations for genes in the identified region are presented in Supplementary Figure 4, available at Carcinogenesis Online, showing that there were multiple strong connections identified in the literature between these DNA repair genes. Notably, the associated GTF2H4 gene has a strong literature-based connection with ERCC2 and MMS19, the previously known NER genes. Most of the keywords describing the functional connections in the pathway analysis were ‘repair’, ‘damage’ and ‘excision’.
Discussion
In this largest lung cancer GWAS meta-analysis among 28998 Europeans, we identified a novel genome-wide significant susceptibility variant rs114596632 in the DNA repair gene GTF2H4 at 6p21.33. In addition, we confirmed two previously reported lung cancer-associated SNPs in DNA repair genes MSH5 and XRCC4 in the combined analysis. Interestingly, The GTF2H4 SNP rs114596632 was found to have an effect on the DRC phenotype in removing BPDE-DNA adducts in cultured cells and the mRNA expression levels of GTF2H4 in the established cell lines. Further GRAIL pathway analysis revealed that GTF2H4 had a strong connection with multiple NER genes. Our findings highlight the significant role of DNA repair genes in the development of lung cancer.
DNA repair is a complicated biological process, consisting of several distinct but often connected pathways, that plays a fundamental role in maintaining genomic stability and integrity. Defects in the complex DNA repair machinery can lead to point mutations as well as chromosomal aberrations, which increase the risk of cancer (36). Several types of cancer, including lung cancer, are characterized by defective DNA repair, indicating the critical role of DNA repair in the pathogenesis and development of lung cancer (37,38).
Previous association studies in candidate genes have explored associations between DNA repair gene SNPs and lung cancer susceptibility, but the results were inconsistent (39,40). Recently, the pathway-based analysis on previously published GWAS data revealed distinct genes and pathways associated with lung cancer risk (41,42). However, their findings were based on only one GWAS in an Asian population, which need to be validated and extended in other populations to evaluate the robustness of the findings. In our current DNA repair pathway analysis that used a large sample size aggregated across six published GWASs, we found that rs114596632 in GTF2H4 was associated with lung cancer risk. We have an adequate statistical power (>80%) to detect the observed association of rs114596632. Also, the effect of rs114596632 on lung cancer risk had the same direction across all of the six GWASs, suggesting that the observed association was consistent and reliable.
GTF2H4, a general transcription factor IIH (TFIIH) subunit 4, is involved in both the NER process of DNA repair and transcription control interacting with variable factors important in carcinogenesis (43). The well-known NER pathway, consisting of at least 25 major genes, that mainly repairs bulky DNA lesions such as pyrimidine dimers and chemical adducts (44). The NER-specific factors can be released from the core TFIIH, thereby promoting the excision of the damaged oligonucleotide (45). Prior studies of genetic variants in GTF2H4 have observed that they are associated with risk of multiple sclerosis and cervical cancer but not aspirin-exacerbated respiratory disease (46–48). The eQTL analysis indicated that rs114596632 T allele was significantly associated with decreased mRNA expression levels of GTF2H4 in normal lymphoblastoid cell lines. Furthermore, bioinformatics prediction revealed that rs114596632 in GTF2H4 might be located in an enrichment of predicted motif ZEB1, an E-box transcriptional repressor known to induce epithelial to mesenchymal transition in lung cancer (49). It is known that reduced protein function due to SNPs in DNA repair genes can result in reduced DRC for carcinogenic adducts and oxidative lesions (29). Notably, we found that the cancer-free individuals with the rs114596632 T risk allele had a diminished DRC, who may be at risk of tobacco-induced lung cancer. With these consistent genotype-phenotype correlations, we proposed that the rs114596632 T allele might disturb the binding efficiency of the motif ZEB1, thereby decreasing the expression of GTF2H4 and cellular DRC and thus contributing to the risk of lung cancer. However, because the exact molecular mechanism underlying the association of rs114596632 in GTF2H4 with lung cancer risk has not been fully understood, further studies including fine-mapping, next generation sequencing and detailed functional analyses are warranted.
A number of studies have demonstrated associations between the HLA region and many types of cancer, including lung cancer (11,50–52). The underlying mechanism for these associations is still unknown; however, our HLA association analysis revealed that rs114596632 was only partially tagged by HLA-B*0801 and HLA-C*0701, suggesting a possible joint effect of the HLA immune system and DNA repair in the pathogenesis and risk of lung cancer (53). Although further sequencing in large lung cancer cases and control subjects is needed and currently under way to determine association between the HLA region and lung cancer risk, additional mechanistic studies of the role played by GTF2H4 in lung carcinogenesis are warranted.
To date, many studies have shown a consistent association between MSH5 and risk of lung cancer (5,24,54). MSH5 represents a strong candidate for lung cancer susceptibility, as it is involved in MMR and meiotic recombination (55). Interestingly, another unreported MSH5 SNP rs3115672 was also found to be associated with risk of lung cancer in the present study, which confirmed the important role of MSH5 in the etiology of lung cancer. However, this newly identified rs3115672 and previously reported rs3131379 are in complete LD in MSH5 (r 2 = 1.00), suggesting a completely consistent susceptibility locus in this region. Interestingly, the SNP rs114596632 in GTF2H4 is located 828kb upstream of MSH5, but the LD and recombination hotspot analyses revealed that rs114596632 was in moderate LD with rs3131379 in MSH5. Further phenotype correlation analysis showed that rs114596632 in GTF2H4, but not rs3131379 in MSH5, was associated with the DRC phenotype and mRNA expression, possibly revealing that GTF2H4 had an independent role of DNA repair capacity that modulates risk of lung cancer.
In the present study, two SNPs in XRCC4 were found to be associated with lung cancer risk. In the non-homologous end joining (NHEJ) pathway, XRCC4 forms a complex with LIG4, stabilizing and stimulating LIG4 activity (56). Deficiency of XRCC4 in mice results in embryonic lethality associated with severe dysfunction of apoptosis of the newborns (57). Two published studies and ours have reported that SNPs in XRCC4 were involved in the susceptibility of lung cancer; importantly, our previous study identified a functional SNP rs2075685 in the XRCC4 promoter, whose variant allele was associated with an increased XRCC4 expression (19). Our two newly identified XRCC4 SNPs rs1056503 and rs2035990 have only a weak LD with rs2075685, suggesting that these XRCC4 SNPs may be independent casual SNPs. MirSNP prediction revealed that rs2035990 located in the 3′-UTR of XRCC4, a microRNA-567 binding region, might result in gene dysregulation (58). Meanwhile, although rs1056503 in XRCC4 is a synonymous SNP, the G allele was associated with both lung cancer risk and lower mRNA expression levels in the present study. Therefore, further studies are needed to investigate biological mechanisms underlying the observed associations between SNPs in XRCC4 and lung cancer risk.
In conclusion, our large meta-analysis of published GWASs among 28998 Europeans identified a new lung cancer susceptibility locus in GTF2H4 and also provide some evidence supporting two previously reported loci in MSH5 and XRCC4. Given our findings of a novel GTF2H4 variant are biologically plausible, our results provide some new insight into genetic architecture and carcinogenesis mechanisms of lung cancer.
Supplementary material
Supplementary Tables 1–6 and Figures 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
As Duke Cancer Institute members, Q.W. acknowledge support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (NIH CA014236). Q.W. was also supported by a start-up fund from Duke Cancer Institute, Duke University Medical Center. TRICL: This work was supported by P30 CA023108 (CA) and the Transdisciplinary Research in Cancer of the Lung (TRICL) Study, U19-CA148127 on behalf of the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network. The SLRI study was supported by Canadian Cancer Society Research Institute (020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award to RH The ICR study was supported by Cancer Research UK (C1298/A8780 andC1298/A8362—Bobby Moore Fund for Cancer Research UK) and NCRN, HEAL and Sanofi-Aventis. Additional funding was obtained from NIH grants (5R01CA055769, 5R01CA127219, 5R01CA133996, and 5R01CA121197). The Liverpool Lung Project (LLP) was supported by The Roy Castle Lung Cancer Foundation, UK. The ICR and LLP studies made use of genotyping data from the Wellcome Trust Case Control Consortium 2 (WTCCC2); a full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Sample collection for the Heidelberg lung cancer study was in part supported by a grant (70–2919) from the Deutsche Krebshilfe. The work was additionally supported by a Helmholtz-DAAD fellowship (A/07/97379 to MNT). The KORA Surveys were financed by the GSF, which is funded by the German Federal Ministry of Education, Science, Research and Technology and the State of Bavaria. The Lung Cancer in the Young study (LUCY) was funded in part by the National Genome Research Network (NGFN), the DFG (BI576/2-1; BI 576/2-2), the Helmholtzgemeinschaft (HGF) and the Federal office for Radiation Protection (BfS: STSch4454). Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum Muenchen. Support for the Central Europe, HUNT2/Tromsø and CARET genome-wide studies was provided by Institut National du Cancer, France. Support for the HUNT2/Tromsø genome-wide study was also provided by the European Community (Integrated Project DNA repair, LSHG-CT- 2005–512113), the Norwegian Cancer Association and the Functional Genomics Programme of Research Council of Norway. Support for the Central Europe study, Czech Republic, was also provided by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101). Support for the CARET genome-wide study was also provided by grants from the US National Cancer Institute, NIH (R01 CA111703 and UO1 CA63673), and by funds from the Fred Hutchinson Cancer Research Center. Additional funding for study coordination, genotyping of replication studies and statistical analysis was provided by the US National Cancer Institute (R01 CA092039). The lung cancer GWAS from Estonia was partly supported by a FP7 grant (REGPOT245536), by the Estonian Government (SF0180142s08), by EU RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure’s Roadmap and by University of Tartu (SP1GVARENG). The work reported in this paper was partly undertaken during the tenure of a Postdoctoral Fellowship from the IARC (for MNT). The Environment and Genetics in Lung Cancer Etiology (EAGLE), the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), and the Prostate, Lung, Colon, Ovary Screening Trial (PLCO)studies and the genotyping of ATBC, the Cancer Prevention Study II Nutrition Cohort (CPS-II) and part of PLCO were supported by the Intramural Research Program of NIH, NCI, Division of Cancer Epidemiology and Genetics. ATBC was also supported by US Public Health Service contracts (N01-CN-45165, N01-RC-45035 and N01-RC-37004) from the NCI. PLCO was also supported by individual contracts from the NCI to the University of Colorado Denver (NO1-CN-25514), Georgetown University(NO1-CN-25522), Pacific Health Research Institute (NO1-CN-25515),Henry Ford Health System (NO1-CN-25512), University of Minnesota(NO1-CN-25513), Washington University(NO1-CN-25516), University of Pittsburgh (NO1-CN-25511), University of Utah (NO1-CN-25524), Marshfield Clinic Research Foundation (NO1-CN-25518), University of Alabama at Birmingham (NO1-CN-75022, Westat, Inc. NO1-CN-25476), University of California, Los Angeles (NO1-CN-25404). The Cancer Prevention Study II Nutrition Cohort was supported by the American Cancer Society. The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 andRO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C) and study coordination at the GENEVA Coordination Center (U01 HG004446)for EAGLE and part of PLCO studies. Funding for the MD Anderson Cancer Study was provided by NIH grants (P50 CA70907, R01CA121197, R01CA127219, U19 CA148127, R01 CA55769, K07CA160753) and CPRIT grant (RP100443). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is funded through a federal contract from the NIH to The Johns Hopkins University (HHSN268200782096C). The Harvard Lung Cancer Study was supported by the NIH (National Cancer Institute) grants CA092824, CA090578, and CA074386.
Supplementary Material
Acknowledgements
We thank the staff and participants of all TRICL studies for their important contributions. We are also grateful to the patients, clinicians and allied health care professions.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- DRC
DNA repair capacity
- FDR
false discovery rate
- GWAS
genome-wide association studies
- HLA
human leukocyte antigen
- LD
linkage disequilibrium
- NER
nucleotide excision repair
- OR
odds ratio
- SNP
single nucleotide polymorphism
- TRICL
Transdisciplinary Research In Cancer of the Lung
References
- 1. Siegel R., et al. (2014) Cancer statistics, 2014. CA. Cancer J. Clin., 64, 9–29. [DOI] [PubMed] [Google Scholar]
- 2. Coté M.L., et al. (2012) Increased risk of lung cancer in individuals with a family history of the disease: a pooled analysis from the International Lung Cancer Consortium. Eur. J. Cancer, 48, 1957–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rooney A. (2003) Family history reveals lung-cancer risk. Lancet. Oncol., 4, 267. [DOI] [PubMed] [Google Scholar]
- 4. Matakidou A., et al. (2005) Systematic review of the relationship between family history and lung cancer risk. Br. J. Cancer, 93, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y., et al. (2008) Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet., 40, 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKay J.D., et al. (2008) Lung cancer susceptibility locus at 5p15.33. Nat. Genet., 40, 1404–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hung R.J., et al. (2008) A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature, 452, 633–637. [DOI] [PubMed] [Google Scholar]
- 8. Amos C.I., et al. (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., et al. (2014) Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat. Genet., 46, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manolio T.A., et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang K., et al. (2010) Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet., 11, 843–854. [DOI] [PubMed] [Google Scholar]
- 12. Ramanan V.K., et al. (2012) Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet., 28, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong H., et al. (2010) Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet., 86, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoeijmakers J.H. (2001) Genome maintenance mechanisms for preventing cancer. Nature, 411, 366–374. [DOI] [PubMed] [Google Scholar]
- 15. Wood R.D., et al. (2001) Human DNA repair genes. Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- 16. Wood R.D., et al. (2005) Human DNA repair genes, 2005. Mutat. Res., 577, 275–283. [DOI] [PubMed] [Google Scholar]
- 17. Hung R.J., et al. (2008) International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol. Biomarkers Prev., 17, 3081–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kazma R., et al. (2012) Lung cancer and DNA repair genes: multilevel association analysis from the International Lung Cancer Consortium. Carcinogenesis, 33, 1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu H., et al. (2011) An analysis of single nucleotide polymorphisms of 125 DNA repair genes in the Texas genome-wide association study of lung cancer with a replication for the XRCC4 SNPs. DNA Repair (Amst)., 10, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qian B., et al. (2011) Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer, 73, 138–146. [DOI] [PubMed] [Google Scholar]
- 21. Doherty J.A., et al. (2013) DNA repair genotype and lung cancer risk in the beta-carotene and retinol efficacy trial. Int. J. Mol. Epidemiol. Genet., 4, 11–34. [PMC free article] [PubMed] [Google Scholar]
- 22. Sakoda L.C., et al. (2012) Germ line variation in nucleotide excision repair genes and lung cancer risk in smokers. Int. J. Mol. Epidemiol. Genet., 3, 1–17. [PMC free article] [PubMed] [Google Scholar]
- 23. Buch S.C., et al. (2012) Genetic variability in DNA repair and cell cycle control pathway genes and risk of smoking-related lung cancer. Mol. Carcinog., 51(suppl. 1), E11–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Timofeeva M.N., et al. (2012) Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet., 21, 4980–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian A., et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U. S. A., 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lange S.S., et al. (2011) DNA polymerases and cancer. Nat. Rev. Cancer, 11, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Z., et al. (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res., 37, W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coetzee S.G., et al. (2012) FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res., 40, e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang L.E., et al. (2013) Genome-wide association study reveals novel genetic determinants of DNA repair capacity in lung cancer. Cancer Res., 73, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei Q., et al. (2003) Repair of UV light-induced DNA damage and risk of cutaneous malignant melanoma. J. Natl. Cancer Inst., 95, 308–315. [DOI] [PubMed] [Google Scholar]
- 31. Wei Q., et al. (1993) DNA repair and aging in basal cell carcinoma: a molecular epidemiology study. Proc. Natl. Acad. Sci. U. S. A., 90, 1614–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. International HapMap Consortium. (2003) The International HapMap Project. Nature, 426, 789–796. [DOI] [PubMed] [Google Scholar]
- 33. Gourraud P.A., et al. (2014) HLA diversity in the 1000 genomes dataset. PLoS One, 9, e97282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raychaudhuri S., et al. (2009) Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet., 5, e1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Candore G., et al. (2002) Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun. Rev., 1, 29–35. [DOI] [PubMed] [Google Scholar]
- 36. Luijsterburg M.S., et al. (2011) Chromatin and the DNA damage response: the cancer connection. Mol. Oncol., 5, 349–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sevilya Z., et al. (2014) Low integrated DNA repair score and lung cancer risk. Cancer Prev. Res., 7, 398–406. [DOI] [PubMed] [Google Scholar]
- 38. Vogelstein B., et al. (2004) Cancer genes and the pathways they control. Nat. Med., 10, 789–799. [DOI] [PubMed] [Google Scholar]
- 39. Li W., et al. (2014) DNA repair pathway genes and lung cancer susceptibility: a meta-analysis. Gene, 538, 361–365. [DOI] [PubMed] [Google Scholar]
- 40. Ricceri F., et al. (2012) Is there evidence of involvement of DNA repair polymorphisms in human cancer? Mutat. Res., 736, 117–121. [DOI] [PubMed] [Google Scholar]
- 41. Lee D., et al. (2013) Pathway-based analysis using genome-wide association data from a Korean non-small cell lung cancer study. PLoS One, 8, e65396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chu M., et al. (2014) A genome-wide gene–gene interaction analysis identifies an epistatic gene pair for lung cancer susceptibility in Han Chinese. Carcinogenesis, 35, 572–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gervais V., et al. (2004) TFIIH contains a PH domain involved in DNA nucleotide excision repair. Nat. Struct. Mol. Biol., 11, 616–622. [DOI] [PubMed] [Google Scholar]
- 44. Leibeling D., et al. (2006) Nucleotide excision repair and cancer. J. Mol. Histol., 37, 225–238. [DOI] [PubMed] [Google Scholar]
- 45. Coin F., et al. (2008) Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell, 31, 9–20. [DOI] [PubMed] [Google Scholar]
- 46. Briggs F.B., et al. (2010) Variation within DNA repair pathway genes and risk of multiple sclerosis. Am. J. Epidemiol., 172, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castle P.E., et al. (2001) An association of cervical inflammation with high-grade cervical neoplasia in women infected with oncogenic human papillomavirus (HPV). Cancer Epidemiol. Biomarkers Prev., 10, 1021–1027. [PubMed] [Google Scholar]
- 48. Kim J.Y., et al. (2012) Lack of association between GTF2H4 genetic variants and AERD development and FEV1 decline by aspirin provocation. Int. J. Immunogenet., 39, 486–491. [DOI] [PubMed] [Google Scholar]
- 49. Gemmill R.M., et al. (2011) ZEB1-responsive genes in non-small cell lung cancer. Cancer Lett., 300, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Madeleine M.M., et al. (2008) Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res., 68, 3532–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu M.S., et al. (2002) Association of HLA-DQB1*0301 and HLA-DQB1*0602 with different subtypes of gastric cancer in Taiwan. Jpn. J. Cancer Res., 93, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hu J., et al. (2012) HLA-DRB1*1501 and HLA-DQB1*0301 alleles are positively associated with HPV16 infection-related Kazakh esophageal squamous cell carcinoma in Xinjiang China. Cancer Immunol. Immunother., 61, 2135–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramnath N., et al. (2006) Is downregulation of MHC class I antigen expression in human non-small cell lung cancer associated with prolonged survival? Cancer Immunol. Immunother., 55, 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Broderick P., et al. (2009) Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res., 69, 6633–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Y.C., et al. (2006) Molecular diagnostic markers for lung cancer in sputum and plasma. Ann. N. Y. Acad. Sci., 1075, 179–184. [DOI] [PubMed] [Google Scholar]
- 56. Critchlow S.E., et al. (1997) Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol., 7, 588–598. [DOI] [PubMed] [Google Scholar]
- 57. Gao Y., et al. (1998) A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell, 95, 891–902. [DOI] [PubMed] [Google Scholar]
- 58. Liu C., et al. (2012) MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics, 13, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.