Supplemental Digital Content is available in the text
Abstract
Breast cancer stem cells (BCSCs) are considered to be responsible for recurrence in breast cancer. The 68 kDa Src-associated protein in mitosis (Sam68) has been linked to the development and progression of breast cancer; however, the posttranscriptional regulation and role of Sam68 in BCSC self-renewal remain unclear.
Sam68 was ectopically overexpressed or knocked down using a siRNA; the self-renewal potential of breast cancer cell lines was assessed using flow cytometry, in vitro mammosphere culture and a xenograft model in NOD/SCID mice. Activation of beta-catenin was assessed by immunohistochemical staining, Western blotting, and luciferase reporter gene assays. The ArrayExpress dataset GSE45666 was used to identify conserved microRNAs downregulated in breast cancer; real-time PCR, Western blotting, luciferase reporter assay, and xenografted tumor model were used to confirm miR-204 regulated Sam68.
We found that endogenous Sam68 expression correlated positively with the self-renewal potential of breast cancer cell lines. Overexpression of Sam68 promoted, whereas knockdown reduced, breast cancer cell self-renewal potential in vitro and tumorigenicity in vivo. The Wnt/beta-catenin pathway was identified as a functional mediator of Sam68-induced self-renewal in SKBR-3 and MCF-7 cells. Furthermore, miR-204 was found to be frequently downregulated in human breast cancer and confirmed to directly target Sam68; miR-204 inhibited the self-renewal of breast cancer cell lines by targeting and suppressing Sam68.
Our study reveals that Sam68 is regulated by miR-204 and may play an important role in the self-renewal of BCSCs via activating the Wnt/beta-catenin pathway. Sam68 may represent a novel therapeutic target for breast cancer.
INTRODUCTION
Breast cancer is the second cause of tumor-related deaths in females, and more than 400,000 women die due to breast cancer worldwide every year.1 Many patients suffer recurrence within 5∼10 years after surgery or chemotherapy.2 Breast cancer stem cells (BCSCs) are considered to be the major source of breast cancer recurrence.3 Cancer stem cells (CSCs) influence tumor growth, metastasis, and recurrence due to their self-renewal potential and multidirectional differentiation ability,3–5 and act as the “seed cells” that continue to produce new cells in tumor tissue.6 Present study indicates that the removal of CSCs is required to completely eliminate a tumor.7 Additionally, high CSC expression of ATP-binding cassette, subfamily G member 2 (ABCG2)8 and ATP-binding cassette, subfamily B, member 1 (ABCB1),9 which can efflux chemotherapy drugs, are associated with chemoresistance, while activation of the Notch pathway10 and Wnt/beta-catenin pathway11 are linked to radioresistance in breast cancer. Characterization of the biological mechanisms leading to the development and self-renewal of BCSCs may have great clinical significance and reveal novel therapeutic targets for breast cancer.
The 68 kDa Src-associated protein in mitosis (Sam68) is a GTPase-activating protein (GAP)-related protein involved in intracellular signal transduction, cell proliferation, apoptosis, and other malignant processes.12,13 The protein expression level and posttranslational regulation of Sam68 may affect tumor progression.12 A recent study in spermatocytes found that alternative splicing of Sam68 affected its interaction with RNA polymerase II 215 kDa subunit (RNAPII) and played major role in spermatogenesis and male fertility.14 Another study showed that Sam68 influences the self-renewal of BCSCs by affecting the oncogene serine/arginine-rich splicing factor 1 (ASF).15 In vitro experiments showed that overexpression of Sam68 in breast cancer cells promoted cell proliferation and decreased the expression of the FOXO family, which is closely related to the self-renewal of stem cells.12 These findings indicate that Sam68 plays an important role in BCSCs; however, it remains unclear whether Sam68 can regulate the self-renewal capacity of BCSCs.
Beta-catenin, a key molecule in the Wnt signal transduction pathway, plays an important role in cell adhesion as well as tumor growth, invasion, and metastasis.16 Activated beta-catenin can inhibit embryonic germ cell differentiation and malignant transformation, and the nuclear levels of beta-catenin increase during the process of tumorigenesis.17 Overexpression of beta-catenin and its downstream target genes c-myc (a proto-oncogene) and cyclin D1 is closely related to the development of breast cancer18 and thyroid cancer.19 Inhibition of beta-catenin expression can block development of the breast and pregnancy-induced mammary gland proliferation, suggesting that beta-catenin is a breast stem cell survival factor.20 Chen et al21 found that the expression of beta-catenin and self-renewal capacity of Sca1(+) mouse breast cancer cells increased after 2 Gy irradiation.
In the present study, we report that Sam68 promoted the self-renewal capacity of breast cancer cells in vitro and in vivo via a mechanism linked to activation of the beta-catenin signaling pathway. Additionally, we identified that miR-204 is frequently downregulated in human breast cancer, directly targets the 3′-untranslated region (3′-UTR) of Sam68 and modifies Sam68-induced self-renewal in breast cancer cells. In vivo xenograft formation assays supported the phenotype observed with miR-204-transfected cells and Sam68 replenished cells. Therefore, Sam68 may play a major role in the self-renewal of BCSCs and represent a novel therapeutic target for breast cancer.
METHODS
Cell Culture and Human Breast Cancer Specimens
The breast cancer cell lines, normal human breast epithelial cells (NBECs), and breast cancer specimens were established as previously described.22
Plasmids and Generation of Stably Engineered Cell Lines
The conserved miR-204 binding site in the full-length sequence of Sam68–3′-UTR is from 1047 base pairs (bp) to 1055 bp. The region of human Sam68–3′-UTR and Sam68–3′-UTR-mutant, from 998 to 1179 was cloned into the pGL3-basic luciferase reporter plasmid (Promega, Madison, WI). pMSCV/Sam68 (with 3′-UTR or without 3′-UTR) overexpressing human Sam68 was constructed as previously described. MiR-204 was cloned into the bgl II/EcoR I sites of the PMSCV plasmid. Plasmids were prepared according to a previously described protocol.23 To knockdown Sam68, our previously reported human Sam68 siRNA sequence (5′-GGACCACAAGGGAATACAATC-3′; synthesized by Invitrogen Co., Carlsbad, CA) was cloned and kept in our lab, and retroviral production and infection were performed as described previously.24
Transient Transfections
The negative control microRNA (miRNA), microRNA-204, microRNA-204 inhibitor or siRNA were transfected into cells cultured in 6-well plates using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The sense strand sequences of the siRNA designed to target human TCF4 was 5′-AAGUCCGAGAAAGGAAUCUGA-3′ and 5′-UCAGAUGUCAACUCCAAACAA-3′ for LEF1.
RNA Extraction, Reverse Transcription, and Real-Time RT-PCR
RNA extraction, reverse transcription, and real-time RT-PCR were performed according to standard methods as previously described.12 PCR primers were as follows: OCT4-forward: 5′-GGTTCTCGATACTGGTTCGC-3′; OCT4-reverse: 5′-GTGGAGGAAGCTGACAACAA-3′; SOX2-forward: 5′-GCTTAGCCTCGTCGATGAAC-3′; SOX2-reverse: 5′-AACCCCAAGATGCACAACTC-3′; cyclin D1-forward: 5′-AACTACCTGGACCGCTTCCT-3′; cyclin D1-reverse: 5′-CCACTTGAGCTTGTTCACCA-3′; TCF4-forward: 5′-GGGAAATTTTTTGCGACTGTACAC-3′; TCF4-reverse: 5′-AGGCACTCAGCCACACATTG-3′; CD44-forward: 5′-ACCCCATCCCAGACGAAGACAGTC-3′; CD44-reverse: 5′-GGGATGAAGGTCCTGCTTTCCTTCG-3′; Nanog-forward: 5′-ATGGAGGAGGGAAGAGGAGA-3′; Nanog-reverse: 5′-GATTTGTGGGCCTGAAGAAA-3′; C-MYC-forward: 5′-TTCGGGTAGTGGAAAACCAG-3′; C-MYC-reverse: 5′-CAGCAGCTCGAATTTCTTCC-3′; GAPDH-forward: 5′-GACTCATGACCACAGTCCATGC-3′; GAPDH-reverse: 5′-AGAGGCAGGGATGATGTTCTG-3′.
Western Blotting Analysis
Western blotting analysis was performed according to standard methods as previously described.12 The following primary antibodies were used: anti-Sam68 (sc-333, dilution, 1:500; Santa Cruz Biotechnology, Delaware Ave Santa Cruz, CA), anti-beta-catenin, anticyclin D1, anti-p21cip1, anti-p27KIP1, anti-p-Rb, antitotal-Rb, anti-p-AKT, antitotal-AKT, anti-c-Myc (1:1000, Millipore, Billerica, MA), anti-P-84, anti-LEF-1 (1:500, Abcam, Cambridge, MA), anti-α-tubulin, anti-TCF-4, anti-LEF1, and anti-GAPDH (1:1000, Sigma, Saint Louis, MO). Nuclear extracts were prepared using the Nuclear Extraction Kit (Active Motif), according to the manufacturer's instructions.
Mammosphere Culture
One thousand cells were seeded in suspension in serum-free DMEM-F12 as described by Song et al.25 Cultures were fed once every 3 days. On day 20, the length and width measurements of the mammospheres were obtained using Zeiss Axiovision software (Carl Zeiss Co. Ltd, Jena, Germany).
Hoechst 33342 Staining and Flow Cytometry
To identify and isolate side population (SP) cells, the cells were dissociated and resuspended at 1 × 106 cells/ml in DMEM supplemented with 5% fetal bovine serum, preincubated at 37°C for 30 minutes without or with 100 mM verapamil (Sigma). If verapamil was used in the subsequent steps, it was included at 50 mM. Cells were labeled with 2.5 mg/ml Hoechst 33342 (Sigma) in staining media at 37°C for 90 minutes, incubated on ice for 10 minutes, washed twice with ice-cold phosphate-buffered saline (PBS), and then analyzed (20,000 cells per experiment with three replication) on a FACStar plus (BDIS) cell sorter equipped with dual Coherent I-90 lasers.
Immunofluorescent Assay
Immunofluorescent (IF) staining was performed on cells cultured on cover slips using an antihuman beta-catenin monoclonal antibody (Santa Cruz Biotechnology; 1:200 dilution). DAPI was used to stain the nuclei. Images were acquired using a laser scanning microscope (Axioskop 2 Plus, Carl Zeiss Co. Ltd).
Luciferase Reporter Assay
Cells were seeded in triplicate in 24-well plates and allowed to settle for 24 hours, then transfected with the indicated plasmids plus 1 ng of the pRL-TK Renilla plasmid using Lipofectamine 2000 Reagent (Life Technologies), and assayed using the Dual-Luciferase Reporter Assay (Promega, Madison, WI) according to the manufacturer's instructions 48 hours later.
Xenograft Tumor Model
NOD/SCID mice (4–5 weeks of age, 18–20 g) were purchased from the Center of Experimental Animal of Guangzhou University of Chinese Medicine. The Institutional Animal Care and Use Committee of Sun Yat-sen University approved all experimental procedures. The mice were randomly divided into groups (10 mice/group). Each mouse was inoculated in situ with SKBR-3-vector (or SKBR3-NC or SKBR-3-miR-204 + Sam68-vector) cells in the left breast and SKBR-3-Sam68 (or SKBR-3-miR-204 or SKBR-3-miR-204 + Sam68-ORF or SKBR-3-miR-204 + Sam68–3′-UTR) cells in the right mammary pad; the different groups of mice were inoculated with different number of cells: 1 × 104 cells/mouse, 5 × 103 cells/mouse, 1 × 103 cells/mouse, 1 × 102 cells/mouse, and 10 cells/mouse. On day 50, the animals were euthanized and the tumors were excised and weighed.
RESULTS
Endogenous Sam68 Expression Positively Correlates With the Self-Renewal Potential of Breast Cancer Cell Lines
The proportion of SP cells was significantly higher in the breast cancer cell lines than the normal breast cell lines NBEC1 and NBEC2 (Fig. 1A). Western blotting was used to assess the total level of Sam68 expressed in each cell line (Fig. 1B). As shown in Figure 1C, a positive correlation was observed between the expression of Sam68 and the proportion of SP cells (r = .634, P = 0.028).
FIGURE 1.
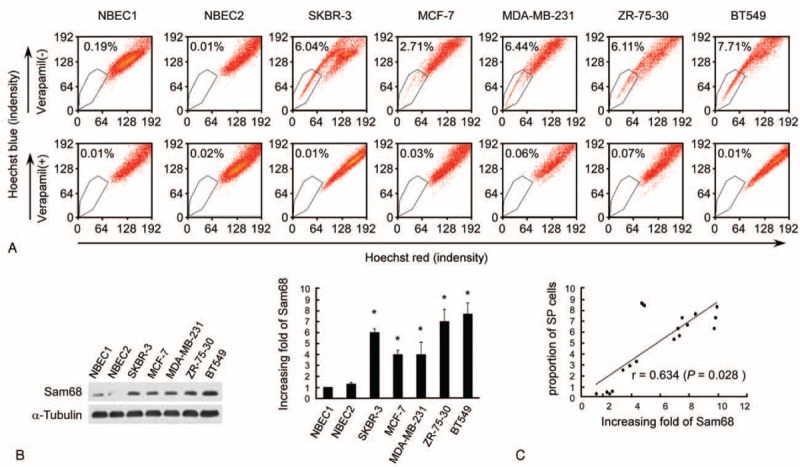
Enhanced expression of Sam68 positively correlates with the stem cell phenotype in breast cancer cell lines. (A) Assessment of the proportion of side population (SP) cells in cancer and noncancer breast cell lines using Hoechst 33342 staining and flow cytometry. Verapamil-treated cells were used as negative controls (lower). Each cell line was assessed in triplicate and the average value is shown in the upper left corner of each plot. (B) Western blot of Sam68 protein expression in breast cell lines (left); α-tubulin was used as a loading control. The fold changes in Sam68 expression relative to the expression of NBEC1were analyzed by densitometry; each cell line was assessed in triplicate and the mean ± SD values are shown (right). ∗P < 0.05 vs. NBEC1, name tested. (C) Correlation between the protein level of Sam68 protein and the proportion of SP cells in breast cancer cells by Student t test. Each cell line had three dots as each experiment was repeated 3 times.
Sam68 Promotes the Self-Renewal Potential of Breast Cancer Cell Lines
To further evaluate the function of Sam68 in vitro, SKBR-3 and MCF-7 breast cancer cells stably overexpressing Sam68 or in which Sam68 was knocked down were established (Fig. 2A). As shown in Figure 2B, overexpression of Sam68 increased, whereas knockdown of Sam68 reduced, the proportion of SP cells. Additionally, Sam68-overexpressing cells formed a higher number of more bulky mammospheres, while Sam68-knockdown cells formed a lower number of smaller mammospheres (Fig. 2C).
FIGURE 2.
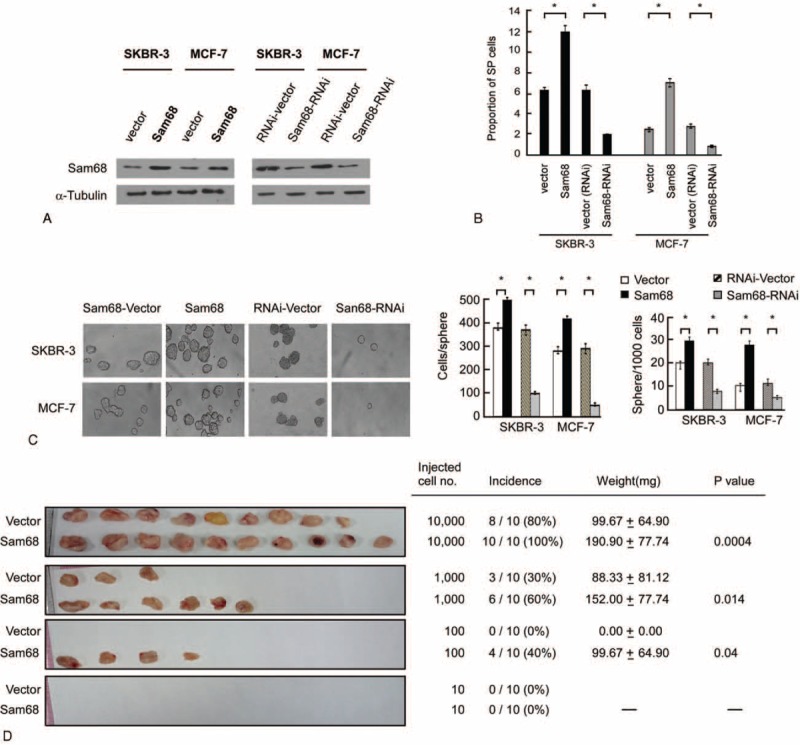
Sam68 is essential for the self-renewal phenotype and proliferation of breast cancer cells. (A) Western blot of Sam68 protein expression in SKBR-3-Sam68-vector, SKBR-3-Sam68, SKBR-3-RNAi-vector, SKBR-3-Sam68-RNAi, MCF-7-Sam68-vector, MCF-7-Sam68, MCF-7-Sam68-RNAi-vector, and MCF-7-Sam68-RNAi cells; α-tubulin was used as a loading control. (B) Assessment of the proportion of side population (SP) cells using Hoechst 33342 staining and flow cytometry. Each cell line was assessed in triplicate and the mean ± SD values are shown. ∗P < 0.05 versus vector or RNAi vector, name tested. (C) Mammosphere formation assay for the indicated cells. The mean number ± SD of cells per sphere and number of spheres formed per 100 cells in 3 independent experiments are shown. (D) Xenograft tumor model in NOD/SCID mice. Each mouse was inoculated with SKBR-3-vector cells in the left breast and SKBR-3-Sam68 cells in the right mammary pad; each group (n = 10 each) was inoculated with different number of cells. P < 0.05 compared to vector or RNAi-vector, name tested.
Furthermore, when subcutaneously implanted into NOD/SCID mice, Sam68-overexpressing SKBR-3 cells formed significantly higher numbers of larger tumors compared to vector-transfected SKBR-3 cells (Fig. 2D); as few as 100 Sam68-overexpressing cells could form tumors in NOD/SCID mice. Taken together, these assays demonstrated that ectopic overexpression of Sam68 enriched the self-renewal potential of breast cancer cells.
Sam68 Activates the Wnt/Beta-Catenin Signaling Pathway in Breast Cancer Cell Lines
Real-time RT-PCR demonstrated that overexpression of Sam68 increased, whereas knockdown of Sam68 reduced, the expression of c-MYC, cyclin D1, MMP-7, TCF4, CD44, Nanog, OCT4, and SOX2 in SKBR-3 and MCF-7 breast cancer cells (Fig. 3A). Subcellular fractionation followed by Western blotting and IF staining assays demonstrated that overexpression of Sam68 resulted in substantial nuclear accumulation of beta-catenin in SKBR-3 and MCF-7 breast cancer cells, while knockdown of Sam68 reduced nuclear translocation of beta-catenin. These results suggested that Sam68 might contribute to activation of the Wnt/beta-catenin signaling pathway (Fig. 3B and C). In agreement with this observation, overexpression of Sam68 markedly increased the transactivation activity of beta-catenin in SKBR-3 and MCF-7 cells, as indicated by beta-catenin reporter gene assays based on the TCF and LEF genes (Fig. 3D). Conversely, knockdown of Sam68 reduced TCF/LEF transcription activity in SKBR-3 and MCF-7 cells (Fig. 3D). These data suggested that Sam68 enhances the nuclear translocation of beta-catenin and consequently promotes transcription of TCF/LEF.
FIGURE 3.
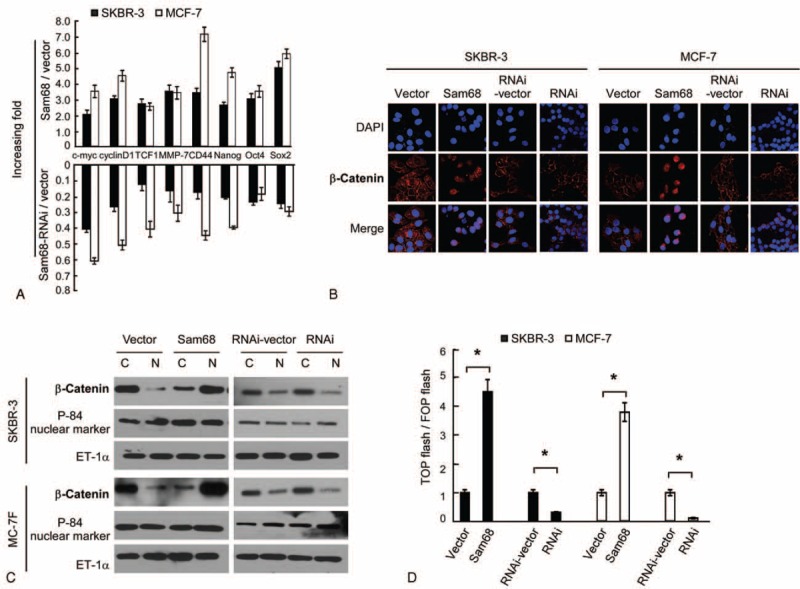
Sam68 activates beta-catenin in breast cancer cells. (A) Real-time PCR quantification of c-MYC, cyclin D1, TCF-1, MMP-7, CD44, Nanog, OCT4, and SOX2 mRNA expression in SKBR-3 and MCF-7 cells. Gene expression levels are normalized to GAPDH and expressed relative to vector-control cells. (B) Subcellular localization of beta-catenin was assessed by immunofluorescent staining in the indicated cells. (C) Western blotting of beta-catenin protein expression in the nuclear (N) and cytoplasmic (C) fraction of SKBR-3-Sam68-vector, SKBR-3-Sam68, SKBR-3-RNAi-vector, SKBR-3-Sam68-RNAi, MCF-7-Sam68-vector, MCF-7-Sam68, MCF-7-Sam68-RNAi-vector, and MCF-7-Sam68-RNAi cells. P-84 was used as a nuclear marker and ET-1a as a loading control. (D) The indicated cells were transfected with TOP flash or FOP flash and Renilla pRL-TK, and subjected to dual luciferase assays 48 hours after transfection. Firefly luciferase reporter gene activity was normalized to Renilla luciferase activity.
The Wnt/Beta-Catenin Pathway Is a Functional Mediator of Sam68-Induced Self-Renewal in Breast Cancer Cell lines
To further validate the role of beta-catenin in the Sam68-induced self-renewal potential of breast cancer cells, we blocked the Wnt/beta-catenin pathway by knocking down TCF4 or LEF1 in Sam68-overexpressing SKBR-3 and MCF-7 cells. Inhibition of beta-catenin signaling not only reduced the protein expression (Fig. 4A) and transcriptional activity of TCF/LEF (Fig. 4B), but also abrogated the self-renewal ability induced by overexpression of Sam68 (Fig. 4C). On the other hand, activation of beta-catenin signaling by ectopically overexpressing TCF4 or LEF1 in Sam68-knockdown cells mimicked the effects of overexpressing Sam68, as it increased the proportion of SP cells, mammosphere formation in suspension culture and transcriptional activity of TCF/LEF (Fig. 4B–D). Taken together, these results indicate that Wnt/beta-catenin signaling is a functional mediator of Sam68-induced self-renewal in breast cancer cells.
FIGURE 4.
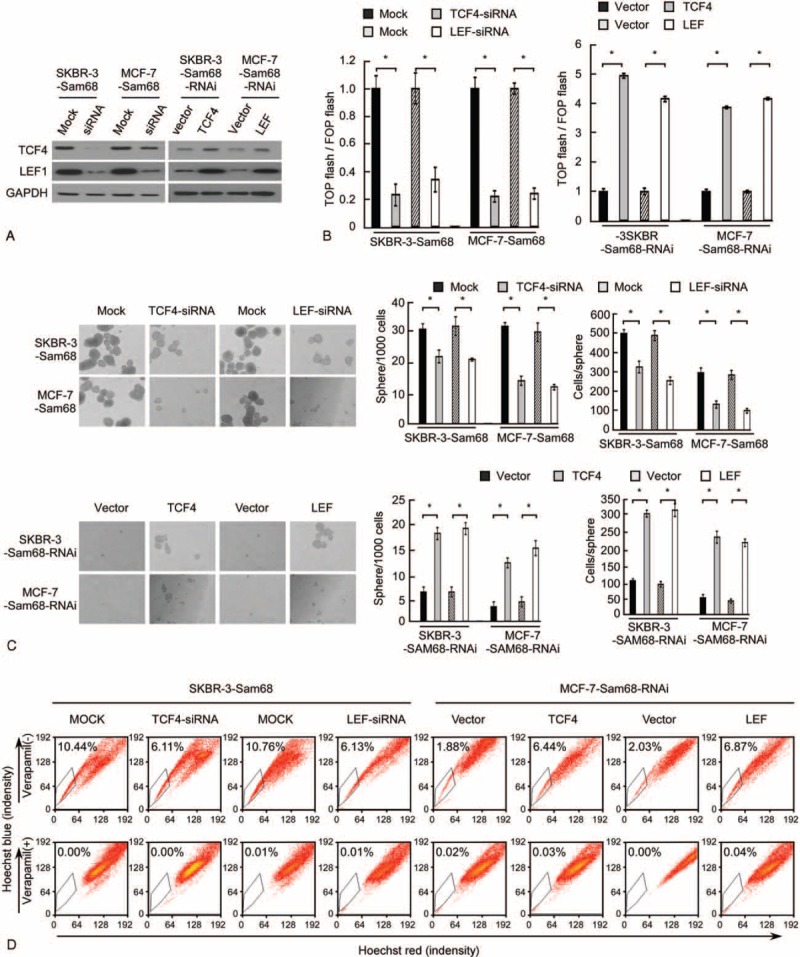
Inhibition of beta-catenin activity attenuates The Sam68-induced breast cancer stem cell renewal phenotype. (A) Western blotting analysis to confirm the knockdown or overexpression of TCF4 or LEF1 in the indicated cells. (B) Luciferase-reporter assay of TCF/LEF transcriptional activity in the indicated cells. Experiments in A to I were repeated at least 3 times with similar results; error bars in A, B, C, and D are mean ± SD. ∗P < 0.05. (C) Representative micrographs (left) and quantification (right) of mammosphere formation. (D) Detection of the proportion of SP cells by flow cytometry.
Overexpression of Sam68 Is Associated With Activation of the Beta-Catenin Pathway in Human Breast Cancer
Immunohistochemical staining was performed to further explore the clinical relevance of these findings in human breast cancer. An association was observed between the Sam68 expression level and beta-catenin nuclear expression in the 158 clinical specimens tested (Fig. 5A). As shown in Figure 5B, breast cancer samples with high Sam68 expression showed higher levels of beta-catenin activation (37/44 samples; 84.1%) than those with low Sam68 expression (40/114 samples; 35.1%; Figure 5B; P < 0.05). These data indicate that overexpression of Sam68 is associated with activation of beta-catenin in human breast cancer.
FIGURE 5.
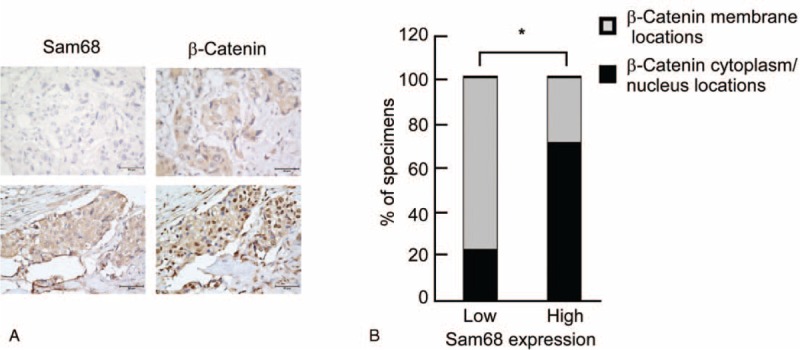
Analysis of Sam68, proteins in the Wnt/beta-catenin pathway and stem cell markers in human breast cancer samples. (A) IHC staining of breast cancer specimens. IHC analyses were performed independently twice on each sample. Two representative cases are shown. Original magnification, ×400. (B) Percentage of specimens showing Sam68 expression based on the cytoplasmic/nuclear or membrane localization of beta-catenin. ∗P < 0.05, name tested.
MicroRNA-204 Is Frequently Downregulated in Human Breast Cancer and Directly Targets the 3′-UTR of Sam68
To investigate the factors that may lead to upregulation of in breast cancer, we investigated whether miRNAs target this gene using TargetScan. Based on analysis of seed region of that are conserved between mammals and vertebrates, we identified several miRNAs that may target Sam68. Using the ArrayExpress dataset GSE45666, we observed that miR-204 was the only miRNA that was significantly downregulated in 101 breast cancer tissues compared to 15 tumor-adjacent normal breast tissues (Fig. 6A and B). The expression of miR-204 was significantly lower in the 15 breast tumor tissues for which the paired adjacent normal tissues were included in the GSE45666 dataset (P < 0.001; Fig. 6C). Additionally, real-time PCR revealed that miR-204 was significantly downregulated in all nine breast cancer cell lines tested, including SKBR-3, MCF-7, MDA-MB-231, ZR-75-30, and BT549 cells, compared with normal breast epithelial cells (NBECs; Fig. 6D). In confirmation of these in vitro analyses, real-time PCR revealed that miR-204 was significantly downregulated in 7 freshly isolated human breast cancer samples compared to the paired adjacent noncancerous tissues from the same patients (Fig. 6E). Taken together, these data indicated that miR-204 is frequently downregulated in human breast cancer.
FIGURE 6.

MicroRNA-204 is downregulated in human breast cancer and reduces the stem cell phenotype in breast cancer via directly targeting the Sam68 3′-UTR. Predicted miR-204 target sequence and mutated miR-204 sequence in the 3′-UTR of Sam68. (B) Expression of miR-204 in 101 breast tumor tissues (tumor) compared with 15 adjacent breast normal tissue samples (adjacent) in the GSE45666 dataset. (C) Expression of miR-204 in 15 breast tumor tissues (tumor) and the paired adjacent normal tissues (adjacent) in the GSE45666 database. (D) Real-time PCR analysis of miR-204 expression in normal breast epithelial cells (NBEC1) and breast cancer cell lines. (E) Expression of miR-204 in 7 freshly isolated breast tumor tissues (tumor) and the paired adjacent normal tissues (adjacent). Average miR-204 expression was normalized to U6. Each column represents the mean of 3 independent experiments. (F) Expression of Sam68 mRNA in the indicated cells. (G) Western blotting analysis the expression of Sam68 and its related proteins in the indicated cells. (H) Subcellular localization of beta-catenin was assessed by immunofluorescent staining in the indicated cells. (I) Luciferase assays on the indicated cells were transfected with the pGL3 control reporter, pGL3-Sam68–3′-UTR reporter, or pGL3-Sam68–3′-UTR-mut reporter and increasing amounts of miR-204 plasmid (20 and 50 nM), as indicated. Error bars represent mean ± SD from three independent experiments. ∗P < 0.05.
The ability of miR-204 to regulate the expression of Sam68 was confirmed using real-time PCR and Western blotting. As shown in Figure 6F and G, both the mRNA and protein expression of Sam68 were significantly reduced in miR-204-transfected cells and significantly increased in miR-204-inhibitor-transfected cells. The critical proteins of the Wnt/beta-catenin pathway and AKT pathway, which were verified to be regulated by Sam68 protein, were also modified by the expression of miR-204. These findings suggest a conclusive effect of miR-204 on the expression of Sam68 (Fig. 6G). IF staining assays demonstrated that the overexpression of miR-204 resulted in the substantial nuclear accumulation of beta-catenin in SKBR3 and MCF7 breast cancer cells, while the inhibition of miR-204 reduced the nuclear translocation of beta-catenin (Fig. 6H). A dose-dependent reduction of luciferase activity of the Sam68–3′-UTR was observed after treatment with miR-204 (Fig. 6I). Taken together, these experiments indicated that miR-204 directly targets the Sam68 3′-UTR (Fig. 6H).
MiR-204-Reduced Self-Renewal Can Be Attenuated by Supplementation of Sam68
To understand the role of miR-204 in self-renewal, the proportion of SP cell was examined in breast cancer cells. As shown in the Figure 7A, transfection of the miR-204 decreased the proportion of SP cells, whereas transfection of the miR-204 inhibitor increased the proportion of SP cells. When miR-204-overexpressing SKBR-3 cells were subcutaneously implanted into NOD/SCID mice, they formed significantly higher numbers of larger tumors compared to NC-transfected SKBR-3 cells (Fig. 7D and E), which supported the inhibitory effect of miR-204 in the self-renewal of breast cancer cells.
FIGURE 7.
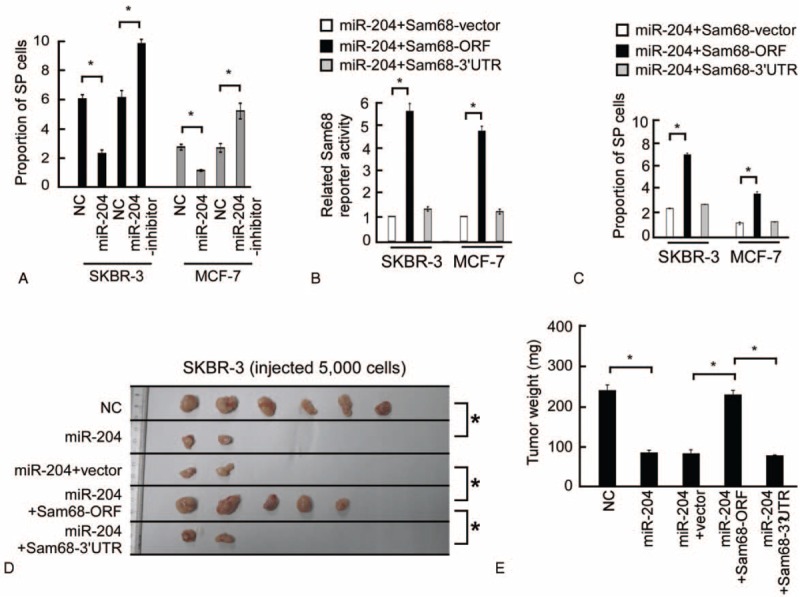
In vivo assays indicated the inhibitory effect of miR-204 and the neutralizing effect of Sam68 on tumorigenesis. (A) The proportion of SP cells decreased in cells transfected with the miR-204 and increased in cells transfected with the miR-204 inhibitor. (B) Related Sam68 reporter activity in the indicated cells. (C) Images of excised tumors 50 days after injection. (D) The weight of excised tumors measured on day 50 by electronic weighing. ∗P < 0.05.
The effect of Sam68 open reading frame (ORF, without the 3′-UTR) and Sam68–3′-UTR (ORF with the 3′-UTR) were examined in the miR-204-overexpressing cells to investigate the effect of Sam68 in miR-204-reduced self-renewal. The luciferase activity of the Sam68 reporter in miR-204-transfected cells could be rescued by overexpressing Sam68-ORF but not by overexpressing the Sam68–3′-UTR (Fig. 7B). Furthermore, the effect of miR-204 to reduce the proportion of SP cells was only abrogated by the supplementation of Sam68-ORF. As shown in Figure 7D and E, a xenograft tumor assay showed that the combination of Sam68 and miR-204 significantly increased the rate and the weight of tumor formation, while the combination of Sam68–3′-UTR and miR-204 had no effect on tumor formation compared with miR-204 alone. Taken together, our results suggest that the suppression of Sam68 plays an important role in miR-204-induced self-renewal.
DISCUSSION
Self-renewal is a crucial stem cell function, as stem cells need to persist for the entire lifetime of the organism.26 Sphere-forming assays under nonadherent cell culture conditions and detection of side-population (SP) cells by flow cytometry are used to evaluate the quantity and activity of CSCs in tumor tissues in vitro.27,28 Researches on the molecular mechanisms of the regulation of BCSCs may lead to the identification of novel molecular targets that could potentially improve breast cancer treatment. A number of BCSC markers have been identified. For example, overexpression of the oncogene erb-b2 receptor tyrosine kinase 2 (ERBB2) at the same time as reducing the expression of the tumor suppressor gene PTEN can enhance expansion of the stem cell population in breast cancer cell lines.29 CD44 was considered to be a acknowledged stem cell marker in many different organs and pathologies,30,31 and may also be a promising therapeutic target for breast cancer.32 In situ hybridization for aldehyde dehydrogenase, cytosolic 1-like (ALDH1) in formalin-fixed, paraffin-embedded specimens identified this stem/progenitor cell marker as a useful prognostic factor for poor clinical outcome in 577 patients with breast carcinoma from 2 different patient populations.29 However, no stem cell marker has yet been identified as a potential target for the elimination of BCSCs. Our previous study found that siRNA-mediated knockdown of endogenous Sam68 expression inhibited the proliferation and tumorigenicity of breast cancer cells in vitro.12 This study indicates that Sam68 plays a significant role in the self-renewal of stem cells in breast cancer, as overexpression of Sam68 increased mammosphere formation and the proportion of SP cells in SKBR-3 and MCF-7 cells; therefore, Sam68 may represent a potential target for elimination of BCSCs during breast cancer therapy.
The molecular mechanisms underlying the self-renewal of BCSCs are not yet fully understood. The WNT pathway, Notch signaling and Hedgehog signaling pathway are known to regulate the self-renewal of normal stem cells.33,34 Sam68 is an RNA binding protein with Src homology (SH) 2 and 3 domain binding sites, and can be coprecipitated with p85 PI3K.35 AKT can activate beta-catenin by inhibiting GSK3beta.36 We previously demonstrated that silencing Sam68 in breast cancer cells was associated with an attenuation of Akt/GSK-3beta signaling.12 The cytoplasmic localization of Sam68 has been observed previously in renal cancer and breast cancer, which was significantly correlated with poor prognosis.12,37 Therefore, Sam68 is involved in multiple biological mechanisms not only in the nuclei, but also in cytoplasm. In this study, we reported that Sam68 promotes the nuclear accumulation of beta-catenin, facilities Wnt/beta-catenin signaling activation and upregulates TCF/LEF transcription activity in breast cancer cells; however, the mechanism that underlies these findings is not clear. Multifunctional kinase GSK-3β is required for the regulatory APC/Axin/GSK-3β complex, which phosphorylates beta-catenin and leads to beta-catenin ubiquitination and proteasomal degradation through the beta-TrCP/Skp pathway.38 In previous studies, Sam68 promotes breast cancer cell proliferation and tumorigenicity through the activation of Akt/FOXO signaling; meanwhile, the phosphorylation level of GSK-3β is enhanced.12,37 Accordingly, phosphorylated inactivated GSK-3β is recruited to the cytomembrane by Wnt receptors with the scaffold proteins APC and Axin. Sequentially stabilized beta-catenin is translocated to the nucleus, where it binds to LEF/TCF transcription factors, displacing corepressors and recruiting additional coactivators to Wnt target genes. Furthermore, the crosstalk between PI3K/Akt and Wnt/beta-catenin signaling pathway through GSK-3β has been reported in colon cancers,39 bladder cancers,40 and gastric cancers.41 Therefore, we speculated that Sam68 could affect the Wnt/beta-catenin signaling pathway. In this study, we investigated how Sam68 regulated the self-renewal potential of breast cancer cells, and observed that overexpression of Sam68 enhanced the nuclear translocation of beta-catenin in both breast cancer cell lines and clinical specimens. Our observations may have potential implications for the critical role of beta-catenin in cell progressions.
Given the potentially important role of Sam68 in BCSCs, we investigated why Sam68 is expressed at low levels in NBECs and frequently overexpressed in breast cancer cells. miRNAs, a class of small noncoding RNAs, inhibit gene translation or facilitate mRNA degradation to repress expression of their target genes.42,43 Using TargetScan, we identified several miRNAs with conserved binding sites in the 3′-UTR Sam68 which could potentially regulate Sam68. Analysis of the ArrayExpress dataset GSE45666 revealed that only miR-204 was significantly downregulated in breast cancer, and we confirmed that the expression of miR-204 was significantly reduced in breast cancer cell lines and freshly isolated human clinical samples. Furthermore, both the mRNA and protein expression and activity of a Sam68 3′-UTR luciferase reporter gene were downregulated by transfection of the miR-204 and upregulated by transfection of the miR-204 inhibitor, demonstrating that miR-204 directly targets the 3′-UTR and regulates the expression of Sam68. Additionally, the miR-204 inhibitor, but not the mutant miR-204 sequence, reduced the proportion of SP cells, indicating that miR-204 may functionally regulate BCSCs via Sam68. Therefore, the present study indicates Sam68 may represent a potential therapeutic target for elimination of BCSCs.
In conclusion, this study demonstrates that Sam68 regulates the enrichment of self-renewing BCSCs via activation of the Wnt/beta-catenin signaling pathway, and that miR-204 directly regulates Sam68. Sam68 and miR-204 may represent candidate therapeutic targets to enable the elimination of BCSCs, which may allow the development of innovative therapies for breast cancer.
Supplementary Material
Footnotes
Abbreviations: 3′-UTR = 3′-untranslated region, BCSC = breast cancer stem cell, CSC = cancer stem cell, IF = immunofluorescent, miRNA = microRNA, mut = mutant, NBEC = normal breast epithelial cell, PBS = phosphate-buffered saline, Sam68 = The 68 kDa Src-associated protein in mitosis, SP = side population, wt = wild-type.
Lan Wang and Han Tian contributed equally to this work.
This work was supported by Guangdong Natural Science Funds for Distinguished Young Scholar (2014A030306023), the Natural Science Foundation of China (no. 81201677, 81272417), National Science and Technique Major Project (201305017).
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Glass AG, Lacey JV, Jr, Carreon JD, et al. Breast cancer incidence, 1980–2006: combined roles of menopausal hormone therapy, screening mammography, and estrogen receptor status. J Natl Cancer Inst 2007; 99:1152–1161. [DOI] [PubMed] [Google Scholar]
- 2.Nishimae K, Tsunoda N, Yokoyama Y, et al. The impact of Girdin expression on recurrence-free survival in patients with luminal-type breast cancer. Breast Cancer 2013; 22:445–451. [DOI] [PubMed] [Google Scholar]
- 3.Clare SE. Breast cancer surgery and angiogenesis: stem cell cycle may explain heterogeneity of recurrence. Int J Surg 2005; 3:286–287. [DOI] [PubMed] [Google Scholar]
- 4.Papadimitriou CA, Topp MS, Serve H, et al. Recombinant human stem cell factor does exert minor stimulation of growth in small cell lung cancer and melanoma cell lines. Eur J Cancer 1995; 31A:2371–2378. [DOI] [PubMed] [Google Scholar]
- 5.Gu Z, Thomas G, Yamashiro J, et al. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene 2000; 19:1288–1296. [DOI] [PubMed] [Google Scholar]
- 6.Di J, Duiveman-de Boer T, Zusterzeel PL, et al. The stem cell markers Oct4A, Nanog and c-Myc are expressed in ascites cells and tumor tissue of ovarian cancer patients. Cell Oncol (Dordr) 2013; 36:363–374. [DOI] [PubMed] [Google Scholar]
- 7.Jasmin C. Tumor stem cells and the curability of early human breast cancer. Recent Results Cancer Res 1993; 127:7–12. [DOI] [PubMed] [Google Scholar]
- 8.Ding XW, Wu JH, Jiang CP. ABCG2: a potential marker of stem cells and novel target in stem cell and cancer therapy. Life Sci 2010; 86:631–637. [DOI] [PubMed] [Google Scholar]
- 9.Bunting KD, Galipeau J, Topham D, et al. Effects of retroviral-mediated MDR1 expression on hematopoietic stem cell self-renewal and differentiation in culture. Ann N Y Acad Sci 1999; 872:125–140.discussion 121–140. [DOI] [PubMed] [Google Scholar]
- 10.Garcia Campelo MR, Alonso Curbera G, Aparicio Gallego G, et al. Stem cell and lung cancer development: blaming the Wnt, Hh and Notch signalling pathway. Clin Transl Oncol 2011; 13:77–83. [DOI] [PubMed] [Google Scholar]
- 11.Gedaly R, Galuppo R, Daily MF, et al. Targeting the Wnt/beta-catenin signaling pathway in liver cancer stem cells and hepatocellular carcinoma cell lines with FH535. PLoS ONE 2014; 9:e99272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L, Wang L, Li Y, et al. Sam68 up-regulation correlates with, and its down-regulation inhibits, proliferation and tumourigenicity of breast cancer cells. J Pathol 2010; 222:227–237. [DOI] [PubMed] [Google Scholar]
- 13.Ramakrishnan P, Baltimore D. Sam68 is required for both NF-kappaB activation and apoptosis signaling by the TNF receptor. Mol Cell 2011; 43:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paronetto MP, Messina V, Bianchi E, et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol 2009; 185:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valacca C, Bonomi S, Buratti E, et al. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol 2010; 191:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyazawa K, Iwaya K, Kuroda M, et al. Nuclear accumulation of beta-catenin in intestinal-type gastric carcinoma: correlation with early tumor invasion. Virchows Arch 2000; 437:508–513. [DOI] [PubMed] [Google Scholar]
- 17.Peng X, Liu T, Wang Y, et al. Wnt/beta-catenin signaling in embryonic stem cell converted tumor cells. J Transl Med 2012; 10:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dakeng S, Duangmano S, Jiratchariyakul W, et al. Inhibition of Wnt signaling by cucurbitacin B in breast cancer cells: reduction of Wnt-associated proteins and reduced translocation of galectin-3-mediated beta-catenin to the nucleus. J Cell Biochem 2012; 113:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishigaki K, Namba H, Nakashima M, et al. Aberrant localization of beta-catenin correlates with overexpression of its target gene in human papillary thyroid cancer. J Clin Endocrinol Metab 2002; 87:3433–3440. [DOI] [PubMed] [Google Scholar]
- 20.Rowlands TM, Pechenkina IV, Hatsell S, et al. Beta-catenin and cyclin D1: connecting development to breast cancer. Cell Cycle 2004; 3:145–148. [PubMed] [Google Scholar]
- 21.Chen MS, Woodward WA, Behbod F, et al. Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J Cell Sci 2007; 120:468–477. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Zhang N, Song LB, et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res 2008; 14:3319–3326. [DOI] [PubMed] [Google Scholar]
- 23.Corsaro A, Thellung S, Russo C, et al. Expression in E. coli and purification of recombinant fragments of wild type and mutant human prion protein. Neurochem Int 2002; 41:55–63. [DOI] [PubMed] [Google Scholar]
- 24.Guan H, Song L, Cai J, et al. Sphingosine kinase 1 regulates the Akt/FOXO3a/Bim pathway and contributes to apoptosis resistance in glioma cells. PLoS ONE 2011; 6:e19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131:1109–1123. [DOI] [PubMed] [Google Scholar]
- 26.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature 2001; 414:105–111. [DOI] [PubMed] [Google Scholar]
- 27.Wang YJ, Bailey JM, Rovira M, et al. Sphere-forming assays for assessment of benign and malignant pancreatic stem cells. Methods Mol Biol 2013; 980:281–290. [DOI] [PubMed] [Google Scholar]
- 28.Yao J, Cai HH, Wei JS, et al. Side population in the pancreatic cancer cell lines SW1990 and CFPAC-1 is enriched with cancer stem-like cells. Oncol Rep 2010; 23:1375–1382. [DOI] [PubMed] [Google Scholar]
- 29.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007; 1:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 2007; 104:973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 2007; 104:10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orian-Rousseau V. CD44, a therapeutic target for metastasising tumours. Eur J Cancer 2010; 46:1271–1277. [DOI] [PubMed] [Google Scholar]
- 33.Takashima S, Mkrtchyan M, Younossi-Hartenstein A, et al. The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature 2008; 454:651–655. [DOI] [PubMed] [Google Scholar]
- 34.Mascaro Cordeiro B, Dias Oliveira I, de Seixas Alves MT, et al. SHH, WNT, and NOTCH pathways in medulloblastoma: when cancer stem cells maintain self-renewal and differentiation properties. Childs Nerv Syst 2014; 30:1165–1172. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Margalet V, Najib S. Sam68 is a docking protein linking GAP and PI3K in insulin receptor signaling. Mol Cell Endocrinol 2001; 183:113–121. [DOI] [PubMed] [Google Scholar]
- 36.Yan D, Avtanski D, Saxena NK, et al. Leptin-induced epithelial-mesenchymal transition in breast cancer cells requires beta-catenin activation via Akt/GSK3- and MTA1/Wnt1 protein-dependent pathways. J Biol Chem 2012; 287:8598–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Yu CP, Zhong Y, et al. Sam68 expression and cytoplasmic localization is correlated with lymph node metastasis as well as prognosis in patients with early-stage cervical cancer. Ann Oncol 2012; 23:638–646. [DOI] [PubMed] [Google Scholar]
- 38.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012; 149:1192–1205. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Pandurangan AK, Lu F, et al. Chemopreventive sphingadienes downregulate Wnt signaling via a PP2A/Akt/GSK3beta pathway in colon cancer. Carcinogenesis 2012; 33:1726–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu K, Fan J, Zhang L, et al. PI3K/Akt to GSK3beta/beta-catenin signaling cascade coordinates cell colonization for bladder cancer bone metastasis through regulating ZEB1 transcription. Cell Signal 2012; 24:2273–2282. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Zhang Y, Xu R, et al. PI3K/Akt-dependent phosphorylation of GSK3beta and activation of RhoA regulate Wnt5a-induced gastric cancer cell migration. Cell Signal 2013; 25:447–456. [DOI] [PubMed] [Google Scholar]
- 42.Krichevsky AM, King KS, Donahue CP, et al. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003; 9:1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawasaki H, Taira K. Hes1 is a target of microRNA-23 during retinoic-acid-induced neuronal differentiation of NT2 cells. Nature 2003; 423:838–842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


