Supplemental Digital Content is available in the text
Abstract
Identification of cancer-associated genes by genomic profiling contributes to the elucidation of tumor development and progression. The methylthioadenosine phosphorylase (MTAP) gene, located at chromosome 9p21, plays a critical role in tumorigenicity and disease progression in a wide variety of cancers. However, the prognostic impact of MTAP in patients with nasopharyngeal carcinoma (NPC) remains obscured. Through data mining from published transcriptomic database, MTAP was first identified as a differentially downregulated gene in NPC. In this study, our aim was to evaluate the expression of MTAP in NPC and to clarify its prognostic significance.
MTAP immunohistochemistry was retrospectively performed and analyzed in biopsy specimens from 124 NPC patients who received standard treatment without distant metastasis at initial diagnosis. The immunoexpression status was correlated with the clinicopathological variables, disease-specific survival (DSS), distant metastasis-free survival (DMFS), and local recurrence-free survival (LRFS). Real-time quantitative polymerase chain reaction (PCR) was used to measure MTAP gene dosage. In some cases, we also performed methylation-specific PCR and pyrosequencing to assess the status of promoter methylation.
MTAP deficiency was significantly associated with advanced tumor stages (P = 0.023) and univariately predictive of adverse outcomes for DSS, DMFS, and LRFS. In the multivariate comparison, MTAP deficiency still remained prognostically independent to portend worse DSS (P = 0.021, hazard ratio = 1.870) and DMFS (P = 0.009, hazard ratio = 2.154), together with advanced AJCC stages III to IV. Homozygous deletion or promoter methylation of MTAP gene were identified to be significantly associated with MTAP protein deficiency (P < 0.001).
MTAP deficiency was correlated with an aggressive phenotype and independently predictive of worse DSS and DMFS, suggesting its role in disease progression and as an independent prognostic biomarker of NPC, which potentially offers new strategy of targeted treatment for patients lacking MTAP expression.
INTRODUCTION
Nasopharyngeal carcinoma (NPC), a common head and neck malignancy in southeastern Asia and Taiwan, is caused by a combination of factors, including Epstein–Barr virus (EBV) infection, environmental influence, and genetic susceptibility.1 In endemic areas, NPC is strongly associated with EBV infection.2 However, little is known about the molecular mechanism underlying NPC pathogenesis. Although advances in concurrent chemoradiotherapy have led to better locoregional control and overall survival, treatment options for advanced disease are still limited.3 Therefore, it would be of great value to work out molecular mechanisms of this cancer and to search for potential prognostic biomarkers. New biomarkers can help to stratify the risk of disease progression and to develop novel therapeutic strategies for NPC patients in a personalized manner.
Mounting evidence has suggested that DNA losses of chromosome 9 are identified in a variety of cancers, particularly 9p21 that harbors several candidate or established tumor suppressor genes, such as CDKN2A (also known as p16INK4A/p14ARF), CDKN2B (also known as p15INK4B), and MTAP (methylthioadenosine phosphorylase).4–10 The MTAP gene, which lies about 100 kb telomeric to CDKN2A, is frequently co-deleted with the CDKN2A and CDKN2B genes in many different cancers.6,7,10–12MTAP encodes a key enzyme in the catabolism of methylthioadenosine (MTA), which is generated during the biosynthesis of polyamines. MTAP is expressed abundantly in a wide range of normal cells and tissues. In normal cells, MTAP cleaves MTA into adenine and 5-methylthioribose-1-phosphate. The latter compound is further metabolized to methionine.13 Adenine and methionine are further metabolized and hence salvaged.
Lack of MTAP expression, either due to homozygous deletion or promoter methylation, is found in numerous hematologic malignancies4,12,14–19 and solid tumors, such as mesothelioma, pancreatic cancer, osteosarcoma, chondrosarcoma, soft tissue sarcoma, gliomas, gastrointestinal stromal tumor, endometrial cancer, esophageal carcinoma, chordoma, biliary tract cancer, malignant melanoma, nonsmall cell lung cancer, etc.5,7,10,20–32 Information on MTAP-deficiency in primary NPC is lacking, and its association with various clinicopathological variables and survival has never been systematically studied. In this study, we first identified deletion of MTAP gene and corresponding downregulation of mRNA in NPC through data mining from array comparative genomic hybridization (aCGH) and gene expression profiling datasets. We further analyzed MTAP gene status, its promoter methylation and immunoexpression in a well-defined cohort of NPC tissues, trying to clarify its prognostic significance and correlation to different clinicopathological parameters.
MATERIALS AND METHODS
Analysis of Public Transcriptomic Datasets of NPC
To identify clinically relevant genes that are critical in the pathogenesis of NPC, we reappraised gene expression profiling datasets for NPC versus non-neoplastic nasopharyngeal tissues from the Gene Expression Omnibus, which contains transcriptome and copy number data (GSE34573) obtained using Affymetrix Human Genome U133 Plus 2.0 and Mapping 250K Nsp SNP Arrays, respectively. The raw CEL files obtained from Affymetrix Human Genome U133 Plus 2.0 and Mapping 250K Nsp SNP Array platforms were imported into Nexus Expression 3 (BioDiscovery, EI Segundo, CA) and Nexus 6 (BioDiscovery) to analyze all probe sets without preselection or filtering, respectively. For the analysis of expression profiling, supervised comparative analysis and functional profiling were performed to identify statistically significant genes that were differentially expressed, with special attention given to the nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process (GO:0006139). Those genes with P < 0.01 and log2-transformed expression fold change >±0.1 were chosen for validation. For the purpose of exploring the copy number alteration of target genes, Mapping 250K Nsp SNP Arrays was also analyzed.
Patient Characteristics and Tumor Samples
In this study, there were 124 NPC patients who received biopsy between January 1993 and December 2002 as previous described.33–37 Patients with distant metastasis were excluded. Adequate paraffin-embedded tissue blocks were obtained from BioBank of Chi-Mei Medical Center, which were approved by the Institutional Review Board of Chi-Mei Medical Center (IRB103-02-013). The slides of tumor tissues were reviewed by 2 pathologists (H-LH and C-FL) who did not know the patients’ clinical information. The identification of histopathologic type of tumors was based on the current WHO classification. The 2 pathologists followed the principle of the 7th American Joint Committee on Cancer (AJCC) system to assess tumor staging.
Immunohistochemical Study
The paraffin-embedded blocks were sectioned at 3-mm thickness onto precoated slides, deparaffinized with xylene and then rehydrated with gradient ethanol. The slides were heated and incubated in a 10 mM citrate buffer (pH 6.0) for 7 min. After inactivating endogenous peroxidase by 3% H2O2, the slides were incubated with primary antibody targeting MTAP (MGC:31876, 1:100, Proteintech, Chicago, IL) for 1 h. Primary antibodies were detected by the use of ChemMate DAKO EnVision kit (DAKO, K5001, Glostrup, Denmark), washed with Tris buffered saline for 15 min and then incubated with a primary antiboarpinteria, CA. Two pathologists (H-LH and C-FL) evaluated the immunostainings of MTAP according to the previous published criteria.6,20,25,38 No cytoplasmic staining or faint cytoplasmic staining in <10% of tumor cells was defined as MTAP deficiency. Moderate or strong cytoplasmic staining in >10% of tumor cells was defined as intact MTAP expression.
Conditions of Real-Time Quantitative PCR Used to Measure MTAP Gene Dosage
In order to isolate specific tumor cells of interest, we performed manual macrodissection in 14 selected tumor samples that contain 90% of tumor cells. The quantity of the DNA extracted was measured by an ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA). The relative fold change of the MTAP gene copy number between normal and tumor tissues was measured by a quantitative polymerase chain reaction (PCR) assay with the use of comparative threshold cycle (Ct) method. The TaqMan assays targeting PIK3R1 (Hs06028467_cn) in 5q13.1 and the exon 8 of the MTAP gene (Hs02079487_cn) in 9p21.3 were purchased from Applied Biosystems. They were separately amplified using the ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). In each well of the PCR reaction contained a total volume of 20 μL, including 1 μL of 20× TaqMan assay probe, 20 ng of genomic DNA, and 10 μL of TaqMan Genotyping MasterMix. The PCR conditions was started at an initial denaturation step of 95°C for 10 min, followed by 40 amplification cycles at 95°C for 15 s, and then extension at 60°C for 1 min. All reactions were conducted in duplicate with a negative control run in parallel.
To measure MTAP gene dosage, we conducted an assay using TaqMan real-time quantitative PCR, with MTAP as the test sequence and PIK3R1 as the reference gene. To obtain gene dosages, the PIK3R1 gene was regarded as a control with the gene copy number of 2, and the normalized ratio of MTAP/PIK3R1 is expected to yield a value of 1 in individuals without deletion. Following our previous defined criteria for formalin-fixed samples, the MTAP/PIK3R1 gene ratio <0.2 was considered as homozygous deletion.25,39 Also, a calibration standard curve was plotted using input templates of 100, 20, 8, and 0.4 ng human genomic DNA (Clontech, Mountain View, CA, USA) to ensure the PCR efficiency and precision in the experiment
Methylation-Specific PCR to Detect MTAP Promoter Hypermethylation
It is known that promoter hypermethylation might be an alternative mechanism for MTAP gene silencing in those with intact or hemizygously deleted MTAP gene. We thus evaluated its promoter methylation for selected cases lacking MTAP immunoexpression but showing no evidence of MTAP homozygous deletion by real-time quantitative PCR. The methylation-specific PCR was performed as previously described.25 In brief, the first-run PCR was performed with primers specific for bisulfite-converted DNA (sense primer, 5′-ATTTTTGAGTTTTGGGTTAAGTTTATTT-3′; antisense primer, 5′-AAAAAACAACACTCCCTACTTAACC-3′), which did not discriminate between methylated (M) and unmethylated (U) sequences. PCR amplification was first run for 30 cycles at the annealing temperature of 59°C, in a reaction mixture of 15 μL containing 15 ng bisulfite-converted DNA, 0.1 μM of each primer, and 0.3 U HotStar Taq DNA polymerase (Qiagen, Gmb Hilden, Germany). The nested PCR for M and U promoters were both performed for 40 cycles at the annealing temperature of 61°C in a reaction mixture of 1 μL of 10-fold diluted, first PCR products, 0.1 μM of each primer, and 0.3 U HotStar Taq DNA polymerase (Qiagen) with the following primer sequences:
M, sense primer, 5′-GTTGTTTAAGAGAATTTTCGTGGTC-3′; antisense primer, 5′-ACTTAACCCAATATTAATAACGTCG-3′.
U, sense primer, 5′-AGTTGTTTAAGAGAATTTTTGTGGTT-3′; antisense primer, 5′-CCCTACTTAACCCAATATTAATAACATCA-3′.
Determination of MTAP Gene Promoter Methylation Using Pyrosequencing
Promoter methylation of MATP was quantified by pyrosequencing to crossly validate that of methylation-specific PCR. For this purpose, after bisulfate treatment and cleanup of DNA by using EpiTect 96 Bisulfite Kit (Qiagen), PCR amplification and sequencing were performed by using Hs_MTAP_01_PM PyroMark CpG assay (Qiagen, Product No. 978746) following our previous protocol.40 Finally, PyroMark Q24 Software was used to quantity cytosine methylation. Methylation was called when the average methylation percentage of all CpG islands tested was higher than 4.56%, which was the mean +3 SD of the average methylation percentage of CpG islands from 10 nontumor nasopharyngeal mucosa, as described previously.41
Treatment and Follow-Up
A complete course of radiotherapy (RT) with a total dose > or =7000 cGy was performed in all 124 patients, following a uniform protocol of RT. In addition, those who with stage II to IV received cisplatin-based chemotherapy. The 124 patients were under regular follow-up after treatment until their last visit or death. The mean duration of follow-up was 59.6 months, ranging from 4 to 117 months.
Statistical Analysis
We used the SPSS 14 software package (SPSS, Inc., Chicago, IL) to perform statistical analysis. The relationships between MTAP expression and various clinicopathological parameters or MTAP gene dosage were assessed by the use of Chi-squared test. This study contained 3 endpoints, including disease-specific survival (DSS), distal metastasis-free survival (DMFS), and local recurrence-free survival (LRFS). We performed Kaplan–Meier method and log-rank test to analyze the survival data. Multivariate analysis was performed using Cox proportional hazards model. For all analyses, the statistical difference was considered significant if the P value was <0.05 under two-sided tests.
RESULTS
Identifying MTAP as a Significantly Differentially Downregulated Gene Associated With Nucleobase, Nucleoside, Nucleotide, and Nucleic Acid Metabolic Process in NPC
From the transcriptomic dataset of 16 NPCs and 4 nontumor nasopharyngeal samples deposited in the public domain, we focused on 151 probes covering 67 genes associated with nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process. The genes with P < 0.01 and log2-transformed expression fold change >±0.1 are listed in Table 1 and Figure 1. Among these statistically significant genes, deficiency of MTAP appeared as the only candidate that can be directly linked to pharmaceutical targeting,25,39 prompting us to characterize the protein expression level in NPC. Since the MTAP gene locus at 9p21.3 has been reported to be frequently deleted in various human malignancies,5,7,10,20–32 we thus evaluated the copy number alterations of MTAP gene by analyzing GSE34573 dataset. Interestingly, MTAP deletion can be identified in 68.8% (11/16) of NPCs including 2 with homozygous and 9 with hemizygous deletion, suggesting a potential role of MTAP gene deletion in its protein deficiency in NPC (Fig. 2).
TABLE 1.
Summary of Differentially Expressed Genes Associated With Nucleobase, Nucleoside, Nucleotide, and Nucleic Acid Metabolic Process (GO:0006139) in the Transcriptome of Nasopharyngeal Carcinoma
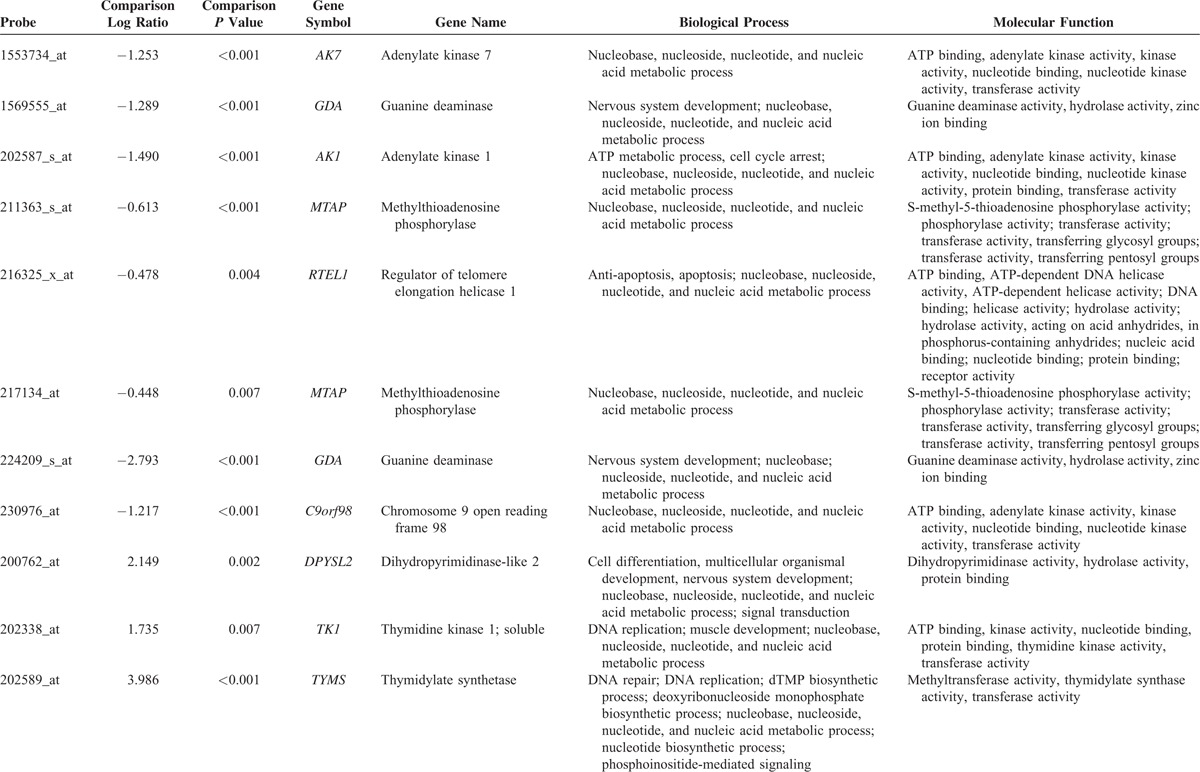
FIGURE 1.
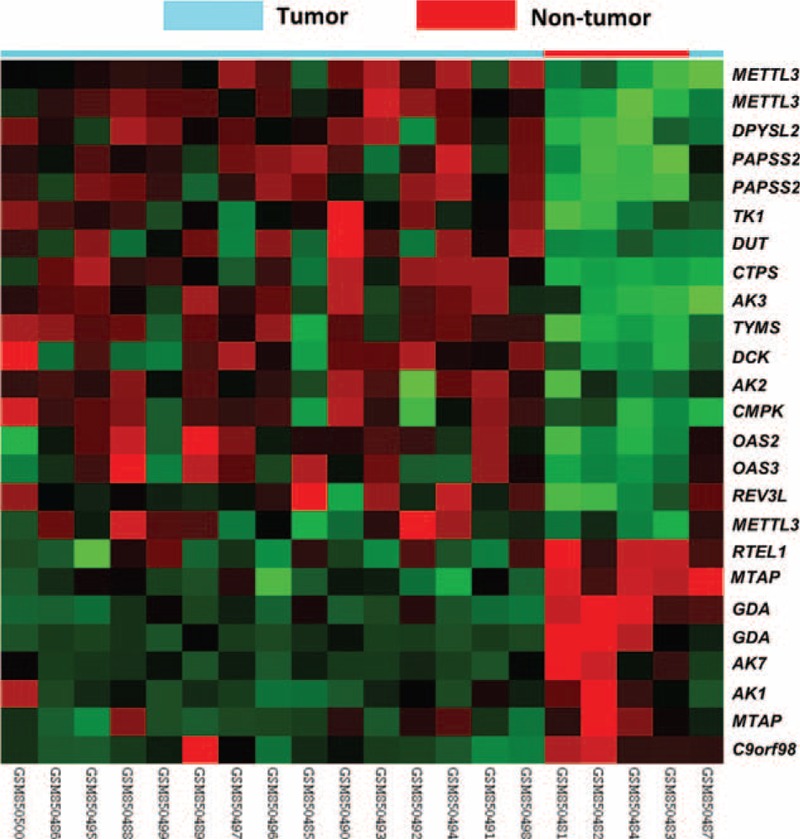
Data mining on public transcriptomic datasets of nasopharyngeal carcinoma. By reappraising the published transcriptomic datasets of nasopharyngeal carcinoma (NPC) versus non-neoplastic nasopharyngeal tissue samples (GSE34573), MTAP is one of the significantly downregulated gene associated with nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process (GO:0006139) in clustering analysis. NPC (blue lines) and non-neoplastic (red lines) tissue specimens were indicated on top of the heatmap, and expression levels of upregulated and downregulated genes were expressed as a series of brightness of red and green colors, respectively, with those unaltered in mRNA expression being coded as black.
FIGURE 2.
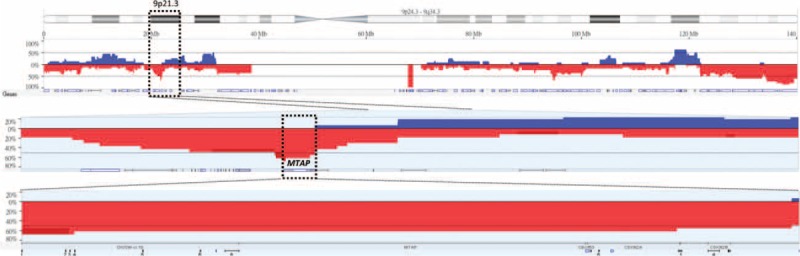
Analysis of copy number alterations of MTAP gene in public transcriptomic datasets of nasopharyngeal carcinoma. Reappraising copy number data in the same public domain dataset (GSE34573) further confirmed that chromosome region 9p21.3 is frequently (68.8%, 11/16) deleted (upper). Of this region, MTAP is located at the most frequently altered gene segment (middle), showing 68.8% (11/16) deletion and 12.5% (2/16) homozygous deletion (lower). MTAP = methylthioadenosine phosphorylase.
MTAP Deficiency and Its Association With Clinicopathological Factors in NPC
The tumor samples consisted of 5 keratinizing squamous cell carcinomas, 54 nonkeratinizing differentiated carcinomas, and 65 nonkeratinizing undifferentiated carcinomas. The study population included 29 women and 95 men. The age of patients ranged from 20 to 83 years (mean, 48.6). MTAP deficiency was identified in 37 (29.8%) cases, which was significantly associated with advanced tumor stage (AJCC stage III to IV, P = 0.023; Table 2). However, there was no association between MTAP deficiency and other clinicopathological factors. In cases with intact MTAP expression, the tumor cells retained moderate or strong cytoplasmic staining, although some nuclear staining was also seen (Fig. 3).
TABLE 1 (Continued).
Summary of Differentially Expressed Genes Associated With Nucleobase, Nucleoside, Nucleotide, and Nucleic Acid Metabolic Process (GO:0006139) in the Transcriptome of Nasopharyngeal Carcinoma
FIGURE 3.
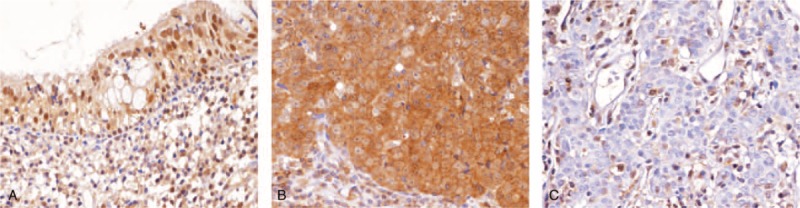
Immunohistochemical staining of MTAP in nasopharyngeal carcinoma. Non-neoplastic nasopharyngeal epithelial tissue (A) and representative low-stage NPC (B) show intact MTAP expression. While representative high-stage NPC (C) is deficient for MTAP expression. In addition, the expression of MTAP is also present either in the lymphocytes (A and C) or stromal cells (B) adjacent to the tumor or non-neoplastic mucosa. MTAP = methylthioadenosine phosphorylase, NPC = nasopharyngeal carcinoma.
Prognostic Significance of MTAP Deficiency in NPC
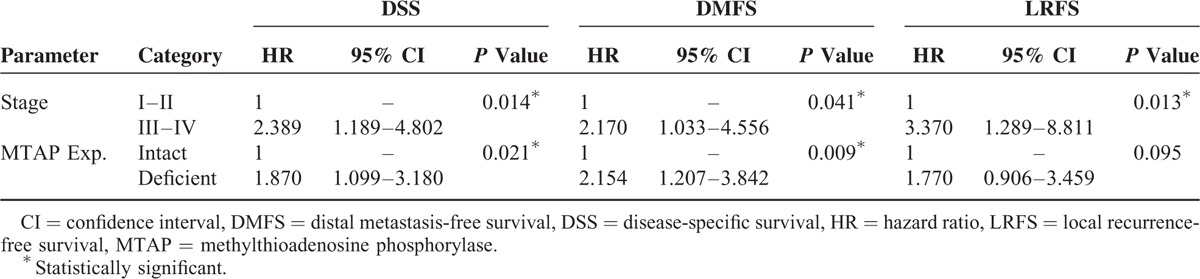
All disease-specific mortality, local recurrence, and distal metastasis were significantly associated with T3 to T4 status (P = 0.029 for DSS; P = 0.009 for DMFS; P = 0.018 for LRFS), N2 to N3 status (P < 0.001 for DSS; P = 0.013 for DMFS; P = 0.016 for LRFS), and stage III to IV (P = 0.002 for DSS; P = 0.007 for DMFS; P = 0.003 for LRFS; Table 3). MTAP deficiency correlated with a significantly shorter DSS (P = 0.002), DMFS (P < 0.001), and LRFS (P = 0.013) in patients with NPC (Fig. 4). In multivariate analysis, MTAP deficiency and stage both independently portended inferior DSS (MTAP deficiency, P = 0.021, hazard ratio [HR] = 1.870) and worse DMFS (MTAP deficiency, P = 0.009, HR = 2.154), respectively (Table 4).
TABLE 1 (Continued).
Summary of Differentially Expressed Genes Associated With Nucleobase, Nucleoside, Nucleotide, and Nucleic Acid Metabolic Process (GO:0006139) in the Transcriptome of Nasopharyngeal Carcinoma
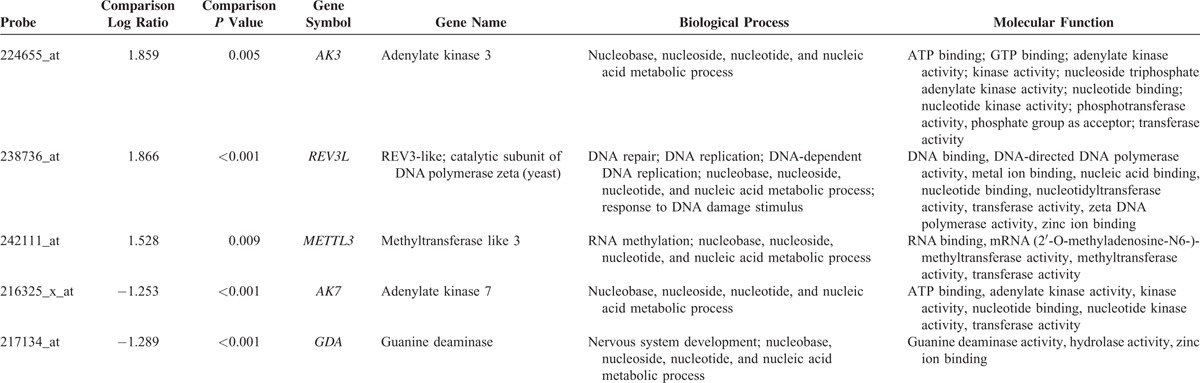
FIGURE 4.
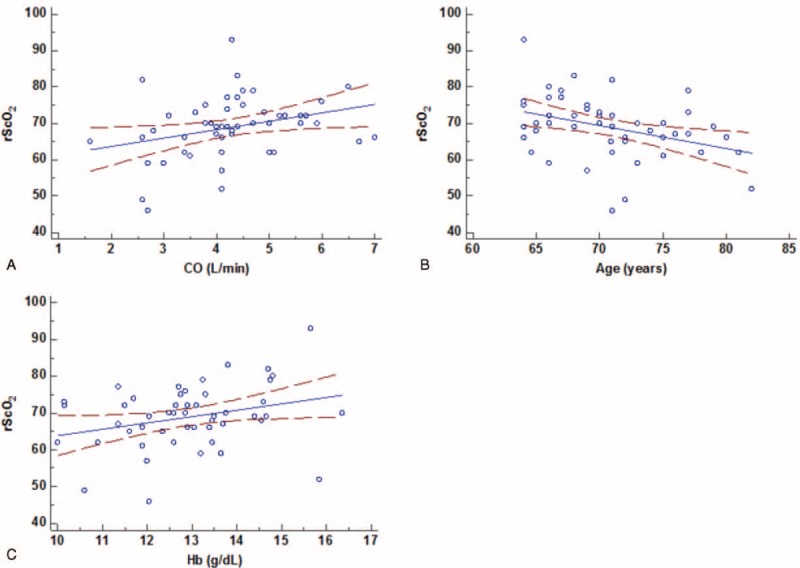
Survival analysis for assessing prognostic significance of MTAP deficiency in nasopharyngeal carcinoma. By using log-rank test, an MTAP deficiency is univariately predictive of inferior disease-specific survival (A), distant metastasis-free survival (B), and local recurrence-free survival (C), MTAP = methylthioadenosine phosphorylase.
TABLE 2.
Associations Between MTAP Expression With Other Important Clinicopathological Variables
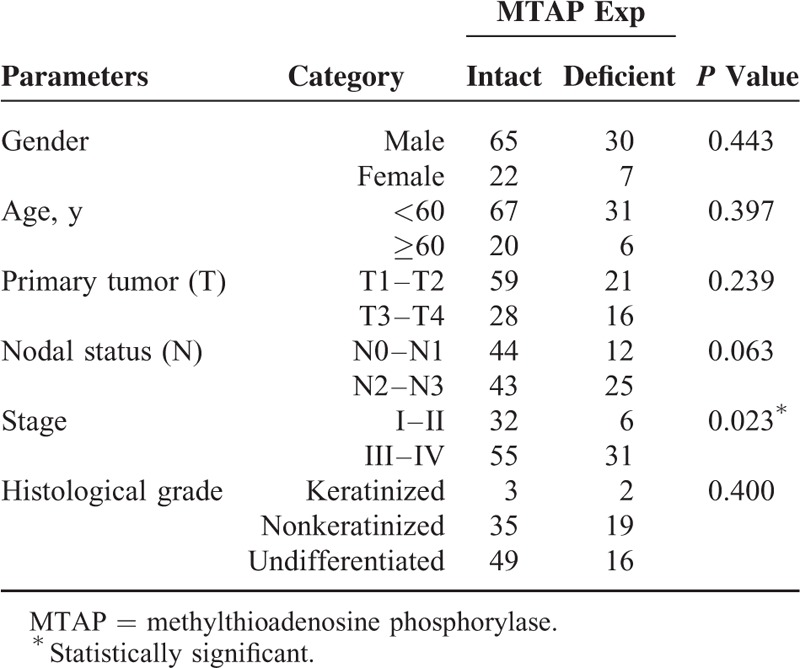
TABLE 3.
Univariate Log-Rank Analyses
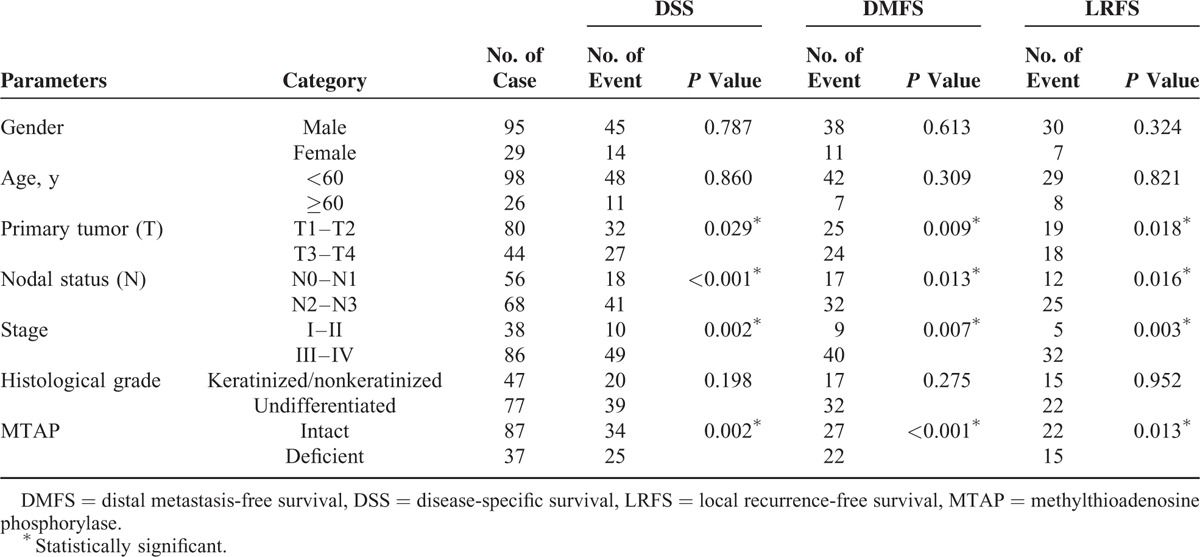
TABLE 4.
Multivariate Survival Analyses
Correlation of MTAP Homozygous Deletion and Promoter Hypermethylation With Immunoexpression
As shown in Table S1, the presence of either MTAP homozygous deletion or promoter hypermethylation had a strong correlation to the loss of protein expression (P < 0.001) in 14 selected NPC samples. In the 14 selected NPC cases, there were 8 cases showing MTAP protein deficiency, including 4 cases with MTAP gene homozygous deletion characterized by MTAP/PIK3R1 gene ratio < 0.2 and another 4 cases with promoter hypermethylation (Fig. 5).
FIGURE 5.
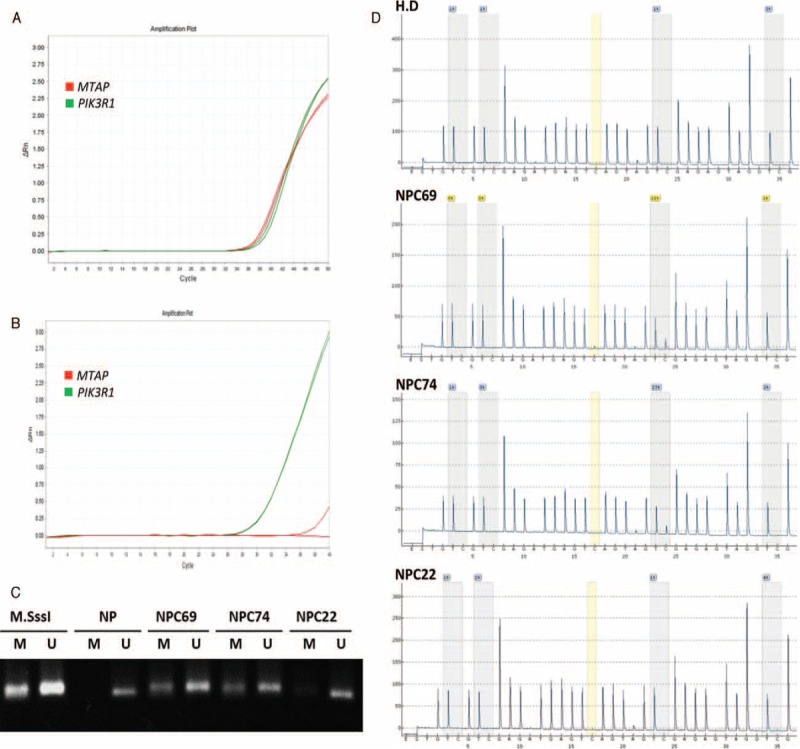
MTAP gene dosage and methylation-specific PCR for analysis of MTAP promoter methylation in nasopharyngeal carcinoma. Representative low-stage NPC with intact MTAP expression has a preserved MTAP gene, with a Ct value similar to that of PIK3R1, both <40 in a quantitative DNA-PCR (A). Representative high-stage and MTAP-deficient NPC has nondetectable MTAP gene, characterized by a Ct value no less than 40 (B). Methylation-specific PCR detects MTAP promoter methylation in MTAP protein-deficient NPCs without homozygous gene deletion (C). PCR products amplified from MTAP promoter with methylation-specific primers can be distinctly visualized in both NPC69 and NPC74, while the MTAP gene promoter of a representative MTAP-intact NPC, NPC22, is unmethylated. Pyrosequencing is performed to quantify the percentage of methylation in specific CpG sites of MTAP (D). The percentages of methylation in NPC69 and NPC74 are 23% and 26%, respectively. While in NPC22, the percentage is 4%, which is less than the cut-off value of methylation and regarded as unmethylated. M = methylation-specific primers, M.SssI = a healthy donor's blood DNA treated with M. SssI methyltransferase, MTAP = methylthioadenosine phosphorylase, NP = DNA from nontumor nasopharyngeal mucosal tissue without treatment of M.SssI methyltransferase, NPC = nasopharyngeal carcinoma, PCR = polymerase chain reaction, U = unmethylation-specific primers.
DISCUSSION
The MTAP gene is located at human chromosomal locus 9p21. Homozygous deletions are not uncommon in several kinds of cancer associated with the loss of tumor suppressor genes as CDKN2A and/or CDKN2B.6,7,10–12 Whether the loss of MTAP activity contributes to tumorigenesis remains debated owing to its frequent codeletion with adjacent tumor suppressor genes. However, mounting evidence has suggested that MTAP may play a role as an independent tumor suppressor in carcinogenesis. First, MTAP deletion may also occur without the loss of neighboring CDKN2A in gliomas and non-small cell lung cancers.10,29 Second, reintroduction of MTAP into MTAP-deleted breast cancer cells (MCF-7) results in dramatic suppression of anchorage-independent growth in vitro and tumorigenicity in vivo.11 Third, in hepatocellular carcinoma, downregulation of MTAP expression is mainly attributed to promoter hypermethylation, rather than genomic loss or mutation.24 Moreover, MTAP deficiency is associated with increased tumorigenicity and induces progression of hepatocellular carcinoma via accumulation of 5′-deoxy-5′-methylthioadenosine (the MTAP substrate).42 As for NPC, the role of MTAP expression is still unclear. In this study, we have first worked out the gene status and protein expression of MTAP at 9p21.3 in NPC and provided convincing evidence that MTAP protein deficiency was correlated with disease progression characterized by worse DSS and DMFS.
Through accumulation of the MTA (the MTAP substrate), MTAP depletion might increase the invasive and vasculogenic capability of melanoma cells by induction of matrix metalloproteinase (MMP) and angiogenic growth factor gene expression. In addition, MTA played a tumor-supporting role in melanoma via activating fibroblasts to construct an ideal microenvironment for tumor growth.43 In hepatocellular carcinoma, Kirovski et al42 also demonstrated similar findings that downregulation of MTAP resulted in MTA secretion from cancer cells, leading to induced expression of MMP and fibroblast growth factors. Furthermore, they found that low MTAP protein expression correlated with advanced tumor stages, which was compatible with our result in NPC. There was evidence suggesting that MTAP depletion may lead to upregulation of ornithine decarboxylase (ODC).44 ODC is the rate-limiting enzyme in the production of polyamines having stimulatory effect on cell proliferation and therefore cancer growth. Introduction of MTAP into MTAP-deleted breast cancer cells (MCF-7) significantly reduced ODC activity, accompanied by reduction in polyamine levels. These findings indicated that other enzyme, in addition to MTAP, in the methionine salvage pathway may also involve in regulating tumor growth.
In normal cells, MTAP cleaves MTA, generated during polyamine biosynthesis, to adenine and 5-methylthioribose-1-phosphate. The latter compound is further metabolized to methionine and adenine is converted to AMP. In MTAP-deficient cells, MTA is unable to be catabolized to generate adenine and methionine. As a consequence, MTAP-deficient cells are more sensitive than MTAP-positive cells to inhibitors of de novo purine synthesis and to methionine deprivation, presenting a possible therapeutic target for cancer treatment.13,45 The inhibitors of purine biosynthesis include methotrexate, 6-mercaptopurine, 6-thioguanine (6-TG), azaserine, and l-alanosine.28,45,46 Both 6-mercaptopurine and 6-TG are rapidly converted to active nucleotide forms in MTAP-deficient cells. 6-TG can be incorporated into DNA, and cells without mismatch repair gene exhibits more resistance to 6-TG treatment.47 In addition, MTA may act as a protective agent. MTA alone, which serves as an anti-inflammatory agent, has been found to possess suppression effect on tumor growth in melanoma and colon cancer.48,49 In recent years, a new strategy of treatment with selective killing of MTAP-deficient tumors has emerged. MTAP-deficient tumors are treated with MTA, followed by high dose of a purine or pyrimidine analog, such as 5-fluorouracil and 6-TG, which needs phosphoribosylation for conversion to its toxic nucleotide. In normal host cells, substantial adenine, generated from MTA, blocks the conversion of the analog to its toxic nucleotide via effective competition for phosphoribosylation by 5-phosphoribosyl-1-pyrophosphate (PRPP). In MTAP-deficient tumor cells, PRPP persists at adequate level due to lack of adenine. And the analog is converted to its toxic nucleotide, selectively killing the tumor cells.50 Thus, it would be of great value to apply this strategy of targeted treatment to NPC in the future.
Finally, there is an interesting issue with regard to the immunolabeling for MTAP which needs to be further elucidated. Although MTAP is originally found to be a cytoplasmic protein, both previous studies and our current one disclose the possibility of its nuclear expression in some kinds of tumors or precancerous lesions, including gastroesophageal high-grade dysplastic lesions, mantle cell lymphomas, and NPCs.17,30 However, little is known about the pathophysiologic process underlying this phenomenon. Further investigation is needed to confirm this expression pattern and to recognize its function as a nuclear protein.
In conclusion, we have found homozygous deletion or promoter hypermethylation of MTAP in a subset of NPCs either through published transcriptomic datasets or in our cases. MTAP deficiency was demonstrated in the immunohistochemical study. Furthermore, MTAP deficiency was associated with advanced tumor stage, as well as worse DSS and DMFS. This result implied that MTAP deficiency plays an important role in tumorigenesis and exhibits an aggressive behavior. It also acts as an independent factor to predict poor survival. This study sheds light on the potential application of target treatment for tumors lacking MTAP activity in NPC.
Supplementary Material
Footnotes
Abbreviations: 6-TG = 6-thioguanine, aCGH = array comparative genomic hybridization, DMFS = distant metastasis-free survival, DSS = disease-specific survival, EBV = Epstein–Barr virus, LRFS = local recurrence-free survival, MMP = matrix metalloproteinase (MMP), MTA = methylthioadenosine, MTAP = methylthioadenosine phosphorylase, NPC = Nasopharyngeal carcinoma, ODC = ornithine decarboxylase, PCR = polymerase chain reaction, PRPP = 5-phosphoribosyl-1-pyrophosphate, RT = radiotherapy.
Conception and design: H-LH, Y-EL, and C-FL; development of methodology: Y-LS, Y-EL, and C-FL; acquisition of data: S-WL; analysis and interpretation of data: H-LH, T-JC, and C-FL; writing and/or revision of the manuscript: H-LH, Y-EL, and C-FL; study supervision: Y-LS and C-FL.
This study is supported by Ministry of Health and Welfare (MOHW103-TD-B-111-05 and DOH102-TD-M-111-102001) and the Biobank at Chi-Mei Medical Center.
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell 2004; 5:423–428. [DOI] [PubMed] [Google Scholar]
- 2.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2006; 15:1765–1777. [DOI] [PubMed] [Google Scholar]
- 3.Jeyakumar A, Brickman TM, Jeyakumar A, et al. Review of nasopharyngeal carcinoma. Ear Nose Throat J 2006; 85:168–170.172–163, 184. [PubMed] [Google Scholar]
- 4.Batova A, Diccianni MB, Nobori T, et al. Frequent deletion in the methylthioadenosine phosphorylase gene in T-cell acute lymphoblastic leukemia: strategies for enzyme-targeted therapy. Blood 1996; 88:3083–3090. [PubMed] [Google Scholar]
- 5.Huang HY, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. J Clin Oncol 2005; 23:548–558. [DOI] [PubMed] [Google Scholar]
- 6.Hustinx SR, Leoni LM, Yeo CJ, et al. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod Pathol 2005; 18:959–963. [DOI] [PubMed] [Google Scholar]
- 7.Illei PB, Rusch VW, Zakowski MF, et al. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res 2003; 9:2108–2113. [PubMed] [Google Scholar]
- 8.Ladanyi M. Implications of P16/CDKN2A deletion in pleural mesotheliomas. Lung Cancer 2005; 49 Suppl 1:S95–S98. [DOI] [PubMed] [Google Scholar]
- 9.Perrone F, Tamborini E, Dagrada GP, et al. 9p21 locus analysis in high-risk gastrointestinal stromal tumors characterized for c-kit and platelet-derived growth factor receptor alpha gene alterations. Cancer 2005; 104:159–169. [DOI] [PubMed] [Google Scholar]
- 10.Schmid M, Malicki D, Nobori T, et al. Homozygous deletions of methylthioadenosine phosphorylase (MTAP) are more frequent than p16INK4A (CDKN2) homozygous deletions in primary non-small cell lung cancers (NSCLC). Oncogene 1998; 17:2669–2675. [DOI] [PubMed] [Google Scholar]
- 11.Christopher SA, Diegelman P, Porter CW, et al. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res 2002; 62:6639–6644. [PubMed] [Google Scholar]
- 12.M'Soka TJ, Nishioka J, Taga A, et al. Detection of methylthioadenosine phosphorylase (MTAP) and p16 gene deletion in T cell acute lymphoblastic leukemia by real-time quantitative PCR assay. Leukemia 2000; 14:935–940. [DOI] [PubMed] [Google Scholar]
- 13.Tisdale MJ. Methionine synthesis from 5′-methylthioadenosine by tumour cells. Biochem Pharmacol 1983; 32:2915–2920. [DOI] [PubMed] [Google Scholar]
- 14.Dreyling MH, Roulston D, Bohlander SK, et al. Codeletion of CDKN2 and MTAP genes in a subset of non-Hodgkin's lymphoma may be associated with histologic transformation from low-grade to diffuse large-cell lymphoma. Genes Chromosomes Cancer 1998; 22:72–78. [PubMed] [Google Scholar]
- 15.Harasawa H, Yamada Y, Kudoh M, et al. Chemotherapy targeting methylthioadenosine phosphorylase (MTAP) deficiency in adult T cell leukemia (ATL). Leukemia 2002; 16:1799–1807. [DOI] [PubMed] [Google Scholar]
- 16.Hori Y, Hori H, Yamada Y, et al. The methylthioadenosine phosphorylase gene is frequently co-deleted with the p16INK4a gene in acute type adult T-cell leukemia. Int J Cancer 1998; 75:51–56. [DOI] [PubMed] [Google Scholar]
- 17.Marce S, Balague O, Colomo L, et al. Lack of methylthioadenosine phosphorylase expression in mantle cell lymphoma is associated with shorter survival: implications for a potential targeted therapy. Clin Cancer Res 2006; 12:3754–3761. [DOI] [PubMed] [Google Scholar]
- 18.Mirebeau D, Acquaviva C, Suciu S, et al. The prognostic significance of CDKN2A, CDKN2B and MTAP inactivation in B-lineage acute lymphoblastic leukemia of childhood. Results of the EORTC studies 58881 and 58951. Haematologica 2006; 91:881–885. [PubMed] [Google Scholar]
- 19.Traweek ST, Riscoe MK, Ferro AJ, et al. Methylthioadenosine phosphorylase deficiency in acute leukemia: pathologic, cytogenetic, and clinical features. Blood 1988; 71:1568–1573. [PubMed] [Google Scholar]
- 20.Behrmann I, Wallner S, Komyod W, et al. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am J Pathol 2003; 163:683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen ZH, Zhang H, Savarese TM. Gene deletion chemoselectivity: codeletion of the genes for p16(INK4), methylthioadenosine phosphorylase, and the alpha- and beta-interferons in human pancreatic cell carcinoma lines and its implications for chemotherapy. Cancer Res 1996; 56:1083–1090. [PubMed] [Google Scholar]
- 22.Chow WA, Bedell V, Gaytan P, et al. Methylthioadenosine phosphorylase gene deletions are frequently detected by fluorescence in situ hybridization in conventional chondrosarcomas. Cancer Genet Cytogenet 2006; 166:95–100. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Castellano JM, Villanueva A, Healey JH, et al. Methylthioadenosine phosphorylase gene deletions are common in osteosarcoma. Clin Cancer Res 2002; 8:782–787. [PubMed] [Google Scholar]
- 24.Hellerbrand C, Muhlbauer M, Wallner S, et al. Promoter-hypermethylation is causing functional relevant downregulation of methylthioadenosine phosphorylase (MTAP) expression in hepatocellular carcinoma. Carcinogenesis 2006; 27:64–72. [DOI] [PubMed] [Google Scholar]
- 25.Huang HY, Li SH, Yu SC, et al. Homozygous deletion of MTAP gene as a poor prognosticator in gastrointestinal stromal tumors. Clin Cancer Res 2009; 15:6963–6972. [DOI] [PubMed] [Google Scholar]
- 26.Hustinx SR, Hruban RH, Leoni LM, et al. Homozygous deletion of the MTAP gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapy. Cancer Biol Ther 2005; 4:83–86. [DOI] [PubMed] [Google Scholar]
- 27.Karikari CA, Mullendore M, Eshleman JR, et al. Homozygous deletions of methylthioadenosine phosphorylase in human biliary tract cancers. Mol Cancer Ther 2005; 4:1860–1866. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Su D, Mizobuchi H, et al. Status of methylthioadenosine phosphorylase and its impact on cellular response to l-alanosine and methylmercaptopurine riboside in human soft tissue sarcoma cells. Oncol Res 2004; 14:373–379. [DOI] [PubMed] [Google Scholar]
- 29.Nobori T, Karras JG, Della Ragione F, et al. Absence of methylthioadenosine phosphorylase in human gliomas. Cancer Res 1991; 51:3193–3197. [PubMed] [Google Scholar]
- 30.Powell EL, Leoni LM, Canto MI, et al. Concordant loss of MTAP and p16/CDKN2A expression in gastroesophageal carcinogenesis: evidence of homozygous deletion in esophageal noninvasive precursor lesions and therapeutic implications. Am J Surg Pathol 2005; 29:1497–1504. [DOI] [PubMed] [Google Scholar]
- 31.Sommer J, Itani DM, Homlar KC, et al. Methylthioadenosine phosphorylase and activated insulin-like growth factor-1 receptor/insulin receptor: potential therapeutic targets in chordoma. J Pathol 2010; 220:608–617. [DOI] [PubMed] [Google Scholar]
- 32.Wong YF, Chung TK, Cheung TH, et al. MTAP gene deletion in endometrial cancer. Gynecol Obstet Invest 1998; 45:272–276. [DOI] [PubMed] [Google Scholar]
- 33.Hsu HP, Li CF, Lee SW, et al. Overexpression of stathmin 1 confers an independent prognostic indicator in nasopharyngeal carcinoma. Tumour Biol 2014; 35:2619–2629. [DOI] [PubMed] [Google Scholar]
- 34.Lan J, Huang HY, Lee SW, et al. TOP2A overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol 2014; 35:179–187. [DOI] [PubMed] [Google Scholar]
- 35.Lee SW, Chen TJ, Lin LC, et al. Overexpression of thymidylate synthetase confers an independent prognostic indicator in nasopharyngeal carcinoma. Exp Mol Pathol 2013; 95:83–90. [DOI] [PubMed] [Google Scholar]
- 36.Win KT, Lee SW, Huang HY, et al. Nicotinamide N-methyltransferase overexpression is associated with Akt phosphorylation and indicates worse prognosis in patients with nasopharyngeal carcinoma. Tumour Biol 2013; 34:3923–3931. [DOI] [PubMed] [Google Scholar]
- 37.Wu LC, Chen LT, Tsai YJ, et al. Alpha-methylacyl coenzyme A racemase overexpression in gallbladder carcinoma confers an independent prognostic indicator. J Clin Pathol 2012; 65:309–314. [DOI] [PubMed] [Google Scholar]
- 38.Miyazaki S, Nishioka J, Shiraishi T, et al. Methylthioadenosine phosphorylase deficiency in Japanese osteosarcoma patients. Int J Oncol 2007; 31:1069–1076. [PubMed] [Google Scholar]
- 39.Li CF, Fang FM, Kung HJ, et al. Downregulated MTAP expression in myxofibrosarcoma: a characterization of inactivating mechanisms, tumor suppressive function, and therapeutic relevance. Oncotarget 2014; 5:11428–11441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang YH, Wu WJ, Wang WJ, et al. CEBPD amplification and overexpression in urothelial carcinoma: a driver of tumor metastasis indicating adverse prognosis. Oncotarget 2015; 6:31069–31084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chanudet E, Huang Y, Ichimura K, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia 2010; 24:483–487. [DOI] [PubMed] [Google Scholar]
- 42.Kirovski G, Stevens AP, Czech B, et al. Down-regulation of methylthioadenosine phosphorylase (MTAP) induces progression of hepatocellular carcinoma via accumulation of 5′-deoxy-5′-methylthioadenosine (MTA). Am J Pathol 2011; 178:1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens AP, Spangler B, Wallner S, et al. Direct and tumor microenvironment mediated influences of 5′-deoxy-5′-(methylthio)adenosine on tumor progression of malignant melanoma. J Cell Biochem 2009; 106:210–219. [DOI] [PubMed] [Google Scholar]
- 44.Subhi AL, Diegelman P, Porter CW, et al. Methylthioadenosine phosphorylase regulates ornithine decarboxylase by production of downstream metabolites. J Biol Chem 2003; 278:49868–49873. [DOI] [PubMed] [Google Scholar]
- 45.Chen ZH, Olopade OI, Savarese TM. Expression of methylthioadenosine phosphorylase cDNA in p16-, MTAP-malignant cells: restoration of methylthioadenosine phosphorylase-dependent salvage pathways and alterations of sensitivity to inhibitors of purine de novo synthesis. Mol Pharmacol 1997; 52:903–911. [DOI] [PubMed] [Google Scholar]
- 46.Hori H, Tran P, Carrera CJ, et al. Methylthioadenosine phosphorylase cDNA transfection alters sensitivity to depletion of purine and methionine in A549 lung cancer cells. Cancer Res 1996; 56:5653–5658. [PubMed] [Google Scholar]
- 47.Swann PF, Waters TR, Moulton DC, et al. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science 1996; 273:1109–1111. [DOI] [PubMed] [Google Scholar]
- 48.Andreu-Perez P, Hernandez-Losa J, Moline T, et al. Methylthioadenosine (MTA) inhibits melanoma cell proliferation and in vivo tumor growth. BMC Cancer 2010; 10:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li TW, Zhang Q, Oh P, et al. S-adenosylmethionine and methylthioadenosine inhibit cellular FLICE inhibitory protein expression and induce apoptosis in colon cancer cells. Mol Pharmacol 2009; 76:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lubin M, Lubin A. Selective killing of tumors deficient in methylthioadenosine phosphorylase: a novel strategy. PLoS ONE 2009; 4:e5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


