Abstract
Large scale, quantitative proteomic studies have become essential for the analysis of clinical cohorts, large perturbation experiments and systems biology studies. While next-generation mass spectrometric techniques such as SWATH-MS have substantially increased throughput and reproducibility, ensuring consistent quantification of thousands of peptide analytes across multiple LC-MS/MS runs remains a challenging and laborious manual process. To produce highly consistent and quantitatively accurate proteomics data matrices in an automated fashion, we have developed the TRIC software which utilizes fragment ion data to perform cross-run alignment, consistent peak-picking and quantification for high throughput targeted proteomics. TRIC uses a graph-based alignment strategy based on non-linear retention time correction to integrate peak elution information from all LC-MS/MS runs acquired in a study. When compared to state-of-the-art SWATH-MS data analysis, the algorithm was able to reduce the identification error by more than 3-fold at constant recall, while correcting for highly non-linear chromatographic effects. On a pulsed-SILAC experiment performed on human induced pluripotent stem (iPS) cells, TRIC was able to automatically align and quantify thousands of light and heavy isotopic peak groups and substantially increased the quantitative completeness and biological information in the data, providing insights into protein dynamics of iPS cells. Overall, this study demonstrates the importance of consistent quantification in highly challenging experimental setups, and proposes an algorithm to automate this task, constituting the last missing piece in a pipeline for automated analysis of massively parallel targeted proteomics datasets.
Introduction
Molecular biology is increasingly becoming a data-driven science which enables researches in biology and medicine to investigate large numbers of biological systems on a genome-wide scale. Underlying this transition is the ability to generate robust, comprehensive and fully quantitative “data matrices” capturing measurements across many samples (first dimension) in a genome-wide fashion (second dimension). In nucleic acid sequencing-based fields, this transition has advanced enough to allow for large-scale inference from thousands of samples in a reproducible and comparable manner [1, 2, 3, 4, 5].
In contrast, in the field of proteomics the transition to high-throughput measurements across large numbers of samples has proven challenging (Supplementary Note 1 and Röst et al. [6]). While discovery-oriented techniques, such as data-dependent acquisition (DDA) [7, 8, 9, 10], have recently allowed the identification of a large part of the human proteome [11, 12], it has become apparent that these methods suffer from poor reproducibility in large scale experiments. Particularly when applied in high throughput to complex protein mixtures, e.g. whole proteomes, the resulting data matrices contain many missing values. To improve reproducibility, alternative approaches based on targeted proteomics were developed which provide high consistency and quantitative accuracy across many experimental conditions due to their deterministic acquisition strategy. Specifically, selected reaction monitoring (SRM) proved to be invaluable for large-scale measurements geared towards systems biology [13] or biomarker discovery [14, 15, 16, 17]. However, while SRM-based targeted proteomics produces highly consistent data matrices, it is limited by low throughput, resulting in output matrices with typically only few tens of quantified proteins per study (Supplementary Note 1, Fig. S1).
Recently, we developed SWATH-MS based on the principle of targeted analysis of data-independent acquisition (DIA) data as a method for massively parallel targeted proteomics [18]. Our targeted analysis of DIA data based on OpenSWATH was able to increase the throughput of targeted proteomics by several orders of magnitude compared to SRM-based approaches, and is, in principle, able to to generate proteome-wide data matrices [19, 6]. However, obtaining consistent and accurate matrices from targeted proteomics data is challenging as most current software was developed for low-throughput SRM data and focused on manual analysis and visualization of the data [20, 21, 22, 23, 24, 25, 26]. Even fully automated software solutions for peak picking and error rate estimation [27, 28, 19] generally only operate on a single MS run at a time and are unable to efficiently integrate experimental information from multiple targeted MS runs. However, a single MS run may not contain sufficient information to confidently select the correct peptide elution time point among multiple detected peak groups of similar quality in a given chromatogram (Fig. 1 a). Analyzing single MS runs in isolation, therefore, cannot ensure consistent peak picking across all the measurements constituting a whole experiment (Supplementary Note 2).
Figure 1. TRIC: Alignment algorithm for targeted proteomics data.
(a) In a targeted proteomics experiment, each run is typically analyzed individually, giving rise to multiple putative peak groups per run that may not be directly mappable due to chromatographic shifts. (b) The TRIC algorithm selects a set of high-confidence “anchor points” (peptides) for pairwise non-linear alignment and chromatographic distance estimation. (c) Based on the chromatographic distance, an optimal guidance tree (I) is computed (nodes are runs, edges are pairwise alignments). Next (II), the algorithm uses a starting point (1) to transfer identification confidence to nearby runs (iterations 2 and 3) using the guidance tree (III). In an optional last step (IV), runs without suitable peakgroups are re-visited to perform optional noise re-quantification (integration of all fragment ion signal at the aligned position is integrated; orange circles). (d) The confidence transfer step uses a starting peakgroup (top run) to select a narrow region in a neighboring run (gray region in second run) from which a peak gets selected. This procedure is repeated across all runs to identify the correct peak or establish peak boundaries in runs without any analyte signal (bottom run). In a real application, the alignment order may not be linear but follow the guidance tree.
Here we describe the TRIC (TRansfer of Identification Confidence) algorithm, an automated method which integrates all information from a targeted proteomics experiment to accurately and consistently determine the correct elution peak in each MS run. The software is designed to exploit the particular structure of chromatographic fragment ion-based peak data found in targeted proteomics, providing quantification and identification in the same algorithm. The TRIC algorithm uses a reference-free alignment approach based on individual, pairwise non-linear retention time (RT) de-warping which allows it to scale to hundreds of targeted proteomics LC-MS/MS runs. Together with the OpenSWATH framework [19], TRIC allows fully automated analysis of next-generation targeted proteomics datasets with high throughput. The software is vendor-independent and provided as an open-source package (Modified BSD Licence) at https://pypi.python.org/pypi/msproteomicstools.
Results
Design and Structure of the TRIC Algorithm
The TRIC algorithm was developed to perform integrative analyses of a large number of targeted MS injections with a focus on robustness, scalability and performance (Supplementary Note 3 and 4). Inherently, the algorithm was designed to correct for non-linear chromatographic distortion between the runs, minimize the effect of outlier runs, and be scalable to hundreds of MS runs. This is achieved through a guidance tree learned from the data which removes the need for a reference run in alignment and by using a locally adaptive retention time tolerance for each pairwise alignment (Supplementary Note 5) [29, 30]. The algorithm does not rely on MS1 measurements but works directly on chromatographic peak identifications from primary identification tools, such as OpenSWATH [19] or PeakView, which usually identify multiple potential peakgroups in an extracted fragment ion chromatogram. Using the fragment ion data, the algorithm is able to boost identification confidence for peaks consistently detected across multiple runs and can provide improved peak boundaries for analyte quantification. Starting from a typical targeted proteomics dataset with a number of individually analyzed and scored runs containing multiple potential peak group candidates for identification (Fig. 1 a), the algorithm performs the following steps:
-
(a)
Alignment: Using a set of high-confidence endogenous peptides, an estimate of the pairwise chromatographic distance between all MS runs is obtained (Fig. 1 b) and used to generate a guidance tree. Then, for each edge in the tree, a pairwise non-linear transformation between the RT domains of the two MS runs (nodes) is computed (Fig. 1 c; step I). Note that TRIC does not rely on spike-in peptides and that the alignment using a guidance tree removes the need for a reference run, leading to an optimal alignment strategy which prevents direct alignment of chromatographically dissimilar runs (Supplementary Note 5, Figs. S10 and S11).
-
(b)
Confidence transfer: Traversal of the global guidance tree starts for each measured peptide (targeted proteomics assay) from a suitable “seed” (Fig. 1 c; step II). During traversal, each node (LC-MS/MS run) of the tree is visited sequentially (step III, iterations 2 and 3) and a confident identification is mapped from one node (run n) to an adjacent node (run m), where our choice of guidance tree ensures that the mapping only occurs between chromatographically similar runs (Supplementary Note 3). During confidence transfer (Fig. 1 d), the identification confidence of all peakgroups in run m within the adaptive retention time window (area indicated in gray) is increased; if the confidence score of the best peakgroup passes the user-defined threshold, it gets added to the final result matrix. The approach automatically adopts the retention time window for different parts of the tree, depending on the quality of the pair-wise alignment, thus increasing robustness and decreasing the influence of outlier runs (Supplementary Note 5, Figs. S12 and S13).
-
(c)
Re-quantification: In the last (optional) step, runs in the guidance tree where no peakgroup passed the confidence filter can be re-visited for re-quantification. In these cases, the software can infer the peak boundaries from the closest neighboring run (node) and quantify the fragment ion signal within those boundaries. These imputed values, however, are not substitutes for quantification events, but merely serve as upper bounds of the analyte signal for the run in question (orange circles, step IV).
In order to control the false discovery rate (FDR), our software also offers the option to perform an error-rate correction based on known false signals (“decoy signals”) at the assay level [31], preventing the accumulation of false positive identifications when analyzing multiple runs. This step can remove individual rows from the data matrix if they do not pass the filter criteria (see Methods). This is achieved by requiring a more stringent quality threshold for the “seed” identification, similar to the way the Mayu software [32] operates.
Technical Validation
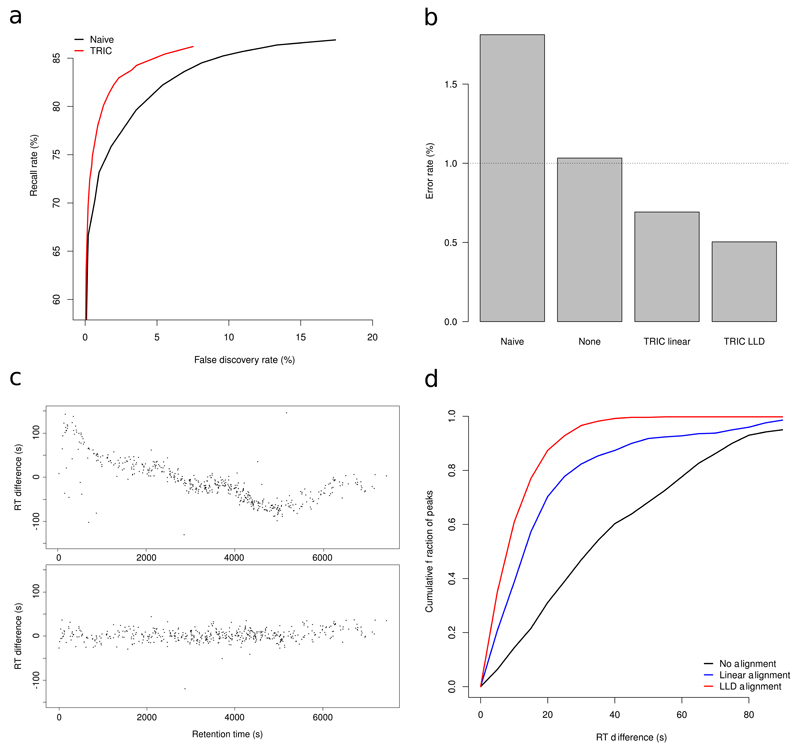
To validate the alignment and FDR control approach implemented in TRIC, we created a manually validated data set of 7,232 chromatograms which were extracted from the Streptococcus pyogenes dataset of Röst et al. [19] (Supplementary Note 4). First, 452 peptides were randomly selected from the data, giving rise to 7,232 chromatograms that were loaded into the Skyline software where the correct elution peak, if present, was identified by visual inspection (Supplementary Table 1). In parallel, we analyzed the same data with TRIC and compared its performance to the current state-of-the-art, that is applying a fixed q-value cutoff in each run individually (“naïve approach”). We found that the TRIC algorithm decreased the error rate substantially compared to the naïve approach, while maintaining high recall for peakgroups across all runs (see FDR-Recall behavior in Fig. 2 a and Supplementary Note 4, Figs. S2–S5). At the same FDR cutoff, TRIC was able to reduce the error rate by more than 3-fold from 1.8 % to 0.5 % (better than the expected 1 %, Fig. 2 a,b). Running TRIC with non-linear RT alignment substantially outperformed linear alignment strategies as well as the naïve approach (Fig. 2 b).
Figure 2. Identification and alignment accuracy of TRIC on manually annotated data.
We used a set of over 7,000 manually validated peakgroups to validate the TRIC algorithm. (a) FDR-Recall plot displaying recall rate versus the false discovery rate allows evaluation of the performance of TRIC compared to the naïve approach of using a fixed q-value cutoff applied to each run individually. As mis-classified peaks cannot be recovered even at high score cutoffs, a recall of 100 % cannot be reached. (b) Error rates at reported FDR cutoffs of 1 % for the naïve approach and TRIC without RT alignment (None), linear alignment (Linear) and non-linear k-nearest neighbor alignment (LLD). (c) The error of reported retention times are plotted without (top) and with (bottom) non-linear alignment on a sample run. (d) The cumulative fraction of peaks having less than a given error in retention time is plotted. TRIC with k-nearest neighbor smoothing (LLD) achieves high peak counts at low RT errors and outperforms linear or no alignment.
Next, we evaluated the accuracy of the fragment ion peak boundaries reported by TRIC in the “re-quantification” step. From a dataset of eight S. pyogenes runs with large chromatographic differences, we removed 506 high-confidence peakgroups to test whether TRIC could recover them. We observed clear non-linearities in the retention times which TRIC was able to correct satisfactorily (Figure 2 c top versus bottom panel). After correction, over 96.6 % of all data fell within ±30 s (that is within 2 chromatographic peak widths and less than 1 % of the chromatographic gradient) around the true retention time, compared to 82 % and 47 % for linear or no alignment (Fig. 2 d and Supplementary Note 4, Fig. S8). Similarly, we found that 80 % of all reported intensity values deviated less than 25 % from the true intensity value (Supplementary Note 4, Fig. S8). We conclude that the alignment and confidence transfer procedure performed by TRIC improves accuracy, reduces the error rate and correctly accounts for large non-linear chromatographic effects.
Application to microbial virulence
We then applied the TRIC algorithm to the full 12 SWATH-MS runs described in Röst et al. [19], comparing cultures of strain SF370 grown in 0 % and 10 % human plasma to study proteomic changes that occur upon vascular invasion of S. pyogenes. TRIC substantially lowered the overall number of missing values as well as the number of incomplete rows generated with each newly added run in the label-free, quantitative proteomics data matrix (Fig. 3 a). The guidance tree created by the alignment mostly reflected the biological condition (case versus control) and not the acquisition order, while chromatographically dissimilar runs were correctly placed at the periphery of the tree, decreasing their influence on the alignment process (Fig. 3 b).
Figure 3. Analysis of a microbial dataset investigating S. pyogenes virulence.
A dataset of 12 runs of S. pyogenes exposed to human plasma was analyzed with TRIC. (a) The data matrix occupancy is higher after alignment with TRIC (fewer missing values are observed). (b) The computed guidance tree captures orthogonal information to injection order (root mean square deviation between runs is indicated for each edge). Control samples are in blue and plasma-exposed samples are in red (note that the tree is substantially different from injection order as samples were shot in three batches: R1, R2 and R3). (c) Number of precursors appearing in a specific number of runs before (left; N=95 685) and after (right; N=120 348) running TRIC; fully aligned precursors increased by 39 % while precursors found in only a single run decreased by 93.7 %. (d) The cumulative number of the number of peptides quantified using a fixed 0.01 q-value cutoff without alignment (left) and after applying TRIC and a minimal q-value cutoff of 0.0015 (right). While TRIC decreased the variance of the number of identifications across runs, the cumulative number of peptides also saturates more quickly indicating less accumulation of false positive identifications.
The final assay-level data matrix (Supplementary Note 6, Fig. S14) was to 87 % populated with quantified peakgroups, compared to 69 % using a “naïve approach” with a fixed q-value cutoff of 0.0015 (Fig. 3 a and Supplementary Note 6). Using the aligned data matrices, we identified 130 S. pyogenes proteins that significantly change upon exposure to human plasma (adjusted p <0.01 and effect size larger than 1.5), up by 37 % from 95 proteins without alignment (Supplementary Tables 2 and 3). The number of assays quantified in all 12 runs increased by 39 % from 4971 to 6914 (Fig. 3 c,d) and substantially fewer peptides were identified in a single run only (down by over 15-fold, compare Fig. 3 c,d); TRIC thus added additional quantitation events to singleton peptides (Supplementary Note 6, Fig. S15).
We then investigated whether our algorithm was able to improve the identification consistency and error rate control across the whole experiment. We found that TRIC increased identification consistency across all runs, and the cumulative number of peptide identifications shows early saturation (Fig. 3 e, f; see Supplementary Note 6, Figs. S16–S18 for data on assay level and protein level). This is consistent with a complete mapping of the expressed peptides, while a continuing increase in cumulative identifications, as seen without alignment, would be consistent with an accumulation of false positive identifications.
Protein turnover analysis using TRIC
Protein degradation rates vary substantially across the proteome, and together with transcription rates are the main determinant of the amount of protein in a cell [33, 34]. Protein degradation plays an important role in many cellular processes including cell-cycle, DNA repair, growth and differentiation, and it has been linked to several diseases [35]. To study protein degradation using targeted proteomics, we performed a pulsed-SILAC experiment on induced pluripotent stem cells (iPSCs) obtained from a healthy human donor, allowing us to measure in vitro protein turnover rates in a personalized fashion: After growing the iPSCs in biological duplicates, we replaced the light medium with heavy labeled medium at timepoint zero, harvested samples after 1.5, 4.5 and 13.5 hours and analyzed their proteomes on an AB Sciex 5600 plus TripleTOF system in SWATH mode (Fig. 4 b). Using a matching spectral library, OpenSWATH quantified 5,484 heavy-light pairs mapping to 1,427 proteins (achieving 87 % library coverage for the light precursors).
Figure 4. Pulsed-SILAC experiment performed on human iPSCs.
A human iPS cell line was exposed to a pulse of heavy amino acids and sampled at four time points in duplicates (see Panel b). (a) The RT difference between the light and heavy signal as a function of the intensity. Aligned values reported by TRIC (in red) have lower intensity and higher RT error (distribution on top only displays values below 104 in intensity) (c) Standard deviation of the RT difference between heavy and light pairs with and without TRIC alignment. For the analysis without alignment, a simple FDR cutoff was applied (naïve approach). Alignment increases the number of quantified SILAC pairs at the cost of slightly higher variance. Pairs from both replicates are aggregated. No heavy-light pairs are expected at t=0 as heavy amino acids were added afterwards. (d) The number of isotopic SILAC pairs quantified per sample increases through the TRIC alignment, especially for the earlier time points with little heavy isotope signal. For each timepoint, average values across two replicates are shown with standard deviation.
We reasoned that this dataset would allow us to evaluate our algorithm on a highly heterogeneous time-course dataset with interesting biological applications. First, this setup allowed us to directly test whether TRIC over-annotated the resulting data-matrices, i.e. by falsely aligning heavy species at time point zero (before heavy amino acids were added). Furthermore, it provided a straight-forward metric to assess the quality of the data by checking the elution time difference error between corresponding heavy and light peptides (the two channels were treated completely independently for the purpose of this analysis). Third, as heavy lysine and arginine get incorporated into the proteome starting from time point zero, the algorithm would have to accurately quantify very low abundant heavy peptide species, as expected in the early time points–a very challenging task.
Applying TRIC increased the number of quantified SILAC pairs by 62 % and 40 % in the time points 1.5 h and 4.5 h, respectively, while only adding few false positive heavy identifications at time point zero (Fig. 4 d and Supplementary Note 7). Similarly, the number of quantified proteins detected in fewer than three samples decreased by a factor of 1.9, while the number of quantified proteins identified in five or more samples increased by 59 %. The additional quantification events reported by TRIC increased the error in heavy/light elution only slightly (Fig. 4 c). When matched within their respective intensity range, the error distributions are very similar (Fig. 4 a, top distribution). Thus, by applying TRIC to a data structure typically encountered in time-course experiments, the number of quantification events increased by up to 60 % without any significant impact on accuracy.
We then used the SILAC ratios to compute the relative isotopic abundance (RIA) for each peptide over time and fitted an exponential decay model as described by Pratt et al. [36] (see Methods). After filtering and correction for dilution, we obtained the median kloss, the rate of loss of light isotope over time, for 1075 proteins (Fig. 5 a). The computed protein-level turnover rates ranged from less than 10 h to several hundred hours with a median protein turnover time of 39.0 h (Fig. 5 b and Supplementary Table 4 and 5). A gene ontology (GO) enrichment analysis on the proteins with the highest and lowest turnover rates using GORILLA [37] identified 20 significantly (q-value <0.05) enriched terms (Supplementary Table 6); with an enrichment of 3.44-fold, the “cell adhesion” GO term was significantly (p <10−7) enriched in the set of proteins with high turnover (Fig. 5 c and Supplementary Note 7, Fig. S19). The enrichment in cell adhesion molecules is both consistent with the hypothesis of a generally faster turnover of these molecules in human cells as well as with the critical involvement of this class of molecules in the regulation of pluripotency (Supplementary Note 7). Thus, TRIC enables the accurate identification of protein turnover rates in human iPSCs in a highly challenging time-series experiment.
Figure 5. Protein turnover rates in human iPSCs.
Targeted proteomics analysis of protein turnover in human iPSCs. (a) Relative isotopic abundance (RIA) is plotted for an example protein, Importin Alpha, with 5 peptides (dashed lines). The median of all decay curves fitted through 1.0 at timepoint zero for all peptides estimates protein-level kloss. (b) Global protein turnover rates are estimated after correction for protein dilution. (c) Proteins in GO category “cell adhesion proteins” show significantly higher turnover than expected (p <10−7). All peptides of the respective proteins exhibit substantially higher degradation rates than the base distribution (shown on the very right). Only proteins with two or more peptides are shown (box indicates first and third quartile with median shown in black; whiskers extend to the most extreme data point which is no more than 1.5 times the length of the box away from the box).
Discussion
The availability of accurate, consistent and complete protein quantification data matrices is crucial for systems biology investigations in the field of proteomics. They are the basic currency of data-driven experiments and their accuracy largely determines the success of downstream analyses [6]. The TRIC algorithm described here is capable of creating consistent targeted proteomics data matrices by performing retention time alignment of fragment ion chromatograms and subsequent identification and quantification. The algorithm is specifically designed for targeted proteomics data and works directly with chromatographic peaks identified by upstream tools on MS2 level [19, 38, 27]. By relying on fragment ion-based identification in all runs, TRIC omits the error-prone step of mapping unidentified MS1 features across runs commonly performed in MS1-based alignment software (Supplementary Note 2) [29, 39, 30, 40, 41, 42]. Instead, the TRIC algorithm employs a “confidence transfer” step where identification confidence (and not the identification itself) is transferred across runs.
The chosen alignment strategy using a globally optimal guidance tree results in minimal alignment error since every alignment step is local and performed between two highly similar runs (as opposed to aligning all runs against a more distant reference run). This makes TRIC scalable to a large number of samples, tolerant to outlier runs and applicable to heterogeneous experimental conditions; which we demonstrate in Supplementary Note 5 using a dataset with over two hundred blood plasma samples [43]. On this dataset, our reference-free strategy has better precision-recall characteristics than a reference-based approach and the data indicate direct benefits of the adaptive retention time windows (Supplementary Note 5, Figs. S10-S13).
Using a validation dataset of 7,232 manually curated ion chromatograms, we find that our algorithm can reach high recall rates while reducing the error rate by a factor of three or more compared to state-of-the-art per-run analyses. TRIC also compares favorably to untargeted data analysis tools such as DIA-Umpire [44], which performs orthogonal peptide identification using demultiplexed pseudo-spectra and subsequent RT alignment (Supplementary Note 4). TRIC achieves higher recall (85 % versus 59 %) at lower error rate (0.3 % versus 3.8 %) than DIA-Umpire, thus highlighting the benefits of using targeted fragment ion information for identification and alignment (Supplementary Note 4, Figs. S6 and S7). Furthermore, when applying TRIC to large-scale microbial and human targeted proteomics datasets, the number of quantified values consistently increases by 30-60 %, which directly leads to an improvement of statistical power and biological information (Supplementary Notes 5, 6 and 7). When applying TRIC to small and large scale data sets (hundreds of injections), we observe consistent performance and scalability to large sample numbers, thus making SWATH-MS amenable to multi-center studies and, through TRIC, achieve consistent and reproducible research data across labs.
Online Methods
Experimental procedures
Cell culture
The iPSC line used in this study (CTL1R-1) was derived and initially cultured as described in Adamo et al. [45]. Cells were growing in single-cell condition in mTeSR-1 and then adapted for 2 passages in a custom medium, composed as follows: DMEM, Knockout Serum Replacement 15 % (Sigma), Pen-Strep 1 %, Non-essential aminoacids 1 %, Glutamine 1 %, Probumin 0.5 % (Millipore), beta-meracptoethnaol 0.1 mM, L-Proline 500 mg/l (Sigma), FGF2 10 ng/ml (Peprotech). The medium was conditioned for 24 hours on a mouse embryonic fibroblast layer inactivated with mitomycin-C and filtered before use. In the SILAC version of the medium a custom DMEM (Lonza) without arginine and lysine was complemented with 84 mg/l 13C6 15N4 Arg10 (Sigma) and 146 mg/l 13C6 15N2 Lys8 (Sigma). Cells were scraped and washed in cold PBS upon reaching 70 % confluence approximately for protein harvest.
Cells were counted using a Bürker chamber with Trypan blue counting 5 fields and averaging. Each count was done in duplicate.
MTS assay
20 µl of CellTiter 96 AQueous One Solution reagent (Promega) was added into each well of a 96 multiwell plate containing 5×103 cells in 100 µl of mTeSR. Plates were incubated for 1 hour at 37 °C and 490 nm absorbance was recorded using Glomax Multi Detection System (Promega).
Protein extraction and in-solution digestion
The iPSC cell pellets were lysed on ice by using a lysis buffer containing 8 M urea (EuroBio), 40 mM Tris-base (Sigma-Aldrich), 10 mM DTT (AppliChem) and complete protease inhibitor cocktail (Roche). The resulted mixture were sonicated in 4 °C for 5 mins using a VialTweeter device (Hielscher-Ultrasound Technology) and centrifuged at 21,130 g and 4 °C for 1 hr to remove the insoluble material. The supernatant protein mixtures were transferred and the protein amount was determined with a Bradford assay (Bio-Rad, Hercules, CA, USA). The protein mixtures were reduced by 5 mM tris(carboxyethyl)phosphine (Sigma-Aldrich) and alkylated by 30 mM iodoacetamide (Sigma-Aldrich). Then 5 volumes of precooled precipitation solution containing 50 % acetone, 50 % ethanol, and 0.1 % acetic acid was added to the protein mixture and kept at -20 °C overnight. The mixture was centrifuged at 20,400 g for 40 min. The pellets were washed with 100 % acetone and 70 % ethanol with centrifugation at 20,400 g for 40 min. The samples were then redissolved by 100 mM NH4HCO3 and were digested with sequencing-grade porcine trypsin (Promega) at a protease/protein ratio of 1:40 overnight at 37 °C [46]. Digests were purified with Vydac C18 Silica MicroSpin columns (The Nest Group Inc.). Peptide amount was determined by using Nanodrop ND-1000 (Thermo Scientific) and about 0.7 µg peptide mixtures were analyzed in each LC-MS run. An aliquot of retention time calibration peptides from iRT-Kit (Biognosys) was spiked into each sample before all LC-MS analysis at a ratio of 1:20 (v/v) for linear RT correction in OpenSWATH [47].
Shotgun measurement
The peptides digested from two biological replicates of iPSC cells at time zero (cells in light medium) were both measured on an AB SCIEX 5600 plus TripleTOF mass spectrometer operated in DDA mode. The mass spectrometer was interfaced with an Eksigent NanoLC Ultra 2D Plus HPLC system as previously described [48, 49]. Peptides were directly injected onto a 20-cm PicoFrit emitter (New Objective, self-packed to 20 cm with Magic C18 AQ 3-µm 200-Å material), and then separated using a 120-min linear gradient of 2 % buffer B to 35 % buffer B (buffer A 0.1 % (v/v) formic acid, 2 % (v/v) acetonitrile, buffer B 0.1 % (v/v) formic acid, 90 % (v/v) acetonitrile) at a flow rate of 300 nL/min. MS1 spectra were collected in the range 360–1,460 m/z. The 20 most intense precursors with charge state 2–5 which exceeded 250 counts per second were selected for fragmentation, and MS2 spectra were collected in the range 50–2,000 m/z for 100 ms. The precursor ions were dynamically excluded from reselection for 20 s.
SWATH-MS measurement
The same LC-MS/MS systems used for shotgun measurements above was also used for SWATH analysis [48, 49]. Specifically, in the present SWATH-MS mode, the AB SCIEX 5600 plus TripleTOF instrument was specifically tuned to optimize the quadrupole settings for the selection of 64 variable wide precursor ion selection windows. The 64-variable window schema was optimized based on a Cal-51 human cell tryptic digest sample, covering the precursor mass range of 400–1,200 m/z. The effective isolation windows can be considered as being 399.5 to 408.2, 407.2 to 415.8, 414.8 to 422.7, 421.7 to 429.7, 428.7 to 437.3, 436.3 to 444.8, 443.8 to 451.7, 450.7 to 458.7, 457.7 to 466.7, 465.7 to 473.4, 472.4 to 478.3, 477.3 to 485.4, 484.4 to 491.2, 490.2 to 497.7, 496.7 to 504.3, 503.3 to 511.2, 510.2 to 518.2, 517.2 to 525.3, 524.3 to 533.3, 532.3 to 540.3, 539.3 to 546.8, 545.8 to 554.5, 553.5 to 561.8, 560.8 to 568.3, 567.3 to 575.7, 574.7 to 582.3, 581.3 to 588.8, 587.8 to 595.8, 594.8 to 601.8, 600.8 to 608.9, 607.9 to 616.9, 615.9 to 624.8, 623.8 to 632.2, 631.2 to 640.8, 639.8 to 647.9, 646.9 to 654.8, 653.8 to 661.5, 660.5 to 670.3, 669.3 to 678.8, 677.8 to 687.8, 686.8 to 696.9, 695.9 to 706.9, 705.9 to 715.9, 714.9 to 726.2, 725.2 to 737.4, 736.4 to 746.6, 745.6 to 757.5, 756.5 to 767.9, 766.9 to 779.5, 778.5 to 792.9, 791.9 to 807, 806 to 820, 819 to 834.2, 833.2 to 849.4, 848.4 to 866, 865 to 884.4, 883.4 to 899.9, 898.9 to 919, 918 to 942.1, 941.1 to 971.6, 970.6 to 1006, 1005 to 1053, 1052 to 1110.6, 1109.6 to 1200.5 (containing 1 m/z for the window overlap). SWATH MS2 spectra were collected from 50 to 2,000 m/z. The collision energy (CE) was optimized for each window according to the calculation for a charge 2+ ion centered upon the window with a spread of 15 eV (equation: slope = 0.0625, intercept= -3.5 eV). An accumulation time (dwell time) of 50 ms was used for all fragment ion scans in high-sensitivity mode and for each SWATH-MS cycle a survey scan in high-resolution mode was also acquired for 250 ms, resulting in a duty cycle of circa 3.45 s.
Peptide identification for shotgun proteomics
Profile-mode wiff files from shotgun data acquisition were centroided and converted to mzML format using the Sciex Data Converter v.1.3 and converted to mzXML format using msconvert v.3.0.4238 from the proteowizard package [50]. The MS2 spectra were queried against the reviewed canonical SwissProt complete proteome database for human (ex_sp_9606, August 2014) appended with reversed sequence decoys [31]. Two types of search engines, X!Tandem [51] and OMSSA [52], were used through iPortal interface for proteomic workflows [53]. The search parameters were: static modifications of 57.02146 Da for cysteines, variable modifications of 15.99491 Da for methionine oxidations. The parent mass tolerance was set to be 50 p.p.m and mono-isotopic fragment mass tolerance was 0.1 Da (which was further filtered to be <0.05 Da for building the spectral library); fully-tryptic peptides and peptides with up to two missed cleavages were allowed. The identified peptides were processed and analyzed through Trans-Proteomic Pipeline 4.5.2 (TPP) [54] and were validated using PeptideProphet [55] (PeptideProphet parameter is –p0 –dDECOY_ –OAPdlIw). All the peptides were filtered at a false discovery rate (FDR) of 1 % (iProphet probability > 0.8790) [56].
Spectral library generation
The raw spectral libraries were generated as described in Schubert et al. [57] from all valid peptide spectrum matches for the shotgun measurement of the light peptides, and then refined into non redundant consensus libraries [49] using SpectraST [58]. For each peptide, the retention time was mapped into the iRT space [47] with reference to a linear calibration constructed for each shotgun run, as previously described [49]. The light MS assays constructed from the 6 most intense y ions (all b ions and other ions removed) with Q1 range from 400 to 1200 m/z excluding the precursor SWATH window were used for targeted data analysis of SWATH maps. This library was subsequently used to generate corresponding assays for all heavy peptides by shifting the y ion transitions in mass corresponding to the number of lysine and arginine contained in the partial sequence. Decoy assay were appended to the final library as described previously [19].
SWATH-MS data analysis and q-value estimation
SWATH-MS raw data was converted from wiff files to mzXML using msconvert included in the open-source proteowizard package [50]. The OpenSWATH analysis workflow was essentially executed as described in Röst et al. [19] but the improved single executable OpenSwathWorkflow was used instead of the multi-step workflow to perform peak-picking and feature detection on all SWATH-MS runs [59]. The experimental data and the assay libraries for the S. pyogenes samples were obtained from previously published analyses (Röst et al. [19]) while the pulsed-SILAC data was analyzed with a sample-specific library as described above. Both libraries contained target assays as well as decoy assays and we used pyprophet [38], an open-source re-implementation of the mProphet algorithm [27], to perform q-value estimation on individual runs after feature detection. The algorithm uses semi-supervised machine learning techniques to optimally separate true target assays and known false “decoy” signals and estimate q-values based on their distributions [27]. The reported features and their associated q-values (used as scores here) comprise the input for the TRIC algorithm.
Decoy based FDR control
In a separate step from the actual alignment, our approach allows for the adjustment of the false discovery rate based on a decoy model. This approach can be applied before or after alignment (for convenience, it has been integrated into the TRIC executable). The algorithm applies a q-value threshold based on the score of the best peakgroup per row (since quantification events are likely not independent, this is the most conservative approach). This approach attempts to control the number of decoy rows in the output matrix by using the estimate of r, the ratio of false positives to decoys as computed by the Storey-Tibshirani method implemented in mProphet and pyProphet [27, 38]. First, this ratio r is used to estimate the number of false positive rows nfp in the data matrix based on the number of decoy rows ndecoy in the matrix: nfp=r⋅ndecoy. The q-value threshold is lowered until the desired number of false positives rows (as estimated with the above formula) is reached, for example until nfp/ntot reaches 0.01 (where ntot represents the total number of target rows). Thus, a more stringent score-cutoff is chosen that limits the number of decoy rows in the final data matrix to the user-defined FDR value. In the examples described here, the q-value cutoffs used to achieve a 1 % FDR were 0.0015 for the S. pyogenes study, 0.0022 for the pulsed-SILAC study and 2.1×10−5 for the blood plasma dataset in the supplementary (generally, the more samples are analyzed, the lower the q-value threshold). When comparing the output of TRIC to the naïve approach with a fixed q-value cutoff, we use these value computed during the FDR procedure also for the naïve data matrix (unless otherwise indicated).
Validation dataset
A random subset of 452 peptides were chosen from the S. pyogenes data described in Röst et al. [19] and 7,232 chromatograms were extracted across 16 LC-MS/MS runs. These 7,232 chromatograms were manually inspected using the Skyline software [20], the correct peak (if present) was marked for each of the 16 runs and then exported. An in-house script compared the results of the manual annotation and the TRIC-based annotation where the peak was considered correct either if its apex differed less than 20 seconds from the manual annotation or of its apex was contained within the manual peak boundaries.
To study the correction of chromatographic distortion, a set of S. pyogenes runs acquired as part of a larger study was chosen for analysis and one run (hroest_K131126_005) was selected as the target and 7 additional runs from the same dataset (hroest_K131126_050 to hroest_K131126_056) were selected for co-alignment (with about 45 other injections between the two acquisitions) to produce realistic alignment conditions. From the target run, half of all high confidence peakgroups (q-value <0.001, 506 peakgroups) were removed. Then the full algorithm was run with the standard settings but –alignment_score changed to 0.001 to allow for more “anchor points”. After running the algorithm on the “training” set, the result was compared to the retention times and intensities of the original 506 high confidence “test” peak groups that were set aside before.
Parameters used
Unless otherwise indicated, the feature_alignment.py (available at https://pypi.python.org/pypi/msproteomicstools) was run with the following settings: LocalMST for –method indicating the use of the MST, 0.0001 for the parameter –alignment_score only allowing highly confident peakgroups with q-value < 0.01 % as “anchor points”, lowess for –realign_method, 0.1 for –max_fdr_quality allowing for the transfer of confidence to peakgroups with a score cutoff of 0.1, a value of 0.01 for –target_fdr indicating 1 % FDR on the data matrix rows, True for –mst:useRTCorrection and 3.0 for –mst:Stdev_multiplier indicating adaptive retention time windows. The optional noise imputation algorithm implemented in requantAlignedValues.py was run with the following settings: singleClosestRun for –method and lowess for –realign_method.
pulsed-SILAC analysis
For the pulsed-SILAC analysis of the iPS cells, TRIC was run as described earlier. The –max_fdr_quality parameter was lowered to 0.05 (to only include higher quality peak groups) and TRIC calculated a m_score cutoff of 0.002198 for the whole dataset. The data was filtered by m_score at the calculated cutoff (0.002198) to produce the data for the “naïve” approach (simple FDR filter) and filtered at 0.025 for the TRIC alignment data. To compare the difference in error between SILAC pairs, only points with an error less than 30 seconds were plotted in Figure 4 (148 high quality pairs and 165 aligned pairs were omitted as they fall outside the plotting window). At each time point, the amount of heavy (IH) and light (IL) precursor was extracted and used to calculated the relative isotopic abundance (RIAt):
| (1) |
analogous to Pratt et al .[36] (with heavy and light switched due to our experimental design being reversed). The value of RIAt is time dependent as unlabelled proteins are replaced with heavy-labelled proteins during the course of the experiment. This is due loss due to dilution of the cells as well as loss due to intracellular protein turnover, where the rate of loss can be modelled as an exponential decay process:
| (2) |
where RIA0 denotes the initial isotopic ratio and kloss the rate of (hourly) loss of unlabelled protein. We assumed RIA0=1 as no heavy isotope was present at t=0, thus the value of RIAt will decay exponentially from initially one to zero after infinite time. As discussed in Pratt et al. [36], these assumptions may reduce measurement error especially at the beginning of the experiment where isotopic ratios are more inaccurate due to the low absolute number of heavy precursor ions. Next, a linear model was fitted per peptide to the logarithmized data to obtain kloss values for all individual peptides. The kloss for each protein was computed as the median of all peptide-level rates. We excluded proteins quantified in a single timepoint only (241 proteins), proteins without a significant correlation (p <0.25 of a linear model) between isotope ratio and time (90 proteins) and increasing isotope ratio over time (21 proteins). After filtering, 1075 proteins were used to compute turnover rates. In order to obtain the protein turnover rate kturnover, we subtracted the dilution ratio D which we obtained by MTS assay from independent experiments on the same cell line at comparable confluence:
| (3) |
Assuming the cells are in steady state and protein synthesis is equal to degradation, the computed kturnover is equal to the degradation rate: kturnover=kdegradation. Please note for the specific purpose of illustrating the TRIC alignment in this paper, we treated heavy and light channels separately, which could be further optimized (e.g., by combining heavy and light assays in the library generation step) for future SILAC experiments.
For gene ontology (GO) term enrichment, we used the Gene Ontology enrIchment analysis and visualization tool (GORILLA) at http://cbl-gorilla.cs.technion.ac.il as described in Eden et al. [37]. We selected the proteins with the highest and lowest turnover rates (10 % and 25 % quantiles) and used all identified proteins as background in GORILLA. We identified 20 significantly (q-value <0.05) enriched GO terms. Most terms enriched for proteins with high turnover (fast degradation) were related to cell signaling, radiation and light response, cell-cell adhesion, extracellular matrix and locomotion (Supplementary Table 6).
Code availability
All source code for TRIC is available at https://github.com/msproteomicstools/msproteomicstools under the 3-clause BSD licence.
Human subjects approval
Approval by the ethics committees of the Genomic and Genetic Disorder Biobank (Casa Sollievo della Sofferenza, San Giovanni Rotondo, Italy) and the University of Perugia (Azienda Ospedaliera–Universitaria “Santa Maria della Misericordia” Perugia, Italy).
Supplementary Material
Acknowledgements
This work was supported by ETH Zurich (grant ETH-30 11-2 to H.R. and R.A.), the Swiss National Science Foundation (SNSF grant P2EZP3_162268 to H.R. and grant 31003A_166435 to R.A.), ERC Proteomics v3.0 (AdvG grant #233226 R.A.), ERC Proteomics4D (AdvG grant #670821 to R.A.), the PhosphonetX project of SystemsX.ch, the ERC DISEASEAVATARS (grant 616441 to R.A. and G.T.), the Telethon Foundation (Grant n. GGP14265 to G.T.) and the Regione Lombardia (Grant Ricerca Indipendente 2012 to G.T.) and the Fondazione Umberto Veronesi (G.D.).
We would like to thank SyBIT project of SystemsX.ch for support and maintenance of the lab-internal computing infrastructure. Specifically, L. Blum from SyBIT contributed substantially to make this software available to the whole lab and provided critical feedback.
Footnotes
Data Accession
All raw data and output generated by the tools described in this manuscript are deposited at the peptide atlas site and can be accessed at http://www.peptideatlas.org/PASS/PASS00788/. Source data for figures presented in this manuscript is provided in Supplemental Data 1.
Author Contributions
H.L.R. designed and wrote the code, performed the data analysis and produced the figures. Y.L., G.D. and M.Z performed the iPS cell experiment and acquired the mass spectrometric data. P.N. and G.R. contributed to the code and provided an initial prototype of the implementation. B.C.C. and L.G. acquired mass spectrometric data and gave critical input. G.T., L.M. and R.A. designed and supervised the study. All authors contributed to the manuscript.
Competing financial interests
The authors declare that they have no competing financial interests.
References
- [1].Hudson TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- [5].International Consortium for Blood Pressure Genome-Wide Association Studies and others. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Röst HL, Malmström L, Aebersold R. Reproducible quantitative proteotype data matrices for systems biology. Mol Biol Cell. 2015;26:3926–3931. doi: 10.1091/mbc.E15-07-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].de Godoy LMF, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- [8].Hebert AS, et al. The one hour yeast proteome. Mol Cell Proteomics. 2014;13:339–347. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Beck M, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Desiere F, et al. The peptideatlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Omenn GS, et al. Metrics for the Human Proteome Project 2015: progress on the human proteome and guidelines for high-confidence protein identification. J Proteome Res. 2015;14:3452–3460. doi: 10.1021/acs.jproteome.5b00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Picotti P, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li X-j, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med. 2013;5:207ra142. doi: 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Drabovich AP, et al. Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma. Sci Transl Med. 2013;5:212ra160. doi: 10.1126/scitranslmed.3006260. [DOI] [PubMed] [Google Scholar]
- [16].Surinova S, et al. Prediction of colorectal cancer diagnosis based on circulating plasma proteins. EMBO Mol Med. 2015a:e201404873. doi: 10.15252/emmm.201404873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Surinova S, et al. Non-invasive prognostic protein biomarker signatures associated with colorectal cancer. EMBO Mol Med. 2015b;7:1153–1165. doi: 10.15252/emmm.201404874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Röst HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- [20].MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martin DB, et al. MRMer, an interactive open source and cross-platform system for data extraction and visualization of multiple reaction monitoring experiments. Mol Cell Proteomics. 2008;7:2270–2278. doi: 10.1074/mcp.M700504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mead JA, et al. MRMaid, the Web-based Tool for Designing Multiple Reaction Monitoring (MRM) Transitions. Mol Cell Proteomics. 2009;8:696–705. doi: 10.1074/mcp.M800192-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Prakash A, et al. Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J Proteome Res. 2009;8:2733–2739. doi: 10.1021/pr801028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Walsh GM, et al. Implementation of a data repository-driven approach for targeted proteomics experiments by multiple reaction monitoring. J Proteomics. 2009;72:838–852. doi: 10.1016/j.jprot.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sherwood CA, et al. MaRiMba: A Software Application for Spectral Library-Based MRM Transition List Assembly. J Proteome Res. 2009;8:4396–4405. doi: 10.1021/pr900010h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bertsch A, et al. Optimal de novo Design of MRM Experiments for Rapid Assay Development in Targeted Proteomics. J Proteome Res. 2010;9:2696–2704. doi: 10.1021/pr1001803. [DOI] [PubMed] [Google Scholar]
- [27].Reiter L, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nature Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- [28].Teleman J, et al. Automated selected reaction monitoring software for accurate label-free protein quantification. J Proteome Res. 2012;11:3766–3773. doi: 10.1021/pr300256x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Prakash A, et al. Signal Maps for Mass Spectrometry-based Comparative Proteomics. Mol Cell Proteomics. 2006;5:423–432. doi: 10.1074/mcp.M500133-MCP200. [DOI] [PubMed] [Google Scholar]
- [30].Mueller LN, et al. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- [31].Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [32].Reiter L, et al. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Doherty MK, Hammond DE, Clague MJ, Gaskell SJ, Beynon RJ. Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J Proteome Res. 2009;8:104–112. doi: 10.1021/pr800641v. [DOI] [PubMed] [Google Scholar]
- [34].Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- [35].Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- [36].Pratt JM, et al. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1:579–591. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- [37].Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Teleman J, et al. DIANA - algorithmic improvements for analysis of data-independent acquisition MS data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu686. [DOI] [PubMed] [Google Scholar]
- [39].Prince JT, Marcotte EM. Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Analytical Chemistry. 2006;78:6140–6152. doi: 10.1021/ac0605344. [DOI] [PubMed] [Google Scholar]
- [40].Kohlbacher O, et al. TOPP–the OpenMS proteomics pipeline. Bioinformatics. 2007;23:e191–197. doi: 10.1093/bioinformatics/btl299. [DOI] [PubMed] [Google Scholar]
- [41].Sturm M, et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- [43].Liu Y, et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol. 2015;11:786. doi: 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tsou C-C, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nature methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Adamo A, et al. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nature Genetics. 2015 Feb;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- [46].Kim SC, et al. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J Proteome Res. 2006 Dec;5:3446–3452. doi: 10.1021/pr0603396. [DOI] [PubMed] [Google Scholar]
- [47].Escher C, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liu Y, et al. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol Cell Proteomics. 2014 Jul;13:1753–1768. doi: 10.1074/mcp.M114.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Collins BC, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nature Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- [50].Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics (Oxford, England) 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- [52].Geer LY, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- [53].Kunszt P, et al. iPortal: the swiss grid proteomics portal: Requirements and new features based on experience and usability considerations. Concurrency and Computation: Practice and Experience. 2015;27:433–445. [Google Scholar]
- [54].Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular Systems Biology. 2005;1:17. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [56].Shteynberg D, et al. iProphet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Schubert OT, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protoc. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- [58].Lam H, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- [59].Rost HL, Aebersold R, Schubert O. Automated SWATH Data Analysis Using Targeted Extraction of Ion Chromatograms. bioRxiv. 2016:044552. doi: 10.1007/978-1-4939-6747-6_20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.