Abstract
The Oxford Nanopore MinION sequences DNA by sensing changes in electrical current flow in real-time as molecules traverse nanopores. Optionally, the voltage across specific nanopores can be reversed, ejecting the DNA molecule. This enables “Read Until”, the selection of specific DNA molecules for sequencing. We use dynamic time warping to match reads to reference, selecting regions of small genomes, individual amplicons, or normalization of the amplicon set. This first demonstration of direct selection of specific DNA molecules in real-time enables many novel future applications.
Nanopore sequencing, represented by Oxford Nanopore Technologies (ONT) MinION sequencer, enables a new paradigm for real-time analysis1. MinION reads are electrical current measurements with values determined by the specific DNA bases in contact with the nanopore (Supplementary Fig. 1). Uniquely the sequencer streams current values from individual nanopores as DNA molecules pass through, enabling analysis of current traces during sequencing. The MinION streams data from all channels simultaneously, with each channel individually addressable and able to reverse the voltage across its pore, rejecting the read. In principle this enables selective sequencing. Molecules sampled by the sequencer can be either sequenced to completion or rejected and an alternative molecule sampled. Rejected reads are unlikely to be sequenced again as the motor protein required for sequencing will already have migrated along the DNA2–4. This type of approach has been described, but not yet applied, by Oxford Nanopore and named “Read Until”.
To maximize selective sequencing, identification must be accomplished before the read completes. This depends on two parameters. Firstly, speed of sequence identification and placement on the reference and secondly, average read length of the library. Read length is limited by input material, with mapped reads reported exceeding 100 kb5. “Read Until” requires matching the shortest fragment of the longest possible read to a specific position within the reference. However, implementation of “Read Until” is complicated by the fact that sequence data streamed by the device is a current squiggle, not a decoded sequence of bases. To date there are no publicly available methods for matching squiggle data directly to a reference.
One approach to “Read Until” would be base calling read fragments, but current implementations are too slow and benefit from subsequent optimizations to maximize base call quality1,6–8. Therefore, we focused on matching current data directly to a reference sequence in squiggle space. The matching of squiggles shares much in common with comparison of audio signals and so we turned to dynamic time warping (DTW), an algorithm first applied to the matching of speech9,10. Guaranteed to find the optimal alignment of two series of time-ordered data, DTW has been previously used in analysis of sequence data11.
We use DTW to match observed short squiggles to a reference in real-time, manipulating the output from the sequencer to demonstrate selective sequencing on the MinION. We apply this approach to two problems. The selection of specific regions from a whole genome library of Lambda and the selective sequencing of individual amplicons from a pooled set of molecules to provide efficient coverage balancing, motivated by recent field experiments with Ebola12. All methods and scripts are freely available under the MIT license from https://github.com/mattloose/RUscripts including methods for post processing reads after sequencing is complete. Jupyter notebooks for figures are also available at https://github.com/mattloose/RUFigs. All read data are available from the European Nucleotide Archive (Supplementary Table 1). The API required for addressing the sequencer is available directly from Oxford Nanopore Technologies on request.
Matching fragments of read to a reference requires converting the reference to a hypothetical current trace. The appropriate model can be extracted from a base called MinION FAST5 file, and an expected squiggle calculated for a reference sequence (Supplementary Fig. 1, Supplementary Table 2). Given two perfect squiggles it is possible to match one to another by visual inspection. This process is complicated by the sequencing environment including voltage changes, noise and interactions between channels. These behaviors are captured by three variables: shift, scale and drift, which together describe the variation between pores and the underlying model (Supplementary Fig. 2). The base caller provided by ONT most likely calculates these values using expectation-maximization but derives the data to do so from the entire read8. For short nanopore reads z-score normalisation can be used which overcomes shift and scale, but not drift. Based on examination of these parameters drift is insignificant and so we z-score normalized both reference and incoming read fragments. Raw current traces from nanopores are subdivided into events, representing specific DNA sequences within the pore, and errors in event detection resulting in event insertions/deletions for identical molecules (Supplementary Fig. 1)8. DTW subsequence search compensates for residual differences in signal between read and reference and potential insertions/deletions in events measuring similarity between temporal sequences varying in amplitude or speed10,11.
Using synthetic data any 8mer squiggle from lambda can be correctly mapped with DTW (Supplementary Fig. 3). On real data longer query sequences are required presumably due to noise. Without normalization, only 40% of data can be mapped correctly. This is improved by z-score normalization and match length. The naive DTW algorithm has O(m×n) complexity where m is reference length and n query length. Reducing query length will improve search time. We chose 250 events as a balance between speed and accuracy, matching 1 read every 0.3 seconds on a single compute core. Over 8 cores, 512 channels were processed in 19.2s suggesting “Read Until” could be attainable using DTW. At current sequencing speeds (70 b/s) 250 events represents 3.5 seconds data collection.
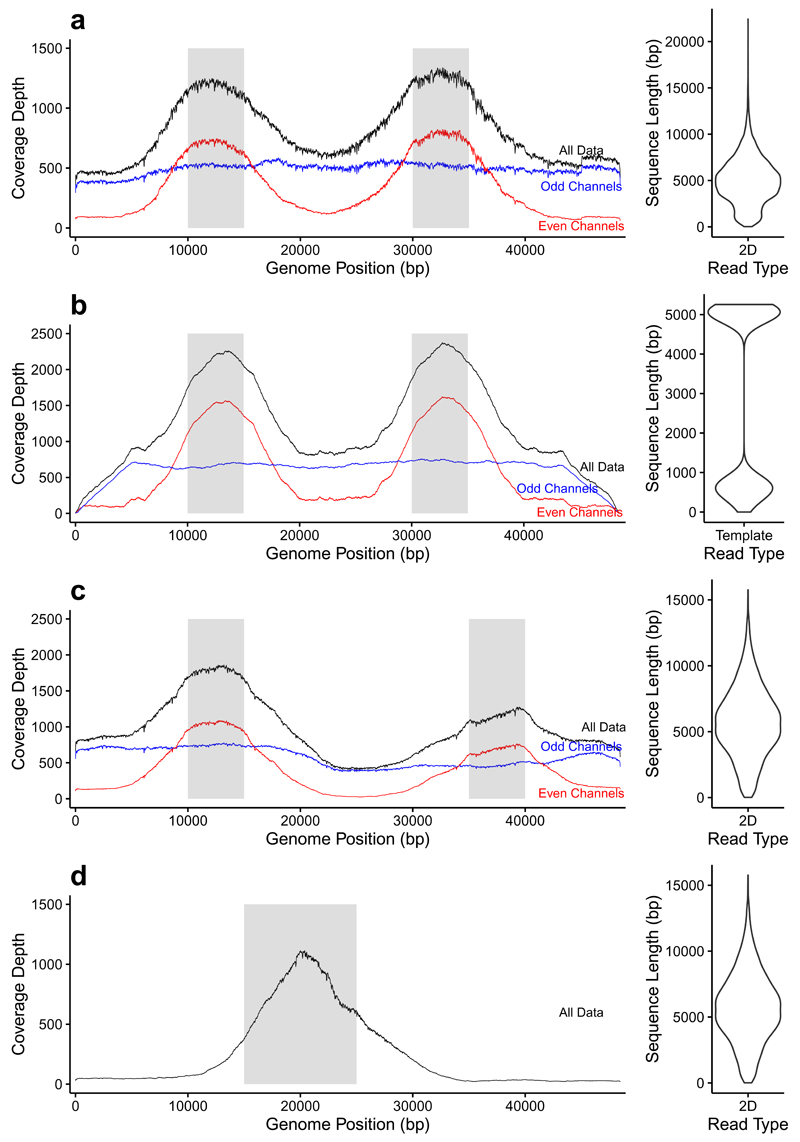
We developed “Read Until” applications using lambda DNA; target enrichment and amplicon sequencing (Supplementary Fig. 4). Lambdas small genome size (48,502 bp) results in a total search space of just 97,004 bp. We enriched two 5kb regions, from 10-15 kb and 30-35 kb, rejecting all other reads. As an internal control, we applied “Read Until” on even numbered channels only. Adjacent channels sample reads from the same pool of molecules allowing for a direct comparison. We sequenced for 47 hours at 30 b/s generating 31,609 reads of which 8,901 were 2D and 8,178 alignable to the reference giving a mean coverage of 840×. Considering all channels, two peaks are observed correlating with the target regions (Fig. 1A). Splitting these data into odd and even channels reveals even channels rejecting reads mapping outside the target regions. In contrast, reads from the entire genome are found within the odd channels. The peak correlates with the mean 2D read length at 5 kb. For comparison, we simulated this experiment using ideal data and uniform long reads revealing sub optimal enrichment in our experiment (Fig. 1B, Supplementary Fig. 5). One contributing factor is likely to be variation in read lengths seen in the real library compared with simulation. Repeating these experiments using 70 b/s SQK6 chemistry, selecting for reads from 10-15 kb and 35-40 kb, shows the same result (Fig. 1C). To validate our ability to be selective we switched approach on this flowcell, running “Read Until” on all channels selecting reads mapping within 15-25 kb (Fig. 1D).
Figure 1.
Targeted sequencing of specific regions of the lambda genome by direct selection using “Read Until”. (a) shows selective enrichment of the lambda genome in two 5kb regions (10-15 kb and 30-35 kb) sequencing with SQK5 chemistry (30b/s). Read Until is only applied to even numbered channels. (b) repeats this experiment on simulated reads under ideal conditions. Note the more consistent read lengths. (c) shows selective sequencing of lambda using SQK6 chemistry (70 b/s) enriching at 10-15 kb and 35-40 kb. Read Until is only applied to even numbered channels. (d) shows selective sequencing on all channels of one 10 kb region (15-25 kb). Violin plots show 2D read length for each library except (b) which is a template only simulation.
This illustrates one use for “Read Until”: regions can be prioritized until a specific goal has been achieved, then switch to a second experiment analyzing alternative regions from the same library. To process data fast enough these experiments were run using 22 cores (Intel(R) Xeon(R) CPU E5-2690 v3 @ 2.60GHz) on a separate server. With refactored code this experiment can be repeated using only 8 cores, processing each read within approximately 250 bases of the data being available to process (i.e. a total sequence of 550 bases, Supplementary Fig. 6).
The speed of match by DTW is constrained by reference length9. In theory, DTW can be applied to genomes up to 5Mb in length although this requires more compute cores than available on a laptop, with 8 cores required to process 64 channels, suggesting 64 cores would be required for a maximally efficient 512 channel flowcell (Supplementary Fig. 6). Hence, we focus on amplicon sequencing on the MinION, with a resulting smaller search space. This approach is ideally suited to the device; the first major field study to use the MinION exploited this to rapidly sequence amplicons from the 19 kB Ebola genome13. However, this requires adequate coverage of every amplicon with as little sequencing as necessary to minimize the resource required and volume of data to transfer for subsequent analysis.
Several optimizations are possible for amplicon matching with “Read Until”. Removing sequence not from the start or end of an amplicon results in a shorter reference squiggle and increase in matching speed. As minION sequencing is 5”-3”, the effective reference can be halved by removing unnecessary complement sequence. Using a 900 bp window around the start sites for an 11 amplicon approach reduces the reference to 19,800 events from 38,000 (Supplementary Fig. 7). This approach scales to larger genomes since the search space depends on amplicon number, not genome size.
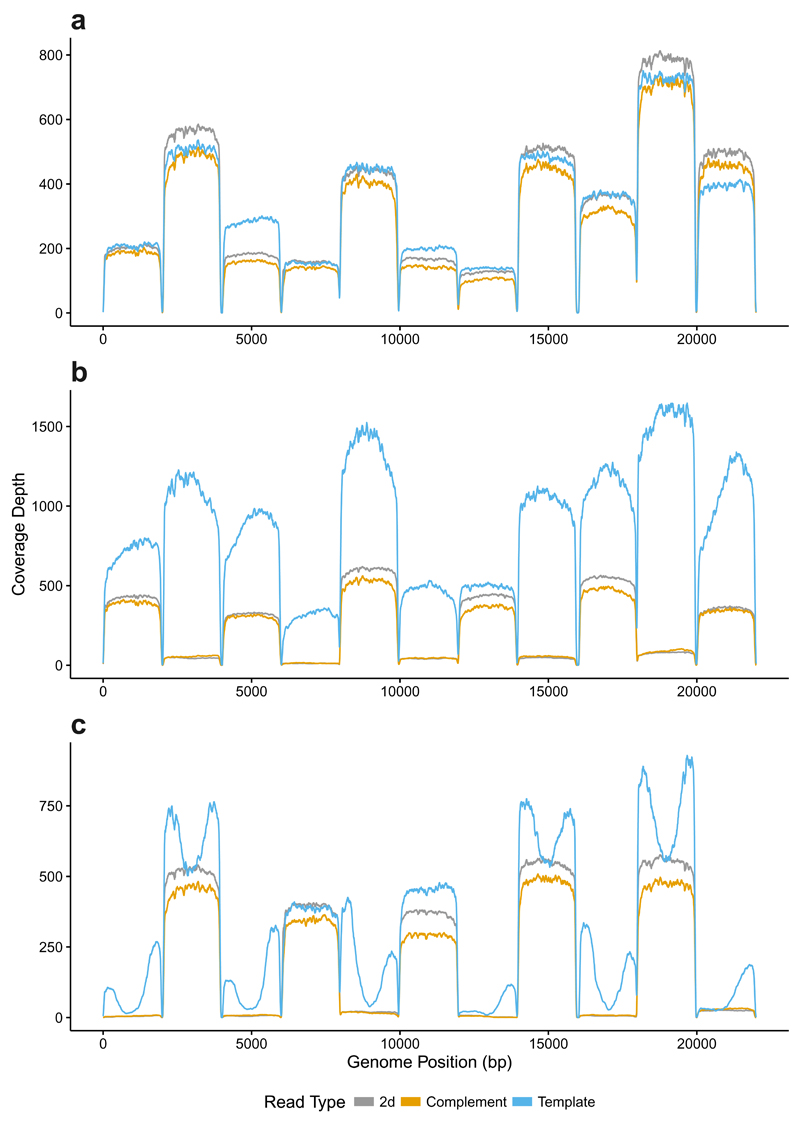
Sequencing Ebola in a lab environment is limited by safety. We therefore used 11 amplicons spanning 22kb of the lambda genome (Supplementary Table 3). Individual amplicons were pooled prior to library preparation at approximately equimolar concentrations and libraries prepared following Quick et al1,12. We performed multiple short duration runs to select for specific amplicons. Fig. 2A shows a control 20 minute run without “Read Until” generating 3,657 reads of which 1,671 were 2D. All 11 amplicons were found, although at different levels. We then applied “Read Until” to exclude even numbered amplicons (Fig. 2B). This was successful, although leakage was observed on rejected amplicons as had been seen with the whole genome selection. Two subsequent repeats of this experiment show significantly improved performance and clear rejection (Supplementary Fig. 8). We then selected the inverse amplicons, successfully blocking sequencing of odd numbered amplicons (Fig. 2C). The “peaks” of template coverage visible correspond to rejected reads, the initial bases of which become template only short reads.
Figure 2.
Selective sequencing of specific individual amplicons from a library using “Read Until”. (a) shows coverage plots for amplicons sequenced without Read Until applied. (b) shows selective sequencing of odd numbered amplicons. 2D reads are absent from even numbered amplicons. Template reads can be seen for all amplicons. (c) shows the inverse relationship where even numbered amplicons are selected for sequencing. The peaks in template coverage seen are the short reads being rejected rapidly by read until. Supplementary Fig. 8 repeats (b) showing this same phenomenon on odd numbered amplicons. Note that the lower quality of complement reads results in lower coverage when mapped to a reference and thus 2D coverage is often higher than either template or complement.
Misplaced reads were characterized by sorting squiggles into individual amplicon groups using DTW. Subsequent alignment of base called data to the reference demonstrates sorting is accurate, although a small proportion of reads are misplaced (Supplementary Fig. 9). This suggests DTW extracts informative matching data as compared with base calling followed by mapping.
An obvious application is ensuring uniform or minimal coverage over each amplicon. Challenges include read counting and overcoming sources of latency within the system. From observation, the MinION generates read starts which do not result in read data being written to disk. These may represent short reads, or non DNA material blocking a pore. Furthermore, reads which have been matched may not include hairpin sequence resulting in only a template and not a 2D read. It is not sufficient to keep track of the observed read starts from the sequencer, it is also necessary to monitor reads written to disk. We therefore track reads from initial signal from the sequencer through to the hard disk. An additional complication resulted from minKnow (the software controlling the minION) buffering writing of reads to disk. This lag introduces a delay between reads completing translocation through the pore and the data being available on disk for analysis.
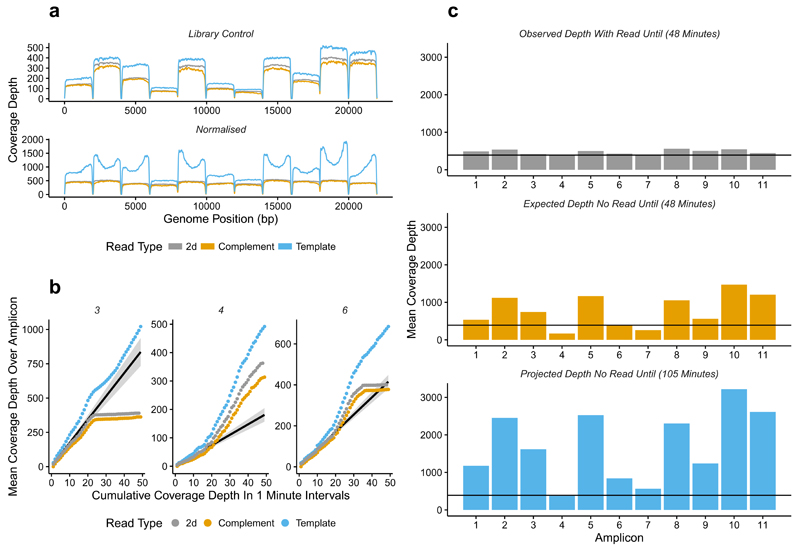
We tested “Read Until” in real-time by obtaining uniform 2D coverage of our 11 amplicon library. 10 minutes of sequencing without “Read Until” showed differential coverage of each amplicon (Fig. 3A). We then restarted the run with normalization and rejected reads once that amplicon had crossed a specified threshold, aiming for at least 200× coverage of each. The “Read Until” script automatically stops the sequencer when set coverage thresholds are surpassed, here after 48 minutes (Fig. 3A). Once base called, the 2D coverage exceeds 200×; consistent with latency within the system rather than leakiness in blocking of individual amplicons and confirmed by investigating mean coverage depth in 1 minute intervals over the run (Fig. 3B, Supplementary Fig. 10). Amplicons with high coverage in the library are rejected first (2,3,5,8,10 and 11), whilst those with lowest coverage continue sequencing (see amplicons 4 and 7). Fig. 3B presents example dynamics observed for each amplicon type. Amplicon 3 accumulates 2D reads for 20 minutes but then rejects all reads. Amplicon 4 accumulates 2D reads at a steady rate until 20 minutes at which point sequencing rate increases. This is due to increased sequencing capacity available for Amplicon 4 as other amplicon coverage thresholds are reached (~350× 2D sequencing). Amplicon 6 follows a similar trajectory although reaching its peak later around 35 minutes.
Figure 3.
Normalization of amplicons in a library using “Read Until”. (a) shows coverage over each of the 11 amplicons without “Read Until” (top pane) and with “Read Until” applied (bottom pane) trying to normalize coverage. Gray - 2D reads, Orange - complement, Blue - Template. (b) shows the cumulative coverage for three representative amplicons (3,4,6) in 1 minute intervals. The black line is predicted 2D coverage for each amplicon calculated by fitting to the first 15 minutes of data. (c) shows the observed 2D coverage after 48 minutes (top pane), the total coverage predicted for each amplicon based on the first 15 minutes of sequencing (middle pane) and the projected depth if the reads were allowed to accumulate without “Read Until” until each amplicon exceeded the minimum coverage threshold, which would take approximately 105 minutes (bottom pane).
Extrapolating from the first 15 minutes of data allows comparison between the observed levels of coverage and the expected coverage depth after 48 minutes without “Read Until” applied (Fig. 3C). The time taken for all amplicons to exceed 200× coverage without “Read Until” can also be calculated: 105 minutes with unnecessary reads slowing subsequent analysis. Simulating these experiments using a library with a 10 fold range of input amplicons and aiming to reach 100× coverage shows that “Read Until” reaches the target coverage in one third the time taken otherwise (Supplementary Fig 11). Increasing the variance in the input library to 50 fold results in a 4 fold faster run.
This is the first demonstration of selective sequencing of specific molecules without prior enrichment during sample preparation. This approach, a basic application of DTW from the mlpy library, is surprisingly accurate14. Although the current implementation is not sufficiently fast for large genomes, the algorithm has the potential to be optimized both algorithmically, using a number of well described approaches including lower bounding, and computationally by exploiting parallel GPU or FPGA processors15,16. Any method for “Read Until”, regardless of genome size or complexity, must be able to match a single read within the reference faster than the read itself is generated. For 512 channels sequencing at 300 b/s, a reasonable goal would be to place a read within 0.02 seconds allowing all channels to be processed within 10 seconds.
Exploiting the real-time nature of the minION, where reads are available for analysis immediately, has been shown by many groups13,17,18. Here we extend this to real-time analysis and selective sequencing on a MinION with “Read Until”, demonstrating this approach can be applied to amplicon sequencing. Given that reads only need map to a subset of the reference, amplicon based selective sequencing can be applied to genomes of any size and up to 50 amplicons on current fast laptops (Supplementary Fig. 11). The anticipated increasing speed of nanopore sequencing (“fast mode” at 300 b/s and beyond) and the scaling up of the MinION to 3,000 channels, and the PromethION with 144,000 channels, will challenge the implementation of “Read Until” in real-time and require algorithmic enhancements and computational power. Yet “Read Until” based approaches will enable new approaches to sequencing, such as exon sequencing without target capture, controlled depth of coverage over entire genomes, counting applications for RNA seq and many applications that have yet to be imagined.
Online Methods
Library Preparation and Sequencing
Lambda whole genome libraries were prepared without PCR amplification according to the standard protocol using either version 5 or version 6 library preparation kits (ONT, SEQ-MAP005 and SEQ-MAP006) supplied by Oxford Nanopore Technologies (ONT). This protocol has been extensively described by Ip et al1. Lambda amplicons were amplified using Platinum Taq DNA polymerase (Thermo Scientific Invitrogen, 10966018). Primers were designed to amplify eleven 2kb regions spanning 22 kb of the Lambda genome (reference strain sequence J02459.1) (see Supplementary Table 3). To prepare a mixed amplicon library, 5ul of each PCR reaction were pooled together followed by two rounds of Agencour Ampure XP purification (Beckman Coulter, A63881). This approach approximates that of Quick et al12 . Libraries were then prepared using ONT version 6 library preparation kits (ONT, SEQ-MAP006) according to the instructions provided by ONT. Libraries were sequenced on both original (Fig. 1A) and Mk1 minIONs (all other data) using either FLO-MAP003 or FLO-MAP103 flow cells respectively. MinKNOW 0.50.2.15 was used to control the minION device using the standard 48 hour run scripts, base calling was performed using the versions of Metrichor available at the time. The "Read Until" API version used was commit b4cd8e1.
Original read data corresponding to individual experiments are available for download with study accession PRJEB12567 from the ENA and are described in Supplementary Table 1.
Code availability
All code is freely available from https://github.com/mattloose/RUscripts with documentation at http://mattloose.github.io/RUscriptsdocs/.
Generation of a reference Squiggle
In order to squiggle match to a reference genome it is necessary to convert the nucleotide sequence to a squiggle. To do this, for each kmer in the reference we use the expected mean current value from the model file. So for the sequence:
ATGCGCGTAGCTGATCGATCG
with a 5mer model file we identify the following overlapping 5mers and assign the appropriate means for the first 6 positions as shown in Supplementary Table 2 which results in a squiggle sequence of:
64.0277060402, 74.1940612355, 72.5600759653, 76.3160881774, 66.6657142728, 61.7330951621
For amplicon sequencing, the reference can be reduced by excluding sequences that do not occur at the start or end of the amplicon regions. This is the suggested running mode for aReadUntil.py with the -c flag reducing the reference sequence to exclude those regions unlikely to appear in the start of individual reads.
““Read Until”” Implementation
Whilst sequencing was in progress, we applied ““Read Until”” using the ONT supplied ““Read Until”” API. In the standard ONT workflow, read data are generated by the minION device under the control of the MinKNOW software and reads are written to a folder on the users hard drive. These reads are then processed by the Metrichor service to be basecalled and the basecalled files returned to a downloads folder. The raw files are moved to an uploads folder (Supplementary Fig. 4a). ““Read Until”” in its current form requires the user to run a second utility whilst minKNOW is running (Supplementary Fig. 4b). This program, ws_event_sampler.exe, streams event data from minKNOW as soon as they are received from the minION device to a second application which will process these data. This second application incorporates a specific API from ONT to communicate with ws_event_sampler.exe and send messages to and from the sequencer requesting reads to be rejected as appropriate. We provide two example applications here, namely gReadUntil.py (Supplementary Fig. 4 ci,ii) and aReadUntil.py (Supplementary Fig. 4 di,ii), for enriching genomic DNA regions or balancing amplicons respectively. These are implemented in python and are freely available from https://github.com/mattloose/RUscripts. ws_event_sampler.exe provides a useful simulation mode to stream example data for testing purposes and provided users have access to the “Read Until” API from ONT, can be used to test “Read Until” (Supplementary Fig. 4 cii, dii).
Detailed use of these scripts are provided in the documentation alongside the GitHub repository (http://mattloose.github.io/RUscriptsdocs/). Use of these scripts are described below. For live running, scripts require an API available from Oxford Nanopore and this varies dependent on the specific version of minKnow being used. We caution those interested in “Read Until” that the API and implementation are currently under development and thus subject to change. The code provided here currently works with all available versions of the “Read Until” API, but only MinKNOW versions 0.48.13-0.51.1.51. Versions of minKNOW from 0.51.3 onwards restore read until functionality. The authors welcome correspondence with anyone wishing to apply “Read Until” to resolve these issues during the early development phase of this technology.
It is important to note that each of these applications depend on the underlying model usually used to decode the sequence for base calling. We do not distribute this information, but rather provide an application, getmodels.py, which will extract the appropriate data from a previously base called file (Supplementary Fig. 4 e). DTW is obviously sensitive to the model being used and users should ensure they are using the most appropriate model for the chemistry in use. This utility exports model files with names based on the model being extracted. This means the script may overwrite a previously generated model file if the read contains the same model. Note that this script will only extract models from reads which have been base called using the Oxford Nanopore Technologies HMM base caller. No model is encoded in reads called with open source base callers or neural network derived approaches. This script should be run each time a chemistry change occurs, typically defined by the introduction of a new nanopore by Oxford Nanopore. Note that reads called using a neural network based base caller will not contain a model file. The necessary model files may be obtained on request from Oxford Nanopore Technologies.
Additional scripts are provided for testing the ability of Dynamic Time Warping to sort reads without access to the ONT API (ampliconSPLIT.py), to simulate genome section (test_gReadUntil.py) and a utility to optimally sort amplicons from a run prior to base calling and allowing offline “balancing” of samples (ampbalance.py).
Dynamic Time Warping
Dynamic Time Warping (DTW) is a Dynamic Programming algorithm that is guaranteed to find the optimal alignment of two sequences of values. It has a "remarkable history of independent discovery and publication"9. We provide a brief summary of the algorithm but refer the reader to the extensive literature on this topic for additional information9,15. DTW is closely related to the Levenshtein Edit Distance for strings of discrete values19, to the Needleman–Wunsch algorithm used to align protein sequences20 and the Smith-Waterman algorithm21. Unlike DTW, these last two algorithms additionally employ substitution matrices and gap penalties in determining the cost of moves.
DTW Description
Let Q be a sequence of length m and R a sequence of length n where m maybe << n.
Let D be an m*n matrix of distances between elements of Q and R with D[i][j] = f(Q[i] , R[j]).
Here, distances may be defined using a range of metrics (f) such as Euclidean distance, squared Euclidean Distance, absolute difference or Manhattan distance.
The optimal alignment of Q and R is obtained by traversing D from D[m][i+m] to D[0][i] where 0 <= i <= n-m choosing, at each step, the lowest cost move; the cost of moving from D[i][j] to a neighbour D[g][h], with (g < i and h==j) or (g==i and h < j) or (g < i and h< j), is determined using a "cost" matrix (C).
The "cost" matrix C is an m*n matrix with the value of C[i][j] determined as follows:
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
| (Eq. 4) |
The basic DTW algorithm has a computational complexity of O(m*n) and the dynamic programming used to determine the cost matrix defeats simple automated vectorisation. Consequently, a number of variations that trade guarantees of optimality against performance have been suggested, for a thorough overview see the survey chapter from Müller15.
An important variation of DTW for searching for an alignment of a relatively small sequence within a much larger sequence, termed subsequence DTW (sDTW), as implemented in the mlpy DTW package, is used in the current work14. Essentially sDTW requires the following small change to Eq. 3 above:
| (Eq. 3a) |
As in https://github.com/lukauskas/mlpy/blob/master/mlpy/dtw/cdtw.c line 411. A detailed description is provided in Muller15.
Running ““Read Until””
The following section briefly describes the scripts and utilities found within the github code repository for this paper (https://github.com/mattloose/RUscripts). Fully worked examples are provided in the online documentation.
getmodels.py
All squiggle matching applications require a reference squiggle to match to. The script getmodels.py extracts the required data to do so from a base called minION read. This script provides the model files that are required for all the “Read Until” applications described here. The model in use by ONT to describe minION data is updated frequently and people should use a read derived from the chemistry and sequencing speed they are actually using for sequencing. getmodels.py outputs two files, a model for the template and a model for the complement. For ““Read Until”” it is the template model that is used. The script will also output the kmer length in the model and the structure of the file for reference. Note that for simulated runs using ws_event_sampler.exe the model file used by the simulator should be used by the "Read Until" scripts.
ampliconSPLIT.py
ampliconSPLIT.py simulates the separation of read data into individual amplicons and can be run on read files pre or post basecalling. The script will process read files, sorting them into individual amplicons and placing them in a subfolder for each amplicon. This script provides a method of testing the ability of DTW to sort reads based on the template strand alone. Line 298 of the code sets the length of the read which is searched and its position from the read. We take 250 events excluding the first 50 events of the read. The depth parameter can be used to limit the number of amplicons returned - this is a basic implementation of balancing. However, we recommend that for balancing of amplicons after a run is complete, the script ampbalance.py (described below) is used.
ampbalance.py
This script builds on the approaches taken for “Read Until” but also includes the complement model. The script will look within a read to identify the template and complement strands from the read, map both to a reference and then determine those reads most likely to give rise to a 2d read.
Live “Read Until” - gReadUntil.py and aReadUntil.py
Within the folder ReadUntil are two scripts, gReadUntil.py and aReadUntil.py. These require access to files from the “Read Until” API provided by Oxford Nanopore and not distributed here. All necessary dependencies for the “Read Until” API should be installed as specified by ONT. Note that changes to the API will most likely break our code as presented here. The API is an alpha release, is under current active development, and so the scripts cannot be guaranteed to work. Full instructions for running these scripts and installing the API are provided in the associated github repository.
General considerations
1. What to map
An important consideration for “Read Until” is which region and length of a prospective read you wish to map to a reference. Reads will begin with a leader sequence derived from the library preparation step which can influence the placement of a read on the reference. As the length of the sample from the read increases, the leader sequence relatively disrupts the mapping less. The “Read Until” API allows the user to specify the offset into a read from which events are sampled and the length of sequence which is sampled with an instruction in the form of:
setup_conditions = {"ignore_first_events": 50, "padding_length_events": 0, "events_length": 250, "repetitions": 1}
These parameters should be tuned for specific applications.
2. What to monitor
For read counting applications it is important to consider what should be counted as a read. The assumption that a read appearing via the API will be written to disk is mistaken. Particularly for short fragments, a read may terminate for a number of reasons and the minKnow software choose to not write this to disk. In the case of 2D sequencing (where template and complement strand are both sequenced), a read cannot have assumed to be 2D just because the start of the read has been seen. Thus it is necessary to monitor reads written to disk. The minKnow software itself has lag in writing reads to disk after they have been generated by the sequencer and so latency throughout the system will impact on counting where knowledge of the final read is required.
gReadUntil.py
This script will select for specific regions of a genome, mapping reads in real time using DTW and rejecting reads appropriately. This script is limited by the length of the query sequence as the time for mapping is proportional to the length of the reference. This code as presented will only process the odd numbered channels for “Read Until”. To change this behaviour, modify the mp_worker function accordingly. Optimal performance on a flow cell will require as many processors as can be dedicated to it.
A second script is provided - test_gReadUntil.py which can take an input collection of reads and filter them based on mapping location in the genome.
aReadUntil.py
This script will map specific amplicons to a reference and demonstrates the ability to select for individual amplicons directly on the sequencer. We recommend a powerful laptop with at least 8 cores for running this alongside the sequencer. Alternatively, the code can be run from a second server. In order to count the number of reads generated, this code looks at reads as they are written to disk by minKnow as well as following the output stream of data in real time. Thus if running on a second server, reads should be synchronised using rsync from the minION laptop to the second server as soon as they are generated. An additional conflict may occur if Metrichor is being run in real time alongside the script. Metrichor itself will move files from the “reads” folder to a subfolder called “uploads”. We suggest that users do not run Metrichor alongside this code at this time to avoid conflicts.
An example “ids” file is provided in the root directory of the repository called “lambda_amplicons.txt” which can be used to map amplicons from the amplicon experiments presented here.
This script implements a number of additional features worth highlighting. It directly interacts with minKnow and, if the -s option is set, will stop the currently running sequencing program when the required depth of sequencing has been reached. This can be set using either the -d flag, which will apply that coverage depth to each amplicon, or the -cd flag which takes a comma delimited list of depths. MinKNOW interaction requires the IP address of the machine running minKnow being supplied to the script. It is important to make sure that localhost is not blocked. This communication with MinKNOW will prevent the script from running if MinKNOW itself is not functioning. The -sim flag will write out illustrative fast5 format files when being used in conjunction with the simulation mode available in ws_event_sampler.exe. This is provided for testing purposes and the files only contain the information necessary for aReadUntil.py.
“Read Until” Efficiency
Simulation of “Read Until” is possible using the ws_event_sampler utility to stream events artificially derived from a reference sequence. Data presented in Supplementary Figures 5,6 and 11 are derived from the use of this simulator in conjunction with the gReadUntil or aReadUntil scripts. For whole genome simulations, the reference sequences used were as described. For amplicon sequencing, reference amplicon sequences were created with different copy numbers of each amplicon as shown in Supplementary Table 4.
Data Analysis
Jupyter notebooks documenting the analysis for each panel from the manuscript are available from https://github.com/mattloose/RUFigs . All requisite files are contained within the repository. For the calculation of coverage plots, BAM files were processed with the genomeCoverageBed tool from BedTools using the -d option to report depth at every position22,23. For clarity of presentation individual coverage traces were smoothed over 50 base pair windows using a simple Perl script (see https://github.com/mattloose/RUFigs/fig2/cov_windows.pl ). BAM files for each time window were generated by extracting timed reads in minute windows using minoTour, which logs sequencing time stamps derived from the raw read files automatically against each aligned file24. Similarly labelled file sets can be obtained using either poretools or poRe25,26.
Supplementary Material
Acknowledgements
This work was supported by BBSRC TRDF award BB/M020061/1. The authors thank T. Evans for comments and discussion on the manuscript and figures, N. Loman and J. Quick for helpful discussions, advice and motivation, and M. Blythe for discussions. We also thank Oxford Nanopore Technologies for access to the Read Until API and helpful discussions and support.
Footnotes
Accession Codes
Author Contributions
ML conceived and designed the project, wrote the code, ran sequencing, performed analysis and wrote the manuscript, MS carried out analysis and wrote and contributed to code and contributed to the manuscript, SM generated libraries, ran sequencing and contributed to the manuscript.
Competing Financial Interests
ML is a member of the MinION access program (MAP) and has received free-of-charge flow cells and kits for nanopore sequencing and travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences.
References
- 1.Ip C, et al. MinION Analysis and Reference Consortium: Phase 1 data release and analysis [version 1; referees: 2 approved] 2015;4 doi: 10.12688/f1000research.7201.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heron A, et al. METHOD FOR ATTACHING ONE OR MORE POLYNUCLEOTIDE BINDING PROTEINS TO A TARGET POLYNUCLEOTIDE. WO patent WO 2015/110813 A1. 2015
- 3.Heron A, et al. MODIFIED HELICASES. US patent US 2015/0191709 A1. 2015
- 4.Moysey R, Heron Andrew J. METHOD OF CHARACTERIZING A TARGET POLYNUCLEOTIDE USING A PORE AND A HEL308 HELICASE. WO patent WO 2013/057495 A3. 2013
- 5.Urban JM, Bliss J, Lawrence CE, Gerbi SA. Sequencing ultra-long DNA molecules with the Oxford Nanopore MinION. bioRxiv. 2015 doi: 10.1101/019281. [DOI] [Google Scholar]
- 6.Timp W, Comer J, Aksimentiev A. DNA base-calling from a nanopore using a Viterbi algorithm. Biophys J. 2012;102:L37–39. doi: 10.1016/j.bpj.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodwin S, et al. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015;25:1750–1756. doi: 10.1101/gr.191395.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 9.Sankoff D, Kruskal JB. Time warps, string edits, and macromolecules : the theory and practice of sequence comparison. Addison-Wesley Pub. Co., Advanced Book Program; 1983. [Google Scholar]
- 10.Miodonska Z, Bugdol MD, Krecichwost M. Dynamic time warping in phoneme modeling for fast pronunciation error detection. Comput Biol Med. 2015 doi: 10.1016/j.compbiomed.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Skutkova H, Vitek M, Sedlar K, Provaznik I. Progressive alignment of genomic signals by multiple dynamic time warping. J Theor Biol. 2015;385:20–30. doi: 10.1016/j.jtbi.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016 doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quick J, et al. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 2015;16:114. doi: 10.1186/s13059-015-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albanese D, et al. mlpy: Machine Learning Python. arXiv. 2012 [Google Scholar]
- 15.Müller M. Information Retrieval for Music and Motion. Springer-Verlag; Berlin Heidelberg: 2007. [Google Scholar]
- 16.Sart D, Mueen A, Najjar W, Keogh E, Niennattrakul V. Data Mining (ICDM), 2010 IEEE 10th International Conference on; pp. 1001–1006. [Google Scholar]
- 17.Ashton PM, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2015;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 18.Greninger AL, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levenshtein VI. Binary codes capable of correcting deletions, insertions and reversals. Soviet Physics Doklady. 1966;10:707–710. [Google Scholar]
- 20.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 22.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11, 12, 11–11, 12, 34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loose M. minoTour - a platform for real-time analysis and management of Oxford Nanopore minION reads.
- 25.Loman NJ, Quinlan AR. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics. 2014;30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson M, et al. poRe: an R package for the visualization and analysis of nanopore sequencing data. Bioinformatics. 2015;31:114–115. doi: 10.1093/bioinformatics/btu590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.