Supplemental Digital Content is available in the text
Keywords: Chinese population, genetic variants, leukoaraiosis (LA), pyrosequencing, restriction fragment length polymorphism (RFLP), sNP
Abstract
Leukoaraiosis (LA) is a frequent neuroimaging finding commonly observed on brain MRIs of elderly people with prevalence ranging from 50% to 100%. Multiple susceptibility genes or genetic risk factors for LA have been identified in subjects of European descent. Here, we report the first replication study on several common and novel genetic variations in the Chinese population. In this study, a total of 244 subjects (201 LA patients and 43 controls) were enrolled according to our new and strict definition for LA. Subsequently, 6 genetic variants at 5 genes, rs3744028 in TRIM65, rs1055129 in TRIM47, rs1135889 in FBF1, rs1052053 in PMF1, and rs1801133 (C677T) and rs1801131(A1298C) in MTHFR, were selected for genotyping using polymerase chain reaction (PCR)-based pyrosequencing and restriction fragment length polymorphism (RFLP) together with capillary electrophoresis (CE) and agarose gel electrophoresis. Finally, Pearson's χ2 and multivariate logistic regression tests were used to examine the associations between the genotypes and LA. Among these candidate polymorphisms, except for rs1052053 and rs1801131, rs1135889 (P = 0.012) showed significant associations with LA in the dominant model, and the other 3 SNPs, rs3744028 (P = 0.043), rs1055129 (P = 0.038), and rs1801133 (P = 0.027), showed significant associations with LA in the recessive model. However, these differences no longer remained significant after adjusting for age, gender, hypertension, and diabetes mellitus and applying Bonferroni correction or Sidak correction for multiple testing. These results suggest that the above-mentioned genetic variants are not associated with LA risk. In summary, the study did not replicate the susceptibility of rs3744028, rs1055129, and rs1135889 at the Chr17q25 locus for LA nor did it find any other significant results for rs1052053, rs1801133, and rs1801131 in the Chinese population. It strongly indicated the ethnic differences in the genetics of LA. However, the associations of rs3744028 (TRIM65), rs1055129 (TRIM47), rs1135889 (FBF1), and rs1801133 (MTHFR) with LA before Bonferroni correction and Sidak correction for multiple testing are worth highlighting. Thus, we believe that a genome-wide association study and candidate gene association studies are needed to reassess the previous findings and screen novel risk genes for LA in China.
1. Introduction
Leukoaraiosis (LA) is mainly characterized by white matter lesions (WMLs) or white matter hyperintensities (WMHs) on brain T2-weighted and fluid-attenuated inversion recovery (FLAIR) magnetic resonance imaging (MRIs) of elderly people (Fig. 1), with the prevalence ranging from 50% to 100% in population-based studies.[1–4] It has been considered to be a putative predictor for the increased risk of gait disturbance and falls, dementia, stroke, and depression, and even death.[4,5] During the past decade, a large number of genetic variants have been identified as risk factors for LA in Western countries.[6–11] However, no mutations or single nucleotide polymorphisms (SNPs) have been established as accepted risk factors for LA among different ethnicities. Moreover, to date, no large replication studies have been performed in the Asian population, especially in the Chinese population.
Figure 1.
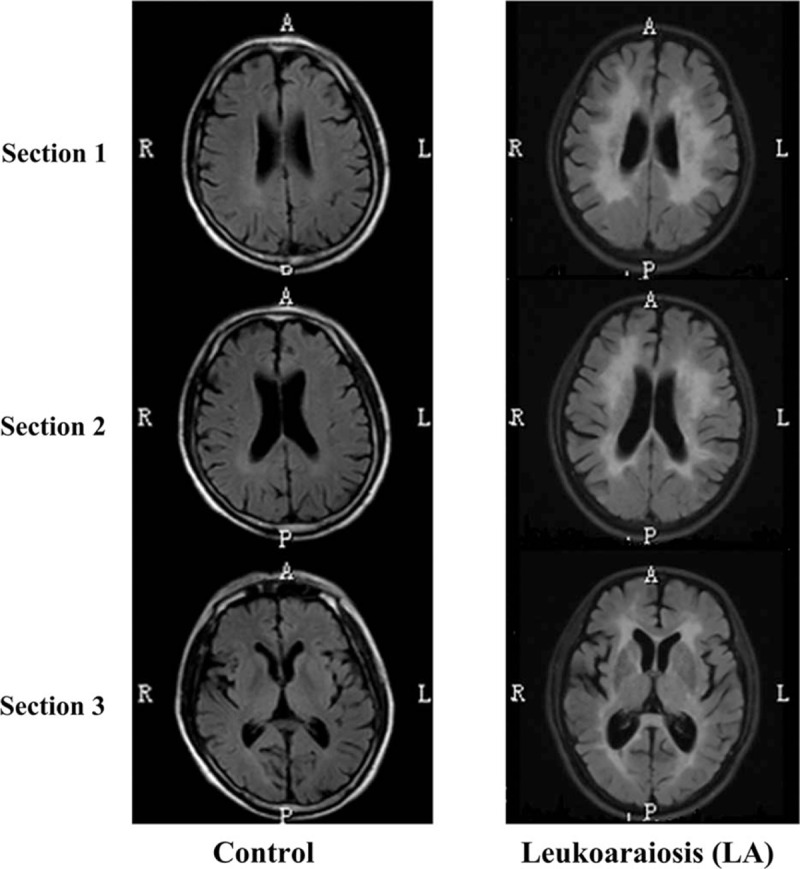
LA in 3 sections of T2-weighted FLAIR MRI. (A) Control without LA represents normal neuroimaging. (B) LA present predominantly in the periventricular and deep/subcortical regions. Sections 1, 2, and 3 represent the centrum semiovale section, lateral ventricles section, and internal capsule section, respectively. R, P, A, and L indicate the right, posterior, anterior, and left on the axial MRI scanning of the brain, respectively. FLAIR = fluid-attenuated inversion recovery, LA = leukoaraiosis, MRI = magnetic resonance imaging.
To confirm the role of these variants in the increased risk of LA, we conducted the first candidate gene association study of LA investigating 6 SNPs in an ethnic Chinese population. Four of these SNPs were selected from the genome-wide association study (GWAS) of Fornage et al,[10] including 2 risk-related SNPs (rs3744028 in TRIM65 and rs1055129 in TRIM47) located in the Ch17q25 locus and 2 missense SNPs (rs1135889 in FBF1 and rs1052053 in PMF1). These 4 SNPs exhibited genome-wide significance (rs3744028 and rs1055129 with P < 1.0 × 10–8) or suggestive association (rs1135889 and rs1052053 with P < 5.0 × 10–5) with WMLs in the GWAS of participants of European descent.[10] Among these SNPs, rs3744028 and rs1055129 were selected from 6 risk SNPs that reached the genome-wide significance threshold of P < 1.0 × 10–8. The other 4 risk-related SNPs, rs9894383, rs11869977, rs936393, and rs3744017, were excluded because they were in strong linkage disequilibrium (LD) with rs3744028 and with each other.
Both rs1135889 and rs1052053 were 2 rare nonsynonymous polymorphisms chosen from 56 highly suggestive risk SNPs with the significance of P < 1.0 × 10–5.[10] They resulted in missense changes and disturbed the structure and function of the protein, suggesting potential importance in the pathogenesis of LA. Recently, rs1052053 has been confirmed as a novel risk factor for LA in subjects from LBC1936.[12] This suggests that those suggestive risk SNPs may be significantly associated with LA risk in other ethnic populations, including the Chinese population. The associations of both rs1135889 and rs1052053 with LA have not been investigated in China.
Two other SNPs, A1298C (rs1801131) and C677T (rs1801133), were selected from the methylenetetrahydrofolate reductase (MTHFR) gene, which acts as a key regulator of homocysteine metabolism and plays a crucial role in the biosynthesis of DNA and RNA.[13–15] Several candidate gene association studies have investigated and found a potential relationship between these 2 mutations and LA in several countries, such as North America,[16] Hungary,[17–19] Germany,[20] and Japan.[21] However, neither SNP has been identified in GWAS of people of European descent,[10] nor have they been detected in the Chinese population. Therefore, rs1801133 (C677T) and rs1801131 (A1298C) were also selected as candidate risk variants in our study.
Thus, the aim of this exploratory study is to investigate the associations of the above-mentioned 6 LA-related SNPs with LA and to recognize predictive factors for the diagnosis of LA in the Chinese population.
2. Methods
2.1. Redefinition and classification of LA
The neuroimaging forms of LA are quite variable. LA is often divided into periventricular WMLs (PVWMLs), including caps around the frontal horns of the lateral ventricles and a pencil-thin lining or a smooth halo along the side of the lateral ventricles, which are all attached to the ventricular system, and deep/subcortical WMLs (DWMLs) occurring as punctate changes or beginning confluent or confluent abnormalities, which are all located apart from the cerebral ventricle in the subcortical white matter.[4] However, the neuroimaging definition of these types of WMLs (PVWMLs and DWMLs) is still unclear. In addition, we found that PVWMLs and DWMLs always coexist in the same LA patient clinically. Therefore, it is difficult for neurologists to clearly diagnose 1 of the 2 types of WMLs, and scientists also feel that comparing the contradictory scientific findings between the different functional studies of LA is laborious because of the different imaging methods and classification schemes. Taking into consideration the difficulty of the clinical LA diagnosis and the previously mentioned shortcomings of the currently predominant LA classification method, our laboratory redefined LA based on the assessment of WMLs in 3 sections (centrum semiovale section, lateral ventricles section, and internal capsule section) of brain T2-weighted and FLAIR MRI neuroimaging (Fig. 1), and we further proposed a new classification scheme to classify LA-associated neuroimaging phenomenon into 3 broad categories: normal, Type I LA, and Type II LA.[22] Subjects with few WMLs in all 3 sections of MRI imaging were considered to be “Normal” without LA. Those subjects with LA in all of the 3 sections were definitely diagnosed as LA patients. Type I LA shows white matter changes including a pencil-thin lining or a smooth halo along the side of the lateral ventricles, small periventricular caps around the frontal horns of the lateral ventricles, and early confluent lesions. Type II LA shows irregular periventricular lesions, largely extending periventricular caps, and severely confluent abnormalities. These 3 neuroimaging categories represented differing levels of severity of LA in the cerebral subcortical white matter on brain MRI (no LA for “normal”, mild LA for “Type I LA,” and severe LA for “Type II LA”). Based on the above-mentioned definition of LA, we included subjects with any degree of WMLs between Type I LA and Type II LA in the LA group, and enrolled those with actually no WMLs in the control group.
2.2. Subjects
There were 244 patients, of which 43 had no LA and 201 had LA, recruited from the First Affiliated Hospital of Xiamen University between 2011 and 2014 (Table 1). They were selected based on the new definition and classification of LA described above. Subjects with any degree of WMLs between Type I LA and Type II LA were included in the LA group, whereas subjects without any white matter changes were included in the control group. All of the subjects met the inclusion criteria, including patients ≥40 years of age and patients with LA or controls without LA. The following patients were excluded: patients <40 years of age, patients suffering from an intracerebral hemorrhage, a subarachnoid hemorrhage, an intracranial infection, a malignant tumor, toxic encephalopathy, Parkinson's disease, ischemic heart disease, multiple sclerosis, or hydrocephalus. The study was approved by the ethics committee of Xiamen University. All study subjects provided written informed consent.
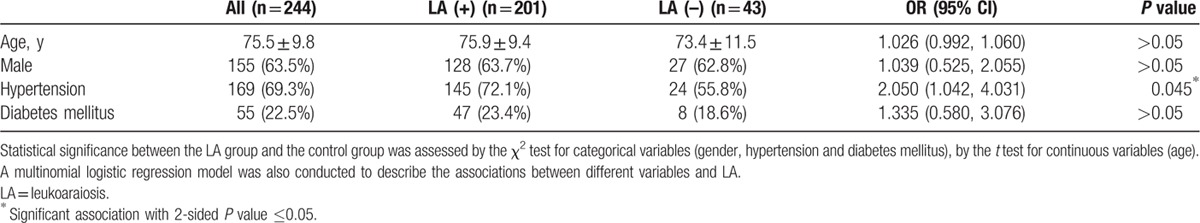
Table 1.
Demographic characteristics of subjects with and without LA.
2.3. Clinical information collection
Clinical data were collected, including gender, age, hypertension, and diabetes. Hypertension was classified according to the 1999 World Health Organization (WHO) Guidelines for the prevention and treatment of hypertension and was considered to be present in patients with a systolic blood pressure ≥140 mmHg and/or a diastolic blood pressure ≥90 mm Hg or in patients requiring treatment with oral antihypertension drugs.[23] Diabetes was determined as a fasting glucose level ≥7.0 mmol/L or 2-hour post glucose load glucose levels ≥11.1 mmol/L, according to the diagnostic criteria of WHO for diabetes.[24]
2.4. Genotyping
Genomic DNA was extracted from frozen whole blood with ethylene diamine tetraacetic acid (EDTA) anticoagulant using the MagCore® Genomic DNA Whole Blood Kit (RBC Bioscience, Taiwan, China). The 3 SNPs rs1052053, rs1801133, and rs1801131 were genotyped using restriction fragment length polymorphism (RFLP). PCR products were digested with the restriction enzyme Hinf I (rs1052053 and rs1801133) and MboII (rs1801131) and analyzed by capillary (BIOptics Qsep100 dna-CE, Taiwan, China) and agarose gel electrophoresis. Three genotypes of rs1052053 were detected: AA (136, 101 bp), AG (136, 101, 82, 54 bp), and GG (101, 82, 54 bp). Three genotypes of rs1801133 (C677T) were detected: CC (369 bp), CT (369, 210, 159 bp), and TT (210, 159 bp). Three genotypes of rs1801131 (A1298C) were detected: AA (204, 29, 28, 22 bp), AC (234, 204, 29, 28, 22 bp), and CC (234, 29, 22 bp) (Supplemental Table 1). The other 3 SNPs without any restriction enzyme sites, rs3744028, rs1055129, and rs1135889, were genotyped using pyrosequencing on the PyroMark Q24 MDx platform (Qiagen, Valencia, CA) (Supplemental Table 1). The primers used in the RFLP and pyrosequencing assays were designed by the Primer Premier 5 software (Premier Biosoft International, Palo Alto, CA) and the Qiagen PyroMark Q24 system, respectively, and the primers were synthesized by Sangon Biotech (Shanghai, China). Some PCR products were sequenced using an ABI 3730 automated DNA sequencing system (Sangon Biotech, Shanghai, China). Details of the PCR and genotyping conditions are described in Supplemental Table 1.
2.5. Statistical analysis
Demographic data was expressed as the mean ± SD for continuous variables and as numbers (percentages) for categorical variables. The differences between the clinical parameters in the LA group and the controls were assessed using the t test and Pearson's χ2 where appropriate. First, each SNP was separately assessed for deviation from the Hardy–Weinberg equilibrium in cases and controls using an exact test as implemented in the Finetti program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). Furthermore, 4 genetic models, the general genetic model, the dominant model, the recessive model, and the multiplicative model (allele test), were used to analyze the influence of every SNP on LA. The minor allele was considered the risk allele of LA. The major allele and major homozygotes were used as the references in the allele test and the genotype test, respectively. The differences in frequencies of the alleles and genotypes between the LA and control groups were analyzed using the χ2 test and Fisher's exact test. A multinomial logistic regression model was also conducted to describe the associations between different genotypes and LA with an adjustment for age and gender. Odds ratios (OR) with 95% confidence intervals (CI) were calculated. A 2-tailed P value ≤0.05 was considered statistically significant in all genetics tests. To exclude or reduce type I errors in the association analysis of each SNP, Bonferroni and Sidak corrections were used to adjust the P value for multiple testing. All statistical analyses were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL).
3. RESULTS
3.1. Population characteristics and epidemiology of LA
The clinical parameters of 244 participants are presented in Table 1. The population is relatively older compared with previous genetic studies of LA.[8,12,25–28] The mean age of the subjects was 75.5 ± 9.8 (45∼97) years and 63.5% were males. There were no significant differences between the LA group and the control group in age (P > 0.05), gender (P > 0.05), and diabetes mellitus occurrence (P > 0.05), but there was a significant difference in the history of hypertension (P = 0.045) between the LA group and the control group. These results suggested that hypertension is the only risk factor for LA, and other clinical variables (age, gender, and diabetes mellitus) did not have much influence on the risk estimation of genetic factors in LA in the present study.
3.2. Association between 6 risk-related polymorphisms and LA
We performed genotyping for rs3744028, rs1055129, and rs1135889 in all of the genomic DNA samples using PCR followed by pyrosequencing (Supplemental Figure 1). The other 3 SNPs, rs1801131, rs1801133, and rs1052053, were tested using the PCR-RFLP method followed by capillary and agarose gel electrophoresis (Fig. 2). Except for rs1801131, which was not detected in our cohort, all of the SNPs were identified in the LA group and the control group, and they met the Hardy–Weinberg criterion of conformity in both groups. Given that rs1801131 was not identified in the present study, we did not include it in the final statistical analysis. The genotype and allele frequencies between patients and controls and the results from the general genetic model, the dominant model, the recessive model, and the multiplicative model are summarized in Table 2.
Figure 2.
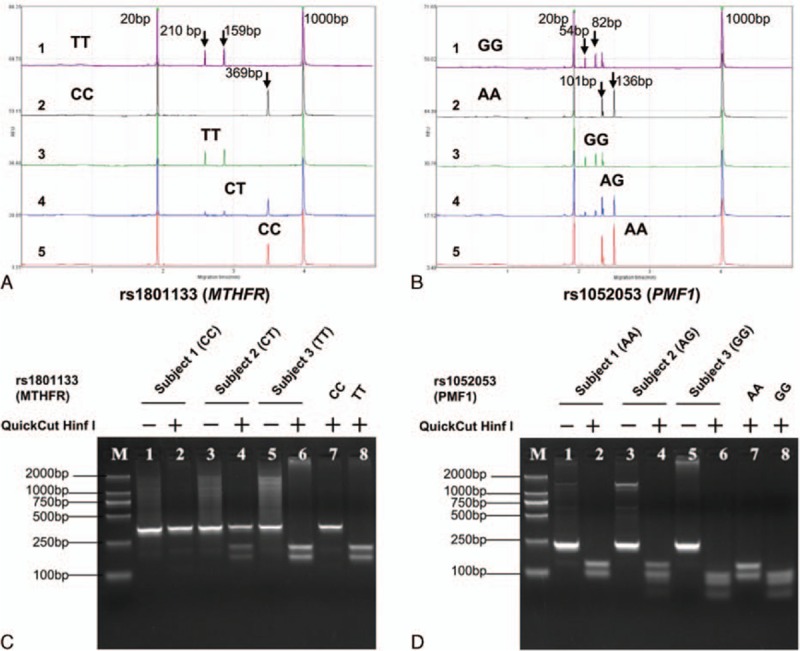
RFLP-based genotyping of rs1801133 (MTHFR) and rs1052053 (PMF1), respectively. A and B show the CE analyses for rs1801133 and rs1052053 after PCR and restriction enzyme digestion, respectively. The number labels shown in A and B represent the positive control (1), negative control (2), minor homozygote (3), heterozygote (4), and major homozygote (5), respectively. C and D show the agarose gel electrophoresis analysis for rs1801133 and rs1052053 after PCR and restriction enzyme digestion, respectively. This experiment aims to validate the results from the CE analysis. CE = capillary electrophoresis, RFLP = restriction fragment length polymorphism.
Table 2.
Statistical analysis of genotypic frequencies among LA subjects and controls.
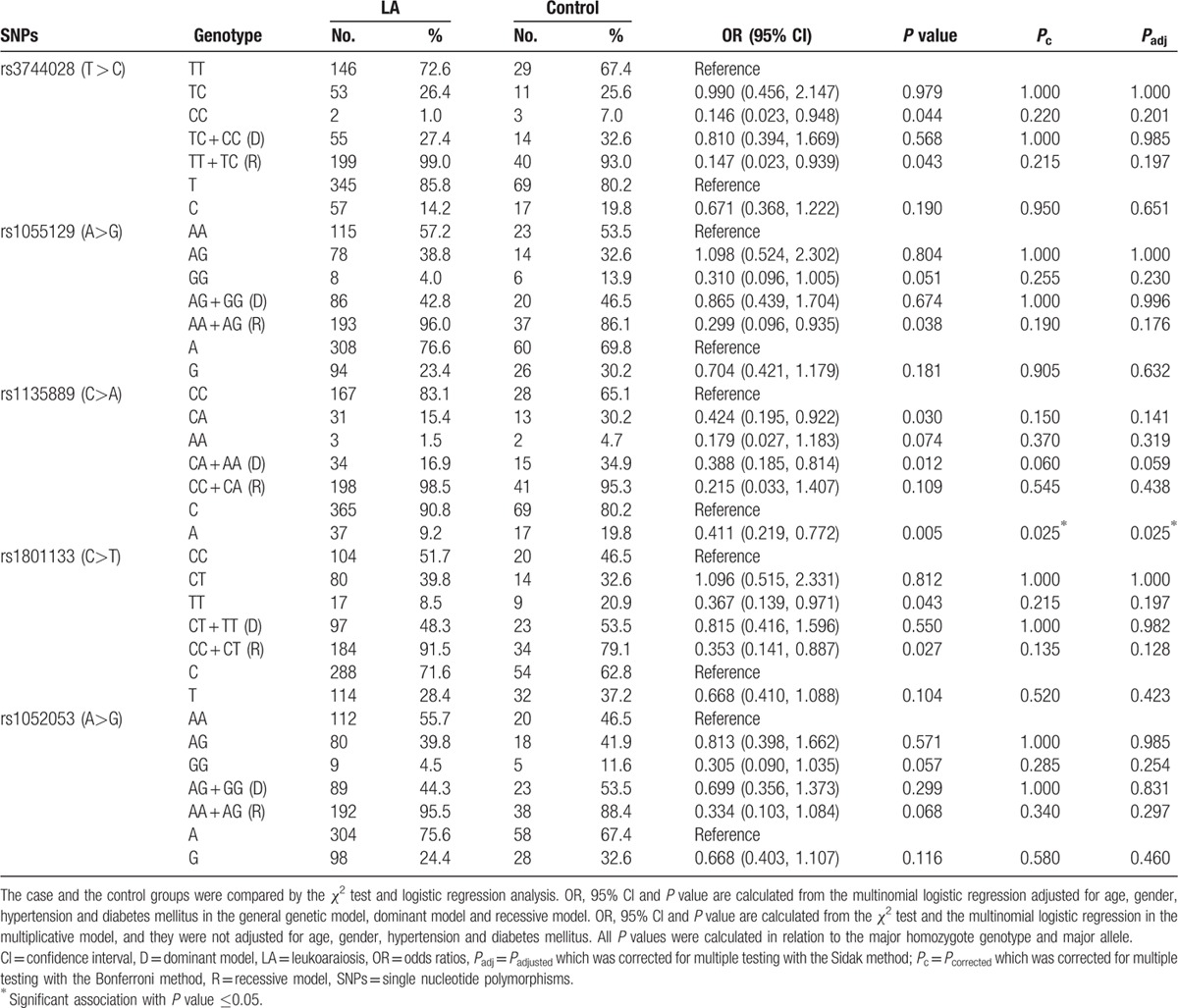
As shown in Table 2. LA was not observed to be significantly associated with rs1055129 and rs1052053, but it was significantly associated with rs3744028, rs1135889, and rs1801133 in the general genetic model. Among these polymorphisms, 2 SNPs, rs3744028 (P = 0.043) and rs1801133 (P = 0.027), together with rs1055129 (P = 0.038) appeared to be significantly associated with LA in the recessive model. Subjects carrying the rs3744028 (CC vs CT + TT: OR 0.147, 95% CI 0.023–0.939, P = 0.043), rs1055129 (GG vs GA + AA: OR 0.299, 95% CI 0.096–0.935, P = 0.038), and rs1801133 (TT vs TC + CC: OR 0.353, 95% CI 0.141–0.887, P = 0.027) homozygous minor alleles had significantly decreased LA risk compared with those carrying 0 minor alleles or 1 minor allele in the recessive genetic model. The other SNP, rs1135889 (P = 0.012), showed to be significantly associated with LA in the dominant model. However, the significant difference of the 4 SNPs between the LA and control groups was lost after Bonferroni correction and Sidak correction for multiple testing. We did not find the frequency distributions of rs3744028 (Pc = 0.215, Padj = 0.197), rs1055129 (Pc = 0.190, Padj = 0.176), rs1135889 (Pc = 0.060, Padj = 0.059), and rs1801133 (Pc = 0.135, Padj = 0.128) to be significantly associated with LA among individuals.
In the multiplicative model, the minor allele of rs1135889 (OR 0.411, 95% CI: 0.219–0.772, Pc = 0.025, Padj = 0.025) was shown to be significantly associated with LA after the adjustment of the p value for 5 tests using the Bonferroni correction method. Together with the trend (Pc = 0.06, Padj = 0.059) shown in the association of rs1135889 with LA in dominant model, this finding suggested that the rs1135889 may be related to LA risk and it could significantly reduce LA risk. Unlike rs1135889, the minor alleles of other 4 SNPs (rs3744028, rs1055129, rs1801133, and rs1052053) did not show significant associations with LA in the multiplicative model.
Taken together, these results suggested that none of the 5 candidate SNPs, rs3744028, rs1055129, rs1801133, rs1052053, and rs1135889, were related to LA in the Chinese population, but the effect of rs1135889 on reducing LA risk was worth highlighting.
4. Discussion
In the present study, we did not confirm the findings from candidate gene association studies and GWAS of LA. We found that rs3744028 in TRIM65, rs1055129 in TRIM47, rs1135889 in FBF1, rs1052053 in PMF1, and rs1801133 and rs1801131 in MTHFR were not associated with LA risk in 244 relatively old Chinese individuals.
Inconsistent with those previous findings from the GWAS in Europeans and the recent multiethnic GWAS,[10,28] as well as the replication studies in Scotland and Japan,[12,27] our results did not show the significant associations of rs3744028 and rs1055129 at the Ch17q25 locus with LA. As observed in studies of African Americans, Hispanics, and Asians (Chinese and Malays),[10,28] the lack of associations of rs3744028 and rs1055129 with LA occurrence should also be interpreted cautiously in the Chinese population. These inconsistent findings may mainly result from ethnic differences (Supplemental Table 2). In addition, the (semi)quantitative scale of LA, which was considered a continuous phenotype in the GWAS by Fornage et al,[10] is different from the visual rating scales used in the present study. This difference also may lead to some differences in the results between the 2 studies.
As shown in the recent multiethnic GWAS by Verhaaren et al,[28] the present study also found that neither rs1135889 nor rs1052053 was significantly associated with LA in the Chinese population. However, the potential association of rs1135889 with LA in dominant model after the adjustment for those clinical variables and the significant effect of the minor allele of rs1135889 on reducing LA risk should be given more attention. Because this observation not only demonstrated the combined influence of age, gender, hypertension, and diabetes mellitus on LA but also indicated the potential effect of rs1052053 on LA risk. Given that there is a suggestive association of rs1135889 in FBF1 with LA and that the FBF1-coding protein acting as a cytoplasmic ligand of the Fas cell surface receptor (TNFRSF6) plays a regulatory role in apoptosis and the immune response through the protein interaction with Fas,[29,30] here, we think that the role of rs1135889 in reducing LA risk is worth highlighting in future studies.
As shown for rs3744028, rs1055129, and rs1135889, rs1801133 was also not shown to be significantly associated with LA after correction for multiple tests. These results are in disagreement with findings from studies of Caucasians from North America and people of European and Japanese descent,[14,16,21] but are consistent with findings from the study of Caucasians from Hungary.[17–19] Although the significant association of rs1801133 TT with LA was missing after correction for multiple tests, the results indicated that rs1801133 TT may also act as a protective factor for LA similarly to rs3744028, rs1055129, and rs1135889, and it could significantly decrease LA risk in the recessive genetic model. This conclusion should be interpreted with caution owing to the ethnic differences described above and the following limitations in the present study. Another polymorphism, rs1801131 (A1298C) in the MTHFR gene, was not identified in any of the Chinese subjects, suggesting that this polymorphism does not exist in the Chinese population. Another possibility may be the low prevalence of rs1801131 in China (Supplemental Table 2).
The limitations of the study are as follows: the small sample size decreases the available statistical power and makes it difficult to examine low-frequency genotypes and also any SNPs of small effect size; although the use of visual rating scales based on the assessment of WMLs in 3 sections of neuroimaging makes the inclusion criteria more strict, it is difficult for scientists to compare with previous studies; and as ischemic stroke was not excluded in the study, it has the potential to confound the observed results.
Because of the above-mentioned limitations and ethnic differences, we could not determine the risk of the candidate SNPs in LA. Therefore, additional genome-wide association studies or large-scale candidate gene association studies (such as exon-capture sequencing studies) are still needed to reassess the identified risk genes and screen for novel risk loci for LA in China. Additionally, we believe that, except for the effect of genetic factors on LA, aberrant gene expression and epigenetic and small RNA regulation may predominantly contribute to increased LA risk or LA pathogenesis. Simpson[31] and Xu et al[32] identified many genes with abnormal mRNA expression in LA by microarray RNA expression analysis in brain tissue and in whole blood, respectively.[33] Our laboratory has performed genome-scale methylation profiling and miRNA expression analysis on LA in whole blood and revealed some specific hyper/hypo-methylated genes and miRNAs associated with LA occurrence and development (unpublished data). Moreover, based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the clusters of potential genes, including the miRNA-targeted genes, together with the pathological demyelination of LA, Lin et al[22] hypothesized that LA may be a neuroinflammatory disorder of the central nervous system (CNS) that is triggered by abnormal DNA methylation and mRNA and miRNA expression-induced inflammatory responses. Much work is required to confirm this hypothesis and to decipher the precise pathogenic genes and signaling pathways underlying the WMLs. We believe that the establishment and study of an LA animal model is of the utmost importance at the moment. Additionally, close collaborations around the world are also required for the study of LA pathogenesis.
5. Conclusion
This is the first genetic association study of LA to explore the risk roles of 6 well-known variants in the occurrence of LA in the Southern Chinese population. In the present study, we did not replicate the susceptibility of rs3744028, rs1055129, and rs1135889 at the Chr17q25 locus and rs1052053 in PMF1 for LA, nor did it find any other significant results for rs1801133 and rs1801131 in MTHFR. These results strongly indicated the ethnic differences in the genetics of LA. However, the associations of rs3744028 (TRIM65), rs1055129 (TRIM47), rs1135889 (FBF1), and rs1801133 (MTHFR) with LA in the dominant and recessive models before Bonferroni correction for multiple testing are worth highlighting. Therefore, additional GWASs or large candidate gene association studies are needed to reassess the previous findings and screen for novel risk genes for LA in China.
Supplementary Material
Acknowledgments
The authors thank all the subjects for participating in this study.
Footnotes
Abbreviations: CI = confidence intervals, DWMLs = deep/subcortical white matter lesions, FLAIR = fluid-attenuated inversion recovery, GWAS = genome-wide association study, LA = leukoaraiosis, MTHFR = methylenetetrahydrofolate reductase, OR = odds ratios, PVWMLs = periventricular white matter lesions, RFLP = restriction fragment length polymorphism, SNPs = single nucleotide polymorphisms, WMHs = white matter hyperintensities, WMLs = white matter lesions.
∗Current address: Translational Medicine Research Center (TMRC), School of Pharmaceutical Sciences, Xiamen University Xiang’an Southern Road, Xindian Town, Xiang’an Dist, Xiamen, Fujian 361102, China.
W-QH and H-MY contributed equally to this work.
Ethical statement and patient consent: This study was approved by the ethical committee of Xiamen University. Written informed consent was obtained from the patient before the publication of this article.
Funding: This study was funded by the Fujian Provincial Science and Technology Grant (No. 2012Y0064), Science and Technology Grant of Xiamen (No. 3502Z20164002), the National Natural Science Foundation of China (Grant No. 81272445, No. 81472031) and CTCTCT.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- 1.Hachinski VC, Potter P, Merskey H. Leuko-araiosis. Arch Neurol 1987; 44:21–23. [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan M. Leukoaraiosis. Practical Neurol 2008; 8:26–38. [DOI] [PubMed] [Google Scholar]
- 3.Launer LJ. Epidemiology of white matter lesions. Top Magn Reson Imaging 2004; 15:365–367. [DOI] [PubMed] [Google Scholar]
- 4.Kim KW, MacFall JR, Payne ME. Classification of white matter lesions on magnetic resonance imaging in elderly persons. Biol Psychiatry 2008; 64:273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debette S, Markus HS. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ 2010; 341:c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atwood LD, Wolf PA, Heard-Costa NL, et al. Genetic variation in white matter hyperintensity volume in the Framingham Study. Stroke 2004; 35:1609–1613. [DOI] [PubMed] [Google Scholar]
- 7.DeStefano AL, Atwood LD, Massaro JM, et al. Genome-wide scan for white matter hyperintensity: the Framingham Heart Study. Stroke 2006; 37:77–81. [DOI] [PubMed] [Google Scholar]
- 8.Paternoster L, Chen W, Sudlow CL. Genetic determinants of white matter hyperintensities on brain scans: a systematic assessment of 19 candidate gene polymorphisms in 46 studies in 19,000 subjects. Stroke 2009; 40:2020–2026. [DOI] [PubMed] [Google Scholar]
- 9.Smith JA, Turner ST, Sun YV, et al. Complexity in the genetic architecture of leukoaraiosis in hypertensive sibships from the GENOA Study. BMC Med Genomics 2009; 2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornage M, Debette S, Bis JC, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol 2011; 69:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assareh A, Mather KA, Schofield PR, et al. The genetics of white matter lesions. CNS Neurosci Ther 2011; 17:525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez LM, Hill WD, Harris SE, et al. Genes from a translational analysis support a multifactorial nature of white matter hyperintensities. Stroke 2015; 46:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueland PM, Hustad S, Schneede J, et al. Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol Sci 2001; 22:195–201. [DOI] [PubMed] [Google Scholar]
- 14.Hassan A, Hunt BJ, O'Sullivan M, et al. Homocysteine is a risk factor for cerebral small vessel disease, acting via endothelial dysfunction. Brain 2004; 127 (pt 1):212–219. [DOI] [PubMed] [Google Scholar]
- 15.Miner SE, Evrovski J, Cole DE. Clinical chemistry and molecular biology of homocysteine metabolism: an update. Clin Biochem 1997; 30:189–201. [DOI] [PubMed] [Google Scholar]
- 16.Hong ED, Taylor WD, McQuoid DR, et al. Influence of the MTHFR C677T polymorphism on magnetic resonance imaging hyperintensity volume and cognition in geriatric depression. Am J Geriatr Psychiatry 2009; 17:847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szolnoki Z, Somogyvari F, Kondacs A, et al. Evaluation of the roles of common genetic mutations in leukoaraiosis. Acta Neurol Scand 2001; 104:281–287. [DOI] [PubMed] [Google Scholar]
- 18.Szolnoki Z, Somogyvari F, Kondacs A, et al. Specific APO E genotypes in combination with the ACE D/D or MTHFR 677TT mutation yield an independent genetic risk of leukoaraiosis. Acta Neurol Scand 2004; 109:222–227. [DOI] [PubMed] [Google Scholar]
- 19.Szolnoki Z, Szaniszlo I, Szekeres M, et al. Evaluation of the MTHFR A1298C variant in leukoaraiosis. J Mol Neurosci 2012; 46:492–496. [DOI] [PubMed] [Google Scholar]
- 20.Linnebank M, Moskau S, Jurgens A, et al. Association of genetic variants of methionine metabolism with methotrexate-induced CNS white matter changes in patients with primary CNS lymphoma. Neurooncology 2009; 11:2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohara K, Fujisawa M, Ando F, et al. MTHFR gene polymorphism as a risk factor for silent brain infarcts and white matter lesions in the Japanese general population: The NILS-LSA Study. Stroke 2003; 34:1130–1135. [DOI] [PubMed] [Google Scholar]
- 22.Lin Q, Huang WQ, Tzeng CM. Genetic associations of leukoaraiosis indicate pathophysiological mechanisms in white matter lesions etiology. Rev Neurosci 2015. [DOI] [PubMed] [Google Scholar]
- 23.1999; 1999 World Health Organization-International Society of Hypertension Guidelines for the Management of Hypertension. Guidelines Subcommittee. J Hypertens. 17:151–183. [PubMed] [Google Scholar]
- 24.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 1998; 15:539–553. [DOI] [PubMed] [Google Scholar]
- 25.Verhaaren BF, de Boer R, Vernooij MW, et al. Replication study of chr17q25 with cerebral white matter lesion volume. Stroke 2011; 42:3297–3299. [DOI] [PubMed] [Google Scholar]
- 26.Adib-Samii P, Rost N, Traylor M, et al. 17q25 Locus is associated with white matter hyperintensity volume in ischemic stroke, but not with lacunar stroke status. Stroke 2013; 44:1609–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabara Y, Igase M, Okada Y, et al. Association of Chr17q25 with cerebral white matter hyperintensities and cognitive impairment: the J-SHIPP study. Eur J Neurol 2013; 20:860–862. [DOI] [PubMed] [Google Scholar]
- 28.Verhaaren BF, Debette S, Bis JC, et al. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet 2015; 8:398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wajant H. The Fas signaling pathway: more than a paradigm. Science 2002; 296:1635–1636. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt T, Karsunky H, Frass B, et al. A novel protein (Fbf-1) that binds to CD95/APO-1/FAS and shows sequence similarity to trichohyalin and plectin. Biochim Biophys Acta 2000; 1493:249–254. [DOI] [PubMed] [Google Scholar]
- 31.Simpson JE, Hosny O, Wharton SB, et al. Microarray RNA expression analysis of cerebral white matter lesions reveals changes in multiple functional pathways. Stroke 2009; 40:369–375. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Stamova B, Jickling G, et al. Distinctive RNA expression profiles in blood associated with white matter hyperintensities in brain. Stroke 2010; 41:2744–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai Z, Stamova B, Xu H, et al. Distinctive RNA expression profiles in blood associated with Alzheimer disease after accounting for white matter hyperintensities. Alzheimer Dis Assoc Disord 2014; 28:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


