Abstract
Background
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the western world. Shorter mean telomere length in leukemic cells has been associated with more aggressive disease. Germline polymorphisms in telomere maintenance genes impact telomere length and may contribute to CLL susceptibility.
Methods
We collected genome-wide data from two groups of patients with CLL (N=273) and two control populations (N=5725). In ancestry-adjusted case-control comparisons, we analyzed eight single nucleotide polymorphisms (SNPs) in genes definitively associated with inter-individual variation in leukocyte telomere length (LTL) in prior genome-wide association studies: ACYP2, TERC, NAF1, TERT, OBFC1, CTC1, ZNF208, and RTEL1.
Results
Three of the eight LTL-associated SNPs were associated with CLL risk at P<0.05, including those near: TERC (O.R.=1.46; 95% C.I.=1.15–1.86; P=1.8×10−3), TERT (O.R.=1.23; 95%C.I.=1.02–1.48; P=0.030), and OBFC1 (O.R.=1.36; 95%C.I.=1.08–1.71; P=9.6×10−3). Using a weighted linear combination of the 8 LTL-associated SNPs, we observed that CLL patients were predisposed to longer LTL than controls in both case-control sets (P=9.4×10−4 and 0.032, respectively). CLL risk increased monotonically with increasing quintiles of the weighted linear combination.
Conclusions
Genetic variants in TERC, TERT, and OBFC1 are associated with both longer LTL and increased CLL risk. Because the human CST complex competes with shelterin for telomeric DNA, future work should explore the role of OBFC1 and other CST complex genes in leukemogenesis.
Impact
A genetic predisposition to longer telomere length is associated with an increased risk of CLL, suggesting that the role of telomere length in CLL etiology may be distinct from its role in disease progression.
Keywords: telomerase, telomere, CST complex, chronic lymphocytic leukemia, OBFC1, TERT, TERC, ACYP2
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is a highly heterogeneous disease with clinical course ranging from indolent to highly progressive(1). Currently, several prognostic markers including somatic hypermutation in the immunoglobulin heavy-chain variable region, ZAP70 and CD38 expression, and TP53/SF3B1/NOTCH1 mutation status are used in combination to estimate disease prognosis(2). Telomere length is also associated with CLL progression, where shorter telomere length is correlated with advanced disease stages, Richter’s transformation(3) and poor patient prognosis(4).
Telomeres are repetitive DNA sequences that cap and protect chromosomes. Telomeric repeats are lost with each somatic cellular division and, when depleted, future mitoses result in the loss of integral genomic DNA. This induces replicative senescence and apoptosis, limiting the replicative potential of somatic cells(5). If adequate oncogenic mutations are acquired before reaching replicative senescence, unlimited proliferation may follow and cancer may result. Thus, long telomeres may increase the risk of cancer by allowing additional time in which a cell can accumulate mutations before reaching this apoptotic checkpoint(6). Following malignant transformation, leukemic cells that proliferate in the absence of an active mechanism of telomere maintenance (e.g. telomerase reactivation) will continue to experience telomere depletion(7, 8). In such tumors, short telomere length would be expected to associate with more advanced and/or highly-proliferative leukemia(9, 10).
Telomere length decreases with age, but can be significantly different between individuals of the same age due to genetic and environmental influences(11). Heritable differences in leukocyte telomere length (LTL) are associated with single nucleotide polymorphisms (SNPs) in eight genes involved in telomere maintenance, including: ACYP2, TERC, NAF1, TERT, OBFC1, CTC1, ZNF208, and RTEL1 (12, 13). Genome-wide association studies (GWAS) indicate that inherited SNPs near the telomerase-component genes TERC and TERT are also associated with increased risk of CLL and implicate telomere biology in leukemia predisposition(14, 15). We therefore hypothesized that inherited variation impacting telomere length may be associated with risk of developing CLL, with the heritable and somatic genetic influences having a net effect on telomere length in tumors.
To determine whether common genetic variants associated with inter-individual variation in telomere length confer risk for CLL, we analyzed eight independent SNPs in 273 patients with CLL and 5725 controls. These SNP have previously been definitively associated with inter-individual variation in LTL in a GWAS of 37,684 individuals of European ancestry conducted by the ENGAGE Consortium Telomere Group(12). In addition to single variant analyses, we also constructed a weighted linear combination of subject genotype at the eight SNPs to create a summary score that quantifies genotypic contributions to differences in LTL across patients and controls. While direct measurement of LTL (e.g. by qPCR) is frequently influenced by differing distributions of potential confounders across patients and controls (e.g. age, sex, chemotherapy), SNP genotypes are present since birth and are not confounded by the effect that these variables may have on both telomere length and cancer risk. Additionally, while the effect of individual alleles in telomere-maintenance genes may be small, the combined effect of numerous such polymorphisms could potentially be quite large and could help identify the “missing heritability” of cancer(16). This Mendelian randomization approach for examining the relationship between telomere length and cancer risk has been previously applied to glioma, melanoma and lung adenocarcinoma, but not to hematologic malignancies (17–19).
MATERIALS AND METHODS
Ethics statement
The genome-wide meta-analysis of mean LTL obtained approval by local ethics committees, as previously outlined(12). CLL patients and associated genotype and phenotype data in the discovery analyses were collected by the Mayo Clinic as part of the Eastern Cooperative Oncology Group (ECOG) 2997 trial and accessed through dbGaP Study Accession phs000621.v1.p1 after review and approval by the NCI Data Access Committee(2). CLL patients and associated genotype and phenotype data in the replication analyses were collected by The Dana-Farber Cancer Institute (DFCI) and accessed through dbGaP Study Accession phs000435.v2.p1 after review and approval by the NHGRI Data Access Committee(1).
Case-control populations
We assessed genetic variants in telomere-associated genes in two independent CLL case-control study groups. The first group included 215 CLL patients from the ECOG 2997 trial (2) and 3390 Illumina iControl subjects. ECOG patients had symptomatic, untreated CLL as defined by NCI 1996 guidelines(20). The second group included 101 non-overlapping CLL patients from DFCI and 2603 controls from the Wellcome Trust Case-Control Consortium (WTCCC) (1, 21). The majority (67%) of DFCI cases were chemotherapy-naïve. Additional case and control data appear in Supplementary Table 1.
Case-control genotyping and quality-control
For ECOG patients, DNA was isolated from peripheral blood, obtained before treatment and shipped overnight at ambient temperature to a central processing laboratory. Mononuclear cells were isolated using Ficoll density-gradient centrifugation and then processed for DNA. All case and control DNA specimens in discovery analyses were genotyped using either the Illumina HumanCNV370-Duo BeadChip or the Illumina HumanHap550 platform. Samples with call rates <95% were excluded from analysis, as were samples with mismatched reported versus genotyped sex. Although all subjects were of self-reported European-ancestry, this was validated using principal components analysis in Eigenstrat(22). Analyzed SNPs had call rates >98% and Hardy-Weinberg equilibrium p-values >0.001 among controls.
For DFCI patients, DNA samples were obtained from normal tissue via either 5mm skin punch biopsies or collection of 2ml of saliva as a source of normal epithelial cells. DNA specimens were genotyped using the Affymetrix 6.0 genotyping array. Genotype data for 2603 European-ancestry control samples genotyped using the Affymetrix 6.0 genotyping array were downloaded from the Wellcome Trust Case-Control Consortium(21). Subjects showing evidence of non-European ancestry, as well as duplicate samples and related subjects (IBD > 0.125) were excluded from analyses. Genome-wide SNP data were used to ensure there was no overlap between ECOG patients and DFCI patients. SNPs with call rates <98% or HWE p-value <0.001 (among controls) were excluded.
To ensure that infiltrating tumor DNA from ECOG patients did not bias allele frequencies in SNP analyses of blood-derived specimens, genome-wide allele frequencies from ECOG patients (blood-derived specimens) were compared to those in DFCI patients (skin biopsy or saliva-derived specimens). With the exception of SNPs on 13q, allele frequencies differed by no more than 5% across patient sets, reflecting the relatively low burden of copy-number alterations in CLL and the comparability of the two CLL case populations(23, 24). Importantly, there was no evidence of copy-number alterations or copy-neutral loss-of-heterozygosity within 250kb of the eight LTL-associated SNPs, as the most common CLL-associated copy-number alterations are located on chromosomes 11,13, 17 (loss) and 12 (gain)(25), while the LTL-associated SNPs are located on chromosomes 2, 3, 4, 5, 10, 17, 19 and 20. As further evidence, allele frequencies in flow-sorted CLL tumor DNA were nearly identical to those in skin biopsy DNA in the eight LTL-associated gene regions among DFCI subjects (Supplementary Figure 1).
SNP imputation
Within the two CLL case-control datasets, we imputed 250kb regions centered on eight SNPs previously associated with LTL in GWAS(12, 13): rs11125529 (ACYP2), rs10936599 (TERC), rs7675998 (NAF1), rs2736100 (TERT), rs9420907 (OBFC1), rs3027234 (CTC1), rs8105767 (ZNF208), and rs755017 (RTEL1). The top LTL SNP was directly genotyped on-array for TERC, TERT and OBFC1 among ECOG cases and the iControls, and was imputed for the other five genes. The top LTL SNP was directly genotyped on-array for ACYP2, TERT, OBFC1, CTC1 and ZNF208 among the DFCI cases and Wellcome Trust controls, and was imputed for the other three genes. Imputation was performed using the Impute2 v2.1.2 software and its standard Markov chain Monte Carlo algorithm and default settings for targeted imputation(26). All 1,000 Genomes Phase I haplotypes were provided as the imputation reference panel(27). All SNPs had imputation quality (info) scores > 0.80 and posterior probabilities > 0.90. Individuals with imputed genotype probabilities <0.70 were excluded to prevent allele misclassification. Imputation was performed separately for the ECOG/iControl case-control set (Illumina array data) and the DFCI/WTCCC dataset (Affymetrix array data).
Statistical analyses
For single locus SNP associations, allele frequencies in all CLL patients were compared to those in the pooled control dataset using logistic regression in SNPTESTv2 under an allelic additive model(28), adjusting for the first 5 ancestry-informative principal components from Eigenstrat and for genotyping array (Illumina vs. Affymetrix). To account for potential errors in imputation, a missing-data likelihood score-test was applied to produce standard errors which account for the additional uncertainty inherent in the analysis of imputed genotypes. P-values <0.05 were considered nominally significant. Sensitivity analyses were conducted wherein cases were compared only to controls genotyped on the same genotyping platform to account for potential differences in genotyping or imputation across arrays.
To investigate the combined effect of the eight LTL-associated SNPs, we created a weighted linear combination by summing the number of “long LTL” alleles that an individual possesses and weighting each allele by its effect size in data from the ENGAGE Consortium Telomere Group(12). The effect size used for weighting was expressed as the number of additional base-pairs of telomere length associated with each allele, adjusted for age and sex, as calculated and previously reported by the ENGAGE Consortium Telomere Group (12, 17). The number of base-pairs was used because the model can be interpreted as the relative difference in estimated LTL across individuals. The weighted model assigns a value of “0” to an individual who possesses 0 of the alleles associated with longer LTL, while an individual possessing all sixteen alleles (two alleles at each of eight SNPs) would have a value of “1215”. These differences in genotypically-estimated LTL were compared between CLL patients and controls using logistic regression, adjusted for the first 5 ancestry-informative principal components from Eigenstrat and genotyping array(22). Odds ratios correspond to the change in cancer risk relative to a one standard deviation (131.9bp) increase in genotypically-estimated LTL, with the standard deviation defined among the pooled controls. P-values <0.05 were considered statistically significant in both discovery and replication analyses. Associations between LTL and tumor stage were tested using the Spearman rank correlation test. All statistical tests presented in the manuscript are two-sided.
RESULTS
After excluding samples with poor call-rates, duplicate samples, cryptically-related individuals, subjects with non-European ancestry, and subjects with imputed genotype probabilities <0.70 for one or more of the eight LTL-associated SNPs, a total of 273 CLL patients (179 ECOG, 94 DFCI) and 5725 controls (3166 Illumina iControls, 2559 Wellcome Trust controls) remained for analyses. The weighted linear combination of the 8 LTL-associated SNPs (16 LTL-associated alleles) ranged from a minimum value of 115bp to a maximum of 1008bp. Per a 31bp/year rate of LTL attrition, this 893bp difference in genotypically-estimated LTL between maximum and minimum values corresponds to an approximately 30 year difference in age-associated telomere attrition. Because the LTL estimates were determined using inherited autosomal SNPs, present since birth, there was no association between the weighted sum and either subject age or sex (Supplementary Figure 2). The mean value of the weighted sum was similar in both control groups (Illumina iControls=541bp, Wellcome Trust controls=544bp).
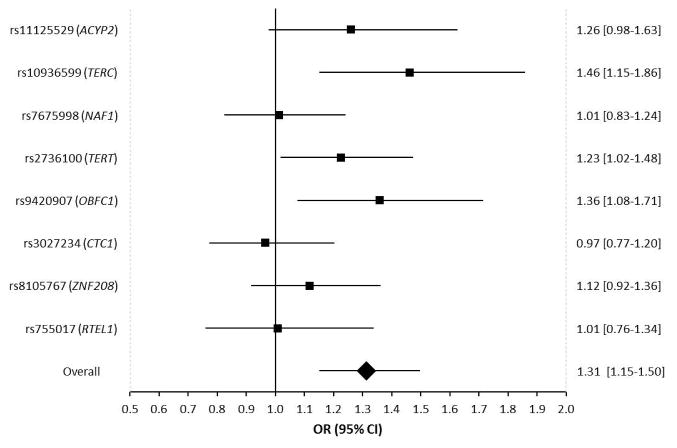
In case-control analyses of the LTL-associated SNPs, the allele associated with longer LTL was associated with increased CLL risk for seven of the eight evaluated SNPs (Figure 1). For three of these SNPs, this association was nominally significant (P<0.05): rs10936599 in TERC (O.R.=1.46; 95% C.I.=1.15–1.86; P=1.8×10−3), rs2736100 in TERT (O.R.=1.23; 95%C.I.=1.02–1.48; P=0.030), and a novel association at rs9420907 in OBFC1 (O.R.=1.36; 95%C.I.=1.08–1.71; P=9.6×10−3) (Table 1, Figure 1). Additionally, a suggestive association was detected at rs11125529 in ACYP2 (OR=1.26; 95%C.I.=0.98–1.63; P=0.075).
Figure 1. Forest plot showing the effect of alleles associated with longer leukocyte telomere length on CLL risk.
Allelic odds ratios are plotted with 95% confidence intervals. The overall estimate is for the combined effect of all 8 SNPs, where the odds ratio relates to the change in CLL risk for one standard deviation increase in genotypically-estimated leukocyte telomere length. Odds ratios are based on combined data from 273 CLL patients and 5725 controls.
Table 1.
Results for each telomere length-associated SNP, including effect on telomere length in the ENGAGE Consortium genome-wide meta-analysis and on CLL risk in the pooled CLL case-control dataset.
| SNPa | Chromosome | Gene | Effect Alleleb | Effect on LTL | Effect on CLL risk | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| EAFc | Beta | BPd | P | EAFe | ORf | 95% CI | P | ||||
|
| |||||||||||
| rs11125529 | 2 | ACYP2 | A | 14% | 0.056 | 66.9 | 4.5×10−8 | 13% | 1.26 | 0.98–1.63 | 0.075 |
| rs10936599 | 3 | TERC | C | 75% | 0.097 | 117.3 | 2.5×10−31 | 76% | 1.46 | 1.15–1.86 | 1.8×10−3 |
| rs7675998 | 4 | NAF1 | G | 78% | 0.074 | 89.7 | 4.3×10−16 | 77% | 1.01 | 0.83–1.24 | 0.90 |
| rs2736100 | 5 | TERT | C | 49% | 0.078 | 94.2 | 4.4×10−19 | 51% | 1.23 | 1.02–1.48 | 0.030 |
| rs9420907 | 10 | OBFC1 | C | 14% | 0.069 | 82.8 | 6.9×10−11 | 14% | 1.36 | 1.08–1.71 | 9.6×10−3 |
| rs3027234 | 17 | CTC1 | C | 79% | 0.021 | 25.3 | 0.020 | 78% | 0.97 | 0.77–1.20 | 0.75 |
| rs8105767 | 19 | ZNF208 | G | 29% | 0.048 | 57.6 | 1.1×10−9 | 29% | 1.12 | 0.92–1.36 | 0.28 |
| rs755017 | 20 | RTEL1 | G | 13% | 0.062 | 74.1 | 6.7×10−9 | 12% | 1.01 | 0.76–1.34 | 0.96 |
All SNPs were associated with LTL in a previous GWAS and replicated at P<0.05 in the ENGAGE Consortium genome-wide meta-analysis(12)
The effect allele is the allele associated with increased leukocyte telomere length.
Effect allele frequency (EAF) calculated in all ENGAGE Consortium subjects (N=37,684)
Base pair (BP) estimates of the per-allele effect on LTL in base pairs calculated from the equivalent age-related attrition in telomere repeat length ratio, as previously described(12)
Effect allele frequency (EAF) calculated in CLL control subjects
Odds ratios (OR) are for each additional copy of the allele associated with longer LTL, calculated in 273 CLL patients and 5725 controls. Odds ratios >1.0 indicate that the “long” allele is more common in CLL patients and the “short” allele is more common in controls.
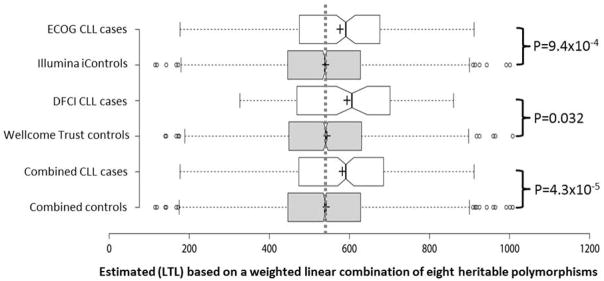
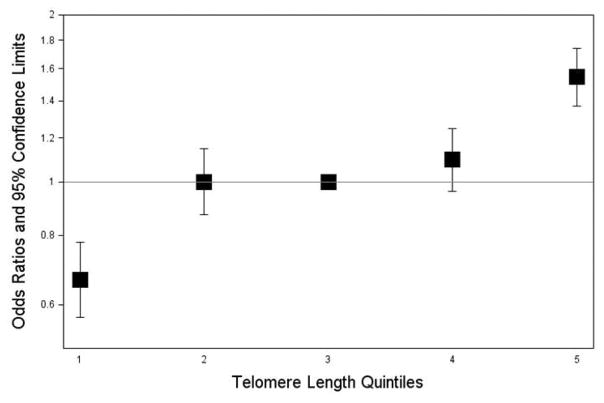
The weighted linear combination of the 8 LTL-associated SNPs was significantly greater in CLL patients than in controls (582bp vs. 542bp, Figure 2), with each standard deviation increase in genotypically-estimated LTL (131.9bp) associated with a 1.31-fold increase in the odds of CLL (95%CI=1.15–1.50; P=4.3×10−5) (Figure 1). Importantly, this statistically significant association was observed in both the ECOG/iControl case-control set (OR=1.31; 95%CI=1.12–1.52; P=9.4×10−4) and the DFCI/WTCCC case-control set (OR=1.32; 95%CI=1.02–1.71; P=0.032) (Figure 2). This provides strong support for both the direction and magnitude of the association. The odds ratio for CLL increased monotonically with increasing quintiles of LTL (Figure 3). Individuals in the highest LTL quintile had a 2.3-fold increased risk of CLL compared to individuals in the lowest LTL quintile (O.R.=2.32; 95% C.I.=1.51–3.58; P=1.3×10−4).
Figure 2. Boxplots of genotypically-estimated leukocyte telomere length (LTL) in CLL patients and controls from the Eastern Cooperative Oncology Group 2997 trial (ECOG) (N=179), Illumina iControls (N=3166), CLL patients from the Dana Farber Cancer Institute (DFCI) (N=94), Wellcome Trust controls (N=2559), and combined analyses (273 CLL patients, 5725 controls).
The vertical dotted line shows the average value in the pooled control sets (542bp). Crosses show samples means; center lines show sample medians. Box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles. Outliers are represented by dots. The notches in boxes are defined as ±1.58*IQR/sqrt(n) and give roughly 95% confidence intervals for sample medians. P-values are adjusted for the first five ancestry-informative principal components and, in the combined analysis, for genotyping platform.
Figure 3. Effect of increasing quintile of genotypically-estimated leukocyte telomere length on CLL risk in combined discovery and replication datasets.
The odds ratios are relative to the median (third) quintile. Vertical bars correspond to 95% confidence intervals. Quintiles were defined among controls and ranges were: quintile one (115bp–432bp), quintile two (433bp–512bp), quintile three (513bp–582bp), quintile four (583bp–652bp), quintile five (653bp–1008bp). Odds ratios are based on combined data from 273 CLL patients and 5725 controls.
To determine if the association between a genetic predisposition to longer telomeres and increased CLL risk was entirely attributable to the previously reported associations between CLL risk and SNPs in TERT and TERC, we re-calculated the weighted linear combination of the 8 LTL-associated SNPs in patients and controls using only the six SNPs located near genes not previously associated with CLL risk (ACYP2, NAF1, OBFC1, CTC1, ZNF208 and RTEL1). Using this reduced 6-SNP estimate of LTL, CLL patients still had a genetic predisposition to significantly longer LTL than controls (P=0.039) (Supplementary Figure 3).
Previous studies indicate that shorter telomere length in CLL tumors is associated with more aggressive disease. We observed a suggestive inverse association between the weighted linear combination and higher tumor stage (P=0.081), indicating that a genetic predisposition to shorter LTL may be associated with risk of more aggressive CLL subtypes.
DISCUSSION
We identify inherited genetic variants in TERC, TERT and OBFC1 that are associated with an increased risk of CLL in case-control analyses. SNPs in these genes have previously been associated with inter-individual variation in LTL(12). By integrating CLL case-control data with data from our previous GWAS of LTL, we observed that CLL risk alleles in TERC, TERT and OBFC1 are associated with longer LTL. While it has previously been reported that inherited SNPs in TERC and TERT are associated with CLL risk(14, 15), our analyses identify a potential mechanism through which these SNPs may impact leukemia risk - by increasing an individual’s LTL. This is supported by our observation that a genetic predisposition to longer LTL is associated with increased CLL risk, even after removing the contributions of TERC and TERT SNPs from the model. Although each of the LTL-associated variants explains only a small proportion of the total variance in telomere length across individuals (12), the summary variable made by combining the eight SNPs accounted for a 893bp difference in LTL. Assuming an annual telomere attrition rate of ~30bp/year in leukocytes, this translates to more than 30 years of age-related telomere attrition.
Telomere length helps cells to regulate the precipitous balance between mitotic competence and cellular senescence and is influenced by both inherited genetic variants and acquired somatic mutations. In studies where telomere length was measured in CLL tumor cells, shorter telomere length was correlated with more aggressive disease(4, 29, 30). Telomere length in cancer tissue can also be affected by patient age, therapeutic regimen, and the rate of tumor proliferation(31). Our analyses use a Mendelian randomization approach to estimate genotypically-determined telomere length in healthy leukocytes, free from the confounding effects of age, sex, therapy, subclinical tumor or disease aggressiveness. Our results complement a recent case-control study, nested within the EPIC cohort, that directly measured LTL in prediagnostic blood specimens from patients with B-cell lymphoma. Similar to our results, they observed that longer LTL was associated with increased cancer risk(32).
Mendelian randomization is an epidemiologic technique that can elucidate the causality of associations(33). As genotypes of individuals are randomized at birth, such studies can eliminate both exogenous confounding factors and the phenomenon of reverse causation. However, factors like linkage disequilibrium, population stratification, and pleiotropy can affect interpretations of Mendelian randomization studies (34). As we analyzed unlinked SNPs that are located on separate autosomes, it is unlikely that linkage disequilibrium would influence the results. Similarly, biases due to population stratification were likely eliminated by carefully excluding individuals with non-European ancestry and adjusting for principal components in all analyses. As genetic variation near TERC and TERT may influence cancer risk independent of its effect on telomere length(35), there remains a possibility that pleiotropy could partially underlie the association between LTL and CLL risk observed in our data. Although our sample size was limited to fewer than 300 cases, we observe statistically significant and extremely concordant effects in both the discovery and replication datasets. Future studies in larger and more ethnically diverse populations appear warranted.
Although an association between CLL risk and both shelterin component genes (POT1) and telomerase component genes (TERC and TERT) has been previously reported, this is the first report of a significant association between CLL risk and a CST complex gene (OBFC1). The human CST complex is encoded by three genes: CTC1, OBFC1 and TEN1. The CST complex competes with shelterin for telomeric DNA and inhibits telomerase-based telomere extension through primer sequestration and physical interaction with the POT1–TPP1 telomerase processivity factor(36). Through binding of the telomerase-extended telomere, CST limits telomerase activity and restricts telomere extension to approximately one event per cell-cycle(36).
An association between longer LTL and increased cancer risk was recently observed in glioma, melanoma and lung adenocarcinoma, where a genetic predisposition to longer telomere length was associated with increased risk of malignancy(17–19). Interestingly, patients diagnosed with CLL are at two-fold to four-fold increased risk of developing melanoma(37, 38). Although this increased risk has been primarily attributed to the immunosuppressive effect of CLL, a common genetic etiology involving telomere lengthening may also contribute. Indeed, a common genetic etiology for melanoma and glioma has already been reported, mediated by shared heritable risk variants in TERC, TERT, and POT1(39).
Several studies have focused on identifying the underlying cause of shorter telomere length in leukemic cells. While a predisposition to longer telomeres at baseline might provide pre-cancerous cells with additional opportunities to acquire mutations and undergo malignant transformation(6, 40, 41), subsequent telomere attrition in aggressive and highly-proliferative tumors could cause leukemic cells to have shorter telomere length than healthy cells(42). This is supported by studies revealing that early-stage CLL (i.e, Binet stage A and Rai stage 0) shows no significant difference between telomere length measured in tumor cells and in healthy cells(7, 29), while telomere length is significantly shorter in tumor cells than healthy cells taken from patients with advanced disease (9, 10). Thus, shorter telomere length in tumor cells may be a marker of high-risk CLL in association with increased tumor burden, perhaps due to an increased rate of telomere attrition in highly proliferative tumors(30).
We observe a strong and consistent association between a genetic predisposition to longer LTL and increased risk of CLL, similar to previous results for lung adenocarcinoma, melanoma and glioma(17–19). Future work is needed to understand the cellular mechanisms underlying this association and to explore the role of OBFC1 and other CST complex genes in leukemogenesis. These studies of CLL initiation will complement existing knowledge about disease progression and can move the field toward better understanding the breadth of factors involved in CLL biology.
Supplementary Material
Acknowledgments
FUNDING:
Work at The University of California, San Francisco was supported by the National Institutes of Health, grant numbers: R01CA52689 (M.R. Wrensch, J.K. Wiencke, J.L. Wiemels, I.V. Smirnov, H.M. Hansen, N.R. Madsen, P.M. Bracci), P50CA097257 (K.M. Walsh, J.K. Wiencke, M.R. Wrensch, N.R. Madsen, J. Ojha), R01CA126831 (M.R. Wrensch, J.K. Wiencke, H.M. Hansen) and R01CA139020 (M.R. Wrensch), The Sontag Foundation (K.M. Walsh), the UCSF Lewis Chair in Brain Tumor Research (M.R. Wrensch), the UCSF Robert Magnin Newman chair in Neuro-Oncology (J.K. Wiencke), and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen (All authors). Work at University of Leicester was undertaken under the framework of European Union Framework 7 ENGAGE Project HEALTH-F4-2007- 201413 (V. Codd, N.J. Samani). N.J. Samani, V. Codd and C.P. Nelson are supported by the British Heart Foundation.
The results published here are, in part, based upon data obtained from dbGaP Study Accession: phs000435.v2.p1 “Whole Exome Sequencing of Chronic Lymphocytic Leukemia”. This work was supported by the National Human Genome Research Institute (5U54HG003067), the Blavatnik Family Foundation, NCI (5R21CA115043-2), the Howard Hughes Medical Institute and the Damon-Runyon Cancer Research Foundation (CI-38-07).
The results published here are, in part, based upon data obtained from dbGaP Study Accession: phs000621.v1.p1 “Genome Wide Association Studies in ECOG 2997 Trial”. Samples and associated genotype and phenotype data were provided by the Mayo Clinic with funding support provided through a cooperative agreement with NCI grant R01CA132780.
This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475. Additional controls were accessed from Illumina’s iControlDB database.
Footnotes
CONFLICT OF INTEREST STATEMENT: The authors report no conflicts of interest.
References
- 1.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grever MR, Lucas DM, Dewald GW, Neuberg DS, Reed JC, Kitada S, et al. Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. J Clin Oncol. 2007;25:799–804. doi: 10.1200/JCO.2006.08.3089. [DOI] [PubMed] [Google Scholar]
- 3.Rossi D, Lobetti Bodoni C, Genuardi E, Monitillo L, Drandi D, Cerri M, et al. Telomere length is an independent predictor of survival, treatment requirement and Richter’s syndrome transformation in chronic lymphocytic leukemia. Leukemia. 2009;23:1062–72. doi: 10.1038/leu.2008.399. [DOI] [PubMed] [Google Scholar]
- 4.Rampazzo E, Bonaldi L, Trentin L, Visco C, Keppel S, Giunco S, et al. Telomere length and telomerase levels delineate subgroups of B-cell chronic lymphocytic leukemia with different biological characteristics and clinical outcomes. Haematologica. 2012;97:56–63. doi: 10.3324/haematol.2011.049874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn EH. Walking the walk from genes through telomere maintenance to cancer risk. Cancer Prev Res (Phila) 2011;4:473–5. doi: 10.1158/1940-6207.CAPR-11-0066. [DOI] [PubMed] [Google Scholar]
- 6.Walsh KM, Rice T, Decker PA, Kosel ML, Kollmeyer T, Hansen HM, et al. Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro Oncol. 2013;15:1041–7. doi: 10.1093/neuonc/not051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoxha M, Fabris S, Agnelli L, Bollati V, Cutrona G, Matis S, et al. Relevance of telomere/telomerase system impairment in early stage chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2014;53:612–21. doi: 10.1002/gcc.22171. [DOI] [PubMed] [Google Scholar]
- 8.Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 9.Lin TT, Norris K, Heppel NH, Pratt G, Allan JM, Allsup DJ, et al. Telomere dysfunction accurately predicts clinical outcome in chronic lymphocytic leukaemia, even in patients with early stage disease. Br J Haematol. 2014;167:214–23. doi: 10.1111/bjh.13023. [DOI] [PubMed] [Google Scholar]
- 10.Dos Santos P, Panero J, Palau Nagore V, Stanganelli C, Bezares RF, Slavutsky I. Telomere shortening associated with increased genomic complexity in chronic lymphocytic leukemia. Tumour Biol. 2015 doi: 10.1007/s13277-015-3556-2. [DOI] [PubMed] [Google Scholar]
- 11.Mirabello L, Yu K, Kraft P, De Vivo I, Hunter DJ, Prescott J, et al. The association of telomere length and genetic variation in telomere biology genes. Hum Mutat. 2010;31:1050–8. doi: 10.1002/humu.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422–7. 7e1–2. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangino M, Hwang SJ, Spector TD, Hunt SC, Kimura M, Fitzpatrick AL, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum Mol Genet. 2012;21:5385–94. doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berndt SI, Skibola CF, Joseph V, Camp NJ, Nieters A, Wang Z, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45:868–76. doi: 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speedy HE, Di Bernardo MC, Sava GP, Dyer MJ, Holroyd A, Wang Y, et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2014;46:56–60. doi: 10.1038/ng.2843. [DOI] [PubMed] [Google Scholar]
- 16.Zaitlen N, Kraft P. Heritability in the genome-wide association era. Hum Genet. 2012;131:1655–64. doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh KM, Codd V, Rice T, Nelson CP, Smirnov IV, McCoy LS, et al. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015;6:42468–77. doi: 10.18632/oncotarget.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iles MM, Bishop DT, Taylor JC, Hayward NK, Brossard M, Cust AE, et al. The effect on melanoma risk of genes previously associated with telomere length. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Doherty JA, Burgess S, Hung RJ, Lindstrom S, Kraft P, et al. Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum Mol Genet. 2015;24:5356–66. doi: 10.1093/hmg/ddv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O’Brien S, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–7. [PubMed] [Google Scholar]
- 21.Genome-wide association study of 14,000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Ouillette P, Collins R, Shakhan S, Li J, Peres E, Kujawski L, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118:3051–61. doi: 10.1182/blood-2010-12-327858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunnarsson R, Mansouri L, Isaksson A, Goransson H, Cahill N, Jansson M, et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica. 2011;96:1161–9. doi: 10.3324/haematol.2010.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouillette P, Malek S. Acquired genomic copy number aberrations in CLL. Adv Exp Med Biol. 2013;792:47–86. doi: 10.1007/978-1-4614-8051-8_3. [DOI] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 29.Bechter OE, Eisterer W, Pall G, Hilbe W, Kuhr T, Thaler J. Telomere length and telomerase activity predict survival in patients with B cell chronic lymphocytic leukemia. Cancer Res. 1998;58:4918–22. [PubMed] [Google Scholar]
- 30.Mansouri L, Grabowski P, Degerman S, Svenson U, Gunnarsson R, Cahill N, et al. Short telomere length is associated with NOTCH1/SF3B1/TP53 aberrations and poor outcome in newly diagnosed chronic lymphocytic leukemia patients. Am J Hematol. 2013;88:647–51. doi: 10.1002/ajh.23466. [DOI] [PubMed] [Google Scholar]
- 31.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;15(17):R109–15. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 32.Hosnijeh FS, Matullo G, Russo A, Guarrera S, Modica F, Nieters A, et al. Prediagnostic telomere length and risk of B-cell lymphoma-Results from the EPIC cohort study. Int J Cancer. 2014;135:2910–7. doi: 10.1002/ijc.28934. [DOI] [PubMed] [Google Scholar]
- 33.Gray R, Wheatley K. How to avoid bias when comparing bone marrow transplantation with chemotherapy. Bone Marrow Transplant. 1991;7(Suppl 3):9–12. [PubMed] [Google Scholar]
- 34.Jansen H, Samani NJ, Schunkert H. Mendelian randomization studies in coronary artery disease. Eur Heart J. 2014;35:1917–24. doi: 10.1093/eurheartj/ehu208. [DOI] [PubMed] [Google Scholar]
- 35.De Semir D, Nosrati M, Li S, Kashani-Sabet M. Telomerase: going beyond the ends. Cell Cycle. 2007;6:546–9. doi: 10.4161/cc.6.5.3980. [DOI] [PubMed] [Google Scholar]
- 36.Chen LY, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–4. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 37.Brewer JD, Shanafelt TD, Call TG, Cerhan JR, Roenigk RK, Weaver AL, et al. Increased incidence of malignant melanoma and other rare cutaneous cancers in the setting of chronic lymphocytic leukemia. Int J Dermatol. 2015;54:e287–93. doi: 10.1111/ijd.12564. [DOI] [PubMed] [Google Scholar]
- 38.Famenini S, Martires KJ, Zhou H, Xavier MF, Wu JJ. Melanoma in patients with chronic lymphocytic leukemia and non-Hodgkin lymphoma. J Am Acad Dermatol. 2015;72:78–84. doi: 10.1016/j.jaad.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 39.Walsh KM, Wiencke JK, Lachance DH, Wiemels JL, Molinaro AM, Eckel-Passow JE, et al. Telomere maintenance and the etiology of adult glioma. Neuro Oncol. 2015;17:1445–52. doi: 10.1093/neuonc/nov082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holt SE, Shay JW, Wright WE. Refining the telomere-telomerase hypothesis of aging and cancer. Nat Biotechnol. 1996;14:836–9. doi: 10.1038/nbt0796-836. [DOI] [PubMed] [Google Scholar]
- 41.Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–5. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sellmann L, de Beer D, Bartels M, Opalka B, Nuckel H, Duhrsen U, et al. Telomeres and prognosis in patients with chronic lymphocytic leukaemia. Int J Hematol. 2011;93:74–82. doi: 10.1007/s12185-010-0750-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.