Supplemental Digital Content is available in the text
Keywords: cord blood, DNA methylation, hepatocyte nuclear factor 4 alpha, melanocortin 4 receptor, metabolic indices, preterm infants, triglyceride
Abstract
The association of preterm birth with obesity and metabolic syndrome later in life is well established. Although the biological mechanism for this association is poorly understood, epigenetic alterations of metabolic-related genes in early life may have important roles in metabolic dysfunction. Thus, we investigated the associations of DNA methylations of melanocortin 4 receptor (MC4R) and hepatocyte nuclear factor 4 alpha (HNF4α) with metabolic profiles in cord blood of term and preterm infants.
We measured metabolic profiles in cord blood samples of 85 term and 85 preterm infants. DNA methylation and mRNA expression levels of MC4R and HNF4α in cord blood cells were quantified using pyrosequencing and real-time PCR. Triglyceride (TG) levels were grouped by percentile as low (<10th percentile), mid (11th–89th percentiles), and high (>90th percentile). A multiple linear regression model was used to assess the differential effects of DNA methylation on metabolic indices in cord blood between term and preterm infants.
The beta-coefficients for associations between TG levels and methylation statuses of MC4R-CpG3 and HNF4α-CpG2 in the P1 promoter differed significantly between term and preterm infants (P = 0.04 and P = 0.003, respectively). DNA methylation statuses of MC4R-CpG3 and HNF4α–CpG2 in the P1 promoter were significantly lower in preterm infants in the high-TG group compared with those in the mid- and low-TG groups (P = 0.01). Notably, preterm infants in the high-TG group had higher TG levels in cord blood than term infants in the high-TG group (60.49 vs 54.57 mg/dL). In addition, MC4R and HNF4α expression levels were higher in preterm infants than in term infants (P < 0.05).
Epigenetic alterations of the newly identified genes MC4R and HNF4α in early life might contribute to metabolic profile changes, especially increased TG levels, in the cord blood of preterm infants.
1. Introduction
Preterm birth (PTB; < 37 weeks gestation) is a major cause of neonatal death and has long-term adverse effects on health.[1] About 28% of early neonatal mortality has been reported to result from PTB.[1,2] The rate of PTB increased from 2.6% in 1993 to 5.9% in 2010 in Korea,[3] and rates are reportedly increased rapidly in developing countries.[4] Children born preterm may have a higher risk of metabolic abnormalities associated with type 2 diabetes (T2D) and cardiovascular disease later in life.[5,6] Changes in lipid metabolism, such as elevated triglyceride (TG) and total cholesterol (TC) levels, in early life may lead to metabolic dysfunction and disease susceptibility.[7] Although the molecular mechanisms underlying the associations remain unknown, epigenetics has recently emerged as a key factor contributing to the development of chronic diseases.[8]
Epigenetics can affect gene activity independent of the DNA sequence and is crucial for development and cellular differentiation.[9,10] Epigenetic alterations induced by environmental stimuli may influence gene expression and may be transmitted across generations, although the mechanisms of epigenetic inheritance remain unclear.[7,11,12] Some of these epigenetic changes appear to be relatively stable and persist throughout life, but others are potentially reversible.[13] Subtle differences in DNA methylation patterns of some genes associated with gestational age (GA) at birth (e.g., DUOX2, TMEM176A, and CASP8) may influence long-term health outcomes.[14,15] Moreover, exposure to prenatal famine was found to lead to altered DNA methylation of genes associated with growth and metabolism (e.g., INSR and CPT1A).[16]
Several previous studies have showed that melanocortin 4 receptor (MC4R), which controls the appetite, and hepatocyte nuclear factor 4 alpha (HNF4α), which regulates lipid homeostasis, are involved in the development of obesity and metabolic-related disorders.[17–21]MC4R signaling regulates insulin sensitivity and glucose utilization.[17] In addition, MC4R activity contributes to increased fat mass by increasing the synthesis of TG and the assembly and secretion of hepatic lipoprotein in the liver.[18] Recently, decreased methylation of MC4R, with corresponding increases in gene expression, was found to lead to obesity in mice fed a high-fat diet.[19]HNF4α affects glucose and lipid metabolism and contributes to the regulation of pivotal metabolic processes in the liver.[20]HNF4α alteration induces impaired insulin signaling, resulting in increased lipid profiles.[21] In animals, maternal chronic exposure to a high-fat diet induced increased HNF4α expression in the fetal liver and contributed to lipid accumulation.[22] In humans, differences in DNA methylation patterns of HNF4α in response to the intrauterine environment have been identified between twins with and without T2D.[23] Most PTB studies have analyzed DNA methylation using cell-free DNA, which can enable the discovery of biomarkers for prediction due to its accessibility and reliability.[24,25] An epigenome-wide association study identified some differentially methylated loci, including the previously known HNF4a, in cord blood of infants with intrauterine growth restriction.[26] On the basis of this evidence, MC4R and HNF4α are plausible biological candidate genes underlying the associations between PTB and metabolic-related disorders.
To our knowledge, no study has found a link between epigenetic alterations of metabolic-related genes and metabolic indices in cord blood. Thus, we investigated the associations between DNA methylations of MC4R and HNF4a and metabolic indices in cord blood of preterm infants, compared with term infants.
2. Methods
2.1. Study population
This case–control study was conducted from 2007 to 2010 at Ewha Womans University Hospital, Seoul, Korea. Pregnant women who received prenatal examinations between 24 and 28 weeks of gestation at the hospital were recruited. The study population included women aged 20 to 42 years, with GAs of 25 to 42 weeks and no complication during pregnancy. A total of 85 preterm infants (GA < 37 weeks) were delivered at the hospital, and 85 term infants (GA 37–42 weeks) were selected randomly from women who delivered healthy infants during the same period as preterm infants. We excluded infants from multiple births and those with major congenital abnormalities or medical problems. All subjects provided clinical data and cord blood samples. Participants gave their written informed consent prior to enrollment in the study. This study was approved by the Institutional Review Board of the Ewha Womans University Hospital.
Trained nurses collected information regarding potential confounding factors, including maternal age, height, weight at delivery, education, mode of delivery, GA, birth weight, height, ponderal index, and Apgar score (AS) 1 and 5 minutes after birth using medical charts. GA was calculated based on the onset of the last menstrual period (LMP). Ultrasonographic estimation of the GA was used when the LMP data were unreliable or when the GA determined by LMP and true fetal size differed by >10 days. The ponderal index was calculated as weight in kilograms divided by cube of height in meters (kg/m3). Maternal education level was dichotomized as high school (<12 years) and university or higher (≥12 years).
2.2. Biochemical assessments
Umbilical cord blood samples were collected into Vacutainer® tubes containing EDTA (BD Biosciences, San Jose, CA) immediately after delivery. The blood samples were centrifuged at 3000 rpm for 10 minutes, and the serum was stored at −80 °C until chemical analysis. Serum glucose, TG, TC, high-density lipoprotein cholesterol (HDL-c), and low-density lipoprotein cholesterol (LDL-c) were measured by automatic analyzer (Model 7180; Hitachi, Tokyo, Japan). The average intraassay coefficients of variation (CVs) and sensitivity limits for the glucose, TG, TC, HDL-c, and LDL-c assays were 1.15% and 2 mg/dL, 1.85% and 9 mg/dL, 1.43% and 4 mg/dL, 2.51% and 3 mg/dL, and 2.71% and 4 mg/dL, respectively. Insulin was analyzed using an immunoradiometric assay kit (Biosource Europe, Nivelles, Belgium) according to the manufacturer's instructions, with an average intraassay CV of 2.40% and a sensitivity limit of 0.2 μU/mL. Insulin resistance levels were determined by the widely used homeostasis model assessment of insulin resistance (HOMA-IR) method, which was calculated as follows: (plasma glucose [mmol/L] × insulin [μU/mL])/22.5.
2.3. DNA methylation analysis by pyrosequencing
Genomic DNA samples were isolated from whole cord blood using the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA), according to the manufacturer's protocol. The total DNA concentration and purity were assessed by spectrophotometer (ND2000; Nanodrop Technologies, Wilmington, DE).
The promoter regions of the MC4R and HNF4α genes were amplified by a primer set designed using the PSQ Assay Design software (Biotage AB, Uppsala, Sweden) (Supplemental Digital Content-Table S1). In MC4R, the 3 CpG sites are located in the promoter region up to 775 to 792 bp upstream of the transcription start site (TSS). The HNF4α gene has 2 promoters (P1 and P2). The 4 CpG sites in the P1 promoter region are located up to 88 to 115 bp, and the 4 CpG sites in the P2 promoter region are located up to 44,903 to 44,920 bp upstream of the TSS (Supplemental Digital Content-Figure S1 and Table S3). Of these, the CpG2 site in the P2 promoter of HNF4α was almost always methylated (>99%). Thus, it was excluded from this study. Methodological details of DNA methylation analysis are reported in our previous study.[27]
2.4. RNA isolation and quantitative real-time PCR
Total RNA was extracted from blood samples of term (n = 38) and preterm (n = 20) infants using the Easy-BLUETM total RNA extraction kit (iNtRON Biotechnology, Sungnam, Korea) according to the manufacturer's protocol. The concentrations and purities of extracted RNA samples were quantitated by bioanalyzer (Agilent Technologies, Santa Clara, CA). Complementary DNA (cDNA) synthesis was performed using 1 μg RNA sample in a 25-μL reaction mixture containing 1 μL of 10 pM oligonucleotide primer, 5 μL 10 × RT buffer, 5 μL 2.5 mM dNTP, 1 μL 40 U RNase inhibitor, and 1 μL 200 U Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) for 60 minutes at 42 °C and 5 minutes at 95 °C. Quantitative real-time polymerase chain reaction (PCR) was carried out in a 20-μL reaction mixture containing 2 μL cDNA, 10 μL SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 1 μL each 10-pM primer, and 6 μL sterile water. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene, was used as an internal control. Amplification was conducted under the following conditions: denaturation at 95 °C for 30 seconds, followed by 40 cycles of denaturation at 95 °C for 15 seconds and annealing/extension at 60 °C for 1 minutes on an ABI PRISM 7000 sequence detection system (Applied Biosystems). All PCR reactions were performed in duplicate, and no template controls were included in any run. We then tested primer specificity by RT-PCR, confirmed using melting curve analysis. Comparative quantitation of each target gene was performed based on the cycle threshold (CT), which was normalized against the CT of GAPDH using the ΔΔCT method. Primer sets and melting temperature for quantitative real-time PCR are described in Supplemental Digital Content-Table S2.
2.5. Statistical analysis
Continuous variables with normal distributions were assessed using Student t test or analysis of variance, followed by the Bonferroni test, and were expressed as means and standard deviations (SD). Categorical variables with nonnormal distributions were assessed using the Wilcoxon rank sum test and expressed as numbers and percentages. To satisfy statistical assumptions for normality, TG, TC, HDL-c, LDL-c, insulin levels, and HOMA-IR data were log transformed. The results for log-transformed data are presented as back-transformed means with 95% confidence intervals. DNA methylation levels of MC4R and HNF4α genes were rescaled using the means and SD to satisfy normality. TG levels were grouped by percentile as low (<10th percentile), mid (11th–89th percentiles), and high (>90th percentile), according to a previous study.[28]
To assess the significant differential effects of DNA methylation statuses of MC4R and HNF4α at each CpG sites on metabolic indices in cord blood between term and preterm infants, a multiple linear regression model was used. The model controlled for maternal height, weight at delivery, education, mode of delivery, GA, ponderal index, and 5-minute AS. The selected confounding variables were marginally related to DNA methylation levels or metabolic indices in all univariate analyses (P < 0.20). Statistical analyses were conducted using the SAS software (ver. 9.3; SAS Institute Inc., Cary, NC). All analyses were 2-tailed, and P values < 0.05 were considered to indicate statistical significance.
3. Results
3.1. General characteristics of study subjects
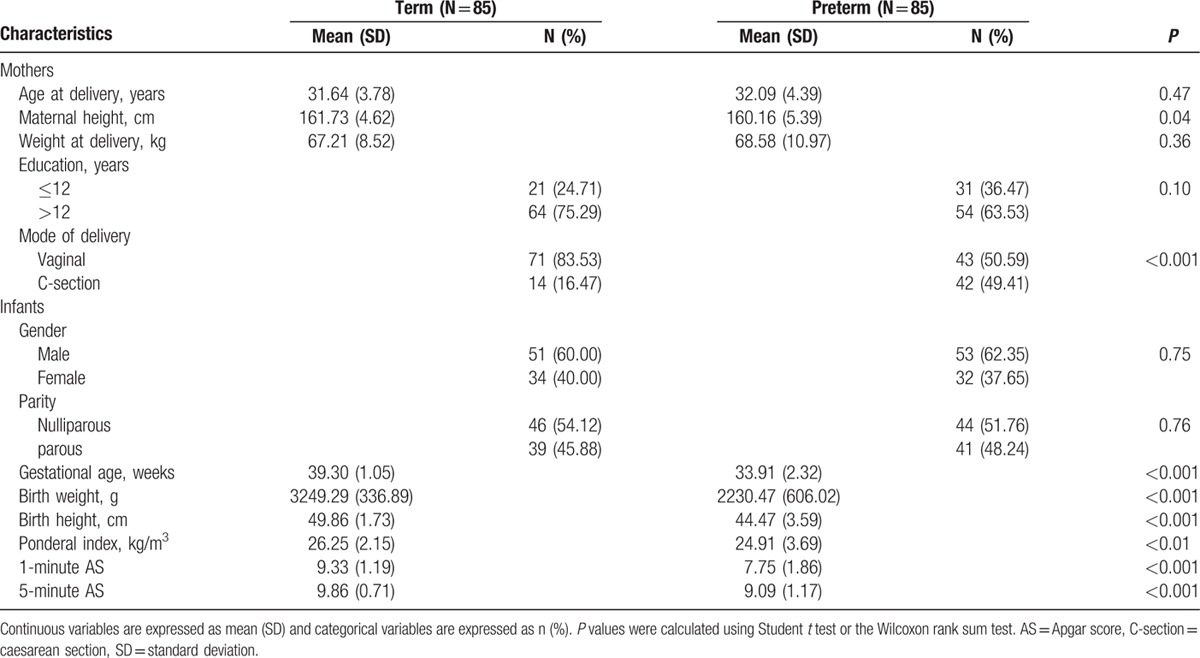
Eighty-five term and 85 preterm infants were included in this study (Table 1). The 2 groups were similar in maternal age, gender, and parity. However, women who had preterm infants were markedly shorter (P = 0.04) and had a higher occurrence of cesarean section (P < 0.001) than women who had term infants.
Table 1.
Demographic characteristics of study subjects.
3.2. Quantitation of DNA methylation and mRNA expression levels in cord blood cells
We measured DNA methylation levels in cord blood cells at 3 CpG sites in the MC4R promoter, and at 7 CpG sites in the 2 HNF4α promoter regions (Supplemental Digital Content-Table S3). To compare differences in methylation patterns in cord blood cells between term and preterm infants, we analyzed the average DNA methylation levels of each CpG sites in the MC4R and HNF4α genes. The median methylation status of the CpG2 and CpG3 sites located within the promoter of MC4R (MC4R-CpG2 and MC4R-CpG3, respectively) was significantly lower in cord blood cells of preterm infants compared with term infants (P < 0.01). In addition, the median methylation status of HNF4α at 4 CpG sites in the P1 promoter was significantly lower in cord blood cells of preterm infants (P < 0.01), whereas the methylation status of the CpG sites within the P2 promoter was similar between the 2 groups (Supplemental Digital Content-Table S4). The MC4R and HNF4α promoter regions contain putative binding sites for transcription factors, including C/EBP alpha and SP1, which are involved in the regulation of lipid metabolism (Supplemental Digital Content-Figure S1).[29,30] The expression levels of MC4R and HNF4α genes were significantly increased in cord blood cells of preterm infants compared with term infants (P = 0.04 and P = 0.048, respectively; Fig. 1).
Figure 1.
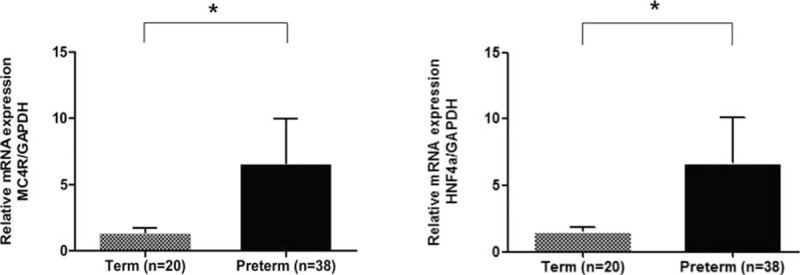
The mRNA expression levels of the MC4R (A) and HNF4α (B) genes in cord blood of preterm infants (n = 20) and term infants (n = 35). MC4R = melanocortin 4 receptor, HNF4α = hepatocyte nuclear factor 4 alpha.
3.3. Metabolic profiles in cord blood from term and preterm infants
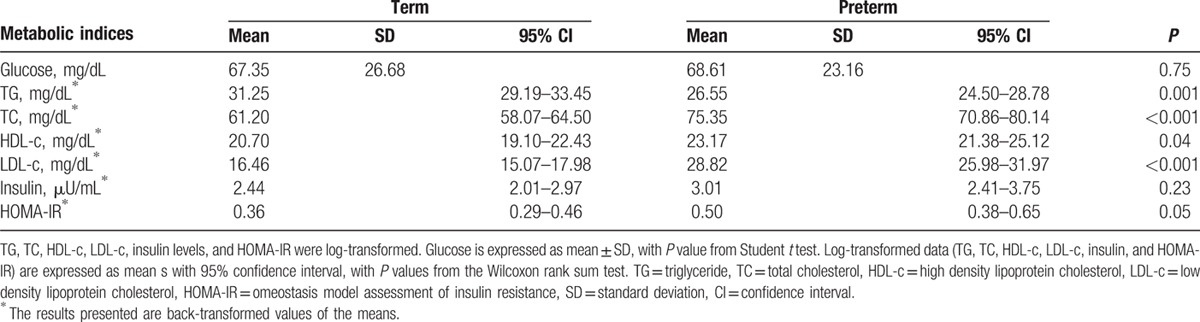
Table 2 shows the average metabolic indices in cord blood of 2 groups. The mean TG level was significantly lower (P = 0.001), whereas the average TC, HDL-c, LDL-c levels, and HOMA-IR were significantly higher in cord blood of preterm infants compared with term infants (P < 0.05). No significant difference in other metabolic indices, such as glucose and insulin levels, was observed between groups.
Table 2.
Metabolic indices in cord blood of study subjects.
3.4. Associations between DNA methylation status of CpGs and metabolic indices in cord blood of term and preterm infants
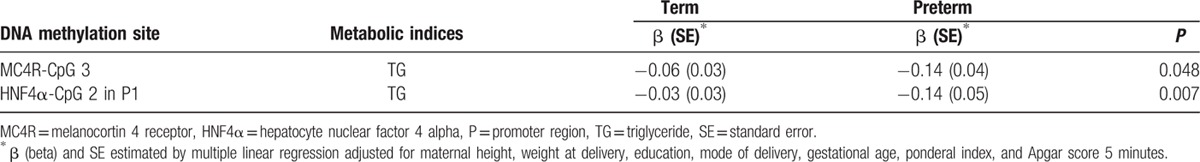
To evaluate the effects of different DNA methylation patterns on metabolic indices between groups, we identified associations between methylation statuses of CpG sites in MC4R and HNF4α and metabolic indices in cord blood of term and preterm infants. We selected 6 CpG sites and 5 metabolic indices that differed significantly between groups. The beta coefficients for associations between log-TG levels and methylation statuses of the MC4R-CpG3 and the CpG2 site located within the P1 promoter of HNF4α (HNF4α-CpG2) in cord blood differed significantly between term and preterm infants (P = 0.04 for MC4R and P = 0.003 for HNF4α; Fig. 2). These associations remained unchanged after adjusting for maternal height, weight at delivery, education, mode of delivery, GA, ponderal index, and 5-minure AS (Table 3).
Figure 2.
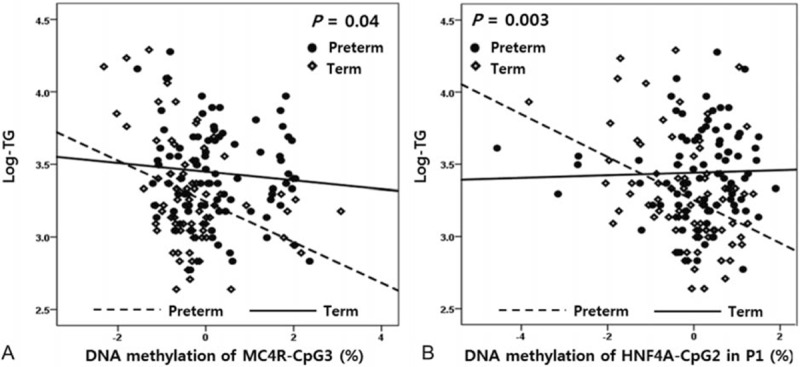
Association between methylation statuses of MC4R and HNF4α at CpG sites and metabolic indices in cord blood of preterm and term infants. DNA methylation of (A) MC4R-CpG3 and TG levels and (B) HNF4α-CpG2 in the P1 promoter and TG levels. Open lines ( - - ) denote preterm infants and closed lines (—) denote term infants. MC4R = melanocortin 4 receptor, HNF4α = hepatocyte nuclear factor 4 alpha, TG = triglyceride.
Table 3.
Associations between methylation status of MC4R and HNF4α at CpG sites and metabolic indices in cord blood of term and preterm infants.
3.5. DNA methylation levels of CpG sites in cord blood cells from term and preterm infants according to TG group
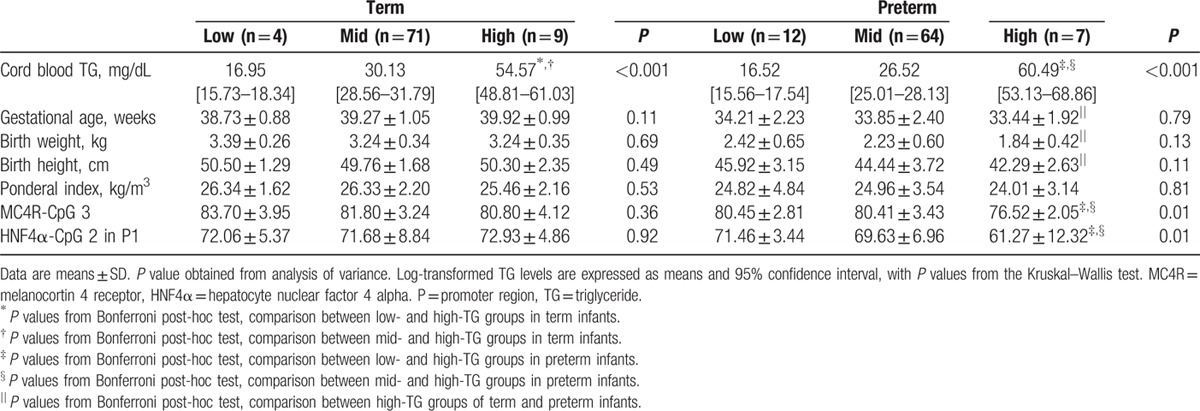
DNA methylation levels of MC4R-CpG3 and HNF4α–CpG2 in the P1 promoter differed significantly among preterm infants according to TG group (P = 0.01; Fig. 3). Methylation statuses of MC4R-CpG3 and HNF4α–CpG2 in the P1 promoter were significantly lower in preterm infants in the high-TG group than in those in the low-TG group (P = 0.04 for MC4R and P = 0.01 for HNF4α). In addition, methylation statuses of MC4R-CpG3 and HNF4α–CpG2 in the P1 were significantly lower in preterm infants in the high-TG group compared with the mid-TG group (both P = 0.01). Notably, preterm infants in the high-TG group were found to have higher TG levels in cord blood than term infants in the high-TG group (60.49 vs 54.57 mg/dL; Table 4). These preterm infants also had significantly lower GA, birth weight, and height than term infants of the high-TG group (P < 0.001). No significant difference in cord blood of term infants was observed according to TG group, except TG level.
Figure 3.
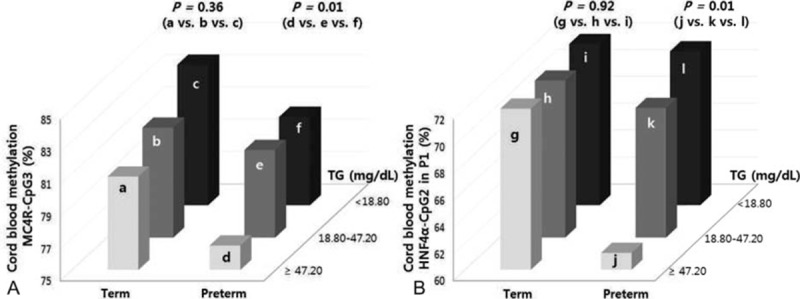
DNA methylation status by TG group of MC4R-CpG3 (A) and HNF4α-CpG2 (B) in the P1 promoter in cord blood cells from term and preterm infants. Preterm infants in the high-TG group show the lowest methylation levels of MC4R-CpG3 and HNF4α-CpG2 in the P1 promoter. P values were calculated using the Bonferroni post-hoc test. a: High-TG group of term infants in A, b: mid-TG group of term infants in A, c: low-TG group of term infants in A, d: high-TG group of preterm infants in A, e: mid-TG group of preterm infants in A, f: low-TG group of preterm infants in A, g: high-TG group of term infants in B, h: mid-TG group of term infants in B, i: low-TG group of term infants in B, j: high-TG group of preterm infants in B, k: mid-TG group of preterm infants in B, and l: low-TG group of preterm infants in B. MC4R = melanocortin 4 receptor, HNF4α = hepatocyte nuclear factor 4 alpha, TG = triglyceride.
Table 4.
DNA methylation status of CpG sites in MC4R and HNF4α in term and preterm infants by TG group.
4. Discussion
This study is the 1st to investigate the associations between DNA methylation statuses of metabolic-related genes and metabolic profiles in cord blood. Our findings showed that DNA methylations of MC4R and HNF4α were associated significantly with TG level in cord blood of preterm infants compared with term infants. Interestingly, preterm infants in the high-TG group were observed to have higher TG levels, despite lower GA and worse birth outcomes, than term infants in the high-TG group. These preterm infants could have disrupted lipid metabolism in utero, and altered lipid profiles may affect metabolic processes and disease susceptibility, which could be mediated partly through epigenetic mechanisms at specific loci.
Epidemiological studies have reported a close link between PTB and the development of metabolic disorders in later life.[31–33] Approximately 35% of patients with T2D are known to have had low birth weight.[34] Impaired fetal growth caused by an adverse prenatal environment leads to metabolic adaptations in the fetus, which could influence long-term risks of metabolic diseases. For instance, a study indicated that extremely low birth weight infants was linked strongly to high TG and low HDL-c levels in 2-year-olds.[35] Young adults who were born prematurely have been observed to have higher blood pressure and LDL-c levels than those who were born at term.[36] In addition, adults who were born preterm with very low birth weight have been shown to have high insulin and glucose levels.[37]
Our findings showed that average TG levels were significantly lower in cord blood of preterm infants compared with term infants, whereas mean TC, HDL-c, and LDL-c levels were significantly higher, consistent with previous studies.[38–41] However, some of these findings are inconsistent with our results that TG, TC, and LDL-c levels in cord blood were higher in late preterm infants than in term infants.[42] Haridas and Acharya reported that preterm infants had higher TG levels than term infants, although the difference was not statistically significant.[43] Notably, the discrepancy in TG levels between the 2 groups may be due to variations in GA (near-full term vs very preterm) of the study subjects.[44] Moreover, this difference may be due to differences in consumption of nutrients, especially fat, associated with higher birth weight.[45,46] During the last trimester of pregnancy, which is accompanied by increased GA and total body fat, TG levels increase and TC levels decrease.[39,45]
Increased TG level is a well-established major risk factor for cardiovascular disease, which is the leading cause of death in many countries. Elevated lipid profiles at birth may affect the regulation of gene expression concerning growth and cell signaling pathways, and contribute to increased risk of diseases.[47] Abnormal lipid profiles in childhood could be predictive of dyslipidemia and cardiovascular disease in adulthood.[48] In addition, altered metabolic profiles triggered by prenatal conditions may be explained partly by epigenetic mechanisms. These epigenetic alterations during prenatal development may persistently affect long-term health outcomes.[15]
Our results provided supporting evidence that higher TG levels were related significantly to decreased methylation of MC4R-CpG3 and HNF4α-CpG2 in the P1 promoter in cord blood of preterm infants, compared with term infants. MC4R, which is relevant to the regulation of energy homeostasis, was found to elevate fatty acid uptake and TG synthesis in adipose tissue.[18,49] Another study implicated that activity of MC4R affects TG levels in cardiovascular patients.[50] To date, very few studies have been conducted to investigate the association between DNA methylation of MC4R and metabolic components in cord blood, although most genetic studies have shown that common variants near MC4R were associated with elevated TG levels and an increased risk of hypertriglyceridemia in obese children and adolescents.[51] In addition, genetic variants of MC4R have been found to lead to substantially higher postprandial response of TG levels in healthy subjects.[52] The active forms of MC4R decrease the activity of stearoyl-coenzyme A desaturase 1, which is involved in TG synthesis and affects LDL levels via the liver, resulting in decreased TG synthesis.[50] Consistent with our findings, the recent study by Widiker et al[19] showed hypomethylation of MC4R and increased expression levels in mice following a high-fat diet, which resulted in obesity. Rats fed a high-fat diet had 43% higher expression levels of MC4R in the paraventricular nuclei than those fed a low-fat diet, which was associated positively with fat mass.[53]
The other gene, HNF4α, is known to be relevant to glucose and lipid metabolism.[54,55] In subjects with hyperlipidemia, variants of HNF4α were involved in the elevation of TG and glucose levels.[56] In offspring of rats fed a high-fat diet, the expression levels of HNF4α were substantially increased in the liver.[22] Many studies have shown that mutations in the P1 promoter of HNF4α are related to the risk of metabolic disorders.[57,58] In a recent study, changes in DNA methylation of CpG sites, including in the known HNF4α, were identified in subjects with T2D.[23] Collectively, we suggest that increased TG levels are associated closely with DNA methylation status of the MC4R and HNF4α loci in cord blood cells.
Of note, our findings identified higher TG levels in high-TG preterm infants compared with term infants, whereas DNA methylation levels of MC4R-CpG3 and HNF4α-CpG2 in the P1 promoter in cord blood cells were significantly decreased in comparison with those with other TG levels. These high-TG preterm infants also had significantly lower GA and birth weight than term infants of the high-TG group. However, no significant association was found in term infants with high TG levels. Cress et al[59] reported that TG levels >65 mg/dL in cord blood were related to fetal distress and adverse perinatal environment. On the basis of this evidence, we suggest that preterm infants with high TG levels may be at greater risk of metabolic-related disorders, although longitudinal studies are needed to clarify these findings.
In this study, we found the associations between DNA methylation of MC4R and HNF4α and metabolic components in cord blood, and differences between term and preterm infants, although in a relatively small sample. Further studies are needed to confirm the differences in methylation of metabolic-related genes as potential contributing factors for the development of metabolic-related diseases, and long-term follow-up studies are also needed.
DNA methylation patterns have tissue and cell-type specificity. However, many studies of DNA methylation have been performed on DNA extracted from whole blood, which contains a heterogeneous cell population; the use of cell-free circulating DNA could enable the discovery of accurate biomarkers due to its accessibility.[24,25,60] Recently, an altered DNA methylation status of HNF4α induced by the intrauterine environment was identified in cord blood from neonates with intrauterine growth restriction.[26]
Overall, this evidence indicates that epigenetic variations of MC4R and HNF4α established in early life may lead to altered metabolic profiles, such as increased TG levels, in cord blood of preterm infants. In particular, high TG levels in preterm infants may represent a greater risk of metabolic-related diseases in adulthood. Particular attention is required for sensitive subjects, such as babies born prematurely, especially those with increased TG levels, although the values are within normal ranges. Additional long-term follow-up studies are needed to gain further information regarding DNA methylation and metabolic components during each period of life.
Acknowledgments
The authors thank Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2013R1A1A2004833) and Ministry of Health & Welfare, Republic of Korea (HI14C0306) through the Korea Health Industry Development Institute (KHIDI) for the support.
Supplementary Material
Footnotes
Abbreviations: AS = Apgar score, GA = gestational age, HDL-c = high-density lipoprotein cholesterol, HNF4α = hepatocyte nuclear factor 4 alpha, HOMA-IR = homeostasis model assessment of insulin resistance, LDL-c = low-density lipoprotein cholesterol, LMP = last menstrual period, MC4R = melanocortin 4 receptor, PTB = preterm birth, T2D = type 2 diabetes, TC = total cholesterol, TG = triglyceride, TSS = transcription start site.
Funding/support: This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF- 2013R1A1A2004833) and Ministry of Health & Welfare, Republic of Korea (HI14C0306) through the Korea Health Industry Development Institute (KHIDI).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- 1.Beck S, Wojdyla D, Say L, et al. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ 2010; 88:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawn JE, Wilczynska-Ketende K, Couspens SN. Estimating the causes of 4 million neonatal deaths in the year 2000. Int J Epidemiol 2006; 35:706–718. [DOI] [PubMed] [Google Scholar]
- 3.Lim JW. The changing trends in live birth statistics in Korea, 1970 to 2010. Korean J Pediatr 2011; 54:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawn JE, Cousens SN, Darmstadt GL, et al. 1 year after The Lancet Neonatal Survival Series–was the call for action heard? Lancet 2006; 367:1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evensen KA, Steinshamn S, Tjonna AE, et al. Effects of preterm birth and fetal growth retardation on cardiovascular risk factors in young adulthood. Early Hum Dev 2009; 85:239–245. [DOI] [PubMed] [Google Scholar]
- 6.Singhal A, Fewtrell M, Cole TJ, et al. Low nutrient intake and early growth for later insulin resistance in adolescents born preterm. Lancet 2003; 361:1089–1097. [DOI] [PubMed] [Google Scholar]
- 7.Menon R, Conneely KN, Smith AK. DNA methylation: an epigenetic risk factor in preterm birth. Reprod Sci 2012; 19:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Cerjak D, Ali O. Finding epigenetic determinants of the syndrome. Austin J Endocrinol Diabetes 2014; 1:1029. [Google Scholar]
- 9.Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J Pathol 2001; 195:97–110. [DOI] [PubMed] [Google Scholar]
- 10.Ginder GD, Gnanapragasam MN, Mian OY. The role of the epigenetic signal, DNA methylation, in gene regulation during erythroid development. Curr Top Dev Biol 2008; 82:85–116. [DOI] [PubMed] [Google Scholar]
- 11.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 2007; 27:363–388. [DOI] [PubMed] [Google Scholar]
- 12.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 2014; 157:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop KS, Ferguson LR. The interaction between epigenetics, nutrition and the development of cancer. Nutrients 2015; 7:922–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder JW, Conneely KN, Cubells JC, et al. Neonatal DNA methylation patterns associate with gestational age. Epigenetics 2011; 6:1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parets SE, Conneely KN, Kilaru V, et al. Fetal DNA methylation associates with early spontaneous preterm birth and gestational age. PLoS One 2013; 8:e67489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobi EW, Goeman JJ, Monajemi R, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 2014; 5:5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 2008; 40:716–718. [DOI] [PubMed] [Google Scholar]
- 18.Nogueiras R, Wiedmer P, Perez-Tilve D, et al. The central melanocortin system directly controls peripheral lipid metabolism. J Clin Invest 2007; 117:3475–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widiker S, Karst S, Wagener A, et al. High-fat diet leads to a decreased methylation of the Mc4r gene in the obese BFMI and the lean B6 mouse lines. J Appl Genet 2010; 51:193–197. [DOI] [PubMed] [Google Scholar]
- 20.Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci U S A 1997; 94:13209–13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miura A, Yamagata K, Kakei M, et al. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J Biol Chem 2006; 281:5246–5257. [DOI] [PubMed] [Google Scholar]
- 22.McCurdy CE, Bishop JM, Williams SM, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest 2009; 119:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One 2012; 7:e51302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levenson VV. DNA methylation as a universal biomarker. Expert Rev Mol Diagn 2010; 10:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parets SE, Bedient CE, Menon R, et al. Preterm birth and its long-term effects: methylation to mechanisms. Biology 2014; 3:498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Einstein F, Thompson RF, Bhagat TD, et al. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS One 2010; 5:e8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo JY, Lee S, Lee HA, et al. Can proopiomelanocortin methylation be used as an early predictor of metabolic syndrome? Diabetes Care 2014; 37:734–739. [DOI] [PubMed] [Google Scholar]
- 28.Genest J, Jr, McNamara JR, Ordovas JM, et al. Lipoprotein cholesterol, apolipoprotein A-I and B and lipoprotein (a) abnormalities in men with premature coronary artery disease. J Am Coll Cardiol 1992; 19:792–802. [DOI] [PubMed] [Google Scholar]
- 29.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev 2006; 86:465–514. [DOI] [PubMed] [Google Scholar]
- 30.Blondet A, Gout J, Durand P, et al. Expression of the human melanocortin-4 receptor gene is controlled by several members of the Sp transcription factor family. J Mol Endocrinol 2005; 34:317–329. [DOI] [PubMed] [Google Scholar]
- 31.Burdge GC, Hanson MA, Slater-Jefferies JL, et al. Epigenetic regulation of transcription: a mechanism for inducing variations in phenotype (fetal programming) by differences in nutrition during early life? Br J Nutr 2007; 97:1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson JR, Hyde MJ, Gale C, et al. Preterm birth and the metabolic syndrome in adult life: a systematic review and meta-analysis. Pediatrics 2013; 131:e1240–e1263. [DOI] [PubMed] [Google Scholar]
- 33.Desai M, Jellyman JK, Ross MG. Epigenomics, gestational programming and risk of metabolic syndrome. Int J Obes 2015; 39:633–641. [DOI] [PubMed] [Google Scholar]
- 34.Boyko EJ. Proportion of type 2 diabetes cases resulting from impaired fetal growth. Diabetes Care 2000; 23:1260–1264. [DOI] [PubMed] [Google Scholar]
- 35.de Jong M, Lafeber HN, Cranendonk A, et al. Components of the metabolic syndrome in early childhood in very-lowbirth-weight infants. Horm Res Paediatr 2014; 81:43–49. [DOI] [PubMed] [Google Scholar]
- 36.Sipola-Leppänen M, Vääräsmäki M, Tikanmäki M, et al. Cardiometabolic risk factors in young adults who were born preterm. Am J Epidemiol 2015; 181:861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hovi P, Andersson S, Eriksson JG, et al. Glucose regulation in young adults with very low birth weight. N Engl J Med 2007; 356:2053–2063. [DOI] [PubMed] [Google Scholar]
- 38.Donegá S, Oba J, Maranhão RC. Concentration of serum lipids and apolipoprotein B in newborns. Arq Bras Cardiol 2006; 86:419–424. [DOI] [PubMed] [Google Scholar]
- 39.Bansal N, Cruickshank JK, McElduff P, et al. Cord blood lipoproteins and prenatal influences. Curr Opin Lipidol 2005; 16:400–408. [DOI] [PubMed] [Google Scholar]
- 40.Nagano N, Okada T, Yonezawa R, et al. Early postnatal changes of lipoprotein subclass profile in late preterm infants. Clin Chim Acta 2012; 413:109–112. [DOI] [PubMed] [Google Scholar]
- 41.Kelishadi R, Badiee Z, Adeli K. Cord blood lipid profile and associated factors: baseline data of a birth cohort study. Paediatr Perinat Epidemiol 2007; 21:518–524. [DOI] [PubMed] [Google Scholar]
- 42.Ghaemi S, Najafi R, Kelishadi R. Cord blood lipoprotein profile in term, preterm, and late preterm newborns. J Res Med Sci 2014; 19:1038–1040. [PMC free article] [PubMed] [Google Scholar]
- 43.Haridas N, Acharya PT. Serum lipid status in neonates. Indian Pediatr 1984; 21:327–334. [PubMed] [Google Scholar]
- 44.Bastida S, Cuesta C, Perea S, et al. Lipid and lipoprotein changes throughout the term-period in neonates from the Toledo Study. Rev Esp Fisiol 1996; 52:23–29. [PubMed] [Google Scholar]
- 45.Lane DM, McConathy WJ. Factors affecting the lipid and apolipoprotein levels of cord sera. Pediatr Res 1983; 17:83–91. [DOI] [PubMed] [Google Scholar]
- 46.Radunovic N, Kuczynski E, Rosen T, et al. Plasma apolipoprotein A-I and B concentrations in growth-retarded fetuses: a link between low birth weight and adult atherosclerosis. J Clin Endocrinol Metab 2000; 85:85–88. [DOI] [PubMed] [Google Scholar]
- 47.Laker RC, Wlodek ME, Connelly JJ, et al. Epigenetic origins of metabolic disease: the impact of the maternal condition to the offspring epigenome and later health consequences. Food Sci Hum Wellness 2013; 2:1–11. [Google Scholar]
- 48.Albisetti M, Chan AK, McCrindle BW, et al. Impaired fibrinolytic activity is present in children with dyslipidemias. Pediatr Res 2004; 55:576–580. [DOI] [PubMed] [Google Scholar]
- 49.Fan W, Boston BA, Kesterson RA, et al. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature 1997; 385:165–168. [DOI] [PubMed] [Google Scholar]
- 50.Brönner G, Sattler AM, Hinney A, et al. The 103I variant of the melanocortin 4 receptor is associated with low serum triglyceride levels. J Clin Endocrinol Metab 2006; 91:535–538. [DOI] [PubMed] [Google Scholar]
- 51.Fernandes AE, de Melo ME, Fujiwara CT, et al. Associations between a common variant near the MC4R gene and serum triglyceride levels in an obese pediatric cohort. Endocrine 2015; 49:653–658. [DOI] [PubMed] [Google Scholar]
- 52.Perez-Martinez P, Garcia-Rios A, Delgado-Lista J, et al. A variant near the melanocortin-4 receptor gene regulates postprandial lipid metabolism in a healthy Caucasian population. Br J Nutr 2011; 106:468–471. [DOI] [PubMed] [Google Scholar]
- 53.Huang XF, Han M, South T, et al. Altered levels of POMC, AgRP and MC4-R mRNA expression in the hypothalamus and other parts of the limbic system of mice prone or resistant to chronic high-energy diet-induced obesity. Brain Res 2003; 992:9–19. [DOI] [PubMed] [Google Scholar]
- 54.Hayhurst GP, Lee YH, Lambert G, et al. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001; 21:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamagata K, Furuta H, Oda N, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 1996; 384:458–460. [DOI] [PubMed] [Google Scholar]
- 56.Weissglas-Volkov D, Huertas-Vazquez A, Suviolahti E, et al. Common hepatic nuclear factor-4alpha variants are associated with high serum lipid levels and the metabolic syndrome. Diabetes 2006; 55:1970–1977. [DOI] [PubMed] [Google Scholar]
- 57.Marcil V, Amre D, Seidman EG, et al. Hepatocyte nuclear factor 4 alpha polymorphisms and the metabolic syndrome in French-Canadian youth. PLoS One 2015; 10:e0117238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonnycastle LL, Willer CJ, Conneely KN, et al. Common variants in maturity-onset diabetes of the young genes contribute to risk of type 2 diabetes in Finns. Diabetes 2006; 55:2534–2540. [DOI] [PubMed] [Google Scholar]
- 59.Cress HR, Shaher RM, Laffin R, et al. Cord blood hyperlipoproteinemia and perinatal stress. Pediatr Res 1977; 11:19–23. [PubMed] [Google Scholar]
- 60.Jacoby M, Gohrbandt S, Clausse V, et al. Interindividual variability and co-regulation of DNA methylation differ among blood cell populations. Epigenetics 2012; 7:1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


