Abstract
Class A G protein coupled receptors (GPCRs) are a large family of membrane proteins that mediate a wide variety of physiological functions (e.g. vision, neurotransmission, and immune response)1–4. Not surprisingly, they are the targets of nearly one-third of all prescribed medicinal drugs5 (e.g. beta blockers, antipsychotics). GPCR activation is facilitated by extracellular ligands, and leads to the recruitment of intracellular G proteins3,6. Structural rearrangements of residue contacts in the transmembrane domain serve as ‘activation pathways’ that connect the ligand-binding pocket to the G protein-coupling region within the receptor. How similar are these activation pathways across different class A GPCRs? Here, we analysed 27 GPCRs from diverse subgroups for which structures of active and/or inactive states are available. We show that despite the diversity in activation pathways between receptors, the pathways converge near the G protein-coupling region. This convergence is mediated by a strikingly conserved structural rearrangement of residue contacts between transmembrane helices 3, 6, and 7 that releases G protein-contacting residues. The convergence of activation pathways may explain how the activation steps initiated by diverse ligands confer GPCRs the ability to bind a common repertoire of G proteins.
Although GPCRs have undergone extensive sequence diversification, they share a conserved structural architecture composed of seven transmembrane (TM) helices. Binding of a ligand to the receptor on the extracellular side results in rearrangement of residue contacts between the TM helices4,7. This leads to large conformational changes in the cytoplasmic side that support receptor activation8–13. The activated receptor binds to and allosterically activates G proteins14–17. In this manner, the TM region of GPCRs plays a pivotal role in transmitting signals across the cell membrane. At the time of performing this study, crystal structures have been determined for 27 class A GPCRs, belonging to seven of the eleven different subgroups (defined based on the endogenous ligand types they bind; www.gpcrdb.org). While most structures correspond to inactive states (generally bound to an antagonist or inverse-agonist), a few structures of receptors in active states (bound to an agonist and with structural rearrangements at the G protein-binding region) have also been solved. There are only five receptors for which the structures of both inactive and active (active or active-intermediate) states are available: the light-activatable rhodopsin8,13, amine-activatable β2 adrenergic receptor (β2AR)7 and M2 muscarinic receptor (M2R)11, nucleoside-activatable A2A receptor (A2AR)12, and peptide-activatable µ-opioid receptor (µOR)10. The remaining structures have been determined only in either inactive or active states. The availability of structures of GPCRs from divergent subgroups (as low as ~20% sequence identity18) that are bound to chemically diverse ligands and known to couple to different G proteins, allowed us to investigate activation pathways across class A GPCRs.
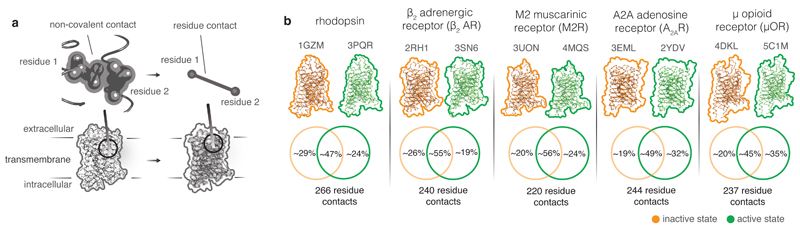
Given that the GPCRs are structurally similar and activate a small set of G proteins, some structural aspects of receptor activation are broadly similar (e.g. contraction of ligand binding site, opening of the cytosolic side due to relocation of TM6). However, receptor activation is mediated by diverse ligands and hence some aspects of ligand-induced GPCR activation must necessarily be receptor-specific. How similar are the activation pathways across different receptors? We carried out a comprehensive comparison of residue contacts of inactive and active state structures. Structural equivalence for residues across the different GPCRs was assigned using the GPCRdb numbering scheme19 from GPCRdb20 (www.gpcrdb.org) (Methods). A contact between a pair of residues is defined to exist if the inter-atomic distance between any two atoms across the residue pair is shorter than the sum of their van der Waals radii plus a cutoff distance4,14 (Fig. 1a; Methods). Analysis of residue contacts in inactive and active state structures of rhodopsin, β2AR, M2R, A2AR and µOR revealed that there are ~220-266 contacts between residues in the seven TM helices and helix 8 for each receptor (Fig. 1b). In every receptor, roughly a half of the residue contacts are reorganized upon activation, whereas the other half does not change upon activation.
Figure 1. Comparison of residue contacts in inactive and active state structures of class A GPCR.
a. Residue contact in a GPCR. Residues are denoted as circles and the non-covalent contacts between residues (residue contacts) are denoted as lines connecting the circles. b. Similarity of residue contacts between inactive and active states of each GPCR. For five class A GPCRs, the comparison of the number of residue contacts between inactive and active states are shown using Venn diagrams.
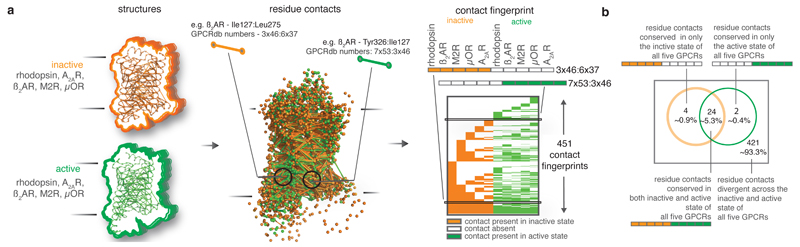
How similar are the contacts that change upon activation among the different receptors? While pairwise structural comparisons of individual receptors reveal some contacts that are reorganized, the combinatorial possibilities of structural comparison of multiple receptors makes it non-trivial to address this question. Therefore, we employed an unbiased, multi-way comparison approach wherein we analyzed the pattern of contacts between structurally equivalent residues across all inactive and active state structures (‘contact fingerprint’; Fig. 2a). Of the 451 contact fingerprints, only 30 (~6.7%) represent contacts that are consistently maintained in all inactive and/or active state structures. The remaining 421 contact fingerprints (~93.3%) represent contacts that are not maintained consistently in the inactive/active state structures across the five GPCRs. This suggests that there is significant diversity in the activation pathways of the different receptors (Fig. 2b).
Figure 2. Patterns of residue contacts across inactive and active state structures of GPCRs.
a. Contact fingerprinting. For every residue contact, the presence (or absence) of a contact between structurally equivalent residues for all structures is computed. The pattern of presence and absence (filled and empty cells respectively) across multiple structures is termed ‘contact fingerprint’. b. Distribution of the contact fingerprints representing the presence of contacts in the five inactive and five active state structures.
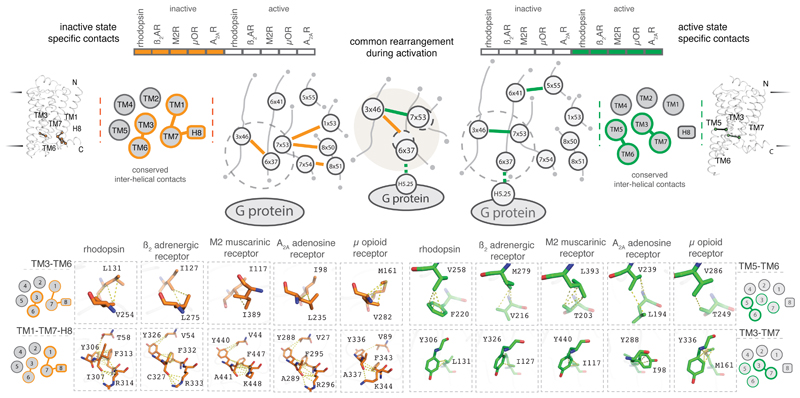
Despite the diversity in the reorganization of residue contacts upon receptor activation, strikingly, four contacts involving seven residues are exclusively maintained in all inactive state structures whilst two contacts involving four residues are maintained exclusively in all active state structures (Fig. 3; Supplementary Table 1). When one considers the role of these contacts in the inactive and active state structures together, a common re-organization of contacts upon activation becomes apparent. This common rearrangement involves residues in TM3 (3x46), TM6 (6x37) and TM7 (7x53) in all the five receptors (Fig. 3). Importantly, the rearranged contacts are proximal to the G protein-coupling region, and distant from the ligand-binding pocket. This highlights that the diverse structural changes among the receptors, that is stabilized by different ligands converges on a common set of rearrangement near the G protein-coupling region.
Figure 3. Conserved rearrangement of residue contacts between inactive and active state structures of GPCRs.
Considering the conserved residue contacts in the inactive (left) and the active (right) states together reveals a conserved rearrangement of residue contacts in all the five receptors. The cartoons show a schematic representation of the contacts at the secondary structure level (inter-helical) and the residue level (inter-residue). TM helices and helix 8 are shown as circles and the presence of inactive and active state specific contacts between the helices are shown as lines. Residues are shown as circles and contacts between residues are shown as lines. Inactive and active state-specific contacts are shown in orange and green respectively. Dotted circles around 6x37 and 7x53 denote the movement of helices 6 and 7 upon activation.
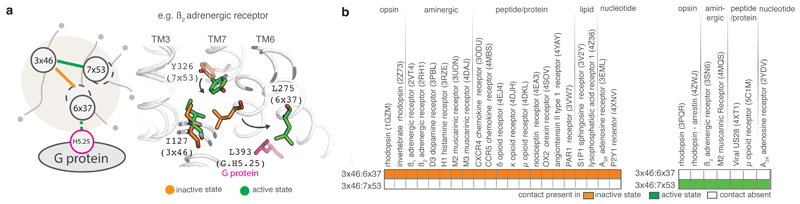
Upon activation, the cytoplasmic side of TM6 moves away from the rest of the transmembrane bundle and several previously inaccessible G protein-coupling residues become accessible7. Many of these residues participate in triggering the conserved allosteric mechanism for GDP release in G proteins14. These include the position 6x37 in the cytoplasmic end of TM6 that upon activation contacts a universally conserved leucine (G.H5.25) of the G protein in the β2AR-Gs structure (Fig. 4a). In the inactive state, the residue at 6x37 is engaged in a contact with a conserved hydrophobic residue at position 3x46 (Fig. 4a). Upon activation, the residue at 3x46 breaks the contact with 6x37 and forms a new contact with Tyr7x53 within the highly conserved NPxxY motif of TM7. In the inactive state, 7x53 is not available to engage with 3x46 because TM helices 7 and 3 are far apart, and TM6 needs to move out for these residues to form a contact. The residues at position 8x50 and 1x53 contact 7x53 in the inactive state and both the contacts are broken upon receptor activation. This contact rearrangement is observed consistently in all the five different GPCRs from diverse subgroups that are activated by different ligands, and couple to diverse G proteins. Thus, despite the diversity in the reorganization of contacts upon activation (Fig. 2b), there exists a common rearrangement of residue contacts near the G protein-coupling site that underlies the activation of diverse class A GPCRs (Fig. 4a).
Figure 4. Conserved rearrangement of residue contacts between inactive and active state structures across diverse class A GPCRs.
a. Illustration of the conserved rearrangement of residue contacts between inactive and active state structures in β2AR. Dotted circles around 6x37 and 7x53 denote the movement of helices 6 and 7 upon activation. b. Conservation pattern of residue contacts involved in the contact rearrangement upon receptor activation in the inactive (left) and active (right) state structures of class A GPCRs.
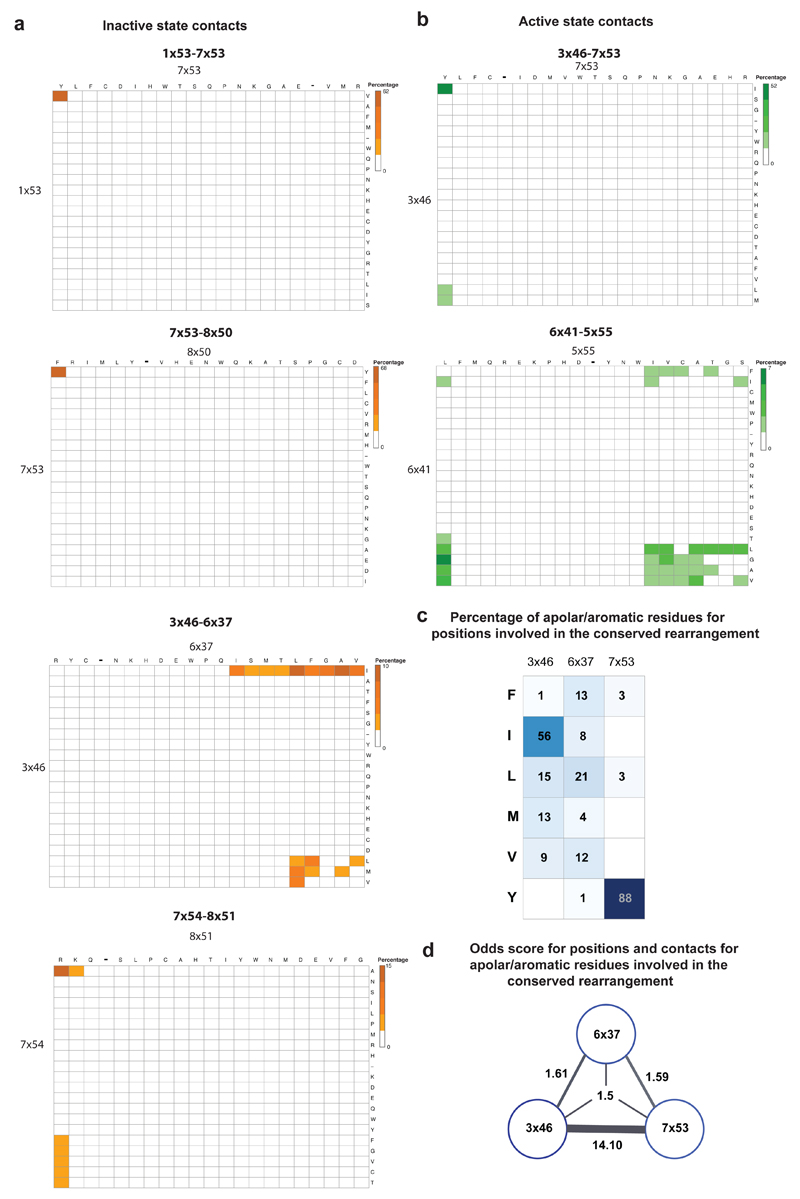
How consistent is the rewiring of contacts between 3x46, 6x37 and 7x53 in other class A GPCRs? We investigated all the other class A GPCR structures that were available only in either inactive or activate state (Methods). These include functionally diverse receptors belonging to seven of the 11 subgroups of class A GPCRs: light-activated opsins, aminergic receptors, peptide-binding receptors, protein-binding receptors, nucleoside-binding receptors, lipid-binding receptors, and other receptors (Fig. 4b). The contact between 3x46 and 6x37 is present consistently in all inactive state structures but in none of the active state structures (Fig. 4b). Similarly, the contact between 3x46 and 7x53 is present consistently in all active state structures but in none of the inactive state structures (Fig. 4b). To investigate the importance of these positions and contacts for class A GPCRs (including those which lack a solved structure), we performed a sequence analysis of all non-olfactory class A GPCRs in human. We find that equivalent positions of both residues that mediate a contact tend to be predominantly large hydrophobic or aromatic residues and hence are likely to fulfill the van der Waals criterion to form a contact (Extended Data Figure 1). These observations collectively suggest that the rearrangement of contacts involving positions 3x46, 6x37 and 7x53 is likely to be conserved, and important for receptor activation in all class A GPCRs.
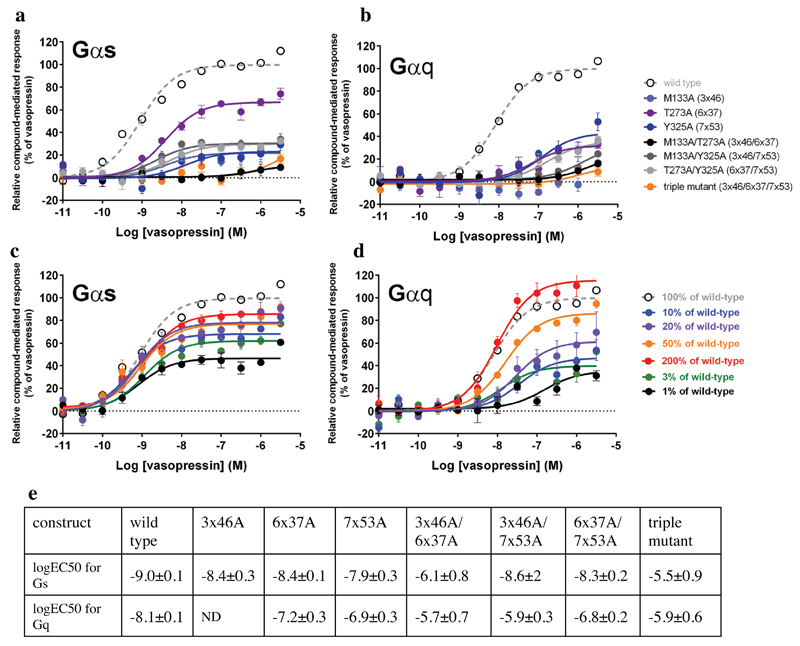
In order to experimentally investigate the importance of the interactions between 3x46, 6x37, and 7x53 in a receptor for which there is no crystal structure available, we examined the signalling profile of the V2 vasopressin receptor using BRET assays21. We created single, double and triple alanine mutants at these positions and measured vasopressin-induced Gs and Gq recruitment to infer receptor function. We observed that the mutations resulted in significantly reduced efficacy and potency of the receptor to activate Gs and Gq when compared to the wild type receptor (Extended Data Figure 2). Furthermore, previous observations from mutagenesis experiments in other GPCRs also highlight the importance of these positions for signaling (Supplementary Table 2). For instance, mutation of 6x37 in α1b-AR results in the reduction of downstream signaling by >70%22. Similarly, mutation of Tyr7x53 in β2AR leads to a significant reduction in G protein activation23. While mutations in these positions affect receptor activation, and downstream signaling, the nature of the substitution is likely to influence how the receptor is affected24. Given their key role, mutation in these positions would likely disrupt functional integrity, which could lead to a pathological condition in a physiological context. Indeed, mutations at these positions in different GPCRs are associated with disease (e.g. hypogonadotropic hypogonadism25; Supplementary Table 3). Thus, given the importance of the positions 3x46, 6x37 and 7x53 in diverse GPCRs, we anticipate that the knowledge about the conserved rearrangement of contacts will guide GPCR modelling, facilitate engineering of GPCRs and help in prioritising disease mutations.
Several studies have provided important insights into distinct mechanisms of activation in individual receptors7–13, and have described networks of contacts that are present in various combinations in different receptors upon activation4,10,26,27. Here, we report the presence of a highly conserved rearrangement of specific residue contacts that mediate the convergence of activation pathways across class A GPCRs. Because the microenvironment (i.e. surrounding residues/second shell residues) in which such rearrangement takes place diverges between receptor families, the detailed mechanism by which this common step is facilitated by diverse ligands is likely to be distinct for different sets of GPCRs. Remarkably, despite such differences, we find that the activation pathways ultimately converge to a common and very specific set of contact rearrangements between topologically equivalent residues near the G protein-coupling region. Future studies aimed at investigating residues at and around these positions can help uncover the unique steps that lead to the triggering of the convergent reorganization in a receptor-specific and ligand-specific manner.
From an evolutionary perspective, uncoupling the structural changes near the ligand-binding pocket from the G protein-coupling region, but still ensuring that the activation pathway converges near the G protein coupling site permits the ligand-binding pocket to evolve independently of the intracellular region that couples to G proteins. In this manner, the convergence of ligand-mediated allosteric activation pathways may have contributed to the evolutionary success of GPCRs to bind and be activated by a plethora of ligands, but still signal via a small repertoire of intracellular signaling proteins inside the cell.
Methods
Dataset
The coordinates of the crystal structures of 27 GPCRs (as of Nov. 2015) were obtained from the Protein Data Bank28 (www.rcsb.org). The structures were classified into inactive and active (active/active-intermediate) states. An inactive state structure is bound to an antagonist or an inverse agonist. An active state structure is bound to an agonist and has undergone structural rearrangements in the G protein-coupling region in comparison to its inactive state structure.
There are inactive state structures for the following GPCRs: rhodopsin (1GZM), invertebrate rhodopsin (2Z73), β1 adrenergic receptor (2VT4), β2 adrenergic receptor (2RH1), D3 dopamine receptor (3PBL), H1 histamine receptor (3RZE), M2 muscarinic receptor (3UON), M3 muscarinic receptor (4DAJ), CXCR4 chemokine receptor (3ODU), CCR5 chemokine receptor (4MBS), κ-opioid receptor (4DJH), μ-opioid receptor (4DKL), nociceptin receptor (4EA3), δ-opioid receptor (4EJ4), orexin 2 receptor (4S0V), angiotensin II type 1 receptor (4YAY), protease activated receptor 1 (3VW7), sphingosine 1P receptor (3V2Y), lysophosphatidic acid receptor 1 (4Z36), adenosine A2A receptor (3EML), and P2Y1 receptor (4XNV).
There are active state structures for the following GPCRs: rhodopsin bound to a peptide resembling the C-terminus of transducin (3PQR) or bound to arrestin (4ZWJ), adenosine A2A receptor (2YDV), β2 adrenergic receptor bound to the G protein Gs (3SN6), M2 muscarinic receptor bound to a G protein mimetic nanobody (4MQS), μ-opioid receptor bound to a G protein mimetic nanobody (5C1M), and viral US28 bound to a G protein mimetic nanobody (4XT1).
There are agonist-bound structures of four GPCRs: β1 adrenergic receptor (2Y01), serotonin 1B receptor (4IAQ), serotonin 2B receptor (4IB4) and neurotensin 1 receptor (4GRV). The β1 adrenergic receptor structure (2Y01) is nearly identical to its inactive state structure in the G protein-coupling region and is thus a case of an agonist-bound inactive conformation. Since there are no antagonist or inverse agonist bound structures of serotonin 1B receptor, serotonin 2B receptor and neurotensin 1 receptor, their agonist bound structures (4IAQ, 4IB4, and 4GRV) cannot be reliably classified as an inactive state or an active state. Hence these structures were excluded.
There are three structures of the P2Y12 receptor (PDBs: 4NTJ, 4PXZ and 4PY0). 4PXZ and 4PY0 are similar to each other, and have a distorted TM6 (in the cytoplasmic side) due to the fusion with BRIL (the helix ‘ends’ right after 6x36, becoming a short unstructured region that links to BRIL). The case of 4NTJ is also unusual. The fusion protein still distorts the cytoplasmic region near 6x36 but, in addition, the ligand binds in a peculiar pose that forces TM6 to ‘move away’ from the transmembrane bundle (at the extracellular side), which modifies the rest of the helix, including the cytoplasmic side. The impact of this peculiar binding pose is so large that most of the extracellular domain is not visible, including the cysteine bridge between TM3 and ECL2. Thus, due to the specific fusion splice points (in 4NTJ, 4PXZ and 4PY0) and the peculiar binding mode of the ligand (in 4NTJ), the cytoplasmic side of TM6 in the P2Y12 receptor crystal structures is distorted and, therefore, these structures were excluded. Similarly, in the case of FFAR1 (PDB: 4PHU), TM7 is shorter than in other class A GPCRs and appears to be truncated. Hence, the structure of FFAR1 was also excluded.
Calculation of residue contacts
We defined the presence of a residue contact between a pair of residues when the distance between any two atoms from the residue pair is less than the sum of their van der Waals radii plus a cut-off distance of 0.5 Å. Contacts involving the backbone atoms between residues that are less than four amino acids apart in the protein sequence were ignored to exclude local contacts4,14. We only considered contacts mediated by the residues of the transmembrane helices (TM1-7) or the amphipathic helix (H8) (based on GPCRdb numbers; www.gpcrdb.org), as (i) ligand binding primarily leads to reorganization in the transmembrane bundle, and (ii) the loops/termini are highly variable in length, structure and intrinsic disorder among the different GPCRs29. While rearrangements of water mediated contacts have been shown to be important for receptor activation10, they are not considered here because not all structures have water molecules resolved in their coordinates. In each structure, residues present in the transmembrane helices (TM1-7) or the amphipathic helix (H8) are treated as nodes and the presence of atomic contacts between pairs of residues are treated as edges connecting the nodes. We then evaluated the presence of residue contacts between topologically equivalent residues across GPCR structures. Structural equivalence was assigned using the GPCRdb numbering scheme19 from GPCRdb20 (www.gpcrdb.org/).
Contact fingerprinting
The functional importance of a given residue contact across a group of proteins can be estimated based on the extent to which topologically equivalent contacts are maintained consistently4,14. For every residue contact within the ensemble, the presence or absence of an equivalent residue contact between topologically equivalent positions across the rest of the GPCR structures was recorded. This information is stored as a bit string of ones (present) and zeros (absent), which are referred to here as ‘contact fingerprints’. Identifying contact fingerprints that represent consistently maintained residues contacts across and between conformational states enabled us to identify conserved rearrangement of residue contacts in class A GPCRs. For the identification of residue contacts that are specific either to the inactive or the active states, we focused only on the GPCRs that had structures determined in both inactive as well as active state. For the analysis of contacts across GPCRs including the inactive and active state only structures (Fig. 4b), the contact distance threshold was computed based on the sum of the van der Waals radii of atoms plus multiple cut-off distance between 0.5 and 0.8 Å, in order to account for variation in the quality of structures.
Sequence analysis
The alignment of 311 non-olfactory, Class A human GPCRs were obtained from GPCRdb20 (www.gpcrdb.org/). 285 positions that spanned all the seven TM segments and H8 were aligned. The percentage of amino acids for residues at positions 3x46, 6x37 and 7x53 were calculated from this alignment. The set of large hydrophobic/aromatic residues was defined to include the following amino acids: V, L, I, M, F, Y, W. The odds of finding large hydrophobic/aromatic residues at both positions that form a contact across Class A GPCRs was calculated as p/(1-p); where p = probability (large hydrophobic/aromatic residues in both positions that mediate a contact).
Structure analysis and visualisation
Computer scripts used for identifying and comparing residue contacts in protein structures were written in Perl. Visualisation of protein structures and residue interaction networks was performed using PyMOL.
Signalling profile of Vasopressin V2 receptor mutants using BRET-based biosensors
Vasopressin V2 receptor construct used in the experiments
The human vasopressin V2 receptor construct included the N-terminal SNAP-tag for easy detection and quantification and Twin-Strep tag for optional purification in pcDNA4/TO vector (SNAP-TS-V2R). The mutations were introduced using a two-fragment PCR approach followed by Gibson assembly as described earlier30 and confirmed by sequencing.
Vasopressin V2 receptor ligands
[Arg8]-Vasopressin (Cys-Tyr-Phe-Gln-Asn-Cys-Pro-Arg-Gly-NH2; disulphide bridge: Cys1-Cys6) was purchased from Genemed Synthesis Inc. (San Antonio, TX, USA). The ligand dilutions were prepared in PBS with 0.1% (w/v) BSA. Stock solutions were stored at -20° C and dilutions were freshly prepared for each experiment.
Biosensor constructs
For the plasmids encoding the RlucII-Gα constructs, Renilla luciferase (RlucII) was inserted into the coding sequence of human Gαs and Gαq constructs at positions 117 and 118, respectively. Gγ1 was N-terminally tagged with GFP10 as described31. Untagged Gβ1 was used for all experiments.
Cell culture and transfection
Human embryonic kidney (HEK) 293SL cells were transiently co-transfected with SNAP-TS-V2R, different RlucII-Gα variants, Gβ1 and GFP10-Gγ1 for G protein activation measurements. Linear 25 kDa polyethyleneimine (PEI) (Polysciences Inc.) was prepared in phosphate-buffered saline (PBS) (Multicell) (PEI:DNA ratio 3:1). The cells were seeded into white Cellstar® PS 96-well cell culture plates (Greiner Bio-One, Germany) at a density of 20,000 cells per well and grown for 48 h at 37° C with 5% CO2.
Biosensor measurements
48 h after transfection the 96-well plates were washed once with 200 µl PBS/well and 90 µl of Tyrode’s buffer (NaCl 137 mM, KCl 0.9 mM, MgCl2 1 mM, NaHCO3 11.9 mM, NaH2PO4 3.6 mM, Hepes 25 mM, glucose 5.5 mM, CaCl2 1 mM, pH 7.4) were added for measurements. The cells were stored at 37°C with 5% CO2 for 2 h prior to the measurement. For the measurement the plates were incubated with 10 µl ligand or vehicle per well for 5 min, then the luciferase substrate, coelenterazine 400a (DeepBlueC), was added to a final concentration of 2.5 μM. After a further 5 min of incubation, luminescence and GFP10 fluorescence were measured at 410 ± 40 nm and 515 nm ± 15 nm respectively, in a Synergy Neo plate reader (Biotek) using 0.4 s integration time. BRET was expressed as a ratio of the emission at 515 nm over the emission at 410 nm.
Quantification of expression levels
SNAP-TS-V2R wild-type and mutant expressing HEK293SL cells, respectively, were labelled with 50 μl 1 μM SNAP-Surface® Alexa Fluor® 647 dye (New England Biolabs Inc.) in complete medium (DMEM, 10% FBS, Penicillin-Streptomycin) for 30 min at 37° C, 5% CO2 in 96-well plates. The wells were washed with 100 μl/well complete medium three times, and once with PBS. For measurements, 90 μl Tyrode’s buffer were added to each well. Fluorescence was measured with 630 nm excitation and 670 nm emission.
Extended Data
Extended Data Figure 1. Sequence analysis of positions involved in the conserved rearrangement during receptor activation.
The alignment of 311 non-olfactory, Class A human GPCRs were obtained from GPCRdb. Percentage of residue pairs making a contact in the inactive state only (a; orange spectrum; five bins) and the active state only (b; green spectrum; five bins). The set of large hydrophobic/aromatic residues was defined to include the following amino acids: V, L, I, M, F, Y, W. c. The percentage of amino acids for residues in the positions (3x46, 6x37 and 7x53) involved in the conserved rearrangement is shown. d. The odds of finding a large hydrophobic/aromatic residue at a pair/triplet of positions that form a contact during the conserved rearrangement. See Methods for details.
Extended Data Figure 2. Signalling profile of Vasopressin V2 receptor mutants using BRET-based biosensors.
Activation of Gs (a) and Gq (b) by Vasopressin V2 receptor wild type and mutants. The positions 3x46, 6x37 and 7x53 were mutated to alanine separately and in combination and their G protein response was measured using BRET-based biosensors. Gs is the primary and Gq is the secondary cognate G protein of the vasopressin V2 receptor. The G protein response was significantly reduced for all mutants, pointing towards a reduced receptor activity. The expression levels of the mutant receptors were quantified using a fluorescent dye bound to the SNAP tag of surface-expressed receptors. While the Y325A (Y7x53A), T273A (T6x37A) and T273A/Y325A (T6x37A/Y7x53A) proteins were expressed at wild-type levels, the expression of M133A (M3x46A) and its combinations showed reduced expression levels. As a comparison, the signalling of reduced levels of WT Vasopressin V2 receptor (% of DNA transfected) for Gs and Gq is shown in panels c and d, respectively. This indicates that the reduction in expression level alone cannot explain the reduced signalling of Y325A (Y7x53A), T273A (T6x37A) and the Y325A (Y7x53A)/T273A (T6x37A) combination. Thus, the reduced signalling activity of Y325A (Y7x53A), T273A (T6x37A) and T273A/Y325A (T6x37A/Y7x53A) is likely due to a change in receptor properties. In the case of receptor mutants (single, double and triple) involving M133A (M3x46A), both the G protein response as well as the expression level of the receptor is reduced. This maybe because the position 3x46 is involved in mediating contacts that are important for both the active and the inactive state, and mutating this position might simultaneously affect receptor biogenesis and downstream response. e. The increased EC50s are consistent with a reduced ability of the receptor to reach the G protein bound active conformation resulting in a reduced potency. This agrees well with a destabilisation of the active state of the mutants.
Supplementary Material
Acknowledgements
We thank C Chothia, G Chalancon, N Latysheva, R Henderson, A Sente, and K Kruse for their comments on this manuscript. We acknowledge LMB International Scholarship (AJV), St. John’s College Benefactor Scholarship (AJV), MRC Centenary Early Career Award (AJV), the MRC (MC_U105185859; MMB, BS, TF; MC U105197215; CGT, GL), Boehringer Ingelheim Fonds (TF), Heptares Therapeutics Ltd (GL), ATIP-Avenir program (GL), Swiss National Science Foundation (310030_153145, GFXS; 31003A_146520, XD; 31003A_159748, DBV), Swiss National Science Foundation Doc.Mobility fellowship P1EZP3_165219 (FMH), the Human Frontiers Science Program (RGP0034/2014, GFXS), the Canadian Institute for Health Research (MOP-10501, MB), H2020 MSCA-ITN-2014-ETN program (X-Probe project, GFXS) and the COST Action CM1207 (GLISTEN, XD, GFXS, CGT, GL). MMB is a Lister Institute Research Prize Fellow. MB holds a Canada research Chair.
References
- 1.Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 3.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 5.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 6.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen SG, et al. Crystal structure of the beta(2) adrenergic receptor-Gs protein complex. Nature. 2011 doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 9.Dror RO, et al. Activation mechanism of the beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, et al. Structural insights into micro-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flock T, et al. Universal allosteric mechanism for Galpha activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manglik A, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 17.Dror RO, et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science. 2015;348:1361–1365. doi: 10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez A, Cordomi A, Caltabiano G, Pardo L. Impact of helix irregularities on sequence alignment and homology modeling of G protein-coupled receptors. Chembiochem. 2012;13:1393–1399. doi: 10.1002/cbic.201200189. [DOI] [PubMed] [Google Scholar]
- 19.Isberg V, et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isberg V, et al. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016;44:D356–364. doi: 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Westhuizen ET, Breton B, Christopoulos A, Bouvier M. Quantification of ligand bias for clinically relevant beta2-adrenergic receptor ligands: implications for drug taxonomy. Mol Pharmacol. 2014;85:492–509. doi: 10.1124/mol.113.088880. [DOI] [PubMed] [Google Scholar]
- 22.Greasley PJ, Fanelli F, Rossier O, Abuin L, Cotecchia S. Mutagenesis and modelling of the alpha(1b)-adrenergic receptor highlight the role of the helix 3/helix 6 interface in receptor activation. Mol Pharmacol. 2002;61:1025–1032. doi: 10.1124/mol.61.5.1025. [DOI] [PubMed] [Google Scholar]
- 23.Gabilondo AM, Krasel C, Lohse MJ. Mutations of Tyr326 in the beta 2-adrenoceptor disrupt multiple receptor functions. Eur J Pharmacol. 1996;307:243–250. doi: 10.1016/0014-2999(96)00247-6. [DOI] [PubMed] [Google Scholar]
- 24.Bohm SK, et al. Identification of potential tyrosine-containing endocytic motifs in the carboxyl-tail and seventh transmembrane domain of the neurokinin 1 receptor. J Biol Chem. 1997;272:2363–2372. doi: 10.1074/jbc.272.4.2363. [DOI] [PubMed] [Google Scholar]
- 25.Tello JA, et al. Congenital hypogonadotropic hypogonadism due to GnRH receptor mutations in three brothers reveal sites affecting conformation and coupling. PLoS One. 2012;7:e38456. doi: 10.1371/journal.pone.0038456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isom DG, Dohlman HG. Buried ionizable networks are an ancient hallmark of G protein-coupled receptor activation. Proc Natl Acad Sci U S A. 2015;112:5702–5707. doi: 10.1073/pnas.1417888112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lans I, Dalton JA, Giraldo J. Helix 3 acts as a conformational hinge in Class A GPCR activation: An analysis of interhelical interaction energies in crystal structures. J Struct Biol. 2015;192:545–553. doi: 10.1016/j.jsb.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatakrishnan AJ, et al. Structured and disordered facets of the GPCR fold. Curr Opin Struct Biol. 2014;27:129–137. doi: 10.1016/j.sbi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Sun D, et al. AAscan, PCRdesign and MutantChecker: a suite of programs for primer design and sequence analysis for high-throughput scanning mutagenesis. PLoS One. 2013;8:e78878. doi: 10.1371/journal.pone.0078878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gales C, et al. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat Struct Mol Biol. 2006;13:778–786. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.