Abstract
Heparan Sulfate (HS) is a glycosaminoglycan (GAG) which forms a key component of the extracellular matrix (ECM). Breakdown of HS is carried out by heparanase (HPSE), an endo-β-glucuronidase of the glycoside hydrolase (GH)79 family. Overexpression of HPSE is strongly linked to cancer metastases - reflecting breakdown of extracellular HS and release of stored growth factors. Here we present crystal structures of human HPSE at 1.6-1.9 Å resolution reveal how an endo-acting binding cleft is exposed by proteolytic activation of latent proHPSE. Oligosaccharide complexes map the substrate-binding and sulfate recognition motifs. These data shed light on the structure and interactions for a key enzyme involved in ECM maintenance, and provide a starting point for design of HPSE inhibitors as biochemical tools and anti-cancer therapeutics.
Introduction
Heparan Sulfate (HS) is a sulfated glycosaminoglycan polysaccharide produced by nearly all animal species1,2. Structurally, HS is comprised of a linear repeating [D-HexUA(β1→4)D-GlcNX(α1→4)]n disaccharide motif, where HexUA is either Glucuronic acid (GlcUA) or its C-5 epimer Iduronic acid (IdoUA), and GlcNX is N-acetyl glucosamine (GlcNAc) or N-sulfo glucosamine (GlcNS). These chains are further elaborated by O-sulfation at O2 of HexUA, O6 of GlcNX and (rarely) O3 of GlcNX. The deacetylation, sulfation and epimerisation reactions involved in biosynthesis of HS are non-templated and do not reach completion, resulting in substantial structural heterogeneity along a chain. Mature HS is modular in composition, with “NS” domains rich in GlcNS, IdoUA and O-sulfated sugars separate from lower O-sulfation, GlcNAc, GlcUA rich “NA” domains (Supplementary Figure 1). This heterogeneity of HS structure provides interaction sites for a large number of different binding partners, and is central to its proper biological function3.
HS occurs in vivo in the form of heparan sulfate proteoglycans (HSPGs), consisting of one or more HS chains covalently linked to a core transmembrane or secreted protein. HSPGs are an important constituent of the extracellular matrix (ECM) which surrounds cells, and perform important structural and signaling functions via HS mediated interactions4. The compositions of HS chains are adapted to their function, and can differ between cells and tissues even when the core HSPG protein is the same. HS chains are also dynamically regulated in response to external stimuli, with turnover by some cells occurring as rapidly as t1/2 = 2.5 h5,6. This turnover is underpinned by a network of enzymes which serve to efficiently synthesize and break down HS in a regulated fashion.
The principal enzyme involved in breakdown of HS is heparanase (HPSE), a member of the Carbohydrate Active Enzymes (CAZy) Glycoside Hydrolase (GH)79 family of carbohydrate processing enzymes7. HPSE catalyzes hydrolysis of internal GlcUA(β1→4)GlcNS linkages in HS, with net retention of anomeric configuration8. HPSE breakdown of HS is not indiscriminate, but instead is restricted to a small subset of GlcUAs reflecting a requirement for specific N- and O-sulfation patterns on neighboring sugars9–11. HPSE present in late endosomes and lysosomes performs an essential housekeeping role in catabolic processing of internalized HSPGs12. In addition, HPSE can be trafficked to the cell surface or released into the ECM in order to effect breakdown of extracellular pools of HS13.
HPSE mediated breakdown of HS in the ECM has several effects on the behavior of nearby cells. Weakening of structural HS networks in the ECM and basement membranes directly facilitates cell motility and extravasation into surrounding tissues14. Latent pools of growth factors stored by HS are released upon breakdown by HPSE, promoting increased cell proliferation, motility and angiogenesis15,16. HS fragments generated by HPSE activity can also activate downstream signaling cascades17. Whilst controlled HPSE activity plays an important role in physiological processing of the ECM, aberrant HPSE expression is associated with inflammation and cancerous growth. The proliferative advantages conferred by HPSE lead to its upregulation by tumors in a variety of tissues, and HPSE overexpression correlates strongly with metastasis and worsened clinical prognoses18–21.
Only one gene with heparanase like catalytic activity has so far been identified in mammals, suggesting that loss of HPSE activity may not easily be compensated for by the cell. Accordingly, HPSE inhibition has attracted intense interest as an anti-cancer strategy, although the efficacy of small molecule inhibitors22,23 has yet to rival those reported for oligosaccharide-like HS mimetics24–26, such as the sulfated phosphomannopentaose derivative PI-88 (currently in phase III clinical trials as an adjuvant therapy for the treatment of viral hepatitis related hepatocellular carcinomas).
Despite intense biological and clinical interest, no 3D structure has yet been reported for human HPSE, although structures of a GH79 exoglucuronidase from Acidobaterium capsulatum (AcaGH79) and, recently, an endo-acting heparanase from Burkholderia pseudomallei (BpHPSE) have helped provide a structural overview for this enzyme family27,28. In order to understand the mechanistic basis of HPSE function, we have determined the crystal structures of human HPSE in apo and ligand bound conformations to illustrate the structural basis of enzyme-substrate binding in HPSE. These data shed light on the structure and function of a key enzyme involved in human HS metabolism, and may be of substantial utility in future efforts to rationally design inhibitors against HPSE.
Results
Tertiary structure of HPSE
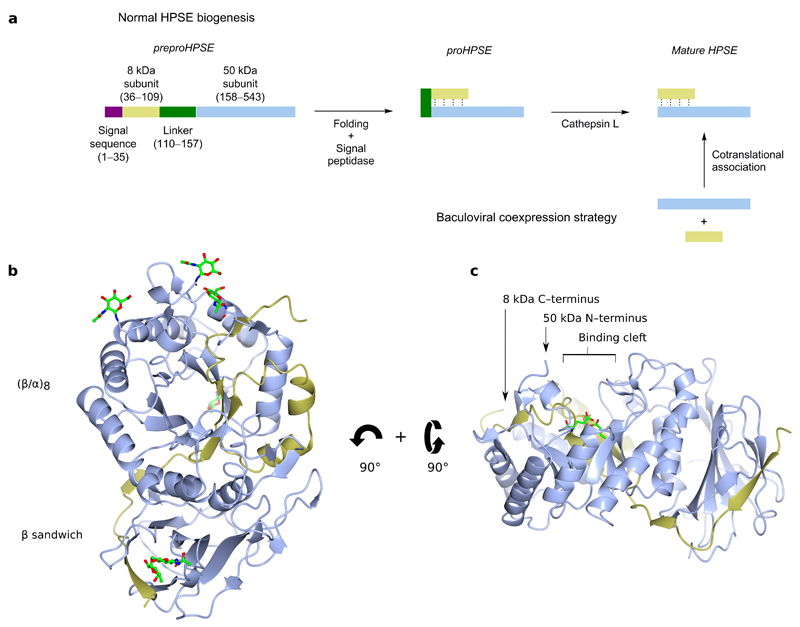
HPSE is initially translated as a pre-proenzyme, containing a signal sequence spanning Met1–Ala35. Cleavage of this signal sequence by signal peptidase leaves an inactive 65 kDa proHPSE, which must undergo further processing for activity. Proteolytic removal by Cathepsin L of a linker spanning Ser110–Gln157 liberates an N-terminal 8 kDa subunit and a C-terminal 50 kDa subunit, which remain associated as a non-covalent heterodimer in mature active HPSE29. For our studies, we expressed HPSE using baculovirus, following a previously described dual expression strategy30. cDNA encoding for the 8 kDa and 50 kDa subunits were placed into a single bacmid under the control separate viral promoters. The two subunits cotranslationally fold into mature heterodimeric HPSE, bypassing the 65 kDa proenzyme form and ensuring expression of only active enzyme (Figure 1a).
Figure 1.
3D structure of apo HPSE. (a) Schematic representation of HPSE biogenesis, and the coexpression strategy utilized in this study. (b) ‘Front’ view of apo-HPSE in ribbon representation; subunits are colored yellow (8 kDa), and blue (50 kDa). 5 sites of N-glycosylation are shown in green. (c) ‘Side’ view of HPSE showing a binding cleft in the (β/α)8 domain in which the catalytic residues (green) reside.
The structure of apo-HPSE refined to 1.75 Å resolution (Table 1) contains a single heterodimer in the asymmetric unit, comprising residues Gln36–Glu109 of the 8 kDa subunit and Lys159–Ile543 of the 50 kDa subunit (numbering based upon full preproenzyme). The domain architecture of HPSE comprises a (β/α)8 domain flanked by a smaller β-sandwich domain. Both 8 kDa and 50 kDa subunits are structurally involved in both domains: the 8 kDa subunit contributes 1 β-sheet to the β-sandwich, and the 1st β-α-β fold of the (β/α)8 domain, with the remaining folds contributed by the 50 kDa subunit (Figure 1b). Overall, the domain architecture of HPSE is superficially similar to that of previously characterized bacterial GH79s27,28, with Cα root mean square difference of 2.35 Å over 392 residues (out of 457) and 2.59 Å over 387 residues for AcaGH79 and BpHPSE respectively (Supplementary Figure 2a).
Table 1.
Crystal data collection and refinement statistics
| Iodide HPSE derivative |
apo HPSE (5e8m) |
M04 S00a complex (5e97) |
M04 S02a complex (5e98) |
M09 S05a complex (5e9b) |
dp4 complex (5e9c) |
|
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Beamline | Diamond I02 | Diamond I02 | Diamond I04 | Diamond I02 | Diamond I03 | Diamond I03 |
| Space group | P21 | P21 | P21 | P21 | P21 | P21 |
| Cell dimensions | ||||||
| a, b, c (Å) | 46.3, 70.6, 78.5 | 46.8, 71.0, 79.0 | 45.9, 70.9, 78.2 | 45.8, 71.3, 77.9 | 46.0, 71.1, 78.3 | 46.2, 71.1, 78.6 |
| α, β, γ (°) | 90.0, 94.6, 90.0 | 90.0, 94.2, 90.0 | 90.0, 94.9, 90.0 | 90.0, 95.1, 90.0 | 90.0, 95.1, 90.0 | 90.0, 95.2, 90.0 |
| Resolution (Å) | 52.42−2.15 (2.21−2.15) | 46.64−1.75 (1.78−1.75) | 40.99−1.64 (1.67−1.64) | 38.42−1.63 (1.67−1.63) | 52.52−1.88 (1.92−1.88) | 52.64−1.73 (1.76−1.73) |
| Rmerge | 0.14 (0.90) | 0.05 (0.88) | 0.05 (0.61) | 0.05 (0.97) | 0.06 (1.01) | 0.06 (1.02) |
| I/σI | 16.4 (2.5) | 12.0 (1.7) | 14.2 (1.7) | 11.7 (1.3) | 12.1 (1.5) | 11.3 (1.1) |
| Completeness (%) | 99.5 (98.9) | 100.0 (99.8) | 99.8 (99.8) | 99.5 (99.8) | 99.8 (99.7) | 99.0 (98.4) |
| Redundancy | 6.1 (5.6) | 4.2 (4.1) | 4.0 (3.6) | 4.0 (3.8) | 4.1 (4.2) | 4.0 (3.5) |
| Refinement | ||||||
| Resolution (Å) | 46.64−1.75 | 40.99−1.64 | 38.42−1.63 | 52.52−1.88 | 52.64−1.73 | |
| No. reflections | 49,329 | 57,972 | 58,820 | 38,830 | 49,793 | |
| Rwork/ Rfree | 0.16/0.20 | 0.17/0.20 | 0.17/0.20 | 0.18/0.22 | 0.17/0.21 | |
| No. atoms | ||||||
| Protein | 3669 | 3652 | 3657 | 3652 | 3660 | |
| Ligand/ion | 63 | 66 | 51 | 60 | 72 | |
| Water | 249 | 397 | 223 | 208 | 228 | |
| B-factors | ||||||
| Protein | 36.3 | 28.4 | 36.3 | 42.1 | 35.5 | |
| Ligand/ion | 47.4 | 41.5 | 45.6 | 59.2 | 68.5 | |
| Water | 44.9 | 37.2 | 41.5 | 45.1 | 42.2 | |
| R.m.s deviations | ||||||
| Bond lengths (Å) | 0.015 | 0.014 | 0.014 | 0.015 | 0.015 | |
| Bond angles (°) | 1.74 | 1.67 | 1.66 | 1.75 | 1.78 |
HPSE contains 6 putative N-glycosylation sites, all residing on the 50 kDa subunit. N-linked GlcNAcs (corresponding to N-glycan trees trimmed by Endoglycosidase (Endo)H during protein preparation) were found in the apo-HPSE structure at Asn162, Asn200, Asn217, Asn238 and Asn459. Additionally a core α1→6 linked Fucose was seen on the GlcNAc linked to Asn459. No noticeable density corresponding to GlcNAc was observed at the N-glycosylation site Asn178, suggesting this position may not be well N-glycosylated during baculoviral expression, or that N-GlcNAc here is not compatible with crystal packing.
The GH79 family belongs to the wider GH-A clan, characterized by a (β/α)8 domain containing the catalytic site7. A clear cleft spanning ~10 Å could be seen in the (β/α)8 domain of apo-HPSE, suggesting the HS binding site was contained within this part of the enzyme (Figure 1c). This cleft contained residues Glu343 and Glu225, previously identified as the catalytic nucleophile and acid-base of HPSE31, required for the retaining catalytic mechanism32. In accordance with the negatively charged nature of its HS substrate, the HPSE binding cleft is lined by basic sidechains contributed by Arg35, Lys158, Lys159, Lys161, Lys231, Arg272, Arg273 and Arg303.
Although not present in mature HPSE, our model indicates that the excised Ser110–Gln157 linker of proHPSE should lie very near or even within the active site cleft of the (β/α)8 domain. This positioning would hinder incoming HS substrates, and is consistent with a previously proposed steric block mechanism for proHPSE inactivation by its own linker33.
Structural basis of HPSE substrate interactions
Interaction of HS with HPSE is influenced by substrate sulfation, and only HS sequences with particular sulfation patterns are hydrolysed. Studies using defined HS oligomers have suggested that HPSE preferentially cleaves a trisaccharide with sulfated GlcNX residues at −2 and +1 positions9,10,34,35 (subsite nomenclature in Ref 36). However, a mechanistic understanding of how HPSE ‘reads’ the sulfation status of HS substrates to select for favorable cleavage sites has so far been lacking.
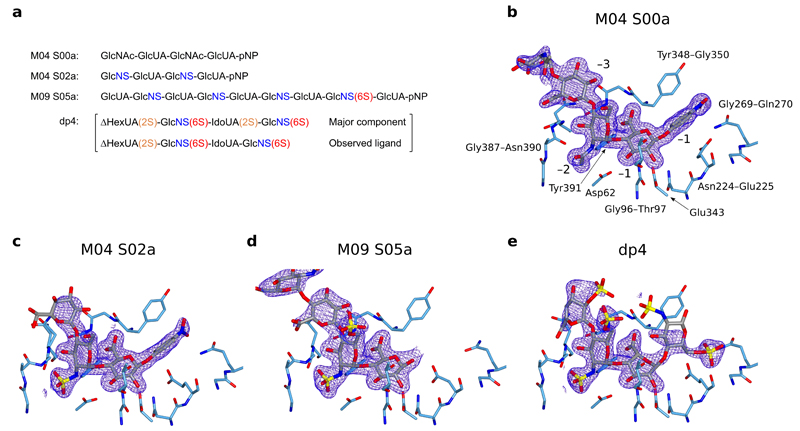
We mapped the substrate binding and sulfate interaction sites of HPSE by obtaining structures of the enzyme in complex with a set of HS analogues. We initially chose three ‘HepMers’; semi-synthetic HS oligomers of defined structure34, to investigate the effect of systematically increasing N- and O-sulfation: M04 S00a (a tetrasaccharide with no sulfation), M04 S02a (a tetrasaccharide with only N-sulfation), and M09 S05a (a nonasaccharide with N-sulfation and a single GlcNS(6S) towards the reducing end) (Figure 2a).
Figure 2.
Active site structures of HPSE in complex with substrate analogues. Densities shown are REFMAC maximum-likelihood/σA weighted 2Fo−Fc syntheses contoured between 0.25 and 0.32 electrons/Å3. (a) Schematic of substrate analogues used in this study, with N-sulfation (blue), 6O-sulfation (red) and 2O-sulfation (orange) highlighted. Structures for sugar monomers are shown in Supplementary Figure 1. pNP refers to paranitrophenol. (b) HPSE active site in complex with M04 S00a, with binding subsites and neighboring amino acids annotated. (c) HPSE in complex with M04 S02a. −3 GlcNS was disordered and has not been modeled. (d) HPSE in complex with M09 S05a. No +1 substituent could be modeled due to poor density, suggesting pNP no longer occupied this subsite. Sugars beyond −4 were disordered and have not been modeled. (e) HPSE in complex with dp4, illustrating interactions made by HPSE to 6O-sulfate at the +1 subsite.
Complexes of HPSE with the three HepMers showed clear electron density for the ligands within the active site cleft, revealing the basis of interaction at the −1 and −2 binding subsites (Figure 2b, c, d). Electron density was progressively more disordered from the −3 sugar onwards as the substrate exited the binding cleft, consistent with HPSE recognizing a trisaccharide spanning the −2, −1 and +1 subsites. For M04 SO0a and MO4 SO2a, electron density for the reducing end paranitrophenol (pNP) moiety was clearly visible at the +1 equivalent position, suggesting that these molecules were poor substrates for HPSE. In contrast, density at +1 for M09 S05a was almost absent.
Binding of GlcUA at the −1 subsite was identical in all HepMer complexes, with the ring in 4C1 conformation and anomeric carbon close to the nucleophile Glu343. Direct H-bonds to HPSE were observed from GlcUA to the sidechains of Asp62, Tyr391, as well as backbone NHs of Thr97, Gly349 and Gly350. A strong network of H-bonds made by the GlcUA C6 carboxylate to Tyr391, Gly349 and Gly350 are similar to that observed in the bacterial GH79s, suggesting a key GH79 motif tuned to recognize GlcUA. Our structures also showed that the presence of GlcUA(2S) or IdoUA(2S) at the −1 subsite cannot be tolerated by HPSE, due to steric clashes between the bulky 2O-sulfate and Asn224.
GlcNX residues at the −2 subsite also adopt a 4C1 conformation in all complexes and illustrate HPSE’s ability to accommodate a variety of GlcNX sugars at this position. Surprisingly, only the N2 position of GlcNX appeared to make any direct H-bonding interactions to HPSE: for both GlcNAc and GlcNS, the amide NH formed an H-bond to the sidechain of Tyr391. For GlcNAc, the amide carbonyl formed an H-bond to both Asn64 and to a structural water molecule, whereas GlcNS made these same interactions as well as H-bonds to a further structural water and the backbone NH of Gly389. For -2 GlcNS(6S), 6O-sulfate was ideally placed to participate in electrostatic interactions with the sidechain of Lys159. Thus GlcNS(6S) at the −2 subsite should be favored over GlcNS or GlcNAc, reflecting additional electrostatic and H-bonding interactions respectively.
Lack of density at the +1 subsite for M09 S05a, in contrast to M04 S00/S02a, suggested that the observed structure reflected several related complexes differing at this position. Inspection of GlcNS(6S) at the −2 subsite also revealed electron density for 6O-sulfate was considerably weaker than that for N-sulfate (Supplementary Figure 2b) indicating this subsite was likely occupied by a mixture of GlcNS and GlcNS(6S). Although M09 S05a contains several GlcNS residues, binding of HPSE far towards the reducing end of M09 S05a is strongly disfavored in crystallo due to the presence of a clashing symmetry molecule at the ‘positive’ end of the binding cleft (Supplementary Figure 2c). Thus we concluded the M09 S05a complex reflected partial HPSE binding only one disaccharide unit further from the reducing end. This would imply GlcNS(6S) occupancy of the +1 subsite, or alternatively, a product complex in which a +1 GlcNS(6S) had been hydrolysed. Consistent with this, we were able to determine kinetic parameters for HPSE hydrolysis of M09 S05a using a colorimetric reducing end assay (KM = 7.70 ± 1.42 µM, kcat = 0.53 ± 0.02 s–1; Supplementary Figure 3), but not for M04 S00a or M04 S02a. Whilst KM for M09 S05a hydrolysis was in the low μM range, comparable to previous studies of HPSE activity (1.64 μM for natural HS37, 46 μM for the synthetic substrate fondaparinux38), kcat for M09 S05a was ~15% of that previously determined for fondaparinux (3.5 s–1), possibly reflecting differences in substrate sulfation or oligosaccharide length.
Because +1 subsite interactions were not resolvable using HepMers, we sought to obtain a structure of HPSE in complex with a heparin-derived tetrasaccharide (hereafter ‘dp4’; Figure 2a; generated by depolymerization of full-length heparin by heparin lyases). Like HS and heparin itself, dp4 is a mixture of structurally related disaccharide units, with the tetrasaccharide ΔHexUA(2S)-GlcNS(6S)-IdoUA(2S)-GlcNS(6S) as the major component. The structure of HPSE with dp4 showed occupancy of the same binding site as for HepMers (Figure 2e), consistent with the overall similarity of heparin to HS. The observed density was not, however, for the predominant dp4 tetrasaccharide, but was better modeled by a molecule containing IdoUA at −1 (instead of IdoUA(2S)), confirming that 2O-sulfate at the −1 subsite is not tolerated. Electron density for dp4 in HPSE was correspondingly weaker than for HepMers across all subsites, reflecting the binding of a minor constituent.
The overall configuration of dp4 across the HPSE active site was similar to that of HepMers, but −1 IdoUA adopted a 2SO conformation, instead of the 4C1 seen for GlcUA in HepMers. In free heparin, IdoUA residues exist in an easily traversable equilibrium between 1C4 and 2SO, with the O2 position held axial or equatorial respectively39. Upon binding to HPSE, H-bonding to Asn224 and the presence of clashing Glu343 at the O2 axial position constrain −1 IdoUA to the 2SO conformation. IdoUA constrained in 2SO may be hindered from undergoing the conformational changes required for hydrolysis40, perhaps explaining why heparin-type ligands containing a IdoUA residues act as competitive inhibitors of HS cleavage.
Although weak, some density at the +1 subsite could be resolved in the dp4 complex, suggesting the presence of +1 6O-sulfate, which we tentatively modelled as GlcNS(6S). In our model, GlcNS(6S) at the +1 subsite adopts an undistorted 4C1 conformation, with 6O-sulfate providing the main interactions to HPSE via H-bonds to the backbone and sidechain NH of Gln270, as well as electrostatic interactions with Arg272. From its positioning in our model, further electrostatic interactions may also exist between +1 N-sulfate and Arg303, although we are unable to confirm this given the poor density for the ligand at this position. Although dp4 contains IdoUA instead of GlcUA at the −1 subsite, both GlcUA 4C1 and IdoUA 2SO conformations involve a similar all-equatorial arrangement of substituents around the sugar ring. Therefore we propose the position of GlcNS(6S) at the +1 subsite is also likely to be relevant for a HS substrate containing GlcUA instead of IdoUA.
HPSE interaction induces distortion of the substrate chain
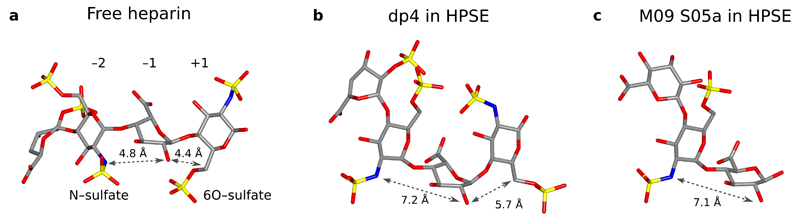
The structure of free heparin has been extensively studied; it adopts a linear right handed helix with a rotation of ~180° and translation of 0.82–0.87 nm per disaccharide unit41–43. Sulfates are presented in clusters on the surface of the sugar chain, with their distribution reflecting whether IdoUA residues are in 1C4 or 2SO conformation. The helical heparin conformation is retained in complex with binding partners, as seen in crystal structures of heparin bound to FGF or antithrombin44–46, which also illustrate the role of sulfation in mediating electrostatic interactions with basic residues on heparin interacting proteins. Conversely, the presence of a bulky negatively charged ‘coat’ surrounding the core glycan may hinder access to nucleophilic protein residues, effectively protecting the sugar residues from enzymatic attack.
Dp4 in complex with HPSE retains an approximately right-handed helical configuration within the active site cleft, similar to free heparin. However, interactions from HPSE to −2 N-sulfate and +1 6O-sulfate introduce a clear bend into the heparin chain across the −2, −1, +1 trisaccharide. Compared to a previous model for an idealized heparin helix42, the −2(N2)→–1(O2) distance for dp4 in HPSE was increased from 4.8 Å to 7.2 Å, and −1(O2)→+1(C6) distance increased from 4.4 Å to 5.7 Å (Figure 3a, b). This ‘bend’ separates the N2 and O6 sulfates adjacent to the −1 anomeric center, allowing the catalytic residues of HPSE to access this position more easily. Although a +1 subsite sugar was not observed in HepMer-HPSE complexes, a similar distortion was seen for HepMers across the −2→–1 subsites (Figure 3c: lengthened −2(N2)→–1(O2) distance of 7.1 Å for M09 S05a), suggesting that the ‘bending’ observed for dp4 is also likely to apply to substrates with a −1 GlcUA instead of IdoUA.
Figure 3.
Oligosaccharide chain distortion is induced by HPSE interaction. (a) Linear helical structure of a free heparin tetrasaccharide, adapted from an NMR derived model of heparin with all IdoUAs restrained in 2SO42. (b) Configuration of dp4 in the HPSE active site, showing a bend around the −2, −1, +1 sugars caused by HPSE interaction with −2 N-sulfate and +1 6O-sulfate. (c) Configuration of M09 S05a in complex with HPSE, showing the same distortion around −2→−1 as observed for dp4.
Our results indicate a dual role for HS sulfation in interactions with HPSE. Not only does sulfation serve as a molecular signal which directs the enzyme to only cleave certain glycan sites, but −2 N-sulfate and +1 O-sulfate moieties also act as mechanistic handles by which HPSE can prize open a substrate HS helix, in order to more effectively access the anomeric center of the −1 sugar.
Discussion
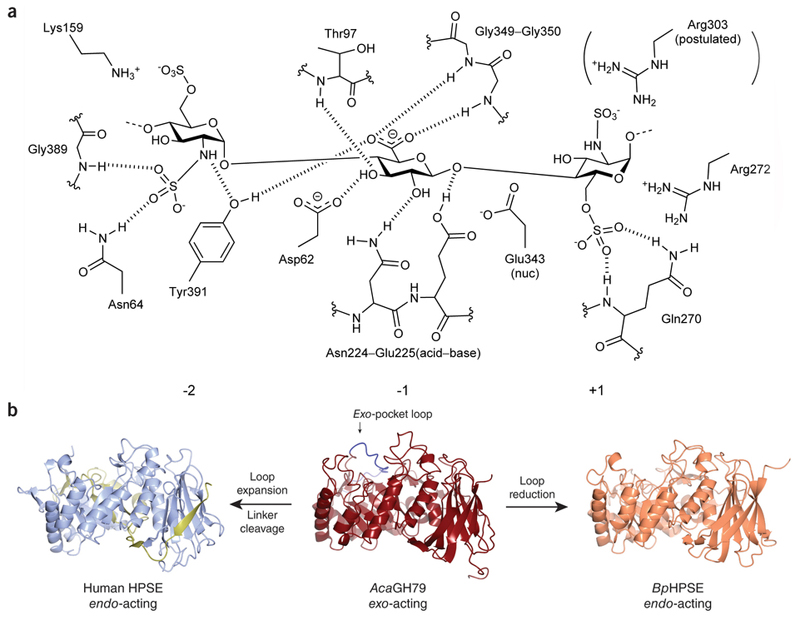
The 3D structure of the human GH79 endoglucuronidase, HPSE, and its interaction with HS substrates (Figure 4) provide a long anticipated structural rationale which tie together the results of numerous biochemical studies on the HS sulfation patterns required for HPSE cleavage9–11. We have confirmed sulfation is key for HPSE interaction with HS, and that the recognized cleavage site is a trisaccharide accommodated into the HPSE binding cleft. Structurally, −2 N-sulfate and +1 6O-sulfate appear to be the main determinants for recognition, as these directly contact the enzyme through H-bonding networks. −2 6O-sulfate and +1 N-sulfate may also further stabilize the HPSE bound trisaccharide through electrostatic interactions to basic residues lining the active site cleft. Our observations are consistent with the major findings of previous biochemical studies.
Figure 4.
Composite summary of HPSE-substrate interactions across the −2, −1 and +1 subsites of the enzyme binding cleft. Interactions as mapped by complexes with HepMers and dp4. The catalytic residues of HPSE have been annotated. Electrostatic interaction with Arg303 is postulated based on the position of N-sulfate in the dp4 model.
Whilst the activity of human (h)HPSE is broadly similar to that of the recently reported bacterial enzyme BpHPSE28, differences exist between the substrates hydrolyzed by each enzyme. BpHPSE was reported to preferentially cleave HS containing GlcNAc residues, whereas GlcNS is highly favored for hHPSE activity. Furthermore, BpHPSE was also found to degrade Chondroitin Sulfate glycosaminoglycans (GAG)s, which are not known to be a substrate for hHPSE. Sequence alignment of several eukaryotic heparanases with BpHPSE and AcaGH79 revealed that whilst residues at the −1 subsite residues (of hHPSE) are well conserved across all species (reflecting absolute specificity for GlcUA) residues at the −2 and +1 subsites show much poorer conservation in the bacterial enzymes, providing a rationale for observed differences in substrate specificity (Supplementary Figure 4). Compared to hHPSE, BpHPSE may represent a more general catabolic enzyme that is used by the bacterium to break down GAGs encountered in its environment.
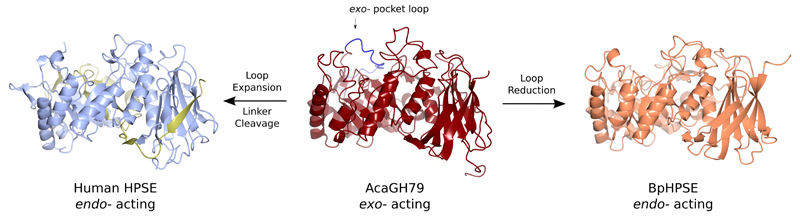
One point of particular interest is the evolutionary relationship between exo- and endo- acting GH79 enzymes; in particular, the sequence corresponding to a loop in AcaGH79 which forms part of its exo-acting substrate binding pocket (Figure 5, Supplementary Figure 5). This loop is substantially reduced in size in endo-acting BpHPSE, converting a binding pocket into a large cleft capable of accommodating long GAG chains. Conversely, the human enzyme appears to have evolved endo specificity though expansion of this loop to form a proteolytically cleavable linker motif. Proteolytically activated proenzymes are abundant in higher eukaryotes, and represent an efficient mechanism to induce enzymatic activity in response to stimuli. However, de novo evolution of proenzymes is extremely unlikely, as it would require the coevolution of both viable enzymatic and proteolyzable sequences. A more plausible scenario would involve expansion of a small loop around the active site of an existing enzyme scaffold, which eventually forms a larger proteolytically cleavable steric block which be removed for activation. This mechanism has already been hypothesized for serine proteases such as trypsinogen/trypsin and chymotrypsinogen/chymotrypsin47. Structural observations on HPSE and related enzymes provide evidence that such evolutionary processes may have also occurred for carbohydrate processing enzymes. We anticipate that further examples of such relationships will become apparent in the near future, as efforts to solve the structures of higher eukaryotic carbohydrate processing enzymes intensify.
Figure 5.
Structural relationships between the active sites of exo- and endo- acting GH79 enzymes. An extended loop in the (β/α)8 domain of AcaGH79 forms part of the exo- acting substrate binding pocket wall. This loop is considerably shortened in BpHPSE, creating an endo- acting binding cleft. Conversely, human HPSE has expanded this loop into a large linker sequence, which is proteolytically removed to produce its endo- acting binding cleft.
In conclusion, our structures of HPSE and its ligand complexes provide insight into the mechanisms of action for a key player in ECM remodeling. The structures we have reported here should be of utility in future efforts to design improved inhibitors of HPSE, as therapeutics and/or chemical biology tools. Indeed, our complex of HPSE with tetrasaccharide dp4 already suggests that introduction of sugars to the −1 subsite, fixed in particular conformations, may be a viable strategy for inhibitor design. More fundamentally, GAGs such as HS play an important role in numerous biological processes both normal and pathological. An understanding of the enzymes involved in their processing will be essential for determining how this important and complex class of carbohydrates is regulated.
Online methods
Production of HPSE expressing baculovirus
DNA was extracted from baculovirus encoding for proHPSE N-tagged with honey bee mellitin signal sequence with a QIAGEN miniprep kit, using 250 µL of virus stock as input. PCR using restriction site tailed primers (Supplementary Table 1) was used to generate cDNA fragments for the mellitin signal peptide, 8 kDa HPSE subunit, and 50 kDa HPSE subunit. N-mellitin-HPSE(8 kDa) was subcloned into the BamHI/PstI sites of pFastBac Dual, under the control of the PolH promoter. N-mellitin-HPSE(50 kDa) was subcloned into the XhoI/KpnI sites of the same vector, under the control of p10. For both fragments, mellitin was ligated to the HPSE gene via an XmaI restriction site, which leaves an extra DPG tripeptide in the protein upon expression and signal peptidase cleavage.
Recombinant bacmid was produced using the Tn7 transposition method in DH10EMBacY (Geneva Biotech)48, and purified using the PureLink miniprep kit (Invitrogen) following standard protocols. V1 baculovirus was produced by transfection of bacmid into low passage adherent Sf21 cells (Invitrogen) using FuGENE HD transfection reagent (ProMega), at a ratio of 2 μg DNA to 4.5 μL FuGENE. V1 → V2 virus amplification was carried out using suspension Sf21 cells, using the YFP marker present in EMBacY baculovirus to determine optimum amplification prior to harvesting (typically ˜60% cells fluorescent). For expression, T. Ni cells (Invitrogen) were infected with V2 baculovirus at a MOI>1, and infection followed using the EMBacY YFP marker to determine optimum timepoint for harvesting (typically 72 h, with > 80 % cells fluorescent). All insect cells used tested negative for mycoplasma contamination.
Expression and purification of HPSE
3L of conditioned media was cleared of cells by centrifugation at 400 g for 15 min at 4°C, followed by further clearing of debris by centrifugation at 4000 g for 60 min at 4°C. DTT (1 mM) and AEBSF (0.1 mM) were added to cleared media, which was then loaded onto a pre-equilibrated HiTrap Heparin HP 5 mL column (GE Healthcare). The heparin column was washed with 10 CV Hep Buffer A (20 mM HEPES [7.4], 100 mM NaCl, 1 mM DTT), and eluted with a linear gradient over 30 CV using Hep Buffer B (20 mM HEPES [7.4], 1.5 M NaCl, 1 mM DTT). HPSE containing fractions were pooled, diluted 10-fold into IEX buffer A (50 mM Phosphate [8.0], 100 mM NaCl, 1 mM DTT), and loaded onto a pre-equilibrated HiTrap Sepharose SP HP 1 mL column at 4°C. Protein was eluted from the SP column with a linear gradient over 30 CV using IEX buffer B (50 mM Phosphate [8.0], 1.5 mM NaCl, 1 mM DTT). HPSE containing fractions were pooled and concentrated to ˜2 mL using a 30 kDa Vivaspin concentrator (GE Healthcare), and treated with 5 μL EndoH (NEB) for 4 h at ambient temperature. Digested protein was purified by size exclusion chromatography (SEC) using a Superdex S75 16/600 column (GE Healthcare) in SEC buffer (20 mM HEPES [7.4], 200 mM NaCl, 1 mM DTT). HPSE containing fractions were concentrated to 10 mg/mL using a 30 kDa Vivaspin concentrator, and buffer exchanged into Hep Buffer A via at least 3 rounds of dilution/reconcentration.
Crystallization of HPSE
HPSE at 10 mg/mL was tested against a range of commercial crystallization screens. Large split crystals were found in several conditions of the Index and PEG/Ion screens, which were taken on for further optimization. Well diffracting single crystals were obtained by the sitting drop vapor diffusion method at 20°C using 100 mM MES [5.5], 100 mM MgCl2, 17% PEG3350, and a protein:well solution ratio of 200:500 nL, with crystals typically appearing after 3 days. apo-HPSE crystals were cryoprotected prior to flash freezing in liquid N2 for data collection, using the mother liquor solution supplemented with 25% ethylene glycol.
Crystal structure solution
HPSE crystal structure was solved by single isomorphous replacement with anomalous scattering (SIRAS) using the apo crystal and an Iodide derivative. The Iodide derivative was prepared by a short soak (~10 s) of an apo crystal in cryoprotectant solution further supplemented with 0.5 M NaI49. Data were collected at beamline I02 of the Diamond Light Source, processed using XDS50, reduced using Aimless51, and phased using the SHELX pipeline52. Initial autobuilding was carried out with Buccaneer53, before subsequent rounds of manual model building and refinement using Coot54 and REFMAC555 respectively. MolProbity56 and Privateer57 were used to assess model validity before deposition to the PDB. Crystal structure figures were generated using ccp4mg58.
Substrate complexes
HPSE complexes were generated by soaking the apo crystal with M04 S00a, M04 S02a, M09 S05a HepMers (Iduron) or dp4 heparin tetrasaccharide (Dextra Laboratories). In each case, a small piece of solid compound was directly dissolved in mother liquor containing the crystal. All soaks were carried out for 20 minutes, before direct flash freezing in liquid N2 for data collection without further cryoprotection. Ligand coordinates were built using jLigand59.
HepMer hydrolysis assays
20 μL assay solutions comprising 50 nM HPSE added to a series of HepMer solutions in 40 mM NaOAc buffer (pH 5.0) were incubated at 37 °C for 20 min, before addition of NaOH and WST-1 dye (Santa Cruz Biotech) to a final volume of 40 μl, and final concentrations of 0.1 M NaOH and 1 mM WST-1. Reactions were developed at 60 °C for 60 min, and the absorbance at 584 nm measured in a 384 well microplate using a POLARstar Optima plate reader (BMG Labtech). A584nm readings were quantified against a D-Glucose standard set up on the same plate. For each compound, a no enzyme control was taken to account for non-enzymatic autohydrolysis of the pNP residue present on the HepMer. Data analysis and curve fitting was done using SigmaPlot 12.5 (Systat Software).
Supplementary Information
Supplementary Figures 1–5 and Supplementary Table 1 are available in the online version of this paper.
Acknowledgements
We thank Diamond Light Source for access to beamlines I02, I03 and I04 (proposal mx-9948) that contributed to the results presented here. DH10EMBacY cells used to generate recombinant bacmids were a gift from I. Berger (University of Bristol). The full HPSE gene was obtained from a baculoviral construct from D. Jarvis (University of Wyoming – US National Institutes of Health grant RR005351). G.J.D. and L.W. acknowledge support from the European Research Council through Advanced Grant Glycopoise (AdG 322942 (G.J.D.)). A.M.B. and C.M.V. thank the UK Medical Research Council for funding (MR/K000179/1 (A.M.B.)).
Footnotes
Accession Codes
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5E8M (apo structure), 5E97 (M04 S00a complex), 5E98 (M04 S02a complex), 5E9B (M09 S05a complex), 5E9C (dp4 complex).
The following structures were referenced in this article: PDB accession codes 3VNY27, 5BWI28.
Author contributions
L.W. and G.J.D designed and interpreted the experiments. L.W. and C.M.V. cloned, expressed and purified proteins with help in eukaryotic protein expression facilities from A.M.B. L.W. carried out kinetics experiments, protein crystallizations, and solved the structures of protein and ligand complexes. L.W. and G.J.D. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Lawrence R, et al. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem. 2008;283:33674–84. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamada S, Morimoto H, Fujisawa T, Sugahara K. Glycosaminoglycans in Hydra magnipapillata (Hydrozoa, Cnidaria): demonstration of chondroitin in the developing nematocyst, the sting organelle, and structural characterization of glycosaminoglycans. Glycobiology. 2007;17:886–94. doi: 10.1093/glycob/cwm051. [DOI] [PubMed] [Google Scholar]
- 3.Rabenstein DL. Heparin and heparan sulfate: structure and function. Nat Prod Rep. 2002;19:312–31. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 4.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iozzo RV. Turnover of heparan sulfate proteoglycan in human colon carcinoma cells. A quantitative biochemical and autoradiographic study. J Biol Chem. 1987;262:1888–900. [PubMed] [Google Scholar]
- 6.Owens RT, Wagner WD. Metabolism and turnover of cell surface-associated heparan sulfate proteoglycan and chondroitin sulfate proteoglycan in normal and cholesterol-enriched macrophages. Arterioscler Thromb. 1991;11:1752–8. doi: 10.1161/01.atv.11.6.1752. [DOI] [PubMed] [Google Scholar]
- 7.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson JC, Laloo AE, Singh S, Ferro V. 1H NMR spectroscopic studies establish that heparanase is a retaining glycosidase. Biochem Biophys Res Commun. 2014;443:185–8. doi: 10.1016/j.bbrc.2013.11.079. [DOI] [PubMed] [Google Scholar]
- 9.Okada Y, et al. Structural recognition by recombinant human heparanase that plays critical roles in tumor metastasis. Hierarchical sulfate groups with different effects and the essential target disulfated trisaccharide sequence. J Biol Chem. 2002;277:42488–95. doi: 10.1074/jbc.M206510200. [DOI] [PubMed] [Google Scholar]
- 10.Pikas DS, Li JP, Vlodavsky I, Lindahl U. Substrate specificity of heparanases from human hepatoma and platelets. J Biol Chem. 1998;273:18770–7. doi: 10.1074/jbc.273.30.18770. [DOI] [PubMed] [Google Scholar]
- 11.Peterson SB, Liu J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 2013;32:223–7. doi: 10.1016/j.matbio.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Goldshmidt O, et al. Human heparanase is localized within lysosomes in a stable form. Exp Cell Res. 2002;281:50–62. doi: 10.1006/excr.2002.5651. [DOI] [PubMed] [Google Scholar]
- 13.Nadav L, et al. Activation, processing and trafficking of extracellular heparanase by primary human fibroblasts. J Cell Sci. 2002;115:2179–87. doi: 10.1242/jcs.115.10.2179. [DOI] [PubMed] [Google Scholar]
- 14.Sasaki N, Higashi N, Taka T, Nakajima M, Irimura T. Cell surface localization of heparanase on macrophages regulates degradation of extracellular matrix heparan sulfate. J Immunol. 2004;172:3830–5. doi: 10.4049/jimmunol.172.6.3830. [DOI] [PubMed] [Google Scholar]
- 15.Elkin M, et al. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661–3. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 16.Ishai-Michaeli R, Eldor A, Vlodavsky I. Heparanase activity expressed by platelets, neutrophils, and lymphoma cells releases active fibroblast growth factor from extracellular matrix. Cell Regul. 1990;1:833–42. doi: 10.1091/mbc.1.11.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodall KJ, Poon IK, Phipps S, Hulett MD. Soluble heparan sulfate fragments generated by heparanase trigger the release of pro-inflammatory cytokines through TLR-4. PLoS One. 2014;9:e109596. doi: 10.1371/journal.pone.0109596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldshmidt O, et al. Cell surface expression and secretion of heparanase markedly promote tumor angiogenesis and metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10031–10036. doi: 10.1073/pnas.152070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlodavsky I, et al. The impact of heparanase and heparin on cancer metastasis and angiogenesis. Pathophysiology of Haemostasis and Thrombosis. 2006;35:116–127. doi: 10.1159/000093553. [DOI] [PubMed] [Google Scholar]
- 20.Vlodavsky I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 21.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. International Journal of Biochemistry & Cell Biology. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Xu YJ, et al. N-(4-{[4-(1H-Benzoimidazol-2-yl)-arylamino]-methyl}-phenyl)-benzamide derivatives as small molecule heparanase inhibitors. Bioorg Med Chem Lett. 2006;16:404–8. doi: 10.1016/j.bmcl.2005.09.070. [DOI] [PubMed] [Google Scholar]
- 23.Courtney SM, et al. 2,3-Dihydro-1,3-dioxo-1H-isoindole-5-carboxylic acid derivatives: a novel class of small molecule heparanase inhibitors. Bioorg Med Chem Lett. 2004;14:3269–73. doi: 10.1016/j.bmcl.2004.03.086. [DOI] [PubMed] [Google Scholar]
- 24.Cassinelli G, et al. Antitumor efficacy of the heparanase inhibitor SST0001 alone and in combination with antiangiogenic agents in the treatment of human pediatric sarcoma models. Biochem Pharmacol. 2013;85:1424–32. doi: 10.1016/j.bcp.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 25.Hammond E, Handley P, Dredge K, Bytheway I. Mechanisms of heparanase inhibition by the heparan sulfate mimetic PG545 and three structural analogues. FEBS Open Bio. 2013;3:346–51. doi: 10.1016/j.fob.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis KD, et al. A phase II study of the heparanase inhibitor PI-88 in patients with advanced melanoma. Invest New Drugs. 2008;26:89–94. doi: 10.1007/s10637-007-9080-5. [DOI] [PubMed] [Google Scholar]
- 27.Michikawa M, et al. Structural and biochemical characterization of glycoside hydrolase family 79 beta-glucuronidase from Acidobacterium capsulatum. J Biol Chem. 2012;287:14069–77. doi: 10.1074/jbc.M112.346288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohlmann L, et al. Functional and structural characterization of a heparanase. Nat Chem Biol. 2015 doi: 10.1038/nchembio.1956. advance online publication. [DOI] [PubMed] [Google Scholar]
- 29.Fairbanks MB, et al. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem. 1999;274:29587–90. doi: 10.1074/jbc.274.42.29587. [DOI] [PubMed] [Google Scholar]
- 30.McKenzie E, et al. Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. Biochem J. 2003;373:423–35. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulett MD, et al. Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry. 2000;39:15659–67. doi: 10.1021/bi002080p. [DOI] [PubMed] [Google Scholar]
- 32.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–9. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 33.Nardella C, et al. Mechanism of activation of human heparanase investigated by protein engineering. Biochemistry. 2004;43:1862–73. doi: 10.1021/bi030203a. [DOI] [PubMed] [Google Scholar]
- 34.Peterson S, Liu J. Deciphering mode of action of heparanase using structurally defined oligosaccharides. J Biol Chem. 2012;287:34836–43. doi: 10.1074/jbc.M112.390161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson SB, Liu J. Unraveling the specificity of heparanase utilizing synthetic substrates. J Biol Chem. 2010;285:14504–13. doi: 10.1074/jbc.M110.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321(Pt 2):557–9. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fairweather JK, Hammond E, Johnstone KD, Ferro V. Synthesis and heparanase inhibitory activity of sulfated mannooligosaccharides related to the antiangiogenic agent PI-88. Bioorg Med Chem. 2008;16:699–709. doi: 10.1016/j.bmc.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 38.Hammond E, Li CP, Ferro V. Development of a colorimetric assay for heparanase activity suitable for kinetic analysis and inhibitor screening. Anal Biochem. 2010;396:112–6. doi: 10.1016/j.ab.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Torri G, et al. Mono- and bidimensional 500 MHz 1H-NMR spectra of a synthetic pentasaccharide corresponding to the binding sequence of heparin to antithrombin-III: evidence for conformational peculiarity of the sulfated iduronate residue. Biochem Biophys Res Commun. 1985;128:134–40. doi: 10.1016/0006-291x(85)91655-9. [DOI] [PubMed] [Google Scholar]
- 40.Speciale G, Thompson AJ, Davies GJ, Williams SJ. Dissecting conformational contributions to glycosidase catalysis and inhibition. Curr Opin Struct Biol. 2014;28:1–13. doi: 10.1016/j.sbi.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atkins ED, Nieduszynski IA. Crystalline structure of heparin. Adv Exp Med Biol. 1975;52:19–37. doi: 10.1007/978-1-4684-0946-8_2. [DOI] [PubMed] [Google Scholar]
- 42.Mulloy B, Forster MJ, Jones C, Davies DB. N.m.r. and molecular-modelling studies of the solution conformation of heparin. Biochemical Journal. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikhailov D, Linhardt RJ, Mayo KH. NMR solution conformation of heparin-derived hexasaccharide. Biochem J. 1997;328(Pt 1):51–61. doi: 10.1042/bj3280051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–20. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 45.Schlessinger J, et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–50. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 46.Dementiev A, Petitou M, Herbert JM, Gettins PG. The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity. Nat Struct Mol Biol. 2004;11:863–7. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- 47.Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation (a review) Proc Natl Acad Sci U S A. 1976;73:3825–32. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bieniossek C, Imasaki T, Takagi Y, Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci. 2012;37:49–57. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dauter Z, Dauter M, Rajashankar KR. Novel approach to phasing proteins: derivatization by short cryo-soaking with halides. Acta Crystallogr D Biol Crystallogr. 2000;56:232–7. doi: 10.1107/s0907444999016352. [DOI] [PubMed] [Google Scholar]
- 50.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallographica Section D-Biological Crystallography. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallographica Section D-Biological Crystallography. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallographica Section D-Biological Crystallography. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D-Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agirre J, et al. Privateer: software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol. 2015;22:833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- 58.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallographica Section D-Biological Crystallography. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lebedev AA, et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallographica Section D-Biological Crystallography. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.