Summary
miR-200b mitigates cell growth and cell cycle progression, and suppresses multiple key cell cycle regulators and Wnt/β-Catenin signaling in ESCC. CDK2 and PAF (a potent oncoprotein) are identified as miR-200b targets, highlighting miR-200b as a promising therapeutic target in ESCC.
Abstract
miR-200b is a pleiotropically acting microRNA in cancer progression, representing an attractive therapeutic target. We previously identified miR-200b as an invasiveness repressor in esophageal squamous cell carcinoma (ESCC), whereas further understanding is warranted to establish it as a therapeutic target. Here, we show that miR-200b mitigates ESCC cell growth by inducing G2-phase cell cycle arrest and apoptosis. The expression/activation of multiple key cell cycle regulators such as CDK1, CDK2, CDK4 and Cyclin B, and the Wnt/β-Catenin signaling are modulated by miR-200b. We identified CDK2 and PAF (PCNA-associated factor), two important tumor-promoting factors, as direct miR-200b targets in ESCC. Correlating with the frequent loss of miR-200b in ESCC, both CDK2 and PAF levels are significantly increased in ESCC tumors compared to case-matched normal tissues (n = 119, both P < 0.0001), and correlate with markedly reduced survival (P = 0.007 and P = 0.041, respectively). Furthermore, CDK2 and PAF are also associated with poor prognosis in certain subtypes of breast cancer (n = 1802) and gastric cancer (n = 233). Although CDK2 could not significantly mediate the biological function of miR-200b, PAF siRNA knockdown phenocopied while restored expression of PAF abrogated the biological effects of miR-200b on ESCC cells. Moreover, PAF was revealed to mediate the inhibitory effects of miR-200b on Wnt/β-Catenin signaling. Collectively, the pleiotropic effects of miR-200b in ESCC highlight its potential for therapeutic intervention in this aggressive disease.
Introduction
microRNAs (miRNAs) are a large class of evolutionarily conserved small non-coding RNAs that modulate gene expression at the post-transcriptional level. miRNAs recognize and bind to the 3′-untranslated region (3′UTR) on their target genes via partially complementary sequences, thereby inducing RNA-induced silencing complex, leading to either target mRNA decay or translation repression (1). Aberrant expression of miRNAs has been implicated in various hallmarks of cancer, including sustaining proliferative signaling, resisting cell death, inducing angiogenesis, evading immune destruction, and activating invasion and metastasis (2,3). Certain tumor types, such as pre-B-cell lymphoma, have been shown to display addiction to specific miRNAs that play a causal role in both cancer initiation and maintenance (4–6). Thus, miRNAs have become attractive targets in cancer therapy. Recently, a number of studies have developed various miRNA delivery strategies for cancer therapeutics in mice and non-human primates, and a miR-34 mimic has become the first miRNA to reach phase I clinical trials (4–7).
Esophageal cancer is the sixth most common cause of cancer-related death worldwide, representing one of the most aggressive cancers with a dismal survival rate of 10–20% (8). Esophageal squamous cell carcinoma (ESCC) is the predominant histological subtype of this disease in Asia and certain regions of Africa, which displays approximately 10% 5-year survival rates (8). Thus, identification of effective therapeutic targets is urgently needed for ESCC treatment. In the past few years, a panel of miRNAs has been identified to play key roles in the pathobiology of ESCC (9). Deregulation of these miRNAs contributes to ESCC stemness, tumorigenesis, invasion, metastasis and chemoresistance, indicating their potential as valuable therapeutic targets (9). Among these candidates, we have previously revealed in ESCC that miR-200b is a very important invasiveness suppressor that impairs the remodeling of cytoskeletal and adhesive machineries (10,11). Loss of miR-200b significantly correlates with adverse clinicopathological characteristics and a shorter survival in ESCC patients (11), which highlights its potential as a valuable therapeutic target. However, the implications of miR-200b in other aspects of ESCC pathobiology besides invasiveness remain unclear.
In this study, we uncovered that miR-200b mediates its tumor suppressive role in ESCC via inducing G2 cell cycle arrest, apoptosis and suppresses cell growth and clonogenic potential. A key cell cycle regulator CDK2 and an oncogenic protein PAF (PCNA-associated factor) (12–15) were identified as direct targets. Thus, our work has revealed how ESCC cells exploit deregulated miR-200b to sustain tumorigenic signals, thereby facilitating unrestricted tumor cell growth.
Materials and methods
Human ESCC cell lines and stable cell clone generation
KYSE150 and KYSE510, two cell line established from primary human ESCC specimens, were characterized previously (16), and both cell lines were provided by Ming-Zhou Guo, Chinese PLA General Hospital, Beijing, China. Human ESCC cell line EC109 was purchased in 2001 from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai, China. All three cell lines were authenticated using short tandem repeat DNA profiling in December 2012. KYSE150 and KYSE510 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), while EC109 cells were maintained in Dulbecco’s modified Eagle’s medium plus 10% FBS. CDK2-DN (dominant negative CDK2) Tetracycline-off stable cell clones were generated as described previously (17), following the instructions from the manufacturer of the Tetracycline-off system (Clontech). Briefly, EC109 and KYSE150 cells were first transfected with the pTet-Off vector that expresses tetracycline-controlled transactivator (Clontech), stable cell clones were selected under 400mg/ml geneticin (Invitrogen). Then, these stable clones were transfected with pUHD-CDK2-DN-HA plasmid, and double stable cell clones were selected using media containing 400mg/ml hygromycin B (Invitrogen). These EC109 and KYSE150 stable clones were cultured in DMEM and RPMI-1640 medium respectively, both supplemented with 10% FBS.
DNA constructs, miRNA mimics, miRNA inhibitors and siRNAs
The pUHD-CDK2-DN-HA plasmid is a gift from Greg Enders, Addgene plasmid # 27651 (18). pcDNA3-PAF-Flag plasmid was provided by Dr. Jae-II Park, The University of Texas MD Anderson Cancer Center. The pTet-Off vector was purchased from Clontech. The pGL3-Control firefly luciferase reporter plasmid and the pRL-TK Renilla luciferase plasmid were obtained from Promega. Construction of the pGL3-Control-CDK2 and pGL3-Control-PAF plasmids was described previously (11), and the corresponding mutants were generated by deleting the ‘seed sequence’ of the miR-200b binding site, i.e. CAGUAUU, which was performed by GenScript. Syn-hsa-miR-200b Mimic, Anti-hsa-miR-200b Inhibitor and negative control scrambled RNA were purchased from Qiagen. A PAF siRNA smart pool was purchased from Dharmacon.
Transfection and chemical treatment
Transfection of plasmids, miRNA mimics, miRNA inhibitors and siRNAs were performed as described previously (11). For DNA and RNA co-transfection, Lipofectamine 2000 was used. Both miRNA mimics and siRNA were transfected at a concentration of 30nM, while miRNA inhibitors were transfected at a dose of 100nM. The CDK2 inhibitor NU6140 was purchased from Tocris, DMSO was used as the vehicle for this chemical.
RNA isolation and real-time PCR
Total RNA was extracted with RNeasy reagent (Qiagen), and real-time PCR was performed as described previously (11). The primers used in this study are CDK2-forward: GCTTTCTGCCATTCTCATCG, CDK2-reverse: GTCCCCAGAGTCCGAAAGAT; PAF-forward: AGCTTTGTTGAACAGGCATTT, PAF-reverse: GGCAGCAGTACAACAATCTAAGC; Axin2-forward: AAGTGCA AACTTTCGCCAAC, Axin2-reverse: ACAGGATCGCTCCTCTTGAA; LEF1-forward: CCGAAGAGGAAGGCGATTTAGCT, LEF1-reverse: GCTCCTGAGAG GTTTGTGCTTGTCT; GAPDH-forward: GCTAGGGACGGCCTGAAG, GAPDH-reverse: GCCCAATACGACCAAATCC.
Flow cytometry
Cell apoptosis was determined by the propidium iodide and Annexin V double staining assay. Briefly, cells were gently dissociated with trypsin and stained with propidium iodide and Annexin V using the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) according to the manufacturer’s instructions. Then, flow cytometry analysis was performed. For cell mitosis evaluation, propidium iodide and phospho-Histone H3S10 double staining was performed. Briefly, Cells were fixed in 70% (v/v) ethanol for 1h, and permeabilized in phosphate-buffered saline (PBS) containing 0.25% (v/v) Triton X-100. After washing with 1% (w/v) bovine serum albumin (BSA) in PBS, cells were incubated with an anti-phospho-Histone H3S10 antibody (Rabbit, Cell Signaling Technology) diluted in 1% (w/v) BSA in PBS for 1.5h at room temperature. Then, cells were washed twice with 1% BSA in PBS, and labeled with Alex 488-conjugated anti-Rabbit antibody diluted in 1% BSA in PBS for 1h at room temperature. After washing with PBS alone, cells were resuspended in PBS containing propidium iodide (20 µg/ml) and RNAse A (200 µg/ml), and incubated at room temperature for 30min before flow cytometry analysis. For cell cycle analysis, cells were firstly fixed for at least 2h in cold 70% (v/v) ethanol diluted in water. Then, cells were incubated 30min at room temperature in PBS containing 20 µg/ml propidium iodide, 200 µg/ml RNAse A and 0.1% (v/v) Triton X-100. The stained cells were filtered to remove cell clumps and kept on ice before flow cytometry analysis. All the flow cytometry data were analyzed using the Flow Jo software.
Western blot
Protein concentration was determined using BCA Protein Assay Kit (Pierce). Proteins were separated on 10% SDS-PAGE and transferred to PVDF membrane (Millipore). The membranes were blocked in 5% non-fat milk and incubated with the following primary antibodies: anti-CDK2 (mouse, Cat#610146), anti-CDK1 (mouse, Cat#610138), anti-Cyclin A (mouse, Cat#611269) and anti-Cyclin B (mouse, Cat#610220) from BD Transduction Laboratories; anti-PARP (rabbit, #9542), anti-phospho-CDK1Y15 (rabbit, #9111), anti-non-phospho β-Catenin (rabbit, #8814), anti-HDAC1 (rabbit, #2062), anti-α-Tubulin (rabbit, #2125), anti-CDK6 (mouse, #3136), and anti-Cyclin D1 (rabbit, #2978) from Cell Signaling Technology; anti-PAF (rabbit, sc-67279) and anti-Actin (mouse, sc-8432) from Santa Cruz Technology. Then, the blots were washed, incubated with anti-Rabbit IgG, HRP-linked antibody (#7074) or anti-mouse IgG, HRP-linked antibody (#7076) from Cell Signaling Technology and detected with ECL Western Blotting Substrate (Pierce).
Cell viability measurement
Cell viability was measured using the CellTiter 96AQueous One Solution Cell Proliferation Assay (Promega), also known as MTS assay, according to the manufacturer’s protocol. Absorbance at 490nm was measured using a microplate reader (BMG Biotech).
Clonogenic assay
After transfection or chemical treatment, cells were plated at a number of 300 or 500 cells/well in 12-well plates and cultured for around 10 days. Then, the colonies were fixed with cold absolute methanol and stained with 1% crystal violet (Sigma) for 30min. Stained colonies were gently washed with water, and dried before images were taken.
Confocal microscopy
ESCC cells transfected with negative control/miR-200b for 36h were fixed with 4% paraformaldehyde for 10min at room temperature. After washing with PBS, the cells were permeabilized in 0.2% Triton X-100 in PBS for 10min at room temperature. After washing with PBS, the cells were blocked for 45min in PBS containing 10% goat serum and 1% BSA at room temperature. After washing with PBS, the cells were incubated with anti-α-Tubulin (mouse, #T9026, Sigma, 1:300 in blocking buffer) and anti-phospho-Histone H3S10 (rabbit, 06-570, Millipore, 1:300 in 1% BSA) for 1h at room temperature. After washing with PBS, the cells were incubated for 1h at room temperature with Alexa Fluor® 488 conjugated goat anti-mouse IgG (H+L) (A-11001) and Alexa Fluor® 594 conjugated goat anti-rabbit IgG (H+L) (A-11037) secondary antibodies (both from ThermoFisher Scientific, 1:300 in 1% BSA). After washing with PBS, nuclei were then counterstained with DRAQ5 (10 µM, Biostatus), and the slides were mounted with FluorSave, and viewed using an inverted confocal microscope (Eclipse Ti-E; Nikon) with 40× and 100× oil-immersion objective lenses.
Luciferase reporter assay
Luciferase reporter assay was performed as described previously (17). Briefly, ESCC cells were transfected in 48-well plates with 200ng firefly luciferase plasmids, 4ng control Renilla luciferase vector pRL-TK, together with other reagents (e.g. miR-200b mimics, PAF siRNA and PAF cDNA constructs). About 36 or 48h after transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Data are presented as ratios between firefly and Renilla luminescence activities.
Statistical analysis
The statistical analyses were performed using the Graphpad Prism 6. Student’s t test was used to determine the differences between two independent groups of samples. Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. Differences were considered significant when the P value was less than 0.05.
Results
miR-200b represses cell growth via inducing apoptosis and G2 cell cycle arrest in ESCC cells
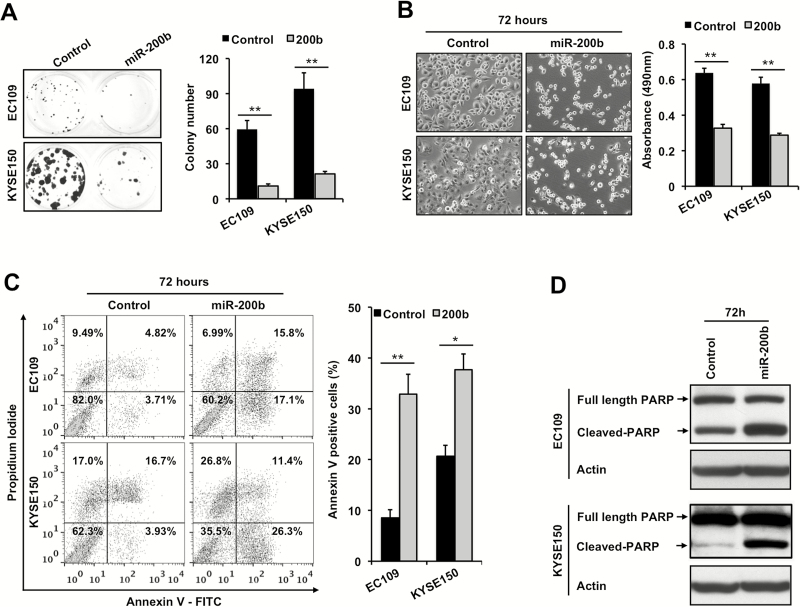
Previous studies have shown that loss of miR-200b promotes invasion in ESCC (10,11), but its involvement in other aspects of ESCC pathobiology remains unclear. To determine the role of miR-200b in ESCC cell growth, clonogenic capacity and apoptosis, we transfected miR-200b mimic in three ESCC cell lines that had reduced expression of miR-200b compared with normal controls, i.e. EC109, KYSE150 and KYSE510 (11). miR-200b mimic transfection was shown to sharply increase miR-200b expression in these cells (11). As shown in Figure 1A and B and Supplementary Figure 1A, available at Carcinogenesis Online, enforced expression of miR-200b markedly blocked their clonogenic potential as well as cell growth (P < 0.01). In keeping with these observations, enforced expression of miR-200b triggered apoptosis at 72h in all three cell lines, as measured by Annexin V positivity and/or PARP cleavage (Figure 1C and D, and Supplementary Figure 1B, available at Carcinogenesis Online). We also determined if inhibition of endogenous miR-200b in KYSE150, a cell line with relatively higher expression of miR-200b (11), could enhance cell growth. Although miR-200b inhibitor could significantly decrease miR-200b expression (11), it did not significantly stimulate cell growth (Supplementary Figure 2A, available at Carcinogenesis Online), probably due to the fact that basal miR-200b expression was already markedly repressed (by ~80%) compared with normal controls (11).
Figure 1.
miR-200b induces apoptosis and inhibits cell growth in ESCC cells. (A) Clonogenic capacity was measured using cells transfected with either a negative control or miR-200b mimics (30nM). Representative images of the colonies are shown on the left panel. (B) Left panel, 72h after transfection, representative images were taken to show the effect of enforced expression of miR-200b on cell growth. Right panel, MTS assay was performed to measured cell growth 72h after transfection. (C, D) Seventy-two hours after transfection, apoptosis was determined by propidium iodide/Annexin V double staining and cleaved-PARP detection. Actin served as a loading control for Western blots. Representative results are shown, and data are presented as mean ± SD. *P < 0.05, **P < 0.01, Student’s t test.
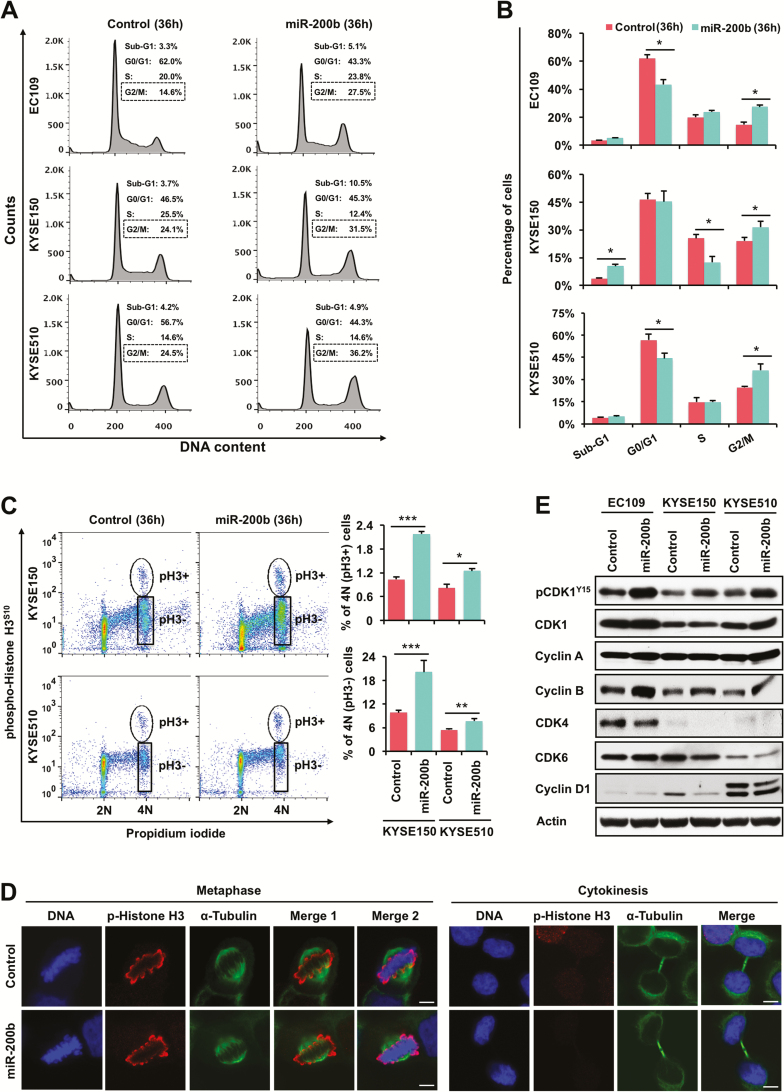
We then investigated whether the clonogenic/growth inhibitory function of miR-200b could be attributed to cell cycle arrest. To this end, those three ESCC cell lines were transfected with either negative control RNA or miR-200b mimics, and cell cycle was determined 36h after transfection before the onset of visible cell death. As shown in Figure 2A and B, miR-200b significantly induced G2/M phase arrest in all three cell lines (P < 0.05). Then, we determined whether miR-200b caused mitotic arrest in these ESCC cells. As shown in Figure 2C, enforced expression of miR-200b significantly increased the proportion of tetraploid cells with both negative phospho-Histone H3S10 staining (G2-phase) and positive phospho-Histone H3S10 staining (late G2 and M phase) in both KYSE150 and KYSE510 cells. However, as shown by the representative microscopic images in Figure 2D, miR-200b overexpression did not cause appreciable mitotic abnormalities, as evidenced by the normal morphology of cells at metaphase and the subsequent cytokinesis. Taken together, these results suggest that miR-200b mainly induces G2-phase arrest in ESCC cells. We also assessed if miR-200b inhibition affects cell cycle in KYSE150, a cell line with relatively higher expression of miR-200b (11). As shown in Supplementary Figure 2B, available at Carcinogenesis Online, miR-200b inhibitor did not alter cell cycle progression, probably due to the same reason mentioned above.
Figure 2.
miR-200b regulates the expression/activation of cell cycle regulators and induces G2-phase arrest in ESCC cells. (A, B) Flow cytometry was performed to determine the effect of miR-200b on ESCC cells 36h after transfection. Control cells were transfected with a non-targeting negative control RNA. (C) ESCC cells transfected with miR-200b mimic/control for 36h were double stained with propidium iodide and phospho-Histone H3S10, then flow cytometry was performed. (D) Confocal microscopy was performed to determine the impact of miR-200b on mitosis. KYSE150 cells transfected with miR-200b mimic/control for 36h were used in this assay, representative images are shown. Scale bars: 5 µm. (E) Western blot assay was performed to determine the impact of miR-200b on the expression of various cell cycle regulators 36h after transfection. Actin served as a loading control. Representative results are shown, and data are presented as mean ± SD. *P < 0.05, ***P < 0.001, Student’s t test.
We then investigated whether miR-200b affected the expression/activation of G2/M cell cycle regulators, such as Cyclin A, Cyclin B, CDK1 and phospho-CDK1Y15 (an inactive form). As shown in Figure 2E, miR-200b mimic transfection consistently increased the expression of inactive phospho-CDK1Y15 in all three cell lines tested, supporting the observed G2-phase arrest. Surprisingly, the expression of Cyclin B, a CDK1 binding partner during G2/M progression, was enhanced by miR-200b (Figure 2E), which might be attributed to CDK1 inactivation and the resulted accumulation in G2-phase arrested cells. The expression of total CDK1 and Cyclin A was not affected by miR-200b in these cells (Figure 2E). We also determined if miR-200b affected G1-phase regulators, such as CDK4, CDK6 and Cyclin D1. As shown in Figure 2E, miR-200b overexpression only decreased the expression of CDK4 but not CDK6 or Cyclin D1. Taken together, these data suggest that miR-200b suppresses ESCC cell growth by inducing G2-phase arrest and apoptosis.
CDK2 and PAF are miR-200b target genes
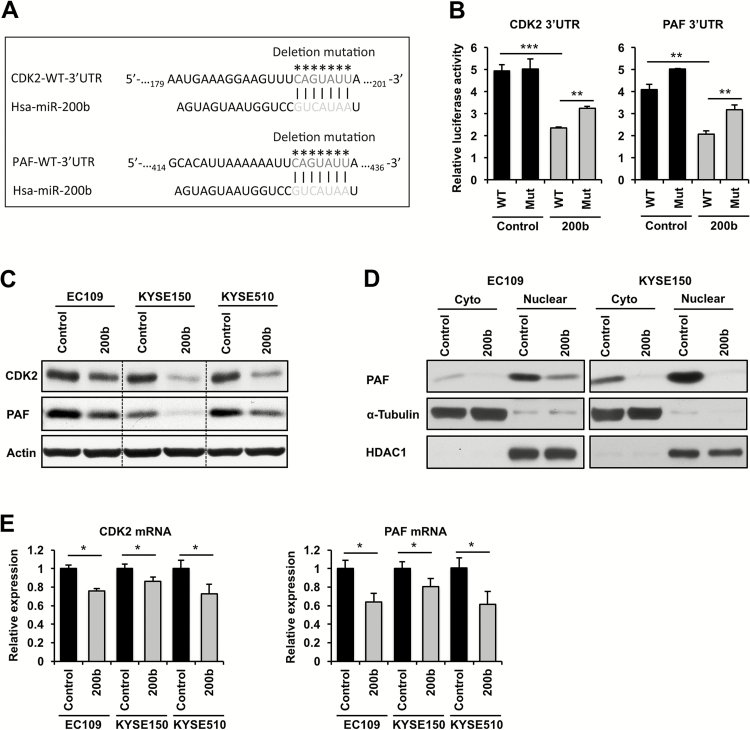
How miR-200b attenuates cell growth and clonogenic potential in ESCC remains unclear. Using quantitative proteomics, we previously identified a list of putative miR-200b targets in ESCC cells (11), among which CDK2 and PAF (PCNA-associated factor) are important regulators of cell cycle progression and/or tumorigenicity (12–15). Here, we determined whether CDK2 and PAF are bona fide targets of miR-200b. To this end, the CDK2 and PAF 3′UTRs containing putative miR-200b binding sites (Figure 3A) as revealed by both TargetScan and miRanda, two well-recognized miRNA target gene prediction databases, were cloned into a luciferase reporter. Mutant reporters that harbor deletions of the miR-200b binding sites were generated to serve as controls (Figure 3A). As shown in Figure 3B, miR-200b mimic transfection dramatically reduced the activity of the luciferase reporters harboring wild-type 3′UTR of CDK2 and PAF (P < 0.01), whereas deletion of miR-200b binding sites in both reporters significantly restored their activities upon miR-200b overexpression (P < 0.01), suggesting that miR-200b directly targets the 3’UTR of both genes. Notably, the activity of the mutant reporters could still be suppressed by miR-200b overexpression, but the decrease was less than that in the wild-type reporters (Figure 3B), suggesting that miR-200b can directly/indirectly target these 3′UTRs via unidentified binding site(s).
Figure 3.
CDK2 and PAF are direct targets of miR-200b. (A) Putative miR-200b binding sites on the 3′UTRs of CDK2 and PAF mRNAs are shown, and the deletion mutation sites of the luciferase reporters are indicated by asterisks. (B) Thirty-six hours after transfection, dual luciferase reporter assay was performed to measure the impact of miR-200b on the 3′UTR of CDK2 and PAF in EC109 cells. WT, wild-type; Mut, mutation. (C–E) ESCC cells were transfected with miR-200b mimics or negative control RNA (30nM), 36h after transfection, the expression of protein and mRNA was determined by Western blot (C and D) and real-time PCR (E), respectively. In Western blot assays, Actin was used as a loading control for whole cell lysates, while α-Tubulin and HDAC1 were used as loading controls for cytoplasmic (Cyto) and nuclear lysates, respectively. GAPDH was used as a loading control for real-time PCR. Representative data are shown, and data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
In support of the 3′UTR reporter data, miR-200b mimic transfection sharply decreased CDK2 and PAF protein expression in all three ESCC cell lines (Figure 3C). Consistent with the major nuclear function of PAF (13–15), we also observed a predominant nuclear localization of PAF in ESCC cells, and enforced expression of miR-200b could dramatically repress nuclear PAF expression (Figure 3D). In contrast to the remarkable changes in protein expression, enforced expression of miR-200b only moderately reduced CDK2 and PAF mRNA expression (Figure 3E), supporting the predominant role of miRNAs in the regulation of translation (1). All together, these results suggest that CDK2 and PAF are bona fide miR-200b targets in ESCC.
CDK2 alone is not a significant functional mediator of miR-200b
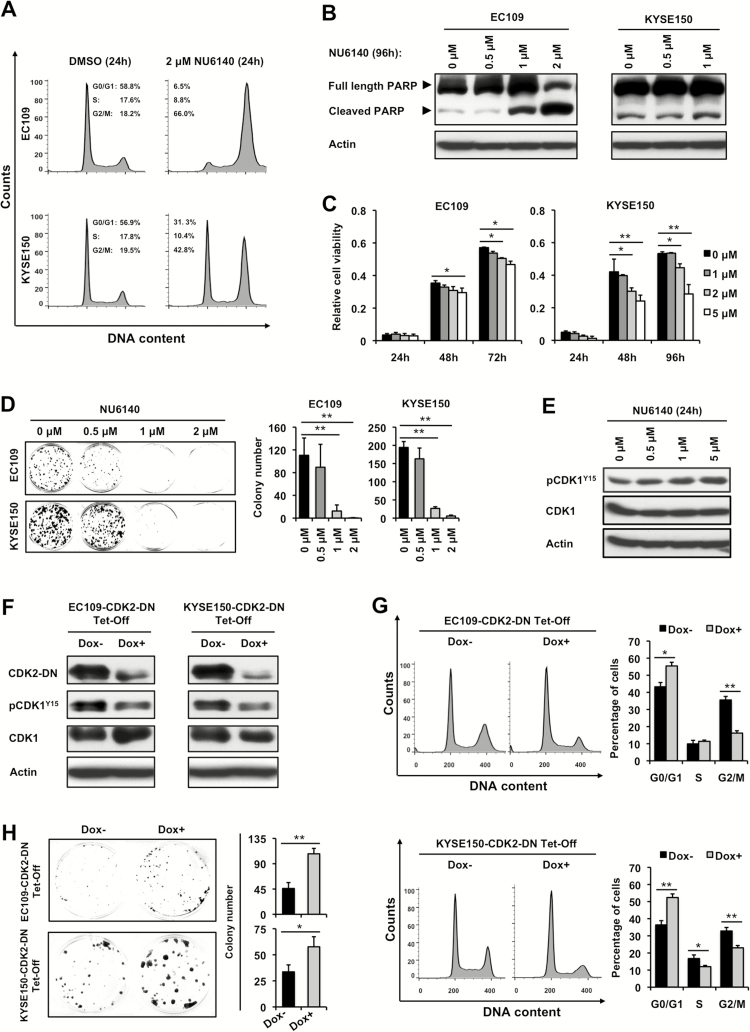
Since enforced expression of miR-200b affected multiple cell cycle regulators besides its direct target CDK2, such as CDK1, CDK4 and Cyclin B (Figures 2E and 3C), we speculated that specific knockdown of CDK2 alone may not phenocopy the biological function of miR-200b. In keeping with this speculation, as shown in Supplementary Figure 3A–D, CDK2 knockdown did not significantly affect colony formation or apoptosis in the two ESCC cell lines tested, and it only slightly increased G0/G1 phase in KYSE150 cells, while no significant cell cycle change was observed in EC109 cells. These data suggest that CDK2 may function synergistically with other cell cycle regulators to mediate the biological effects of miR-200b. To test this hypothesis, NU6140, a CDK2 inhibitor that also inhibits the activity of CDK1 and CDK4 (19), which better mimics the biochemical effects of miR-200b, was used to phenocopy the biological function of miR-200b. As shown in Figure 4A–D and Supplementary Figure 4, available at Carcinogenesis Online, NU6140 markedly induced G2/M arrest, apoptosis, and inhibition of cell growth and clonogenic capacity, which remarkably resembled the biological function of miR-200b. Moreover, as expected, this CDK2 inhibitor also biochemically mimicked the inhibitory effects of miR-200b on CDK1 activity, as shown by the increased expression of inactive phospho-CDK1Y15 (Figure 4E).
Figure 4.
CDK2 does not function alone to mediate the biological effects of miR-200b. (A–E) ESCC cells were treated with a CDK2 inhibitor NU6140 or DMSO vehicle, then cell cycle analysis (A), Western blot assay (B, E), MTS cell growth assay (C) and clonogenic assay (D) were performed. (F–H) Using ESCC cells conditionally expressing a CDK2 dominant negative mutant (CDK2-DN), the effects of CDK2-DN were determined by Western blot assay (F), cell cycle analysis (G) and clonogenic assay (H). Doxycycline (Dox) was used to block the expression of CDK2-DN in these cells. Actin served as a loading control for Western blots. Representative results are shown, and data are presented as mean ± SD, *P < 0.05, **P < 0.01, Student’s t test.
To further validate that miR-200b functions through synergistically blocking multiple cell cycle regulators, a dominant negative CDK2 (CDK2-DN) mutant that has been shown to block the activity of both CDK2 and CDK1, was used to mimic the biological effects of miR-200b (18). EC109 and KYSE150 stable clones conditionally expressing the CDK2-DN mutant under the control of a Tet-Off system were used as cell models (Figure 4F). As shown in Figure 4G and H, enforced expression of CDK2-DN in both cell lines could significantly induce G2/M arrest, increase phospho-CDK1Y15 expression, and inhibit clonogenic growth. Notably, the impact of CDK2-DN on CDK1 was not mediated via blocking CDK2, as specific CDK2 knockdown could not appreciably alter phospho-CDK1Y15 expression in EC109 and KYSE150 cells (Supplementary Figure 3A, available at Carcinogenesis Online). Taken together, these data suggest that CDK2 does not function alone to mediate the biological effects of miR-200b in ESCC, and the exact mechanisms underlying its regulation on cell cycle progression needs further investigation.
The biological function of the miR-200b/PAF axis in ESCC
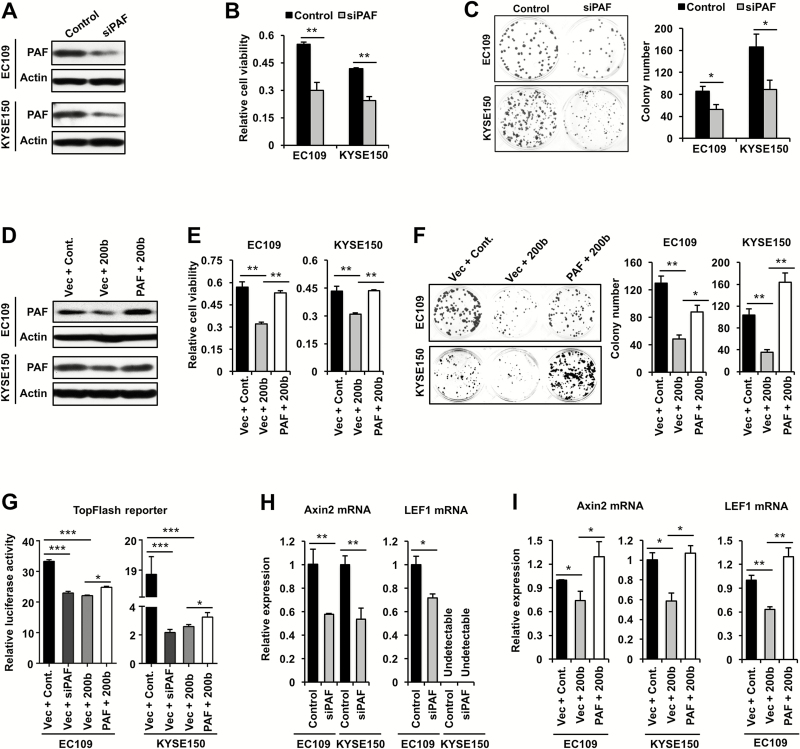
We then proceeded to investigate if PAF can mediate the biological effects of miR-200b in ESCC. Our findings were in support of this notion. First, as shown in Figure 5A–C, siRNA knockdown of PAF phenocopied the impact of miR-200b mimic transfection in both EC109 and KYSE150 cells, i.e. significantly inhibited cell growth and their clonogenic potential (P < 0.01 or P < 0.05). However, PAF knockdown did not appreciably induce apoptosis or cell cycle arrest in these cells (Supplementary Figure 3C and 5), suggesting that PAF mediates miR-200b’s biological function via other mechanisms. Second, as shown in Figure 5D–F, re-expression of PAF expression in miR-200b transfected cells using a PAF construct lacking its 3′UTR significantly restored cell growth and colony formation in both EC109 and KYSE150 cells (P < 0.01 or P < 0.05).
Figure 5.
The biological function of the miR-200b/PAF axis in ESCC cells. (A–C) EC109 and KYSE150 were transfected with PAF siRNA or a negative control siRNA, then Western blot assay was conducted 48h after transfection (A), cell growth assay was measured using MTS assay 72h after transfection (B) and clonogenic assay was performed 48h after transfection (C). (D–F) EC109 and KYSE150 were co-transfected with the indicated plasmids and miR-200b mimics or a negative control miRNA (Cont.), then Western blot assay was conducted 48h after transfection (D), cell growth assay was measured using MTS assay 72h after transfection (E) and clonogenic assay was performed 48h after transfection (F). (G) Dual luciferase reporter assay was performed to measure the TopFlash reporter activity. (H, I) Real-time PCR assay was conducted to measure the mRNA expression of Axin2 and LEF1. GAPDH was used as a loading control for real-time PCR, and Actin was used as a loading control for Western blots. Representative results are shown, and data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
To further support that miR-200b exerts its function via targeting PAF, we determined if miR-200b regulates Wnt/β-Catenin signaling, a pathway that can be potentiated by PAF during intestinal carcinogenesis (14). As shown in Figure 5G, both miR-200b overexpression and PAF knockdown markedly suppressed the Wnt/β-Catenin signaling in both EC109 and KYSE150 cells, as measured by TopFlash reporter assays. Notably, re-expression of PAF in miR-200b mimic transfected cells slightly but significantly restored the TopFlash reporter activity in both cell lines (Figure 5G). Thus, the miR-200b/PAF axis is an important regulator of this pathway, while the incomplete restoration of the TopFlash reporter activity by PAF re-expression suggests that miR-200b may repress this pathway via additional mechanisms. To further validate the role of the miR-200b/PAF axis in Wnt/β-Catenin signaling, we determined its impact on the expression of Axin2 and LEF1, two canonical downstream targets of this pathway (20,21). In line with our hypothesis, as shown in Figure 5H–I, both PAF knockdown and miR-200b overexpression significantly blocked the mRNA expression of Axin2 and LEF1, and restoration of PAF could significantly abolish the inhibitory impact of miR-200b (P < 0.01 or P < 0.05). We next tested whether miR-200b affected β-Catenin activation, a pivotal effector of Wnt/β-Catenin signaling (14). As shown in Supplementary Figure 6, available at Carcinogenesis Online, miR-200b mimic transfection did not appreciably alter the expression of non-phospho (active) β-Catenin, indicating that miR-200b may not regulate the upstream activation of Wnts. Instead, our data suggest that miR-200b may repress this signaling via blocking the potentiating effects of PAF on the transcription activity of β-Catenin complex as shown in a recent study (14).
CDK2 and PAF are frequently increased in ESCC and correlate with poor prognosis in multiple cancers
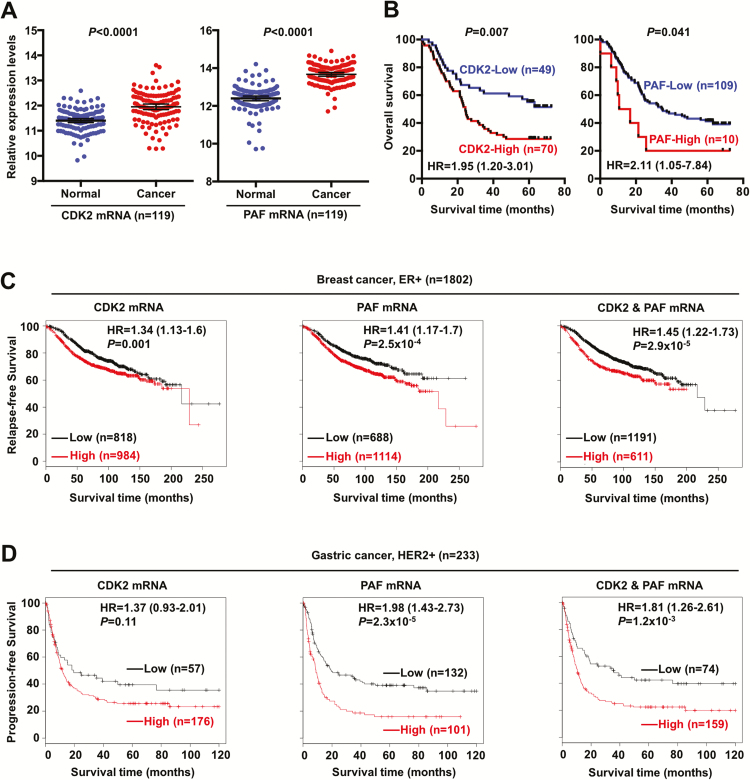
Our previous studies have shown that miR-200b was frequently reduced in ESCC compared with normal controls (11), which prompted us to determine if the newly identified miR-200b targets CDK2 and PAF are aberrantly expressed in ESCC tumors. A recent study has performed transcriptome analyses in 119 cases of paired ESCC tumor and normal tissues (22), using the publicly available data in Gene Expression Omnibus (accession number GSE53625), we analyzed the expression levels of CDK2 and PAF in these case-matched samples. As shown in Figure 6A, both CDK2 and PAF were significantly upregulated in ESCC tumors compared with normal issues (n = 119, both P < 0.0001). Further clinical analysis revealed that patients carrying tumors with high-expression of CDK2 had a markedly reduced 5-year survival rate (Figure 6B, 28 versus 56%, P = 0.007). Similarly, patients carrying tumors with high-expression of PAF also displayed a significantly lower 5-year survival rate (Figure 6B, 20 versus 41%, P = 0.041).
Figure 6.
CDK2 and PAF are unfavorable prognostic markers in multiple cancers. (A) CDK2 and PAF mRNA expression was significantly increased in ESCC tumors compared with case-matched normal control tissues (n = 119, Gene Expression Omnibus accession number GSE53625). (B) Kaplan–Meier analysis was performed to determine the prognostic significance of CDK2 and PAF in the same cohort of ESCC tumors (n = 119, Gene Expression Omnibus accession number GSE53625). Optimal cutoff values of gene expression were used to stratify the samples, and Log-rank test was used in the statistical analyses. HR represents hazard ratio, and 95% confidence interval values of HR are shown. (C, D) Using a publicly available database (http://kmplot.com/analysis/) (49), the prognostic significance of CDK2 and PAF was assessed in large cohorts of ER+ breast tumors (n = 1802) and HER2+ gastric tumors (n = 233). Follow-up data from the 2014 version database was used, and optimal cutoff values of gene expression were used to stratify the samples. Log-rank test was used in the statistical analyses. HR, hazard ratio and 95% confidence interval values of HR are shown.
Given the important role of miR-200b in cell cycle regulation in ESCC cells (Figure 2A–E), we also determined the expression status and prognostic values of other major cell cycle regulators, i.e. CDK1, CDK4 and CDK6, in the same cohort of ESCC patients. As shown in Supplementary Figure 7, available at Carcinogenesis Online, all three CDKs were significantly increased in ESCC compared with case-matched normal tissues (n = 119, P < 0.0001). Kaplan–Meier survival analysis revealed that high-expression of all three CDKs was associated with unfavorable prognosis, i.e. CDK1 (HR = 2.77, P = 0.007); CDK4 (HR = 1.32, P = 0.242); CDK6 (HR = 2.16, P = 0.002). These data highlight the importance of these cell cycle regulators in the pathobiology of ESCC, and the inhibitory effect of miR-200b on their expression/activity, including CDK2 (direct target), CDK1 and CDK4, underlines its significant tumor suppressor function in ESCC.
Apart from ESCC, miR-200b has also been shown to play a tumor suppressor role in other cancers, including breast cancer and gastric cancer (23–26), hence we set out to test whether the two newly identified targets are prognostic markers in these cancers. Publicly available tumor databases were used for this aim. As shown in Figure 6C, both CDK2 and PAF were significantly associated with shorter relapse-free survival in ER-positive breast cancer patients (n = 1802, P = 0.001 and P = 2.5×10−4, respectively). Further analysis revealed that patients carrying tumors with high expression of both CDK2 and PAF had an increased risk (HR = 1.45) than those with CDK2-high alone (HR = 1.34) and PAF-high alone (HR = 1.41). However, as shown in Supplementary Figure 8A, available at Carcinogenesis Online, when all subtypes of breast cancers were included in the analysis, only PAF was a significant prognostic marker (n = 3554, P = 1.2×10−5). In gastric cancer patients, as shown in Figure 6D, the expression of both CDK2 and PAF correlated with reduced progression-free survival in the HER2-positive subtype (n = 233, P = 0.11 and P = 2.3×10−5, respectively), but CDK2 and PAF joint analysis did not increase the prognostic significance (P = 1.2×10−3). When all subtypes of gastric cancers were included in the analysis, as shown in Supplementary Figure 8B, available at Carcinogenesis Online, only PAF was significantly associated with a poor prognosis (n = 641, P = 2.0×10−8). Altogether, these clinical data present compelling evidence to support that deregulation of the miR-200b-CDK2/PAF axis is involved in the pathobiology of ESCC, and possibly in other types of cancers as well.
Discussion
Over the past decade, mounting evidence has highlighted the key roles played by miRNAs in various diseases, most notably cancer (2,3). Using genome-wide miRNA profiling, we previously identified a large set of deregulated miRNAs in ESCC tumors compared with case-matched normal tissues (27). Further investigation revealed miR-200b as a significant prognostic marker and a potent invasiveness suppressor in ESCC (10,11). However, its pathobiological function in ESCC remains less understood. The current study uncovers a role of miR-200b in promoting apoptosis, suppressing cell cycle progression and cell growth in ESCC. A key cell cycle regulator CDK2 and an oncogenic factor PAF (12–15) are identified as targets and functional mediators of miR-200b. The miR-200b-CDK2/PAF regulatory axis is clinically relevant in ESCC patients, as supported by their collective deregulation in ESCC tumors and significant prognostic values.
Although the involvement of miR-200b in epithelial–mesenchymal transition and invasion/metastasis in cancers has been well-established, their function in other traits of cancer cells, such as cell cycle progression, apoptosis and clonogenic growth, remain less understood or controversial (28). While a number of studies found that enforced expression of miR-200b had no significant impact on tumor growth (26,29), a study reported that miR-200b could promote cell growth in cancer cell lines (30), whereas other studies observed significant reductions in tumor growth upon miR-200b overexpression (23,25,31–33). Similarly, the function of miR-200b in cell cycle regulation is also controversial, which is largely cell context-dependent (31–33). Specifically, although miR-200b has been shown to induce G2/M phase arrest in breast cancer cells, it was reported to promote S phase entry in HeLa cells (31–33). The function of miR-200b in cancer cell apoptosis is less understood, to our knowledge, only one study has documented that miR-200b promotes apoptosis in breast cancer cells (33). The present study provides compelling evidence to show that miR-200b is a significant inducer of G2-phase arrest, apoptosis and a suppressor of cell growth in ESCC.
One of the major characteristics of tumor cells is unlimited cell growth, which can result from deregulated and unrestrained cell cycle checkpoints (34). A number of well-known tumor suppressors that are frequently mutated/deregulated in human cancers are cell cycle regulators, such as RB, p53, p14ARF, p15INK4b, p16INK4a (35,36). Inactivation of these factors unleashes tumor cells from the restriction of cell cycle checkpoints, thereby facilitating tumor outgrowth. Notably, the biological function of these proteins are tightly regulated or mediated by cyclin-dependent kinases (CDKs), a family of kinases that directly drive cell cycle progression, which are also commonly deregulated in human cancers (12). Among these CDKs, CDK2 has been shown to play critical roles during cell transformation induced by oncogenes, such as KRAS, MYCN and c-myc (37–40). Specifically, inhibition or genetic depletion of CDK2 was demonstrated to trigger senescence in cells overexpressing Myc, induce anaphase catastrophe and apoptosis in lung cancers with KRAS mutation, and be synthetically lethal to MYCN-driven cancer cells (37–40). Given the important role of CDK2 in tumorigenesis, various pharmacological CDK2 inhibitors have been developed, and numerous chemicals have been shown to have significant anti-cancer efficacy in animals or in clinical trials (12). However, the role of CDK2 in ESCC remains elusive, to our knowledge, no study has specifically assessed the expression status of CDK2 in ESCC tumors or reported its function in ESCC. Our study represents the first to show that CDK2 expression is significantly upregulated in ESCC tumors and correlates with a shorter survival in ESCC patients. Our data also indicate that frequent loss of miR-200b (11), may contribute to CDK2 overexpression in ESCC.
CDK2 is a well-established regulator of G1 and S phase during cell cycle progression (12), thus the effects of miR-200b on G2-phase arrest could not be attributed to the suppression of CDK2. Indeed, specific CDK2 knockdown could not phenocopy the biological function of miR-200b in ESCC cells, including G2-phase arrest, apoptosis and clonogenicity inhibition. However, we cannot exclude the possibility that CDK2 is a significant functional mediator of miR-200b. Since miR-200b regulates the expression/activity of multiple cell cycle regulators besides the direct target CDK2, such as CDK1 and CDK4, we believe that these effects of miR-200b in ESCC are collectively mediated by these factors. It is also possible that CDK2 and these factors are synthetically lethal. Recently, CDK2 has been shown to be synthetically lethal to neuroblastoma cells with MYCN amplification, CDK2 knockdown alone could not induce apoptosis in cells without MYCN amplification, whereas it triggered massive apoptosis in MYCN overexpressed cells (39). In the context of ESCC, although CDK2 knockdown alone could not phenocopy those functions of miR-200b, CDK2 can be synthetically lethal to ESCC cells with aberrant activation/expression of other CDKs caused by miR-200b deregulation, and only simultaneous blockade of them may resemble miR-200b’s functions. Our findings are in support of this concept. Specifically, NU6140, a CDK2 inhibitor (IC50 = 0.41 μM) that can also inhibit CDK1 (IC50 = 6.6 μM) and CDK4 (IC50 = 5.5 μM) (19), was able to phenocopy those biological functions of miR-200b. Similarly, a dominant negative CDK2 mutant (CDK2-DN) that can also block CDK1 activity (via inducing CDK1Y15-phosphorylation), as shown by both our study and others (18), could mimic the role of miR-200b in inducing G2-phase arrest and clonogenicity inhibition. Given that CDK2 knockdown did not appreciably alter the expression of phospho-CDK1Y15 (Supplementary Figure 3A, available at Carcinogenesis Online), the mechanism underlying CDK1 inhibition by CDK2-DN or miR-200b remains unclear. Besides, it should be noted that the non-specificity of NU6140 and CDK2-DN could not conclusively indicate that the function of miR-200b was mediated via those canonical cell cycle regulators. Taken together, these findings suggest that CDK2 alone cannot mediate the biological effects of miR-200b in ESCC, and the mechanism behind how miR-200b regulates cell cycle progression awaits further research. Nevertheless, the impact of miR-200b on those cell cycle regulators and cell cycle progression highlights its importance in the pathobiology of ESCC.
PAF, also known as p15PAF and KIAA0101, was initially identified as a PCNA (Proliferating cell nuclear antigen)-associated factor that was markedly increased in various tumor types compared with normal tissues (41). Later studies showed that PAF could promote DNA damage repair and cell cycle progression (42,43). Importantly, PAF has been shown to be oncogenic, which can transform mouse fibroblasts and induce intestinal and pancreatic ductal neoplasia in mice (13–15). Overexpression of PAF was found to correlate with unfavorable clinical outcome in various cancers including ESCC, breast cancer, lung cancer and hepatocellular carcinoma (44–48). However, the mechanism underlying the deregulation of PAF in cancer remains incompletely understood. To our knowledge, only two studies have suggested that PAF expression can be repressed by E2F factors, possibly downstream of the p53-p21-RB pathway (13,47). The present study is the first to uncover that aberrantly high expression of PAF can be driven by loss of miR-200b in cancer. Our data support the tumor-promoting function of PAF, i.e. enhancing cell growth and clonogenic potential in ESCC. However, the well-established role of PAF in cell cycle progression (43,47) was not observed in ESCC cells, as evidenced by both our study and a recent study (48). The tumor-promoting function of PAF was also not mediated by suppressing apoptosis, as PAF knockdown did not induce appreciable apoptosis in two ESCC cell lines tested. These observations suggest that PAF alone cannot fully mediate the biological function of miR-200b, which we believe is attributed to the fact that miRNAs can simultaneously target large numbers of genes, and PAF may function in concert with other targets to mediate these functions.
PAF has been shown to be oncogenic in colon cancer via hyperactivating Wnt/β-Catenin signaling. The mechanism is that PAF recruits EZH2 to facilitate the association between the RNA polymerase II transcriptional machinery and β-Catenin/TCF complex, thereby enhancing Wnt/β-Catenin target gene transactivation (14). Our data provide compelling evidence to show that miR-200b regulates Wnt/β-Catenin signaling via targeting PAF in ESCC. However, we did not observe appreciable alteration in the expression of non-phospho (active) β-Catenin, a central effector of the canonical Wnt signaling. Thus, the impact of miR-200b on this pathway may be mainly mediated via attenuating the potentiating effects of PAF on the transcriptional activity of the β-Catenin/TCF complex, rather than mitigating the upstream activation of the canonical Wnt signaling. The significance of the Wnt/β-Catenin signaling downstream of the miR-200b/PAF axis requires further investigation in cancers.
In conclusion, this study has uncovered that miR-200b, a frequently deregulated tumor suppressive miRNA in a wide spectrum of cancers, suppresses multiple key cell cycle regulators and mitigates the Wnt/β-Catenin signaling. CDK2 and PAF, two significant prognostic markers in several types of cancers, are identified as direct targets of miR-200b. We believe that CDK2 and PAF function in concert with numerous other targets to fully mediate the pathobiological functions of miR-200b, this research highlights miR-200b as a promising therapeutic target in ESCC.
Supplementary material
Supplementary Figures 1–8 can be found at http://carcin.oxfordjournals.org/
Funding
This work was financially supported by grants from the Natural Science Foundation of China-GuangDong Joint Fund (U1301227) to Dr. Li-Yan Xu, the National Science Foundation of China (81472613) to Dr. En-Min Li, and the Canadian Institutes of Health Research (CIHR) awarded to Dr. Raymond Lai. This study was also supported by the Department of Education, Guangdong Government under the Top-tier University Development Scheme for Research and Control of Infectious Diseases. Hai-Feng Zhang is a recipient of the Li Ka Shing scholarship.
Supplementary Material
Acknowledgement
We thank the technical assistance of Jingzhou Huang in the Flow Cytometry Lab at the Department of Experimental Oncology, Cross Cancer Institute, University of Alberta.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BSA
bovine serum albumin
- ESCC
esophageal squamous cell carcinoma
- miRNA
microRNA
- PBS
phosphate-buffered saline
References
- 1. Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ruan K., et al. (2009) MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett., 285, 116–126. [DOI] [PubMed] [Google Scholar]
- 3. Hayes J., et al. (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med., 20, 460–469. [DOI] [PubMed] [Google Scholar]
- 4. Medina P.P., et al. (2010) OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature, 467, 86–90. [DOI] [PubMed] [Google Scholar]
- 5. Babar I.A., et al. (2012) Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. USA, 109, E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng C.J., et al. (2015) MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature, 518, 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Agostini M., et al. (2014) miR-34: from bench to bedside. Oncotarget, 5, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jemal A., et al. (2010) Cancer statistics, 2010. CA. Cancer J. Clin., 60, 277–300. [DOI] [PubMed] [Google Scholar]
- 9. Sugihara H., et al. (2015) Noncoding RNA Expression Aberration Is Associated with Cancer Progression and Is a Potential Biomarker in Esophageal Squamous Cell Carcinoma. Int. J. Mol. Sci., 16, 27824–27834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H.F., et al. (2015) Loss of miR-200b promotes invasion via activating the Kindlin-2/integrin β1/AKT pathway in esophageal squamous cell carcinoma: An E-cadherin-independent mechanism. Oncotarget, 6, 28949–28960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H.F., et al. (2014) miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting Kindlin-2. Carcinogenesis, 35, 292–301. [DOI] [PubMed] [Google Scholar]
- 12. Chohan T.A., et al. (2015) Cyclin-dependent kinase-2 as a target for cancer therapy: progress in the development of CDK2 inhibitors as anti-cancer agents. Curr. Med. Chem., 22, 237–263. [DOI] [PubMed] [Google Scholar]
- 13. Hosokawa M., et al. (2007) Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res., 67, 2568–2576. [DOI] [PubMed] [Google Scholar]
- 14. Jung H.Y., et al. (2013) PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Mol. Cell, 52, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jun S., et al. (2013) PAF-mediated MAPK signaling hyperactivation via LAMTOR3 induces pancreatic tumorigenesis. Cell Rep., 5, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shimada Y., et al. (1992) Characterization of 21 newly established esophageal cancer cell lines. Cancer, 69, 277–284. [DOI] [PubMed] [Google Scholar]
- 17. Zhang H.F., et al. (2016) The Opposing Function of STAT3 as an Oncoprotein and Tumor Suppressor Is Dictated by the Expression Status of STAT3β in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res., 22, 691–703. [DOI] [PubMed] [Google Scholar]
- 18. Hu B., et al. (2001) S and G2 phase roles for Cdk2 revealed by inducible expression of a dominant-negative mutant in human cells. Mol. Cell. Biol., 21, 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pennati M., et al. (2005) Potentiation of paclitaxel-induced apoptosis by the novel cyclin-dependent kinase inhibitor NU6140: a possible role for survivin down-regulation. Mol. Cancer Ther., 4, 1328–1337. [DOI] [PubMed] [Google Scholar]
- 20. Jho E.H., et al. (2002) Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol., 22, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Filali M., et al. (2002) Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J. Biol. Chem., 277, 33398–33410. [DOI] [PubMed] [Google Scholar]
- 22. Li J., et al. (2014) LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut, 63, 1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang H., et al. (2013) miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin. Cancer Res., 19, 5602–5612. [DOI] [PubMed] [Google Scholar]
- 24. Iliopoulos D., et al. (2010) Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell, 39, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye F., et al. (2014) miR-200b as a prognostic factor in breast cancer targets multiple members of RAB family. J. Transl. Med., 12, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X., et al. (2014) MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene, 33, 4077–4088. [DOI] [PubMed] [Google Scholar]
- 27. Wu B.L., et al. (2011) MiRNA profile in esophageal squamous cell carcinoma: downregulation of miR-143 and miR-145. World J. Gastroenterol., 17, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang H.F., et al. (2014) A family of pleiotropically acting microRNAs in cancer progression, miR-200: potential cancer therapeutic targets. Curr. Pharm. Des., 20, 1896–1903. [DOI] [PubMed] [Google Scholar]
- 29. Sun L., et al. (2012) MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene, 31, 432–445. [DOI] [PubMed] [Google Scholar]
- 30. Hyun S., et al. (2009) Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell, 139, 1096–1108. [DOI] [PubMed] [Google Scholar]
- 31. Xia W., et al. (2010) MicroRNA-200b regulates cyclin D1 expression and promotes S-phase entry by targeting RND3 in HeLa cells. Mol. Cell. Biochem., 344, 261–266. [DOI] [PubMed] [Google Scholar]
- 32. Uhlmann S., et al. (2010) miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene, 29, 4297–4306. [DOI] [PubMed] [Google Scholar]
- 33. Yao Y., et al. (2015) MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J. Cell. Mol. Med., 19, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evan G.I., et al. (2001) Proliferation, cell cycle and apoptosis in cancer. Nature, 411, 342–348. [DOI] [PubMed] [Google Scholar]
- 35. Giacinti C., et al. (2006) RB and cell cycle progression. Oncogene, 25, 5220–5227. [DOI] [PubMed] [Google Scholar]
- 36. Krimpenfort P., et al. (2007) p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature, 448, 943–946. [DOI] [PubMed] [Google Scholar]
- 37. Campaner S., et al. (2010) Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat. Cell Biol., 12, 54–59. [DOI] [PubMed] [Google Scholar]
- 38. Hydbring P., et al. (2010) Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc. Natl. Acad. Sci. USA, 107, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Molenaar J.J., et al. (2009) Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc. Natl. Acad. Sci. USA, 106, 12968–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu S., et al. (2015) CDK2 Inhibition Causes Anaphase Catastrophe in Lung Cancer through the Centrosomal Protein CP110. Cancer Res., 75, 2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu P., et al. (2001) p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene, 20, 484–489. [DOI] [PubMed] [Google Scholar]
- 42. Turchi L., et al. (2009) ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ., 16, 728–737. [DOI] [PubMed] [Google Scholar]
- 43. Emanuele M.J., et al. (2011) Proliferating cell nuclear antigen (PCNA)-associated KIAA0101/PAF15 protein is a cell cycle-regulated anaphase-promoting complex/cyclosome substrate. Proc. Natl. Acad. Sci. USA, 108, 9845–9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yuan R.H., et al. (2007) Overexpression of KIAA0101 predicts high stage, early tumor recurrence, and poor prognosis of hepatocellular carcinoma. Clin. Cancer Res., 13(18 Pt 1), 5368–5376. [DOI] [PubMed] [Google Scholar]
- 45. Kais Z., et al. (2011) KIAA0101 interacts with BRCA1 and regulates centrosome number. Mol. Cancer Res., 9, 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kato T., et al. (2012) Overexpression of KIAA0101 predicts poor prognosis in primary lung cancer patients. Lung Cancer, 75, 110–118. [DOI] [PubMed] [Google Scholar]
- 47. Chang C.N., et al. (2013) p15(PAF) is an Rb/E2F-regulated S-phase protein essential for DNA synthesis and cell cycle progression. PLoS One, 8, e61196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng Y., et al. (2013) Expression of KIAA0101 protein is associated with poor survival of esophageal cancer patients and resistance to cisplatin treatment in vitro . Lab. Invest., 93, 1276–1287. [DOI] [PubMed] [Google Scholar]
- 49. Györffy B., et al. (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat., 123, 725–731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.