Abstract
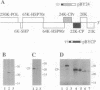
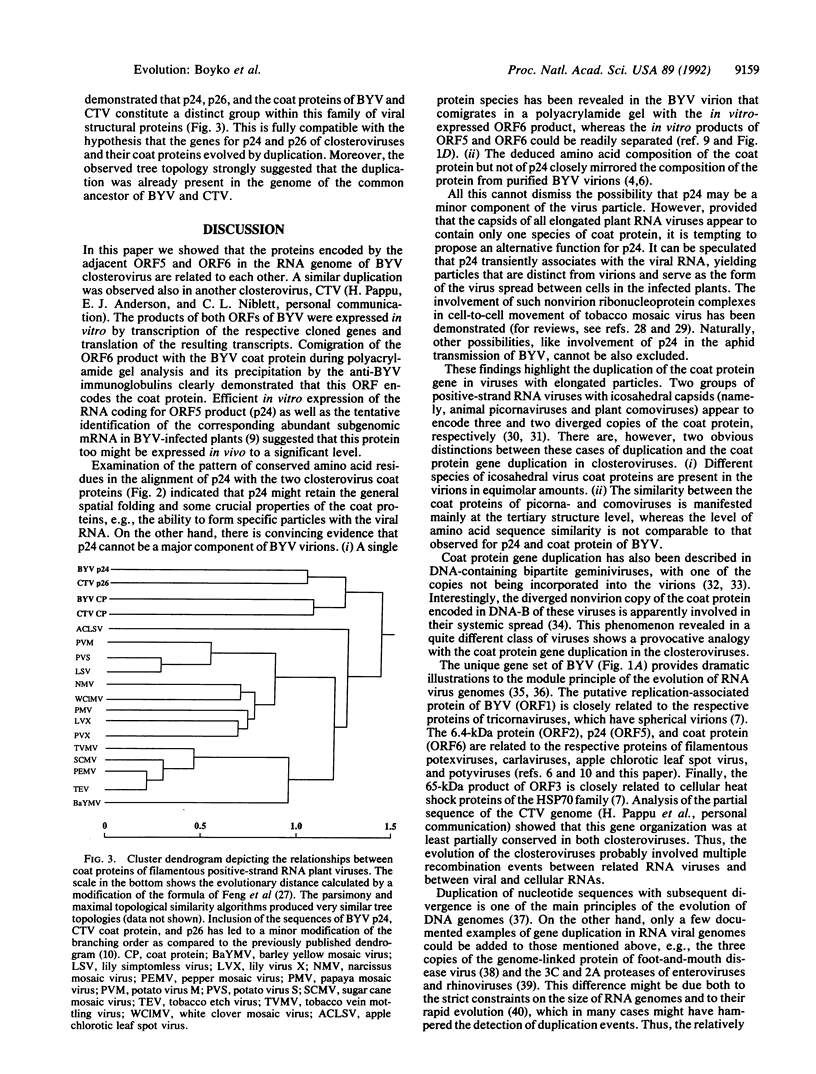
Computer-assisted analysis revealed a striking sequence similarity between the putative 24-kDa protein (p24) encoded by open reading frame (ORF) 5 of beet yellows closterovirus and the coat protein of this virus encoded by the adjacent ORF6. Both of these proteins are closely related to the homologous proteins of another closterovirus, citrus tristeza virus. It is hypothesized that the genes for coat protein and its diverged tandem copy have evolved by duplication. Phylogenetic analysis using various methods for tree generation suggested that the duplication was already present in the genome of the common ancestor of the two closteroviruses. The genes for p24 and coat protein of beet yellows closterovirus were cloned, transcribed, and translated in vitro yielding products of the expected size. It was shown that p24 is translated starting from the first of the two alternative AUG codons located near the 5' terminus of ORF5. The presence of a single protein species in beet yellows closterovirus virions and the near identity of the amino acid composition of this protein with the composition of the ORF6 but not the ORF5 product indicated that p24 is not a major virion component. Most of the amino acids that are conserved in the coat proteins of filamentous viruses of plants are retained also in p24. These observations suggest that p24 may share some structural and functional features with the coat protein but probably fulfills a distinct function in virus reproduction.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agranovsky A. A., Boyko V. P., Karasev A. V., Koonin E. V., Dolja V. V. Putative 65 kDa protein of beet yellows closterovirus is a homologue of HSP70 heat shock proteins. J Mol Biol. 1991 Feb 20;217(4):603–610. doi: 10.1016/0022-2836(91)90517-a. [DOI] [PubMed] [Google Scholar]
- Agranovsky A. A., Boyko V. P., Karasev A. V., Lunina N. A., Koonin E. V., Dolja V. V. Nucleotide sequence of the 3'-terminal half of beet yellows closterovirus RNA genome: unique arrangement of eight virus genes. J Gen Virol. 1991 Jan;72(Pt 1):15–23. doi: 10.1099/0022-1317-72-1-15. [DOI] [PubMed] [Google Scholar]
- Atabekov J. G., Dorokhov YuL Plant virus-specific transport function and resistance of plants to viruses. Adv Virus Res. 1984;29:313–364. doi: 10.1016/s0065-3527(08)60412-1. [DOI] [PubMed] [Google Scholar]
- Biou V., Gibrat J. F., Levin J. M., Robson B., Garnier J. Secondary structure prediction: combination of three different methods. Protein Eng. 1988 Sep;2(3):185–191. doi: 10.1093/protein/2.3.185. [DOI] [PubMed] [Google Scholar]
- Brunstedt J., Moseley J., Hull R. Nucleotide sequence of cDNA encoding the coat protein of beet yellows virus. Virus Genes. 1991 Jul;5(3):267–271. doi: 10.1007/BF00568976. [DOI] [PubMed] [Google Scholar]
- Carpenter J. M., Kassanis B., White R. F. The protein and nucleic acid of beet yellows virus. Virology. 1977 Mar;77(1):101–109. doi: 10.1016/0042-6822(77)90410-x. [DOI] [PubMed] [Google Scholar]
- Davies J. W., Stanley J. Geminivirus genes and vectors. Trends Genet. 1989 Mar;5(3):77–81. doi: 10.1016/0168-9525(89)90030-9. [DOI] [PubMed] [Google Scholar]
- Dolja V. V., Boyko V. P., Agranovsky A. A., Koonin E. V. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology. 1991 Sep;184(1):79–86. doi: 10.1016/0042-6822(91)90823-t. [DOI] [PubMed] [Google Scholar]
- Feng D. F., Johnson M. S., Doolittle R. F. Aligning amino acid sequences: comparison of commonly used methods. J Mol Evol. 1984;21(2):112–125. doi: 10.1007/BF02100085. [DOI] [PubMed] [Google Scholar]
- Forss S., Schaller H. A tandem repeat gene in a picornavirus. Nucleic Acids Res. 1982 Oct 25;10(20):6441–6450. doi: 10.1093/nar/10.20.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Blinov V. M., Donchenko A. P., Koonin E. V. An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J Mol Evol. 1989 Mar;28(3):256–268. doi: 10.1007/BF02102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuno R., Toh H., Hayashida H., Miyata T. Sequence similarity between putative gene products of geminiviral DNAs. Nature. 1984 Apr 5;308(5959):562–562. doi: 10.1038/308562a0. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Mushegian A. R., Ryabov E. V., Dolja V. V. Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J Gen Virol. 1991 Dec;72(Pt 12):2895–2903. doi: 10.1099/0022-1317-72-12-2895. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H. A., Chow K. C., Mickel F. S., Moss K. A., Kern H. F., Scheele G. A. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987 Jan;6(1):43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnichenko N. A., Karpova O. V., Morozov SYu, Rodionova N. P., Atabekov J. G. Translation arrest of potato virus X RNA in Krebs-2 cell-free system: RNase H cleavage promoted by complementary oligodeoxynucleotides. FEBS Lett. 1988 Jul 4;234(1):65–68. doi: 10.1016/0014-5793(88)81304-8. [DOI] [PubMed] [Google Scholar]
- Morozov SYu, Dolja V. V., Atabekov J. G. Probable reassortment of genomic elements among elongated RNA-containing plant viruses. J Mol Evol. 1989 Jul;29(1):52–62. doi: 10.1007/BF02106181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov S. Y., Miroshnichenko N. A., Zelenina D. A., Fedorkin O. N., Solovijev A. G., Lukasheva L. I., Atabekov J. C. Expression of RNA transcripts of potato virus X full-length and subgenomic cDNAs. Biochimie. 1990 Sep;72(9):677–684. doi: 10.1016/0300-9084(90)90051-h. [DOI] [PubMed] [Google Scholar]
- Rao J. K. Sequence homology among the coat proteins of gemini viruses. Biochem Biophys Res Commun. 1985 Jul 31;130(2):892–896. doi: 10.1016/0006-291x(85)90500-5. [DOI] [PubMed] [Google Scholar]
- Reanney D. C. The evolution of RNA viruses. Annu Rev Microbiol. 1982;36:47–73. doi: 10.1146/annurev.mi.36.100182.000403. [DOI] [PubMed] [Google Scholar]
- Sekiya M. E., Lawrence S. D., McCaffery M., Cline K. Molecular cloning and nucleotide sequencing of the coat protein gene of citrus tristeza virus. J Gen Virol. 1991 May;72(Pt 5):1013–1020. doi: 10.1099/0022-1317-72-5-1013. [DOI] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]