Abstract
In oncolytic virotherapy, the ability of the virus to activate the immune system is a key attribute with regard to long-term antitumor effects. Vaccinia viruses bear one of the strongest oncolytic activities among all oncolytic viruses. However, its capacity for stimulation of antitumor immunity is not optimal, mainly due to its immunosuppressive nature. To overcome this problem, we developed an oncolytic VV that expresses intracellular pattern recognition receptor DNA-dependent activator of IFN-regulatory factors (DAI) to boost the innate immune system and to activate adaptive immune cells in the tumor. We showed that infection with DAI-expressing VV increases expression of several genes related to important immunological pathways. Treatment with DAI-armed VV resulted in significant reduction in the size of syngeneic melanoma tumors in mice. When the mice were rechallenged with the same tumor, DAI-VV-treated mice completely rejected growth of the new tumor, which indicates immunity established against the tumor. We also showed enhanced control of growth of human melanoma tumors and elevated levels of human T-cells in DAI-VV-treated mice humanized with human peripheral blood mononuclear cells. We conclude that expression of DAI by an oncolytic VV is a promising way to amplify the vaccine potency of an oncolytic vaccinia virus to trigger the innate—and eventually the long-lasting adaptive immunity against cancer.
Introduction
Vaccinia viruses (VV) have been widely used as vaccines1 and as gene therapy vectors2 for decades and are thus relatively well characterized and bear a solid safety profile.3 Vaccinia virus has also been used as an oncolytic agent in many preclinical and clinical studies3–5 thanks to its fast replication cycle3 and high tropism to cancer tissues.6
In oncolytic virotherapy, the ability of the virus to activate the immune system against tumors is nowadays generally understood to be a key mechanism in full eradication of cancer and for long-term antitumor effects.7,8 In fact, the best results so far in VV-based oncolytic virotherapy have been achieved by using viruses that express immunostimulatory transgenes.4,5,9–11
Vaccinia viruses are immunosuppressive viruses by nature encoding many proteins to evade host immune responses.12 To overcome this problem, we developed an oncolytic VV that expresses intracellular pattern recognition receptor DNA-dependent activator of IFN-regulatory factors (DAI) to obtain a self-immuno-boosting system to activate immune responses in the tumor.
DAI, also known as ZBP1 (Z-DNA-binding protein 1) or DLM-1 is a cytosolic dsDNA sensor that is a potent activator of innate immune responses.13 Activation of DAI increases the production of type I interferons and other cytokines that attract immune cells to the site of activation (infection).14 Lladser et al.14 showed that a DAI-expressing plasmid can be efficiently used as a genetic cancer vaccine inducing memory and effector tumor-specific T-cells. Similarly, myeloid differentiation factor 88 (MyD88), a ubiquitous Toll-like receptor adaptor molecule, has been used as a genetic adjuvant to harness intrinsic immune-stimulating properties of a DNA vaccine15 and a MyD88-encoding adenovirus was shown to potentiate local and systemic antitumor immunity.16
Since vaccinia virus is a dsDNA virus whose entire replication cycle takes place in the cytosol,12 we hypothesized that DAI has a role in innate immune sensing of VV. Moreover, we thought that expression of DAI by a rapidly replicating viral backbone would boost innate immune activation and counteract the immunosuppressiveness of the virus12 and the tumor microenvironment,17 resulting in long-lasting adaptive immunity towards cancer.
Results
DAI-armed oncolytic vaccinia virus displays unaltered oncolytic activity
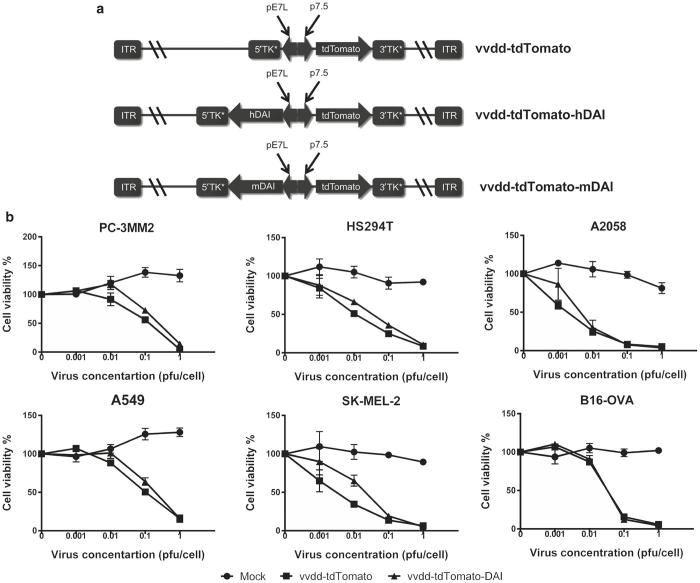
To improve the immunogenicity of oncolytic vaccinia virus, we generated a virus expressing DAI. Separate viruses coding for human (vvdd-tdTomato-hDAI) and mouse DAI (vvdd-tdTomato-mDAI) were constructed (Figure 1a). We used a Western Reserve vaccinia virus strain with two deletions, one in the thymidine kinase gene and another in the vaccinia growth factor gene.9,18,19 These gene deletions render the virus cancer-specific since their expression is necessary for replication in normal cells but not in cancer cells where the corresponding pathways are already active.20,21 Both versions of DAI-expressing viruses and the control virus vvdd-tdTomato express a tdTomato fluorophore for in vitro and in vivo imaging purposes and biosafety concerns.9,22
Figure 1.
Characteristics of the double deleted oncolytic vaccinia virus expressing DAI. (a) Schematic figures of the virus constructs. Two novel armed vaccinia viruses were generated expressing human (vvdd-tdTomato-hDAI) or murine DAI (vvdd-tdTomato-mDAI). Both viruses express also tdTomato fluorescent protein as a reporter gene and are deleted in thymidine kinase and vaccinia growth factor genes. (b) DAI-expression does not hinder virus replication and cell killing efficacy. MTS cell viability assays were performed in several cell lines 5 days postinfection. Virus expressing human DAI was used for human cell lines and mDAI-virus for the mouse cell line B16-OVA.
vvdd-tdTomato-hDAI, vvdd-tdTomato-mDAI, and their relative controls were tested for cancer cell killing potency in several tumor cell lines. The expression of DAI was shown not to hinder cancer cell killing efficacy compared to the control virus without the transgene in all tested cell lines (Figure 1b).
Infection with a DAI-armed virus promotes expression of genes involved in activation of the immune system
Next we wanted to study the overall impact of the DAI-expressing virus on the transcriptome of cancer cells and immune cells. To this end, we performed a wide-genome expression profile for two different cell types, human melanoma cells (HS294T), and human monocyte cells (THP-1) that were infected with the hDAI-virus or a control virus that does not express DAI.
Differentially expressed genes were plotted in a two-dimensional chart to assess the rigorousness of our analysis; this representation shows the consistency of the effects promoted by DAI-armed virus in the different cell lines (Supplementary Figure S1). Then we used the BACA representing tools23 to concisely report the biological processes associated with the significantly downregulated and upregulated genes. The BACA plot analysis revealed a similar effect of the viruses in the melanoma cell line HS294T but a very different one in the monocyte cell line THP-1 (Figure 2). This indicates that DAI-VV can have a very different effect depending on the nature of the cell line. In the THP-1 cell line, there were a lot more upregulated genes than in the melanoma cell line. To deepen our understanding on what those differently expressed genes were and what is the effect that the DAI-armed virus has on the immunological processes, we performed ingenuity pathway analysis on differentially expressed genes between vvdd-tdTomato-hDAI and vvdd-tdTomato in THP-1 cells. Interestingly, the most upregulated networks encompassed pathways related to immune system activation such as dendritic cell maturation, communication between innate and adaptive immune systems, and role of pattern recognition receptors in recognition of viruses (Supplementary Figure S2).
Figure 2.

Infection of THP-1 monocytes with DAI-armed vaccinia virus results in altered expression pattern of genes involved in biological processes. HS294T melanoma cells and THP-1 monocytes were infected with vvdd-tdTomato-hDAI, vvdd-tdTomato, or phosphate-buffered saline, and the wide genome gene expression profile was analyzed. A BACA clustering representation of the differentially expressed biological processes induced by vvdd-tdTomato and vvdd-tdTomato-hDAI is shown in the two different cell lines. The upregulated genes are marked with red dots and down-regulated genes with green dots.
As expected, we also found about sevenfold upregulation of DAI in both cell lines (7.4 and 7.2 in THP1 and HS294T, respectively) treated with DAI-expressing virus compared with the unarmed VV.
In conclusion, we showed that infection with vvdd-tdTomato-hDAI virus promotes upregulation of several genes in monocyte cells and those genes are involved in important immunological pathways.
DAI expression by oncolytic vaccinia virus enhances cancer eradication in vivo by inducing antitumor T-cell responses in a syngeneic melanoma mouse model
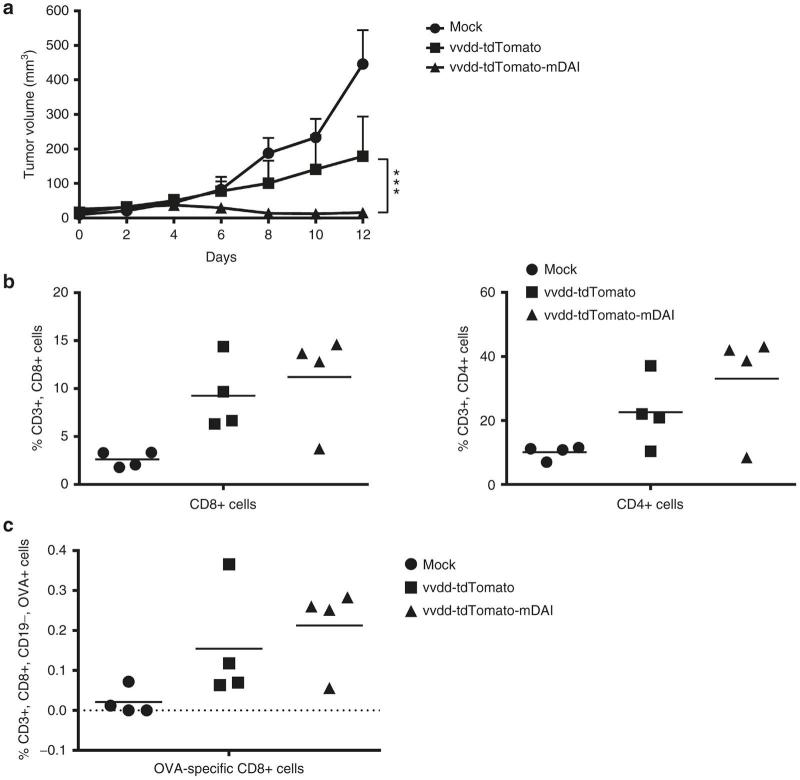
The immunogenic potency of the DAI-virus was tested in vivo using an aggressive syngeneic melanoma model. Subcutaneous B16-OVA tumors were grown in C57BL/6 mice (12 mice/group, one tumor/mouse). The mice were treated intratumorally on days 0 and 2. We observed that tumor growth was significantly (P < 0.001) reduced in the group of mice treated with vvdd-tdTomato-mDAI when compared to the group treated with vvdd-tdTomato (Figure 3a), and in fact most of the tumors treated with the DAI-virus were completely cured. Four mice per group were euthanized on day 12 after the first virus treatments and tumors were collected and prepared for flow cytometric analysis of immune cells. We found that enhanced cancer-eradicating efficacy of the vvdd-tdTomato-mDAI was associated with increased (nonsignificant) infiltration of CD8+, CD4+ (Figure 3b) and more importantly tumor-specific (OVA-specific) CD8+ T-cells into the tumors (Figure 3c).
Figure 3.
In vivo efficacy and antitumor immunity elicited after vvdd-tdTomato-mDAI treatment in an immunocompetent syngeneic melanoma model. B16-OVA melanoma tumors were established by injecting 1 × 105 B16-OVA cells subcutaneously into other flank of C57BL/6J mice (N = 12 per group). Established tumors (one tumor per mouse, ~4 × 4 mm in diameter) were injected intratumorally with viruses in volume of 50 μl on day 0 with 3 × 106 pfu/tumor and on day 2 with 1 × 106 pfu/tumor. Mock tumors were injected with phosphate-buffered saline only. (a) Tumor growth was measured over time (day 0 = day when the first virus treatment was given). (b) General CD8+ and CD4+ T-cells and (c) tumor-specific (OVA peptide-specific) CD8+ T-cells were analyzed by flow cytometry from the tumors of treated mice 12 after the first virus treatment.
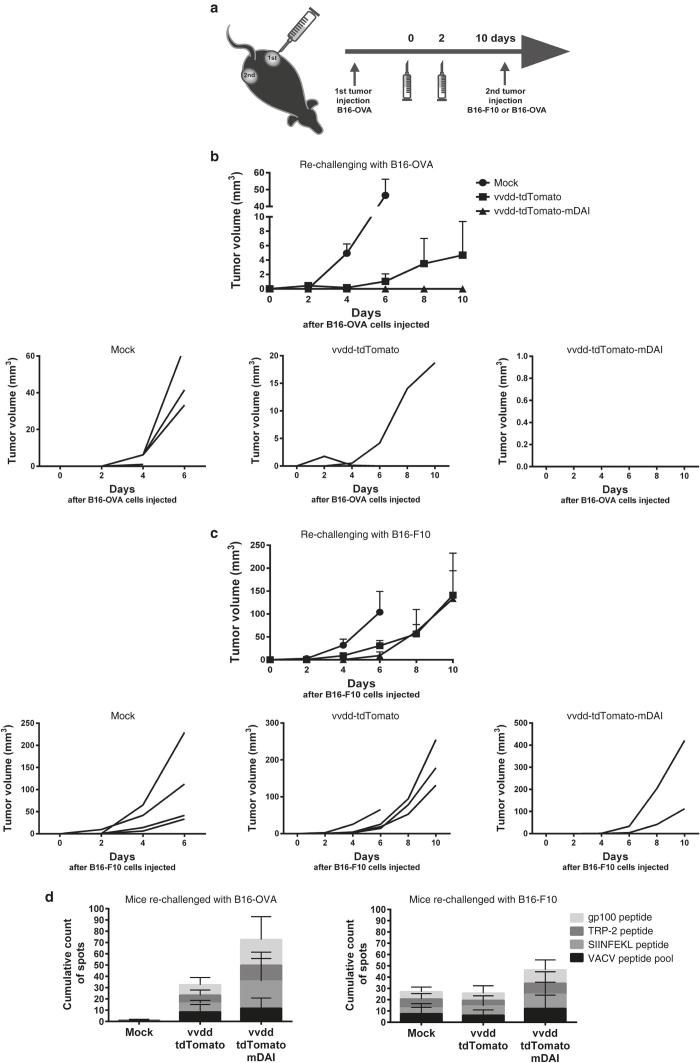
To further study the role of antitumor immunity induced by vvdd-tdTomato-mDAI treatments, we performed a rechallenging experiment using the same mouse melanoma cell line, B16. Ten days after the first treatment with viruses, four mice per group received another injection of tumor cells from the same (B16-OVA) or different cell line without ovalbumin expression (B16-F10) (Figure 4a). This allowed assessment whether the immune response was only directed to OVA or also towards other tumor-specific antigens. The second tumor was not treated, only tumor growth was followed over time. The mice that had been treated with vvdd-tdTomato-mDAI virus completely rejected the growth of the second B16-OVA tumor (Figure 4b), whereas the nontreated mice and the mice treated with the control virus did not. All mice treated with the vvdd-tdTomato control virus or phosphate-buffered saline (PBS), which received injection of non-OVA-expressing tumor cells, allowed growth of a second tumor. In contrast, only half of the mice treated with vvdd-tdTomato-mDAI grew a B16-F10 tumor (Figure 4c).
Figure 4.
DAI-virus boosts tumor-specific immunity without boosting anti-viral immunity in mice bearing syngeneic melanoma tumors. (a) Schematic of the experiment; B16-OVA melanoma tumors were established by injecting 1 × 105 B16-OVA cells subcutaneously into other flank of C57BL/6J mice (N = 12 per group). Established tumors (one tumor per mouse, ~4 × 4 mm in diameter) were injected intratumorally with viruses in volume of 50 μl on day 0 with 3 × 106 pfu/tumor and on day 2 with 1 × 106 pfu/tumor. Mock tumors were injected with phosphate-buffered saline only. The mice were re-challenged with another tumor of the same background: the mice received either B16-OVA cells or B16-F10 cells 3 × 105 cells/tumor, 10 days after the first virus treatment. The second tumor was not treated, only tumor growth was followed. (b) Four mice per group were rechallenged with the same tumor cell line (B16-OVA) and (c) four mice per group were rechallenged with a cell line with the same origin but which does not express ovalbumin (B16-F10). The mean tumor growth (above) and the growth of individual tumors (below) are presented after the second tumor implantation (day 0 = day when second tumor was injected). IFN-γ producing T-cells were assessed from spleens 22 days after virus treatments by ELISpot. Splenocytes were cultured with tumor-associated or virus-associated peptides for 36 hours and spots were counted. (d) Cumulative spot counts of the ELISpot wells of B16-OVA rechallenged mice (left) and B16-F10 rechallenged mice (right).
At the end of the experiment, 22 days after the first virus treatments and 10 days after tumor cell challenge, the spleens of the mice were collected and antitumor and antiviral T-cells were analyzed with ELISpot (Figure 4d and Supplementary Figure S3). Interestingly, the anti-viral T-cell responses were relatively similar in both vvdd-tdTomato and vvdd-tdTomato-mDAI treated mice, but the antitumor responses were increased in DAI-virus-treated mice (no statistical significance, though), especially when the mice were rechallenged with exactly the same tumor cell line, B16-OVA. Intriguingly, we did not only see T-cell responses against the immunogenic OVA peptide, but also against more clinically relevant melanoma antigens TRP-2 and gp100.
These findings indicate that vvdd-tdTomato-mDAI is able to induce a tumor-specific immune response.
vvdd-tdTomato-hDAI controls growth of human tumors in humanized mice and increases infiltration of human immune cells into tumors
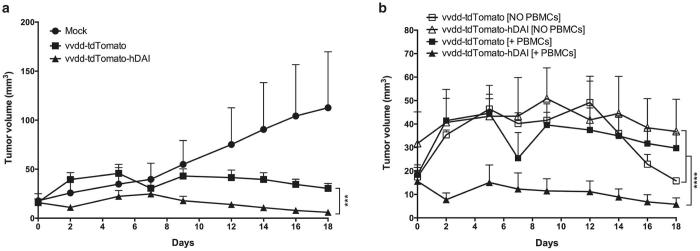
To even better illustrate the immunogenicity and to demonstrate the translational potency of our virus, we performed studies in humanized mice bearing human tumors and a human immune system. Treatments with vvdd-tdTomato-hDAI resulted in significant control of tumor growth (P < 0.001) compared to treatments with the control virus in presence of the human immune system (Figure 5a). Interestingly, the enhanced antitumor effect of the DAI-virus was seen only in mice that received human peripheral blood mononuclear cells (PBMCs), while the effect was absent in mice without white blood cells (Figure 5b), which indicates the important role of immune system activation as a mechanism-of-action of the DAI-virus.
Figure 5.
In vivo efficacy and immunogenicity of the DAI-virus in a peripheral blood mononuclear cell (PBMC) humanized melanoma mouse model. Human melanoma HS294T cells were injected subcutaneously into both flanks of the NSG mice, 5 × 106 cells/flank. When tumors reached injectable size, human PBMCs were injected intravenously to the tail vein in 200 μl of phosphate-buffered saline, 2 × 107 cells/ mouse. Two days later virus treatments were started. Tumors were treated on days 0 and 6 with 1 × 106 pfu/ tumor in volume of 50 μl, mock mice were treated with phosphate-buffered saline only. Parts of the mice were left without PBMCs to have a study control lacking the immune system. (a) Growth of human melanoma HS294T tumors in PBMC humanized NSG mice after virus treatments on days 0 and 6 (N of tumors/group=8). Day 0 = day when the first virus treatment was given. (b) Comparison of the tumor growth in mice humanized with PBMCs (N of tumors/group = 8) and mice that did not receive PBMCs (N of tumors/group = 4).
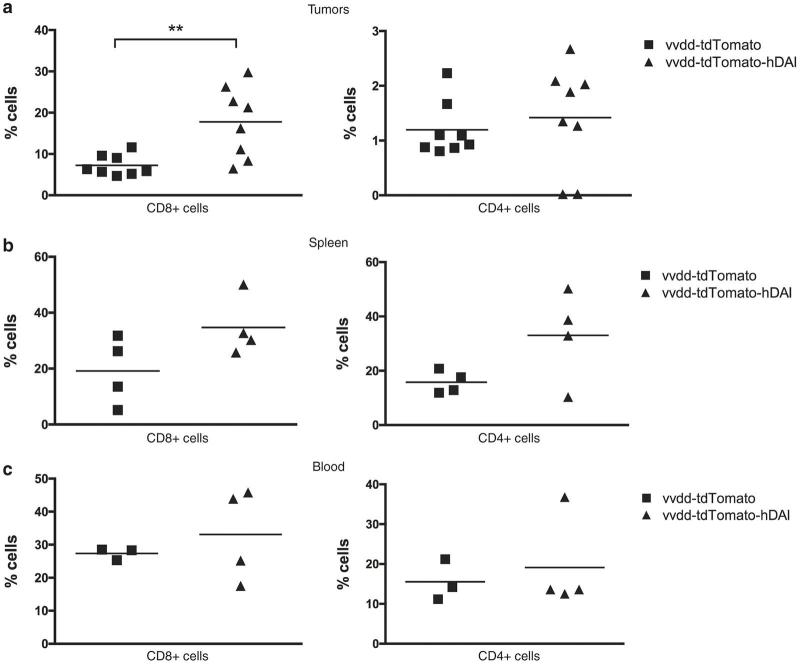
After the experiment tumors, spleens and blood of the mice were collected for flow cytometry analyses. We showed a significant (P < 0.01) increase in the number of human CD8+ T-cells in the tumors of DAI-virus-treated mice compared to mice treated with the control virus (Figure 6a). There was a trend (not significant) toward an increased amount of CD8+ and CD4+ T-cells also in the spleens and blood of DAI-virus-treated mice (Figure 6b,c).
Figure 6.
Oncolytic vaccinia virus expressing human DAI induces infiltration of human T-cells into infected human melanoma tumors in PBMC-humanized mice. Human CD8+ and CD4+ T-cells were analyzed by flow cytometry from (a) tumors, (b) spleens, and (c) blood of mice 18 days after the first treatment.
Discussion
The most promising attempts at oncolytic virotherapy have encompassed use of viruses expressing cytokines or other immunostimulatory molecules to turn on the immune system against the tumor.24,25 However, the efficacy of the viruses to boost the immune system and to elicit a tumor-specific immune response remains modest in preclinical models and cancer patients.24,26 This may be due to the use of only one single immunostimulatory molecule, which may in some cases have limited targets in the tumor site due to downregulation of many receptors.16 By stimulation of a receptor that regulates and induces production of a number of different immunostimulatory cytokines and chemokines would generate a more robust immunological response.12,14 Takaoka et al.13 reported that pattern recognition receptor DAI is a cytosolic sensor for DNA that can induce robust innate immune responses once activated. Few years later, Lladser and colleagues showed DAI to be an efficient genetic adjuvant for DNA vaccines, which can also induce antitumor T-cell responses.14 These intriguing results encouraged us to use DAI in order to potentiate the vaccine effect of an oncolytic vaccinia virus. Therefore, we generated a novel oncolytic vaccine for cancer immunovirotherapy; our approach uses an oncolytic vaccinia virus platform that rapidly replicates specifically in tumor cells and expresses DAI at the infection site. Since the virus is expressing a receptor that senses itself (VV DNA), we have generated a self-amplifying system that can boost viral immunogenicity and innate immune responses at the tumor site regardless of cellular expression of DAI.
In this report, we demonstrate that the oncolytic potency of our new virus construct is not hindered compared to an oncolytic VV control and thus serves as a rapid and effective platform for the immunoboosting molecule DAI. We observed that our DAI-expressing oncolytic VV can activate many relevant pathways including those belonging to dendritic cell maturation and signaling between innate and adaptive immune systems. This is intriguing, since it has been shown by us and others that virus-induced oncolysis releases tumor antigens to the microenvironment for detection by dendritic cells, which eventually can result in activation and maturation of adaptive, tumor-specific immunity.27,28 We showed in this study that the DAI-armed vaccinia virus is able to induce T-cell infiltration into treated melanoma tumors of C57BL/6 mice accompanied with significantly reduced tumor growth. Similarly, activation of the adaptive immune system and antitumor responses were also shown by Lladser et al.14 with a DAI-encoding plasmid. In their approach, they used a coadministration of plasmid expressing tumor antigens to stimulate an antigen-specific response. In our case, probably because oncolysis is able to release epitopes from tumor cells, and the virus platform is more immunogenic, we observed antigen-specific immune responses without the need of additional administration of the antigen. Our rechallenging experiments also showed that we were able to obtain a systemic antitumor response that could control the growth of distant nontreated tumors.
It is known that as cancer progresses, it develops several mechanisms to evade the host immune system.17,29,30 These tumor immune escape mechanisms constitute the immunosuppressive tumor microenvironment that creates challenges for immunotherapies. Likewise, vaccinia viruses have evolved to resist clearance by encoding many genes dedicated to protection of the virus from the host immune system.12 Our results together with other studies done with DAI14 demonstrate that by using DAI as an adjuvant, we are actually able to convert the tumor microenvironment from immunosuppressive into a more favorable variant compatible with antitumor immune responses. In fact, we showed that antitumor T-cell responses are more prominent than antiviral T-cell responses. Importantly, boosting recognition of viral DNA by the expression of DAI, we can also overcome the natural immune evasion mechanisms of the vaccinia virus. This is an intriguing finding because Harrington et al.31 have shown that anti-VV responses are remarkably stronger than the responses against other epitopes in vaccinated mice.
In conclusion, our results suggest that a DAI-armed vaccinia virus as a self-sensing and immuno-boosting system can alter the immunosuppressive tumor microenvironment and would be a way to improve oncolytic and other vaccination strategies.
Materials and Methods
Cell lines
Human lung cancer cell line A549 (ATCC no. CCL-185), human melanoma cell lines HS294T (ATCC no. HTB-140), A2058 (ATCC no. CRL-11147), SK-MEL-2 (ATCC no. HTB-68), human monocyte cell line THP-1 (ATCC no. TIB-202), monkey kidney cell line CV-1 (ATCC no. CCL-70), and mouse melanoma cell line B16-F10 (ATCC no. CRL-6475) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). A highly metastatic human prostate cancer cell line PC-3MM2 was generously obtained from Isaiah J. Fidler, MD Anderson Cancer Center (Houston, TX). The human bladder cancer EJ cells were provided and authenticated by A.G. Eliopoulos, University of Crete Medical School and Laboratory of Cancer Biology (Heraklion, Crete, Greece). B16-OVA, a mouse melanoma cell line expressing chicken ovalbumin, was kindly provided by Professor Richard Vile at Mayo Clinic (Rochester, MN). Jurkat cells, immortalized human T-lymphocytes, were kindly provided by Dr. Pentti Tienari, Program of Molecular Neurology, University of Helsinki (Helsinki, Finland). All cell lines were cultured under recommended conditions.
Viruses
Oncolytic Western Reserve strain vaccinia virus selective for epidermal growth factor receptor pathway mutations (vaccinia growth factor) and tumor-associated hypermetabolism (thymidine kinase) was armed with either human or murine DAI and a tdTomato fluorophore. The control virus vvdd-tdTomato has been described elsewhere.32 For generation of the vvdd-tdTomato-mDAI and vvdd-tdTomato-hDAI, the mDAI and hDAI cDNA was inserted under the control of the pE/L promoter of pSC65-tdTomato32,33 to create pSC65-tdTomato-mDAI and pSC65-tdTomato-hDAI. These shuttle plasmids were cotransfected with vvdd-luc in CV-1 cells. The viruses were propagated on A549 cells and purified by general method described in refs. 18,34. The infectious viral particle concentration was determined by standard crystal violet staining assay on A549 cells.
In vitro tumor cell killing assay
Tumor cells were seeded at 1.0 × 104 cells per well on 96-well plates. On the next day cells were infected for 1 hour at 37 °C in 2% FCS containing medium and then incubated in 5% medium for 5 days. Cell viability was determined by MTS according to the manufacturer’s protocol (Cell Titer 96 AQueous One Solution Cell Proliferation Assay; Promega, Nacka, Sweden).
Whole-genome gene expression profiling
Human HS294T and THP-1 cells were treated with vvdd-tdTomato-hDAI, vvdd-tdTomato, or PBS as a noninfected control. Total RNA was purified from tumor cells after infection with 0.1 pfu/cell of virus for 16 hours. RNeasy Plus Mini kit (Qiagen, Venlo, NL) was used according to manufacturer’s instructions. Independent pools of two RNA samples each (total of 600 ng) were labeled using T7 RNA polymerase amplification method (Low Input Quick Amp Labeling Kit, Agilent Technologies, Santa Clara, CA), according to the instructions of the manufacturer. cRNAs were then labeled with Cy3 and Cy5 dyes (Agilent Technologies) and hybridized to the Agilent 2-color 60-mer oligo arrays (Agilent SurePrint G3 Human GE 8x60K). The slides were washed and scanned with Agilent Microarray Scanner G2505C (Agilent Technologies) and the raw intensity values were obtained with the Feature Extraction software, version 11.0.1.1 (Agilent Technologies). Raw data was quality checked according to the Agilent standard procedures. Data processing and analysis of differential gene expression were done in R. First, the probe profile expression data were normalized using quantile normalization and corrected for batch processing effects using ComBat. Next, after mapping from probsets to Ensemble gene IDs, the differentially expressed genes (DE genes) between pre- and post-treatment samples were identified using limma. For the analysis using the limma package, genes were defined as being differentially expressed after satisfying a minimum fold-change of ±1.5 and a maximum Benjamini-Hochberg adjusted P value of 0.01. Finally, functional categorization of DE genes was performed using a novel R-based package namely BACA. It queries the DAVID knowledgebase and builds a chart showing multiple enrichment analysis results across different conditions/treatments. Each annotation in the chart is represented as a circle (or bubble) that has a size, indicating how many genes in a list of DE genes are associated with it, and a color indicating whether the genes are down- (default color is green) or up- (default color is red) regulated.
Animal experiments
All animal experiments were reviewed and approved by the Experimental Animal Committee of the University of Helsinki and the Provincial Government of Southern Finland. Health status of the mice was frequently monitored and as soon as signs of pain or distress were evident they were euthanized.
For the study of the syngeneic melanoma model, mice were obtained from Harlan Laboratories (Indianapolis, IN) at 8 weeks of age and quarantined for 2 weeks before the study. B16-OVA melanoma tumors were established by injecting 1 × 105 B16-OVA cells subcutaneously into other flank of 10-week-old female C57BL/6J mice (N = 12 per group). Established tumors (one tumor per mouse, ~4 × 4 mm in diameter) were injected intratumorally with viruses in volume of 50 μl on day 0 with 3 × 106 pfu/tumor and on day 2 with 1 × 106 pfu/tumor. Mock tumors were injected with PBS only. In the rechallenging experiment, mice received another tumor, either B16-OVA cells or B16-F10 cells 3 × 105 cells/tumor, 10 days after the first virus treatment.
For the study with PBMC humanized mice, the highly immunodeficient NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were received from The Jackson Laboratory (Bar Harbor, ME) at 7 weeks of age and quarantined for 3 weeks before the study. Human melanoma HS294T cells were injected subcutaneously into both flanks of the NSG mice, 5 × 106 cells/ flank. When tumors reached injectable size, human PBMCs were injected intravenously to the tail vein in 200 μl of PBS, 2 × 107 cells/ mouse (protocol modified from35). Two days later virus treatments were started. Tumors were treated on days 0 and 6 with 1 × 106 pfu/ tumor in volume of 50 μl; mock mice were treated with PBS only. Parts of the mice were left without PBMCs to have a study control lacking the immune system.
The formula (length × (width)2 × 0.5) was used to calculate the tumor volumes. Mice were euthanized when a tumor reached an average diameter of 15 mm.
Isolation of PBMCs for the humanized mice experiment
Human PBMCs were isolated from a buffy coat received from Finnish Red Cross Blood service. Buffy coat blood was diluted 1:1 with PBS and PBMCs were isolated by Ficoll-Paque PLUS (GE Healthcare, Little Chalfont, UK) gradient centrifugation according to manufacturer´s instructions.
HLA typing of tumor cell lines and PBMCs
Genomic DNA was extracted from the HS294T tumor cell line and human PBMCs for HLA typing. Qiagen DNeasy blood and tissue kit (Qiagen, Venlo, NL) was used to extract DNA by manufacturer´s instructions and the HLA genotyping and analysis was carried out in an EFI (European Federation for Immunogenetics) accredited HLA Laboratory. The genotyping of HLA-A, -B, and -C genes were performed using sequence specific primers (Olerup SSP HLA-A-B-C Combi Tray, Applied Biosciences, Stockholm, SE). The reactions were performed according to manufacturers’ instructions giving at least four-digit resolution (for example, HLA-A*01:01). PCR reactions from agarose-gel were evaluated manually and the alleles were called with SCORE software. The HLA alleles were assessed using HLA nomenclature release 3.5.0 (IMGT/HLA database) and carefully interpreted by two persons.
Immune cell analysis
Tumors, spleens, and lymph nodes of treated mice were collected, smashed through a 70 μm cell strainer and cultured over night in 10% RPMI-1640 medium. Single cell suspensions were stained with fluorochrome-conjugated monoclonal antibodies and analyzed using a FACSAria flow cytometer (BD Biosciences) or Gallios flow cytometer (Beckman Coulter, Brea, CA) and FlowJo 7.6 software (TreeStar, Ashland, OR). Anti-mouse FITC-conjugated CD8 (Proimmune), PE-Cy7-conjugated CD19 (BioLegend), APC-conjugated CD4 (BioLegend), PE-Cy7-conjugated CD3e (BioLegend), PE-conjugated IFNgamma (BioLegend) and APC-conjugated SIINFEKL MHC-I pentamer (Proimmune, Oxford, UK) were used for immunocompetent mouse experiment. Anti-human FITC-conjugated CD45RA (BioLegend), PE-conjugated CD197 (BioLegend), PerCP-conjugated CD8 (BioLegend), PE-TexasRed-conjugated CD4 (LifeTechnologies), FITC-conjugated IFNgamma (BioLegend), PE-conjugated IL-6 (BioLegend), and APC-conjugated TNFα (BioLegend) were used for humanized mouse experiment.
IFN-γ ELISpot assays
The amount of activated, interferon-γ-producing T-cells was studied from spleens of treated mice using a Mouse IFN-γ ELISpotPLUS (ALP) kit (Mabtech, 3321-4APT) according to manufacturer´s instructions. Splenocytes were pulsed with PepMix VACV (vaccinia virus) peptide pool (MVA093L, JPT), ovalbumin peptide SIINFEKL (P93, Proimmune), gp100 peptide KVPRNQDWL (P1333, Proimmune) or TRP-2 peptide SVYDFFVWL (P185, Proimmune) in concentration of 50 ng/well. Concanavalin A (Sigma Aldrich) was used as positive control (500 ng/well). ELISpot plates were read and evaluated by ZellNet Consulting (Fort Lee, NJ).
Statistical analysis
Statistical significance was determined by unpaired, two-tailed Student’s t-test using GraphPad Prism 6 (GraphPad Software, La Jolla, CA). Differences in tumor growth were analyzed using two-tailed Student’s t-test (for immunocompetent mouse experiments) or with the nonparametric Mann–Whitney test (for immunocompromised mouse experiments). Results are presented as mean ± SD or as mean ± SEM for tumor growth. All P values were two-sided and considered statistically significant when P ≤ 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****).
Acknowledgments
The authors thank Sirkka-Liisa Holm, Åse Karttunen, Saija Kaikkonen, and Sirpa Hyttinen for technical support. The authors also thank Marja-Liisa Lokki and the HLA Laboratory of Haartman Institute for the HLA-typing. A.H. is Jane and Aatos Erkko Professor of Oncology at the University of Helsinki. This work was supported by University of Helsinki, Marie Curie FP7-PEOPLE-IRG-2008, K. Albin Johansson Foundation, Finnish Cancer Organizations, HUCH Research Funds (EVO), Sigrid Juselius Foundation, Biocentrum Helsinki and Biocenter Finland. M.H. was supported by Finnish Cultural Foundation, Biomedicum Helsinki Foundation, Emil Aaltonen Foundation, Finnish Cancer Foundations and Maud Kuistila Memorial Foundation. A.V. was supported by CREME grant. L.K. was supported by Marie Curie Innovative Training Network (ITN) grant ADVance (FP7-290002).
Footnotes
A.H. is employee and shareholder in TILT Biotherapeutics Ltd. and shareholder in Oncos Therapeutics, Ltd. The other authors declare no potential conflict of interest.
References
- Sánchez-Sampedro, L, Perdiguero, B, Mejías-Pérez, E, García-Arriaza, J, Di Pilato, M and Esteban, M (2015). The evolution of poxvirus vaccines. Viruses 7: 1726–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, B and Flexner, C (1987). Vaccinia virus expression vectors. Annu Rev Immunol 5: 305–324. [DOI] [PubMed] [Google Scholar]
- Zeh, HJ and Bartlett, DL (2002). Development of a replication-selective, oncolytic poxvirus for the treatment of human cancers. Cancer Gene Ther 9: 1001–1012. [DOI] [PubMed] [Google Scholar]
- Breitbach, CJ, Thorne, SH, Bell, JC and Kirn, DH (2012). Targeted and armed oncolytic poxviruses for cancer: the lead example of JX-594. Curr Pharm Biotechnol 13: 1768–1772. [DOI] [PubMed] [Google Scholar]
- Guse, K, Cerullo, V and Hemminki, A (2011). Oncolytic vaccinia virus for the treatment of cancer. Expert Opin Biol Ther 11: 595–608. [DOI] [PubMed] [Google Scholar]
- Peplinski, GR, Tsung, AK, Casey, MJ, Meko, JB, Fredrickson, TN, Buller, RM et al. (1996). In vivo murine tumor gene delivery and expression by systemic recombinant vaccinia virus encoding interleukin-1beta. Cancer J Sci Am 2: 21–27. [PubMed] [Google Scholar]
- Prestwich, RJ, Harrington, KJ, Pandha, HS, Vile, RG, Melcher, AA and Errington, F (2008). Oncolytic viruses: a novel form of immunotherapy. Expert Rev Anticancer Ther 8: 1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, AW, Senzer, N, Cerullo, V, Templeton, NS, Hemminki, A and Nemunaitis, J (2011). Oncolytic viruses for induction of anti-tumor immunity. Curr Pharm Biotechnol 13: 1750–1760. [DOI] [PubMed] [Google Scholar]
- Parviainen, S, Ahonen, M, Diaconu, I, Hirvinen, M, Karttunen, Å, Vähä-Koskela, M et al. (2014). CD40 ligand and tdTomato-armed vaccinia virus for induction of antitumor immune response and tumor imaging. Gene Ther 21: 195–204. [DOI] [PubMed] [Google Scholar]
- Autio, K, Knuuttila, A, Kipar, A, Ahonen, M, Parviainen, S, Diaconu, I et al. (2014). Anti-tumour activity of oncolytic Western Reserve vaccinia viruses in canine tumour cell lines, xenografts, and fresh tumour biopsies. Vet Comp Oncol (epub ahead of print). doi: 111/vco.12119. [DOI] [PubMed]
- Chan, WM and McFadden, G (2014). Oncolytic Poxviruses. Annu Rev Virol 1: 119–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, GL, Benfield, CT, Maluquer de Motes, C, Mazzon, M, Ember, SW, Ferguson, BJ et al. (2013). Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol 94(Pt 11): 2367–2392. [DOI] [PubMed] [Google Scholar]
- Takaoka, A, Wang, Z, Choi, MK, Yanai, H, Negishi, H, Ban, T et al. (2007). DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505. [DOI] [PubMed] [Google Scholar]
- Lladser, A, Mougiakakos, D, Tufvesson, H, Ligtenberg, MA, Quest, AF, Kiessling, R et al. (2011). DAI (DLM-1/ZBP1) as a genetic adjuvant for DNA vaccines that promotes effective antitumor CTL immunity. Mol Ther 19: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullas, PT, Desai, A and Madhusudana, SN (2014). Immunogenicity and efficacy of a plasmid DNA rabies vaccine incorporating Myd88 as a genetic adjuvant. Clin Exp Vaccine Res 3: 202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, ZC, Osada, T, Glass, O, Yang, XY, Lei, GJ, Lyerly, HK et al. (2010). Ligand-independent toll-like receptor signals generated by ectopic overexpression of MyD88 generate local and systemic antitumor immunity. Cancer Res 70: 7209–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaud, C, John, LB, Westwood, JA, Darcy, PK and Kershaw, MH (2013). Immune modulation of the tumor microenvironment for enhancing cancer immunotherapy. Oncoimmunology 2: e25961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCart, JA, Ward, JM, Lee, J, Hu, Y, Alexander, HR, Libutti, SK et al. (2001). Systemic cancer therapy with a tumor-selective vaccinia virus mutant lacking thymidine kinase and vaccinia growth factor genes. Cancer Res 61: 8751–8757. [PubMed] [Google Scholar]
- Guse, K, Sloniecka, M, Diaconu, I, Ottolino-Perry, K, Tang, N, Ng, C et al. (2010). Antiangiogenic arming of an oncolytic vaccinia virus enhances antitumor efficacy in renal cell cancer models. J Virol 84: 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne, SH, Hwang, TH, O’Gorman, WE, Bartlett, DL, Sei, S, Kanji, F et al. (2007). Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J Clin Invest 117: 3350–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller, RM, Chakrabarti, S, Cooper, JA, Twardzik, DR and Moss, B (1988). Deletion of the vaccinia virus growth factor gene reduces virus virulence. J Virol 62: 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, NC, Lin, MZ, McKeown, MR, Steinbach, PA, Hazelwood, KL, Davidson, MW et al. (2008). Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortino V, Alenius H, Greco D (2015). BACA: bubble chArt to compare annotations. BMC Bioinformatics;16:37. [DOI] [PMC free article] [PubMed]
- Pol, J, Bloy, N, Obrist, F, Eggermont, A, Galon, J, Cremer, I et al. (2014). Trial Watch:: Oncolytic viruses for cancer therapy. Oncoimmunology 3: e28694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, SJ, Peng, KW and Bell, JC (2012). Oncolytic virotherapy. Nat Biotechnol 30: 658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähä-Koskela, MJ, Heikkilä, JE and Hinkkanen, AE (2007). Oncolytic viruses in cancer therapy. Cancer Lett 254: 178–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, CG and Fraser, NW (2003). Requirement of an integrated immune response for successful neuroattenuated HSV-1 therapy in an intracranial metastatic melanoma model. Mol Ther 7: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamori, M, Fu, X, Rousseau, R, Chen, SY and Zhang, X (2004). Destruction of nonimmunogenic mammary tumor cells by a fusogenic oncolytic herpes simplex virus induces potent antitumor immunity. Mol Ther 9: 658–665. [DOI] [PubMed] [Google Scholar]
- Schreiber, RD, Old, LJ and Smyth, MJ (2011). Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331: 1565–1570. [DOI] [PubMed] [Google Scholar]
- Igney, FH and Krammer, PH (2002). Immune escape of tumors: apoptosis resistance and tumor counterattack. J Leukoc Biol 71: 907–920. [PubMed] [Google Scholar]
- Harrington, LE, Most Rv, Rv, Whitton, JL and Ahmed, R (2002). Recombinant vaccinia virus-induced T-cell immunity: quantitation of the response to the virus vector and the foreign epitope. J Virol 76: 3329–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviainen, S, Ahonen, M, Diaconu, I, Kipar, A, Siurala, M, Vähä-Koskela, M et al. (2015). GMCSF-armed vaccinia virus induces an antitumor immune response. Int J Cancer 136: 1065–1072. [DOI] [PubMed] [Google Scholar]
- Chakrabarti, S, Sisler, JR and Moss, B (1997). Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques 23: 1094–1097. [DOI] [PubMed] [Google Scholar]
- Earl, PL, Cooper, N, Wyatt, LS, Moss, B and Carroll, MW (2001). Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Mol Biol. Chapter 16:Unit16.16. [DOI] [PubMed]
- Pearson, T, Greiner, DL and Shultz, LD (2008). Creation of “humanized” mice to study human immunity. Curr Protoc Immunol Chapter 15: Unit 15.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.