Abstract
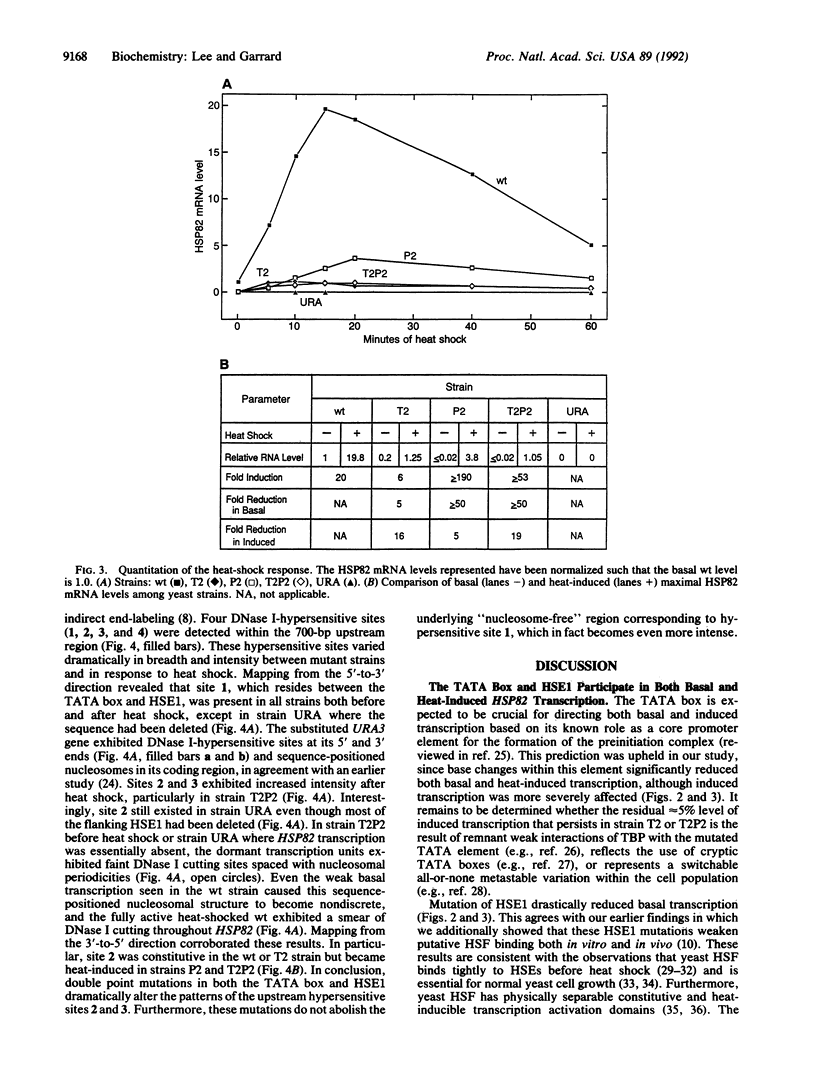
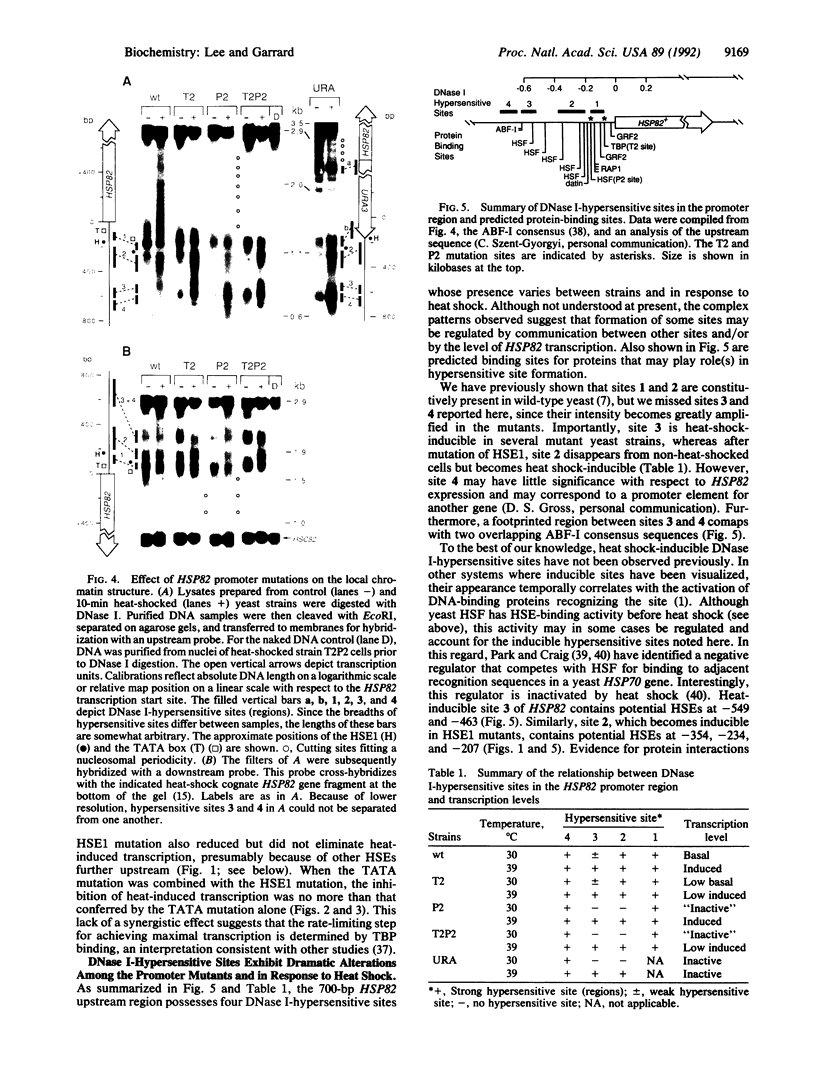
DNase I-hypersensitive sites represent "nucleosome-free" regions in chromatin where the underlying DNA sequence is highly accessible to trans-acting proteins. Here we demonstrate that it is possible to uncouple gene activity from hypersensitive site formation. Point or substitution mutations were introduced into the promoter of the yeast chromosomal HSP82 gene, encoding the 83-kDa heat shock protein (HSP), via site-directed integration. Mutating either the TATA box or heat shock element 1 (HSE1) significantly reduced basal and heat-induced transcription while mutating both essentially inactivated expression. Dormant transcription units exhibited arrays of sequence-positioned nucleosomes; nevertheless, the inactivated genes still retained a hypersensitive site within their mutated promoters. In addition, all yeast strains maintained a heat-inducible hypersensitive site at -600 base pairs (bp), while several mutant strains converted a constitutive hypersensitive site at -300 bp into a heat-inducible one. Thus, mutations in cis-acting elements within a promoter can inactivate transcription without eliminating nucleosome-free regions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amin J., Ananthan J., Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988 Sep;8(9):3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989 Sep;9(9):3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Lue N. F., Buchman A. R., LaPointe J. W., Lorch Y., Kornberg R. D. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 1990 Apr;4(4):503–514. doi: 10.1101/gad.4.4.503. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan J., Manley J. L. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 1992 Feb;6(2):304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne D. C. Regulation of genes controlling synthesis of the galactose pathway enzymes in yeast. Genetics. 1966 Sep;54(3):911–916. doi: 10.1093/genetics/54.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fascher K. D., Schmitz J., Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990 Aug;9(8):2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Finkelstein D. B., Strausberg S. Heat shock-regulated production of Escherichia coli beta-galactosidase in Saccharomyces cerevisiae. Mol Cell Biol. 1983 Sep;3(9):1625–1633. doi: 10.1128/mcb.3.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990 Nov 16;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Gross D. S., English K. E., Collins K. W., Lee S. W. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol. 1990 Dec 5;216(3):611–631. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen B. K., Pelham H. R. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol. 1988 Nov;8(11):5040–5042. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Garrard W. T. Positive DNA supercoiling generates a chromatin conformation characteristic of highly active genes. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9675–9679. doi: 10.1073/pnas.88.21.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Garrard W. T. Transcription-induced nucleosome 'splitting': an underlying structure for DNase I sensitive chromatin. EMBO J. 1991 Mar;10(3):607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Z., Sherman F. Two types of TATA elements for the CYC1 gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):666–676. doi: 10.1128/mcb.11.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Hopper J. E. The relationship of regulatory proteins and DNase I hypersensitive sites in the yeast GAL1-10 genes. Nucleic Acids Res. 1985 Dec 9;13(23):8409–8423. doi: 10.1093/nar/13.23.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Buchman A. R., Kornberg R. D. Activation of yeast RNA polymerase II transcription by a thymidine-rich upstream element in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(2):486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel D., Caplan A. J., Lee M. S., Adams C. C., Fishel B. R., Gross D. S., Garrard W. T. Basal-level expression of the yeast HSP82 gene requires a heat shock regulatory element. Mol Cell Biol. 1989 Nov;9(11):4789–4798. doi: 10.1128/mcb.9.11.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Sotelo J., Wiederrecht G., Okuda A., Parker C. S. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell. 1990 Aug 24;62(4):807–817. doi: 10.1016/0092-8674(90)90124-w. [DOI] [PubMed] [Google Scholar]
- Park H. O., Craig E. A. Positive and negative regulation of basal expression of a yeast HSP70 gene. Mol Cell Biol. 1989 May;9(5):2025–2033. doi: 10.1128/mcb.9.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. O., Craig E. A. Transcriptional regulation of a yeast HSP70 gene by heat shock factor and an upstream repression site-binding factor. Genes Dev. 1991 Jul;5(7):1299–1308. doi: 10.1101/gad.5.7.1299. [DOI] [PubMed] [Google Scholar]
- Pavlović B., Hörz W. The chromatin structure at the promoter of a glyceraldehyde phosphate dehydrogenase gene from Saccharomyces cerevisiae reflects its functional state. Mol Cell Biol. 1988 Dec;8(12):5513–5520. doi: 10.1128/mcb.8.12.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. E. In vivo DNA-binding properties of a yeast transcription activator protein. Mol Cell Biol. 1987 Sep;7(9):3260–3267. doi: 10.1128/mcb.7.9.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Sorger P. K. Heat shock factor and the heat shock response. Cell. 1991 May 3;65(3):363–366. doi: 10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Lewis M. J., Pelham H. R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987 Sep 3;329(6134):81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 1987 Oct;6(10):3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988 Sep 9;54(6):855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Sorger P. K. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990 Aug 24;62(4):793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Györgyi C., Finkelstein D. B., Garrard W. T. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987 Jan 5;193(1):71–80. doi: 10.1016/0022-2836(87)90628-0. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Thomas G. H., Elgin S. C. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988 Jul;7(7):2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J. M. Regulatory DNA-binding proteins in yeast: an overview. Yeast. 1990 Jul-Aug;6(4):271–297. doi: 10.1002/yea.320060402. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Seto D., Parker C. S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988 Sep 9;54(6):841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- Winter E., Varshavsky A. A DNA binding protein that recognizes oligo(dA).oligo(dT) tracts. EMBO J. 1989 Jun;8(6):1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Xiao H., Lis J. T. Germline transformation used to define key features of heat-shock response elements. Science. 1988 Mar 4;239(4844):1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- Zimarino V., Tsai C., Wu C. Complex modes of heat shock factor activation. Mol Cell Biol. 1990 Feb;10(2):752–759. doi: 10.1128/mcb.10.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]





