Abstract
In eukaryotes the regulation of gene expression plays a key role in controlling cell cycle progression. Here, we demonstrate that a forkhead transcription factor, Fkh2, regulates the periodic expression of cdc15+ and spo12+ in the M and G1 phases of the cell division cycle in the fission yeast Schizosaccharomyces pombe. We also show that Fkh2 is important for several cell cycle processes, including cell morphology and cell separation, nuclear structure and migration, and mitotic spindle function. We find that the expression of fkh2+ is itself regulated in a cell cycle-dependent manner in G1 coincident with the expression of cdc18+, a Cdc10-regulated gene. However, fkh2+ expression is independent of Cdc10 function. Fkh2 was found to be phosphorylated during the cell division cycle, with a timing that suggests that this posttranslational modification is important for cdc15+ and spo12+ expression. Related forkhead proteins regulate G2 and M phase-specific gene expression in the evolutionarily distant Saccharomyces cerevisiae, suggesting that these proteins play conserved roles in regulating cell cycle processes in eukaryotes.
Forkhead transcription factors in eukaryotes have been implicated in a number of cellular processes including embryogenesis, development, and the cell division cycle. Forkhead proteins contain a conserved region spanning a monomeric DNA binding (FKH) domain (for reviews, see references 26 and 31). Indeed, a core DNA consensus sequence has been identified to which many forkhead proteins bind (27). Mutations in genes encoding forkhead proteins have been linked with several pathological conditions including thyroid agenesis (11), cleft palate (11), glaucoma (38), speech development defects (30), and tumorigenesis (16, 32, 39).
In eukaryotes the periodic expression of genes plays an important role in the regulation of the cell division cycle. Global approaches in the budding yeast Saccharomyces cerevisiae have provided insight into the groups of genes that are expressed in this manner and also the transcription factors responsible for this cell cycle-dependent expression (10, 44, 47). The MBF (MluI binding factor), consisting of Mbp1 and Swi6, and the related SBF (Swi4/Swi6 cell cycle box binding factor), consisting of Swi4 and Swi6, transcription factor complexes are important for regulation of genes that are expressed early in the cell division cycle (see reference 51 for a review). In contrast, the forkhead proteins Fkh1 and Fkh2 regulate cell cycle-dependent expression of a group of genes, named the CLB2 gene cluster, that includes CLB1 and CLB2, encoding mitotic cyclins (18, 48), in the G2 and M phases (28, 29, 40, 53). Regulation of the CLB2 gene cluster involves the formation of a ternary complex between Fkh2 and the MADS-box protein, Mcm1, and the coactivator Ndd1 (28, 29, 40, 53; see references 8, 17, and 25 for reviews). The altered expression of the CLB2 gene cluster in a fkh1Δ fkh2Δ mutant is associated with defects in the structure of the mitotic spindle, nuclear migration, cell morphology, and cell separation. Recently, a single homologue of FKH1 and FKH2, CaFKH2, was identified in the pathogenic yeast Candida albicans and was shown to play a role in cellular morphogenesis (6). Moreover, the levels of several RNAs that were apparent homologues of genes regulated by Fkh1 and Fkh2 in S. cerevisiae were also affected in a CaFKH2 mutant (6).
In many cases, the molecular details underlying the cyclical transcriptional changes that occur during the cell division cycle are poorly understood. However, recent studies have revealed that the temporal activation of the Mcm1-Fkh2-Ndd1 complex involves a forkhead-associated (FHA) domain located in Fkh2. FHA domains act as phosphothreonine binding motifs that participate in regulated protein-protein interactions (reviewed in reference 15). Cell cycle-dependent phosphorylation of Ndd1 by the Cdc28-Clb2 cyclin-dependent kinase (Cdk) complex promotes interaction with the FHA domain of Fkh2 (13, 41). Fkh2 is also phosphorylated in a cell cycle-dependent manner, but the role of this modification in regulation is unknown (40).
Comparative studies between S. cerevisiae and the distantly related yeast Schizosaccharomyces pombe allow the identification of conserved mechanisms of cell cycle control. In S. pombe a transcription factor complex, MBF (DSC1), related to MBF in S. cerevisiae, that is composed of Res1/Sct1 (9, 50), Res2/Pct1 (35, 54), Rep2 (37), and Cdc10 (33), is responsible for periodic expression early in the cell division cycle. Interestingly, a forkhead transcription factor, Sep1, has also been linked to the regulation of the cell division cycle of S. pombe. In particular, sep1 mutants display defects in cell shape and cell separation (42, 45). Moreover, Sep1 is involved in regulating the periodic expression of the cdc15+ gene in the M and G1 phases (55). Recently, Anderson et al. identified a sequence element in the promoters of several genes including cdc15+ and spo12+, termed a PCB element, that confers M- and G1-phase-specific transcription (3). Furthermore, a transcription factor complex, termed PBF, whose components have not been identified, was found to bind to the PCB element. It was possible that PBF contained Sep1, but no link could be established (3).
To elucidate the mechanisms of cell cycle control of gene expression in S. pombe, we have searched the genome sequence for potential homologues of Fkh1 and Fkh2 in S. cerevisiae. Four potential forkhead-encoding genes were identified, of which two, sep1+ (see above) and mei4+, have been previously characterized. Mei4 is a meiosis-specific transcription factor which regulates the expression of the spo6+ gene (23). However, the remaining two potential forkhead-encoding genes had not been previously characterized. One of these, the open reading frame SPAC1142.08, encodes a protein with strong similarity to Fh11 from S. cerevisiae, which has been linked to the control of RNA processing, possibly by influencing the expression of genes involved in this process (14). Interestingly, the second of these, the open reading frame SPBC16G5.15c, encodes a protein that has the strongest similarity to Fkh2 of S. cerevisiae, suggesting that it may be involved in regulation of the cell division cycle. Based on this homology, we have named SPBC16G5.15c the fkh2+ gene. In this study we examined the potential role of Fkh2 in the regulation of the cell division cycle of S. pombe. We found that the fkh2+ gene is periodically expressed, with a timing similar to that of the Cdc10-regulated gene cdc18+. However, the cell cycle-dependent expression of fkh2+ occurs independently of Cdc10. We also demonstrated that alterations of the levels of Fkh2 in S. pombe cause growth defects in several cell cycle processes, similar to those observed in an fkh1Δ fkh2Δ mutant of S. cerevisiae. Consistent with these phenotypes, we found that Fkh2 is important for cell cycle-dependent gene expression in the M and G1 phases. Moreover, the Fkh2 protein was found to be regulated at the posttranslational level by cell cycle-dependent phosphorylation and by the alteration of protein levels. Collectively, these data suggest that cell cycle events in very distantly related organisms are regulated by a conserved class of forkhead transcription factor.
MATERIALS AND METHODS
Yeast strains and growth conditions.
S. pombe strains used in this study are as follows: CHP428 (h+ leu1-32 ura4-D18 his7-366 ade6-M210) and CHP429 (h− leu1-32 ura4-D18 his7-366 ade6-M216) were from C. Hoffman; NT5 (h− leu1-32 ura4-D18 ade6-M216) was from N. Jones; cdc10-C4 (h− leu1-32 cdc10-C4) and cdc25-22 (h+ cdc25-22 leu1-32 ura4-D18 ade6-M210) were from S. Whitehall; and RB1 (h+ cdc25-22 leu1-32 ura4-D18 ade6-M210 fkh2+::3xPkC::ura4+) was from this study.
All S. pombe strains were grown at 30°C unless otherwise stated. Standard procedures for transformation and growth media were used (1, 36). YE5S medium was used for nonselective growth, and minimal EMM medium was used for selective growth.
To delete the fkh2+ gene, a ura4+ disruption cassette, obtained by PCR using the oligonucleotide primers PFkh2-1 (CCGTGAACCGCTGGCAACCGGAAGGGTAAACGAATGTTGGACAACCAACAAACGAGTGTAAATAAACACACACCAACAGCACAATTGCTGTAGAAAGGCTTAGCTACAAATCCCACT) and PFkh2-2 (CCGTTTGCACAAGAATAGGTCAAAGTCGAAAATAATTAAATTTCGAAGATCATAAGAGTGGGAGAGGGATAAGATATTAAACAAGGTGAATTTGTTGGAACACCAATGTTTATAACCAAG) andplasmid pREP42 (4) as template was introduced into the 428/429 diploid. The heterozygous fkh2+/Δfkh2 diploid was then sporulated, and Δfkh2 haploids were obtained.
Strain RB1, expressing a version of the Fkh2 protein that was tagged at the C terminus with 3×PkC epitopes from the normal chromosomal locus, was created by introducing pREP42-f2Pk linearized with NdeI into cdc25-22 cells. Ura+ colonies were isolated, and tagging was confirmed by PCR and Western blotting.
Plasmids.
To create pREP41MNHfkh2+, the fkh2+ gene was amplified from genomic DNA isolated from CHP429 with oligonucleotide primers Spfkh2-7 (ACGCATGTCGACTATGACTGTTCGCAGACTCG) and Spfkh2-8 (ACGCATGTCGACTTAAGCACTACTTTTAACATTAG), digested with SalI, and ligated into SalI-digested pREP41MNH vector (12). To create pREP42-f2Pk, the C-terminal ∼800 bp of the fkh2+ gene was amplified from genomic DNA isolated from CHP429 using oligonucleotide primers P1PK (AAAACTGCAGTCCTAAAGATGTGGGCTCACC) and P2PK (CGCGGATCCAGCACTACTTTTAACATTAGATTG). The PCR product was then digested with BamHI and PstI and ligated with BamHI- and PstI-digested pREP42PkC (46).
Cell synchronization.
Cells containing the cdc25-22 allele were synchronized at the G2/M transition by shifting a mid-log-phase culture from 25°C (permissive temperature) to 36-37°C (nonpermissive temperature) for 3 to 4 h. The cultures were then shifted back to 25°C, and the growth rate and percent septation were analyzed. Alternatively, cultures were synchronized in early S phase by hydroxyurea (HU) treatment based on a previously described approach (52). HU was added (final concentration, 11 mM) to mid-log-phase cultures growing in YE5S. After a 2.5-h incubation, the cells were collected by filtration, washed with fresh medium, and finally resuspended in fresh YE5S. For fluorescence-activated cell sorter analysis, the cells were stained with propidium iodide as described previously (49). The DNA content of stained cells was then analyzed using a FACScan machine (Becton Dickinson FACScan).
Protein extraction and Western blot analysis.
Protein extracts were made essentially as previously described (46), using lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Nonidet P-40, 10 mM imidazole, 0.1% leupeptin, 0.1% pepstatin A, 1% aprotinin, 1% phenylmethylsulfonyl fluoride, 0.2% Na3VO4, 5% NaF). For samples which were to be phosphatase treated, Na3VO4 and NaF were left out of the lysis buffer. These samples were then treated with 400 U of λ phosphatase (NEB) for 15 min at 37°C prior to Western blot analysis. Protein extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE and analyzed by Western blotting using the Pk antibody (Serotec).
RNA isolation and Northern analysis.
RNA was isolated and Northern blotting was performed as described previously (19). Gene-specific probes (PCR-generated fragments or restriction fragments) were labeled with [α-32P]dCTP (3,000 Ci/mmol). The invariant his3+ transcript was used as a loading control. The oligonucleotide primer pairs for specific gene probes were as follows: cdc15+, Cdc15-F (CTTCTATTGAGCGTGAGTACG) and Cdc15-B (GTGATGATACAACTTCCGATG); htb1+, H2B-Fwd (CGGTAAGGCTCCTAGGGATACC) and H2B-Rev (GTGCGATTCGCTTACTGAGC); spo12+, SPO12-N1 (ATTCAGAATTCATATGTCAGAAACTCAAGCTGACTC) and SPO12-C1 (TTACAGAGCTCGGATCCTATTCTTTTTCTGTCTGGA), respectively. The cdc18+ and his3+ gene-specific probes have been described previously (5), and the fkh2+ gene-specific probe was the SalI-digested PCR product generated using oligonucleotide primers Spfkh2-7 and Spfkh2-8 (see above). Probed membranes were autoradiographed with Fuji medical X-ray film. Membranes were quantified using a phosphorimager (bio-imaging analyzer Fujifilm Bas-1500) and Tina 2.0 software (Raytest).
Light and fluorescence microscopy.
Unfixed cells were observed by light microscopy using a Zeiss Axioscope fluorescence microscope set up for differential interference contrast (DIC) with a 63× oil immersion objective and Axiovision digital imaging system.
Immunofluorescence of microtubules was carried out essentially as described previously (20). A 1:15 dilution of TAT1 antibody (Cancer Research UK) and a 1:50 dilution of fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibodies (Sigma) were used as specified by the manufacturers. Nuclei were observed by first spreading cells onto polylysine-coated slides, which were allowed to dry. The slides were then incubated in ice-cold methanol for 6 min and ice-cold acetone for 30 s, allowed to air dry, and finally mounted with VECTASHIELD mounting medium (Vector Laboratories Inc.) containing 4′,6-diamidino-2-phenylindole (DAPI; Sigma). To view septa, calcofluor white (Sigma) was added to cultures at a final concentration of 1 mg/ml and the cultures were incubated for 5 min at room temperature. The cells were then washed twice with water, and 5 μl of cell suspension was then spotted onto polylysine-coated slides and allowed to dry. The slides were treated with methanol and then acetone and finally mounted as described above. DAPI-, calcofluor- and FITC-treated cells were visualized by excitation at ∼365 nm (DAPI and calcofluor) or 450 to 490 nm (FITC) under a Zeiss Axioscope fluorescence microscope with a 63× oil immersion objective and Axiovision digital imaging system.
RESULTS
Fkh2 is required for normal cell growth and division in S. pombe.
Analysis of the S. pombe genome sequence database revealed the presence of four open reading frames, sep1+, mei4+, SPBC16G5.15c, and SPAC1142.08, containing homology to the 110-amino-acid forkhead DNA binding motif (FKH). Of these, the uncharacterized SPBC16G5.15c open reading frame encodes a protein that is most similar to the architecture and sequence of the Fkh2 protein, which regulates cell cycle-dependent expression of the CLB2 gene cluster in S. cerevisiae (Fig. 1A to C). Hence, we have named this gene fkh2+.
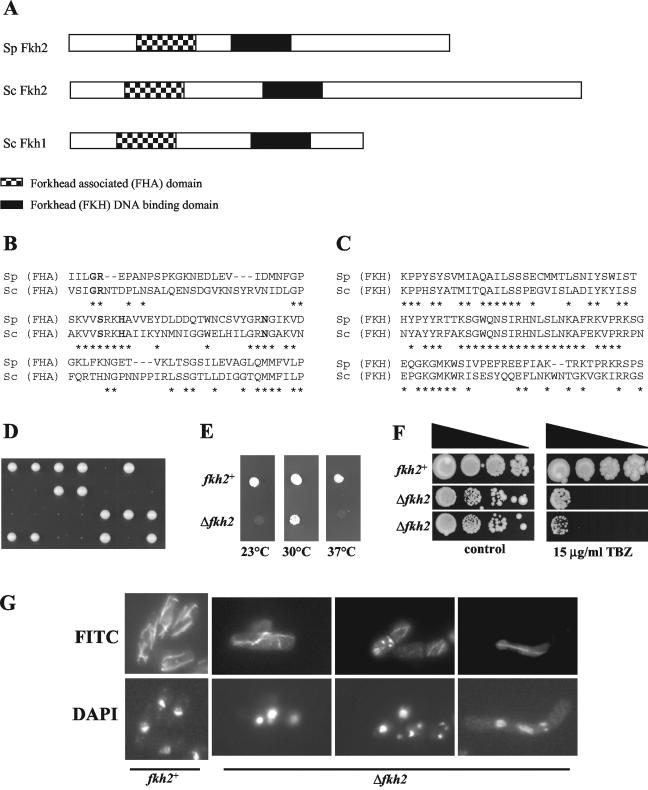
FIG. 1.
Fkh2 is important for normal growth of S. pombe. (A) Schematic representation of Fkh2 (SPBC16G5.15c) in S. pombe and Fkh1 and Fkh2 in S. cerevisiae. The positions of the predicted FKH and FHA domains are indicated. (B and C) Comparisons of the predicted FHA (B) and FKH (C) domains of Fkh2 from S. cerevisiae (Sc) and Fkh2 from S. pombe (Sp). Identical amino acids are indicated by asterisks. Invariant amino acids in FHA domains are indicated in bold type. (D) fkh2+/Δfkh2 heterozygous diploid cells were sporulated, and the resulting tetrads were dissected onto YE5S medium. Plates were photographed after 5 days of incubation at 30°C. (E) Dilutions of mid-log-phase cultures (5 μl) of the wild-type (CHP429) and Δfkh2 haploid strains were plated on YE5S medium and incubated at 23, 30, or 37°C for 3 days. (F) Serial dilutions (10-fold) of mid-log-phase cultures of the wild-type strain (CHP429) and two independent Δfkh2 haploid strains were plated on YE5S medium (control) and YE5S medium containing 15 μg of thiabendazole (TBZ) per ml and incubated at 30°C for 10 days. (G) Δfkh2 cells display aberrant microtubule structures. Cells of the wild-type (NT5) and Δfkh2 strains were grown to mid-log phase in YE5S medium. Microtubules and nuclei were visualized by immunofluorescence using the TAT1 antibody and FITC-conjugated secondary antibody (microtubules) and DAPI staining (nuclei), respectively. Some examples of aberrant Δfkh2 cells are shown.
To investigate the cellular role of Fkh2, one copy of the fkh2+ locus was replaced with the ura4+ selectable marker in a wild-type diploid strain. Sporulation and dissection of this strain generated tetrads that contained two slow-growing Ura+ colonies (Fig. 1D). Moreover, propagation of Δfkh2 haploids revealed that deletion of the fkh2+ gene affects cell growth (Fig. 1F, control plate and data not shown). Δfkh2 cells were also found to be cold sensitive and temperature sensitive (Fig. 1E) and demonstrated increased sensitivity to thiabendazole, a drug that destabilizes microtubules, causing damage to the mitotic spindle (Fig. 1F). These data suggest that the fkh2+ gene is important for normal cell growth and is required for the function of the mitotic spindle. Indeed, aberrant microtubule structures and nuclear organization were detected within a proportion of Δfkh2 cells (Fig. 1G and data not shown). Interestingly, further analysis of Δfkh2 mutant cells revealed that a large proportion (∼80%) display other aberrant phenotypes. Some examples include branched cells with phenotypes such as fragmented DNA and/or multiple septa (Fig. 2A, panel 1), cells containing misplaced nuclei (panel 2), misshapen cells with phenotypes including multiple septa and/or multiple nuclei (panel 3), and cells that were abnormally small and round (panel 4).
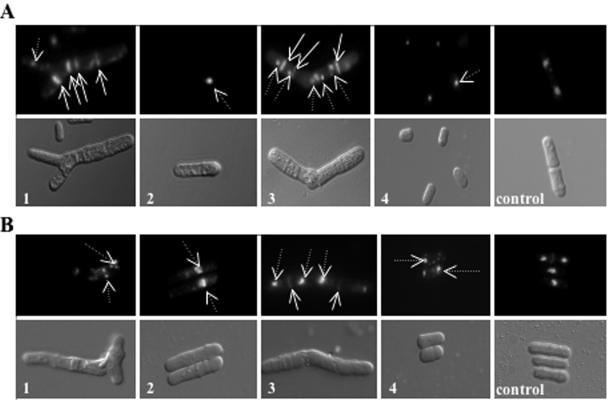
FIG. 2.
Fkh2 is involved in several cell cycle-dependent processes. (A) Cells of mid-log-phase cultures of Δfkh2 (panels 1 to 4) and wild-type (CHP429; panel control) strains were analyzed by DIC and fluorescence microscopy (DAPI staining for nuclei [dotted arrows] and calcofluor staining for septa [solid arrows]). Several different abnormal phenotypes were observed, and some examples are shown. 1, branched cells with phenotypes such as fragmented DNA and/or multiple septa; 2, cells containing misplaced nuclei; 3, misshapen cells with phenotypes including multiple septa and/or multiple nuclei; 4, cells that were abnormally small and round. (B) Cells of a mid-log-phase culture of the CHP429 wild-type strain containing pREP41MNHfkh2+ (panels 1 to 4) or pREP41 vector (panel control) were grown in EMM medium in the absence of thiamine for ∼48 h and then analyzed by DIC and fluorescence microscopy (DAPI staining for nuclei [dotted arrows] and calcofluor staining for septa [solid arrows]). Several examples of phenotypes observed in the Δfkh2 mutant are shown.
We next examined whether expression of fkh2+ (pREP41MNHfkh2+) from the medium-strength thiamine-repressible nmt41 promoter on a multicopy vector (12) affects the growth of wild-type (CHP429) cells. In the absence of thiamine, the growth rate and viability of cells carrying pREP41MNHfkh2+ were reduced (data not shown). Furthermore, after prolonged (48-h) growth in thiamine-free media, many cells shared phenotypes observed in Δfkh2 cells. Some examples of similar phenotypes are shown in Fig. 2B.
Taken together, these data suggest that Fkh2 regulates several cell cycle-dependent processes including septum formation and the function of the mitotic spindle.
Fkh2 affects cell cycle-dependent gene expression.
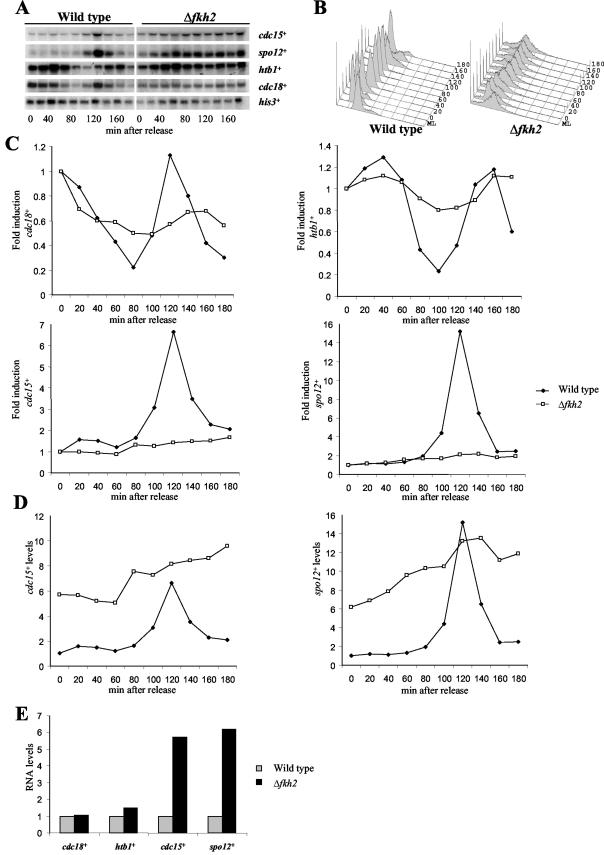
Previously, the expression of several genes such as cdc15+ and spo12+ was shown to be coordinately expressed in the M and G1 phases in S. pombe (43). In contrast, genes such as cdc18+ are expressed in G1 whereas genes such as those encoding histones are expressed later in the cell division cycle in the S phase. We next examined whether we could detect a role for Fkh2 in cell cycle-dependent gene expression. To obtain RNA from synchronized cells, we attempted to construct a Δfkh2 cdc25-22 strain. However, this was unsuccessful, indicating that this combination of mutant alleles is synthetically lethal, further emphasizing the connection between Fkh2 activity and the cell division cycle (data not shown). It was also impractical to use centrifugal elutriation to synchronize cultures of the Δfkh2 mutant, since loss of Fkh2 function causes multiple defects in cell size and shape (Fig. 2A). Hence, to obtain RNA from synchronized cells, wild-type (CHP429) and Δfkh2 mutant strains were blocked at S phase by using HU and RNA was isolated at various times after release from the block (52) (Fig. 3A). Cell synchrony was confirmed by fluorescence-activated cell sorter analysis (Fig. 3B). In wild-type cells, levels of cdc18+ RNA were high at the block and then rapidly dropped before peaking at 120 min. In contrast to cdc18+, peak expression of htb1+, encoding histone H2B, occurred during S phase following the peak of cdc18+ expression, and, in agreement, htb1+ RNA levels were high at several time points after the release of wild-type cells from the HU block and did not peak again until 140 min (Fig. 3A and C). Furthermore, cdc15+ and spo12+ RNA levels were low at the HU block in wild-type cells and then increased after the inhibition of htb1+ expression following release from the block, consistent with the periodic expression of the cdc15+ and spo12+ genes in the M and G1 phases (Fig. 3A and C). It is difficult to assess the synchrony of cultures of the Δfkh2 mutant since many of the cells were abnormal (Fig. 1 and 2). However, our fluorescence-activated cell sorter analysis suggested that the majority of Δfkh2 cells were arrested by HU (Fig. 3B). In Δfkh2 cells the levels of cdc18+ and htb1+ RNA at the HU block, and the kinetics of induction and inhibition of these genes in the first cycle following release from the HU block, displayed profiles similar to those observed in wild-type cells (Fig. 3C and E). However, consistent with reduced synchrony following release from the HU block, the amplitude of these changes in Δfkh2 cells was not as great as that of the changes observed in wild-type cells (Fig. 3C). Furthermore, the induction of cdc18+ and htb1+ RNA levels in the second cycle was delayed in comparison with that in the wild type (Fig. 3C), consistent with the slower growth of Δfkh2 cells (Fig. 1). These studies are consistent with previous analysis of cell cycle-dependent gene expression in a fkh mutant strain of S. cerevisiae (53). Hence, although the phenotypes associated with Δfkh2 cells prevented other analyses of synchrony, these data suggest that the loss of Fkh2 function does not affect the expression of htb1+ and cdc18+ (Fig. 3C and E). In contrast to cdc18+ and htb1+, the expression of the cdc15+ and spo12+ genes was aberrant in Δfkh2 cells (Fig. 3C to E). In particular, RNA levels of cdc15+ and spo12+ were very much increased at the HU block (Fig. 3D and E), in contrast to cdc18+ and htb1+ (Fig. 3E). Furthermore, the expression of cdc15+ and spo12+ appeared to have lost normal cell cycle periodicity (Fig. 3C and D). Collectively, these data suggest that Fkh2 is required for both activation and repression of periodic expression during the cell division cycle and are consistent with the link between Fkh2 function and several cell cycle processes.
FIG.3.
Fkh2 affects cell cycle-dependent gene expression. (A) Northern blot analysis of RNA isolated from cells of the wild-type (CHP429) and Δfkh2 strains that had been synchronized with HU at S phase. Cell pellets were collected every 20 min following release from the HU block. RNA levels of cdc15+, spo12+, cdc18+, htb1+, and his3+ were examined with gene-specific probes. (B) The DNA content of HU-synchronized wild-type and Δfkh2 cells was analyzed by flow cytometry. Samples from mid-log-phase cultures of both strains (ML) were included as controls to allow analysis of the degree of synchronization by HU. (C to E) RNA levels were quantified relative to the his3+ invariant loading control. (C) The fold induction of RNA levels was calculated relative to the time-zero sample of each individual strain. (D) As RNA samples were analyzed on the same membrane in panel A, RNA levels were calculated relative to time zero of the wild-type strain which was set at unity. (E) Comparisons of RNA levels, quantified as described for panel D, at the HU block at time zero.
The expression of fkh2+ is regulated in a cell cycle-dependent manner.
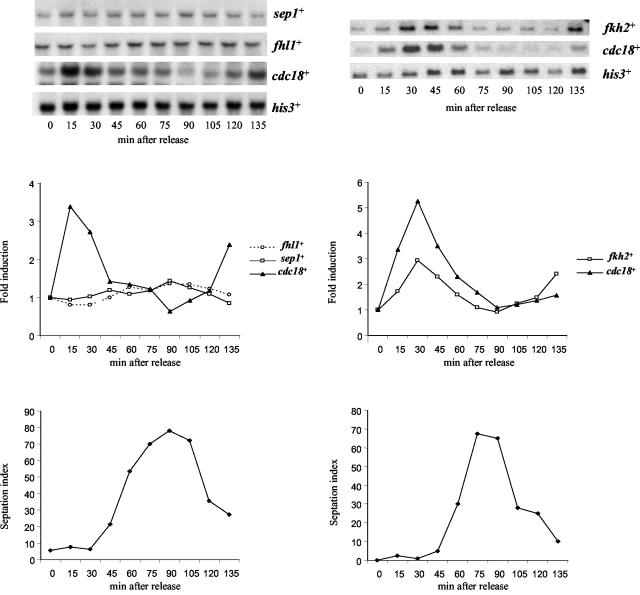
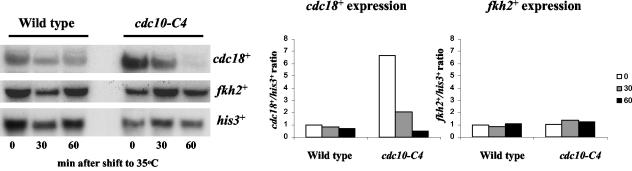
One mechanism of regulation of cell cycle-dependent gene expression in S. cerevisiae involves the cyclical expression of genes encoding specific transcription factors (22). Hence, the expression of fkh2+, sep1+, and SPAC1142.08, which we have named fhl1+ due to the similarity of the predicted protein to Fh11 in S. cerevisiae, was investigated by Northern blot analysis using RNA isolated from a synchronized cdc25-22 mutant (Fig. 4). The expression of mei4+ has previously been shown to be induced in meiosis (23, 24). The expression of the fhl1+ gene varied little during the cell division cycle (Fig. 4). Previous studies of sep1+ expression during the cell division cycle have been contradictory (42, 55). In this study we found that sep1+ RNA levels were unaffected by cell cycle position (Fig. 4). However, in contrast, the levels of fkh2+ RNA were found to fluctuate during the cell division cycle, with peak expression coinciding with the expression of the Cdc10-regulated cdc18+ gene (Fig. 4). We next investigated whether fkh2+ expression is also dependent on Cdc10. RNA was isolated from wild-type (CHP429) and cdc10-C4 mutant cells grown at 25°C (permissive temperature for the cdc10 mutant) or 35°C (nonpermissive temperature for the cdc10 mutant). In agreement with the results of previous studies, the levels of cdc18+ RNA were higher in the cdc10-C4 mutant than in the wild-type control when cells were grown at the permissive temperature (Fig. 5) (34). cdc18+ expression was strongly inhibited by growth of the cdc10-C4 mutant at the nonpermissive temperature in comparison with cdc18+ expression in the wild-type strain (Fig. 5). However, in contrast to cdc18+, fkh2+ RNA levels were relatively unaffected in the cdc10-C4 mutant grown at either the permissive or the nonpermissive temperature (Fig. 5). We also examined the expression of cdc18+ and fkh2+ in a strain containing cdc10-129, a different temperature-sensitive allele of cdc10+. Consistent with our analysis of the cdc10-C4 mutant, cdc18+ RNA levels, but not fkh2+ RNA levels, were inhibited when the cdc10-129 mutant was grown at the nonpermissive temperature (data not shown).
FIG. 4.
fkh2+ expression is regulated during the cell division cycle. The figure shows the results of a Northern blot analysis of RNA isolated from cells of the cdc25-22 strain that had been synchronized at the G2/M phase transition and then returned to 25°C. Cells were harvested every 15 min following the return to 25°C, and cell synchrony was determined by counting individual cell septa. RNA levels were examined with the indicated probes. To examine cell cycle-dependent changes in RNA levels, the data were quantified relative to the his3+-invariant loading control and the fold induction was calculated relative to the time-zero sample.
FIG. 5.
The cell cycle-dependent expression of fkh2+ is independent of Cdc10 function. The figure shows the results of a Northern blot analysis of RNA isolated from cells of the wild-type (CHP429) and cdc10-C4 strains that had been grown to mid-log phase at 25°C and then placed at 35°C for 0, 30, or 60 min. RNA levels were examined with the indicated probes. The RNA levels were quantified relative to the his3+-invariant loading control, and the fold induction was calculated relative to the time-zero sample of the wild-type strain.
These data further link Fkh2 to the regulation of the cell division cycle. Moreover, although fkh2+ expression was regulated at a time in the cell division cycle similar to that of cdc18+, the regulation of fkh2+ was Cdc10 independent.
Fkh2 is regulated during the cell division cycle.
Our data indicate that Fkh2 activity is important for cell cycle-dependent gene expression. Furthermore, the effect of loss of Fkh2 on cdc15+ and spo12+ expression suggests that Fkh2 functions throughout the cell division cycle to regulate these genes. Consistent with these results, indirect-immunofluorescence analysis of the cellular location of 3×PkC-tagged Fkh2, expressed from the normal chromosomal locus, demonstrated that the protein is present in the nucleus of cells at all stages of the cell division cycle (data not shown).
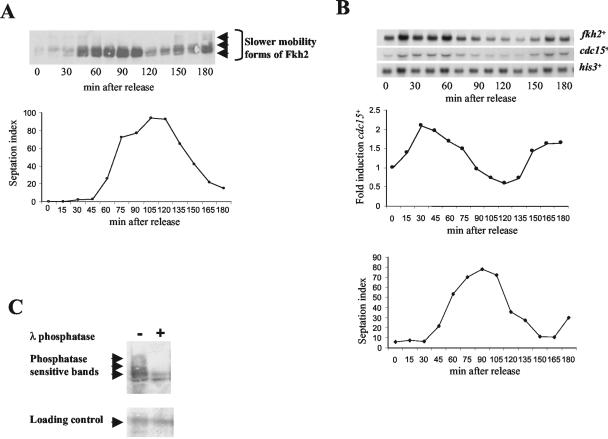
To examine whether the mobility and/or levels of the Fkh2 protein were regulated during the cell division cycle, a cdc25-22 strain (RB1) was constructed that expressed 3×PkC-tagged Fkh2 from the normal chromosomal locus. Western blot analysis of extracts isolated from cells of the RB1 strain and an untagged control strain revealed the presence of specific bands of the predicted mobility for tagged Fkh2 (Fig. 6 and data not shown). An exponentially growing culture of RB1 was synchronized at the G2/M transition, and protein extracts were obtained every 15 min following shift of the culture to the permissive temperature (25°C). Western blot analysis, using the anti-Pk antibody, revealed that slower-mobility forms of Fkh2 were present at different times during the cell division cycle. The slowest-mobility bands were present up to 45 to 60 min following release, prior to the peak of septation (Fig. 6A). Slower-mobility bands then reappeared between 135 and 180 min (Fig. 6A). Our RNA analysis demonstrated that Fkh2 was involved in the regulation of cdc15+ and spo12+ expression during the cell division cycle (Fig. 3). Importantly, we found that the appearance of these slower-mobility shifts in Fkh2 correlated with increased expression of cdc15+, suggesting that they may be linked to the regulation of gene expression (Fig. 6B).
FIG. 6.
The Fkh2 protein is regulated during the cell division cycle. (A) Western blot analysis using the anti-Pk antibody of extracts isolated from cells of RB1 (cdc25-22 fkh2+::3xPkC) synchronized at the G2/M phase transition and then returned to 25°C. Cells were harvested every 15 min following the return to 25°C, and cell synchrony was determined by counting individual cell septa. Slower-mobility forms of Fkh2 are indicated. (B) Northern blot analysis of RNA isolated from cells of the RB1 (cdc25-22 fkh2+::3xPkC) strain synchronized at the G2/M phase transition and then returned to 25°C. Cells were harvested every 15 min following the return to 25°C, and cell synchrony was determined by counting individual cell septa. RNA levels of cdc15+ and fkh2+ were examined with gene-specific probes and were quantified relative to the his3+-invariant loading control. Fold induction was calculated relative to the time-zero sample. (C) Protein extracts, isolated at 30 min following synchronization, were either untreated or treated with λ phosphatase and analyzed by Western blotting using the anti-Pk antibody. Slower-mobility forms of Pk epitope-tagged Fkh2 that are present in the absence of phosphatase are indicated. Loading was determined by copper staining of a nonspecific band (loading control).
Equal amounts of total protein were analyzed by Western blotting, and the results suggested that the total amount of Fkh2 protein varied during the cell division cycle (Fig. 6A and data not shown). This correlates with our finding that the expression of the fkh2+ gene was regulated in a cell cycle-dependent manner (compare Fig. 6A and B). However, the Fkh2 protein could be detected in all protein extracts, and this is consistent with the presence of the Fkh2 protein in the nucleus throughout the cell division cycle (data not shown). Interestingly, the increased levels of Fkh2 correlated with the disappearance of the slower-mobility forms of the protein, suggesting that new expression of the gene may play an important role in the regulation of Fkh2 (Fig. 6A).
It is likely that the cell cycle-dependent alteration in the mobility of Fkh2 represents posttranslational modification(s). To test whether any of these modifications were caused by phosphorylation, a protein sample containing slower-mobility forms of Fkh2 was treated with λ phosphatase (Fig. 6C). This treatment was found to resolve the Fkh2 protein back to faster-mobility forms of the protein, revealing that several of the slower-mobility forms were likely caused by phosphorylation.
Thus, we have identified a forkhead protein, Fkh2, that is involved in the regulation of several processes during the cell division cycle. Furthermore, Fkh2 is modified by phosphorylation in a cell cycle-dependent manner, with a timing suggesting that this is important for regulating gene expression.
DISCUSSION
In this paper we describe the identification and characterization of a forkhead transcription factor, Fkh2, which acts in the cell division cycle of S. pombe. The expression of fkh2+ fluctuates during the cell division cycle, coinciding with that of the cdc18+ gene. In addition, Fkh2 influences several cell cycle processes including cell morphology and cell separation, mitotic spindle structure, and the proper structure and movement of nuclei. Consistent with these roles, Fkh2 was found to be important for cell cycle-dependent gene expression in the M and G1 phases.
The periodic expression of cdc18+ is regulated by the MBF (DSC1) transcription factor complex, suggesting that fkh2+ expression was also regulated by this complex. However, fkh2+ expression is not dependent on the activity of Cdc10, which is a component of MBF (DSC1) (33). These data suggest the presence of an unidentified transcription factor(s) that regulates periodic expression early in the cell division cycle.
The Fkh2 protein in S. cerevisiae is a critical component of Swi Five Factor (SFF), which forms a ternary complex with the MADS-box protein Mcm1 to regulate the CLB2 cluster of genes in the G2 and M phases (28, 29, 40, 53). In addition, a closely related forkhead protein, Fkh1, acts to regulate the cell division cycle independently of Mcm1 (7, 21). Fkh2 in S. pombe is most closely related to Fkh2 and also Fkh1 in S. cerevisiae. Consistent with this relationship, we found that similar cell cycle processes (Fig. 1 and 2) were defective in Δfkh2 cells of S. pombe and fkh1Δ fkh2Δ cells of S. cerevisiae. In S. cerevisiae, these phenotypes have been linked to deregulated expression of the CLB2 gene cluster. Our characterization of a Δfkh2 mutant of S. pombe indicated that Fkh2 also functions to regulate late cell cycle-dependent gene expression. We demonstrated that deletion of the fkh2+ gene resulted in the loss of activation and repression of periodic expression of the cdc15+ and spo12+ genes in the M and G1 phases. S. cerevisiae and S. pombe are very divergent in evolution. Hence, the observation that forkhead proteins play a role in similar cell cycle processes in these yeasts strongly suggests that this is a key function of these proteins in eukaryotes.
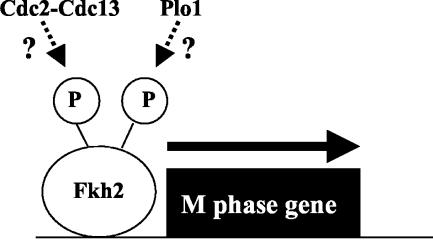
We investigated possible mechanisms of regulation of Fkh2 activity during the cell division cycle. Both fkh2+ mRNA and protein levels change in a cell cycle-dependent manner, suggesting that the levels of Fkh2 are important for normal cell cycle progression. Furthermore, the expression of the fkh2+ gene from a heterologous promoter causes cell cycle defects. However, alteration of protein levels is likely not the only form of regulation of Fkh2. The Fkh2 protein could be detected in the nucleus at all stages of the cell division cycle. Moreover, our data suggested that Fkh2 was phosphorylated on multiple sites in a cell cycle-dependent manner, with the peak of phosphorylation coinciding with increased levels of cdc15+ expression. Given the effects of loss of Fkh2 activity on cdc15+ and spo12+ expression, it is tempting to speculate that phosphorylation of Fkh2 regulates repressor and/or activator functions of the protein. Which kinase(s) is responsible for this phosphorylation? We have found that in S. cerevisiae the Cdc28 Cdk in association with Clb cyclin phosphorylates Fkh2 (A. D. Sharrocks and B. A. Morgan, unpublished data). The timing of phosphorylation of Fkh2 in S. pombe is consistent with a role for the Cdc2 Cdk in association with the B-type cyclin Cdc13 (Fig. 7). Furthermore, we have identified several potential Cdk phosphorylation sites in Fkh2. Recently, Anderson et al. revealed that the polo-like kinase Plo1 was involved in the activation of expression of a group of genes in M and G1 phases, including cdc15+ and spo12+ (3). Thus, it is possible that Plo1 is also responsible for phosphorylation of Fkh2 (Fig. 7).
FIG. 7.
Fkh2 regulates gene expression in the cell division cycle. Fkh2 functions to regulate gene expression in the M and G1 phases of the cell division cycle, and this activity is correlated with cell cycle-dependent phosphorylation of Fkh2.
Based on recent studies with S. cerevisiae, it is likely that the FHA domain of Fkh2 plays a role in the regulation of gene expression in S. pombe. In particular, these studies revealed that the FHA domain of Fkh2 interacts with Cdc28-Clb2-phosphorylated Ndd1, resulting in the activation of gene expression (13, 41). However, analysis of the S. pombe genome database has revealed no obvious homologue of Ndd1. Hence, it is possible that the FHA domain of Fkh2 is involved in intramolecular interactions with a phosphorylated residue(s) within the protein. Alternatively, the FHA domain may interact with other as yet unrecognized coactivator proteins.
Recently, Anderson et al. identified a binding activity that bound to the promoters of several genes, including cdc15+ and spo12+, that are expressed in the M and G1 phases (3). Their analysis also identified a consensus binding site, which they named a PCB element, in these promoters. Interestingly, this sequence is related, although not identical, to the forkhead DNA binding consensus sequence to which forkhead proteins from higher eukaryotes bind (27). Previously, Sep1 was shown to play a role in the periodic expression of cdc15+ (55). However, PBF binding was unaffected in a sep1 mutant (3). Fkh2 affects cdc15+ and spo12+ expression and so it is possible that Fkh2 is a component of PBF. However, to date we have been unable to detect direct binding of Fkh2 to the cdc15+ promoter (data not shown). At present we cannot exclude the possibility that Fkh2 does not bind the cdc15+ promoter directly but, rather, regulates the expression of a transcriptional regulator of cdc15+. Studies of the components of PBF, and of the functions of Sep1 and Fkh2, are important for elucidation of the relationships between these proteins in regulating gene expression.
In summary, we have identified a transcription factor that regulates cell cycle-dependent gene expression in S. pombe. Furthermore, our data suggest that forkhead proteins play conserved roles in regulating cell cycle processes. Indeed, recent studies of metazoans have also linked forkhead proteins with the regulation of cell cycle-dependent gene expression (2). Therefore, it is likely that the elucidation of mechanisms of regulation of forkhead proteins in the cell division cycle of divergent yeasts will yield important insights into the regulation of these key proteins in eukaryotes.
Acknowledgments
We thank Jürg Bähler and Elizabeth Veal for critically reading the manuscript.
The work was funded by Cancer Research UK.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Alvarez, B., A. C. Martinez, B. M. T. Burgering, and A. C. Carrera. 2001. Forkhead transcription factors contribute to the execution of the mitotic programme in mammals. Nature 413:744-747. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, M., S. S. Ng, V. Marchesi, F. H. MacIver, F. E. Stevens, T. Riddell, D. M. Glover, I. M. Hagan, and C. J. McInerny. 2002. plo1+ regulates gene transcription at the M-G1 interval during the fission yeast mitotic cell cycle. EMBO J. 21:5745-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 5.Baum, B., J. Waurin, and P. Nurse. 1997. Control of S-phase periodic transcription in the fission yeast mitotic cell cycle. EMBO J. 16:4676-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boros, J., F. L. Lim, Z. Darieva, A. Pic-Taylor, R. Harman, B. A. Morgan, and A. D. Sharrocks. 2003. Molecular determinants of the cell-cycle regulated Mcm1p-Fkh2p transcription factor complex. Nucleic Acids Res. 31:2279-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breeden, L. L. 2000. Cyclin transcription: timing is everything. Curr. Biol. 10:R586-R588. [DOI] [PubMed] [Google Scholar]
- 9.Caligiuri, M., and D. Beach. 1993. Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell 72:607-619. [DOI] [PubMed] [Google Scholar]
- 10.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65-73. [DOI] [PubMed] [Google Scholar]
- 11.Clifton-Bligh, R., J. M. Wentworth, P. Heinz, M. S. Crisp, R. John, J. H. Lazarus, M. Ludgate, and K. Chatterjee. 1998. Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atrasia. Nat. Genet. 19:399-401. [DOI] [PubMed] [Google Scholar]
- 12.Craven, R. A., D. J. F. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221:59-68. [DOI] [PubMed] [Google Scholar]
- 13.Darieva, Z., A. Pic-Taylor, J. Boros, A. Spanos, M. Geymonat, R. J. Reece, S. G. Sedgwick, A. D. Sharrocks, and B. A. Morgan. 2003. Cell cycle-regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr. Biol. 13:1740-1745. [DOI] [PubMed] [Google Scholar]
- 14.Denmat, S. H., M. Werner, A. Sentenac, and P. Thuriaux. 1994. Suppression of yeast RNA polymerase III mutations by FHL1, a gene encoding for a forkhead protein involved in rRNA processing. Mol. Cell. Biol. 14:2905-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durocher, D., and S. P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58-66. [DOI] [PubMed] [Google Scholar]
- 16.Epstein, J. A., P. Lam, L. Jepeal, R. L. Maas, and D. N. Shapiro. 1995. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J. Biol. Chem. 270:11719-11722. [DOI] [PubMed] [Google Scholar]
- 17.Futcher, B. 2000. Microarrays and cell cycle transcription in yeast. Curr. Opin. Cell Biol. 12:710-715. [DOI] [PubMed] [Google Scholar]
- 18.Ghiara, J. B., H. E. Richardson, K. Sugimoto, M. Henze, D. J. Lew, C. Wittenberg, and S. I. Reed. 1991. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell 65:163-174. [DOI] [PubMed] [Google Scholar]
- 19.Greenall, A., A. P. Haderoft, P. Malakasi, N. Jones, B. A. Morgan, C. S. Hoffman, and S. K. Whitehall. 2002. Role of fission yeast Tup1-like repressors and Prr1 transcription factor in response to salt stress. Mol. Biol. Cell 13:2977-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagan, I., and K. R. Ayscough. 2000. Fluorescence microscopy in yeast, p. 179-206. In V. J. Allan (ed.), Protein localization by fluorescence microscopy. Oxford University Press, New York, N.Y.
- 21.Hollenhorst, P. C., G. Pietz, and C. A. Fox. 2001. Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev. 15:2445-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horak, C. E., N. M. Luscombe, J. Qian, P. Bertone, S. Piccirrillo, M. Gerstein, and M. Snyder. 2002. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 16:3017-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horie, S., Y. Watanabe, K. Tanaka, S. Nishiwaka, H. Fujioka, H. Abe, M. Yamamoto, and C. Shimoda. 1998. The S. pombe mei4+ gene encodes a meiosis-specific transcription factor containing a DNA-binding domain. Mol. Cell. Biol. 18:2118-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iino, Y., Y. Hiramine, and M. Yamamoto. 1995. The role of cdc2 and other genes in meiosis in Schizosaccharomyces pombe. Genetics 140:1235-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorgensen, P., and M. Tyers. 2000. The fork'ed path to mitosis. Genome Biol. 1:1022.1-1022.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann, E., and W. Knochel. 1996. Five years on the wings of forkhead. Mech. Dev. 57:3-20. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann, E., D. Muller, and W. Knochel. 1995. DNA recognition site analysis of Xenopus winged helix proteins. J. Mol. Biol. 248:239-254. [DOI] [PubMed] [Google Scholar]
- 28.Koranda, M., A. Schleiffer, L. Endler, and G. Ammerer. 2000. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406:94-98. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, R., D. M. Reynolds, A. Shevchenko, A. Shevchenko, S. D. Goldstone, and S. Dalton. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896-906. [DOI] [PubMed] [Google Scholar]
- 30.Lai, C. S., S. E. Fisher, J. A. Hurst, F. Vargha-Khadem, and A. P. Monaco. 2001. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519-523. [DOI] [PubMed] [Google Scholar]
- 31.Lai, E., K. L. Clark, S. K. Burley, and J. E. Darnell, Jr. 1993. Hepatocyte nuclear factor 3/fork head or ‘winged helix’ proteins: a family of transcription factors of diverse biologic function. Proc. Natl. Acad. Sci. USA 90:10421-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, J., and P. K. Vogt. 1993. The retroviral oncogene qin belongs to the transcription factor family that includes the homeotic gene fork head. Proc. Natl. Acad. Sci. USA 90:4490-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowndes, N. F., C. J. McInerny, A. L. Johnson, P. A. Fantes, and L. H. Johnston. 1992. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+. Nature 355:449-453. [DOI] [PubMed] [Google Scholar]
- 34.McInerny, C. J., P. J. Kersey, J. Creanor, and P. A. Fantes. 1995. Positive and negative roles for cdc10 in cell cycle gene expression. Nucleic Acids Res. 23:4761-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto, M., K. Tanaka, and H. Okayama. 1994. res2+, a new member of the cdc10+/SW14 family, controls the “start” of mitotic and meiotic cycles in fission yeast. EMBO J. 13:1873-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 37.Nakashima, N., K. Tanaka, S. Sturm, and H. Okayama. 1995. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle “start” function of Res2-Cdc10. EMBO J. 14:4794-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura, D. Y., R. E. Swiderski, W. L. M. Alward, C. C. Searby, S. R. Patil, S. R. Bennet, A. B. Kanis, J. M. Gastier, and V. C. Sheffield. 1998. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat. Genet. 19:140-147. [DOI] [PubMed] [Google Scholar]
- 39.Parry, P., Y. Wei, and G. Evans. 1994. Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer 11:79-84. [DOI] [PubMed] [Google Scholar]
- 40.Pic, A., F. L. Lim, S. J. Ross, E. A. Veal, A. L. Johnson, M. R. Sultan, A. G. West, L. H. Johnston, A. D. Sharrocks, and B. A. Morgan. 2000. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19:3750-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds, D., B. J. Shi, C. McLean, F. Katsis, B. Kemp, and S. Dalton. 2003. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 17:1789-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribár, B., A. Grallertc, É. Oláhb, and Z. Szállásia. 1999. Deletion of the sep1+ forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 263:465-474. [DOI] [PubMed] [Google Scholar]
- 43.Samuel, J. M., N. Fournier, V. Simanis, and J. B. Millar. 2000. spo12 is a multicopy suppressor of mcs3 that is periodically expressed in fission yeast mitosis. Mol. Gen. Genet. 264:306-316. [DOI] [PubMed] [Google Scholar]
- 44.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 45.Sipiczki, M., B. Grallert, and I. Miklos. 1993. Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J. Cell Sci. 104:485-493. [DOI] [PubMed] [Google Scholar]
- 46.Smith, D. A., W. M. Toone, D. Chen, J. Bahler, N. Jones, B. A. Morgan, and J. Quinn. 2002. The Srk1 protein kinase is a target for the Sty1 stress-activated MAPK in fission yeast. J. Biol. Chem. 277:33411-33421. [DOI] [PubMed] [Google Scholar]
- 47.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Surana, U., H. Robitsch, C. Price, T. Schuster, I. Fitch, A. B. Futcher, and K. Nasmyth. 1991. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65:145-161. [DOI] [PubMed] [Google Scholar]
- 49.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14:6193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka, K., K. Okazaki, N. Okazaki, T. Ueda, A. Sugiyama, H. Nojima, and H. Okayama. 1992. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+ and SW14 gene products. EMBO J. 11:4923-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toone, W. M., B. L. Aerne, B. A. Morgan, and L. H. Johnston. 1997. Getting started: regulating the initiation of DNA replication in yeast. Annu. Rev. Microbiol. 51:125-149. [DOI] [PubMed] [Google Scholar]
- 52.Tournier, S., and J. B. A. Millar. 2000. A role for the START gene-specific transcription factor complex in the inactivation of cyclin B and Cut2 destruction. Mol. Biol. Cell 11:3411-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, G., P. T. Spellman, T. Volpe, P. O. Brown, D. Botstein, T. N. Davis, and B. Futcher. 2000. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406:90-94. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, Y., T. Takeda, K. Nasmyth, and N. Jones. 1994. pct1+, which encodes a new DNA-binding partner for p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 8:885-898. [DOI] [PubMed] [Google Scholar]
- 55.Zilahi, E., E. Salimova, V. Simanis, and M. Sipiczki. 2000. The S. pombe sep1+ gene encodes a nuclear protein that is required for periodic expression of the cdc15+ gene. FEBS Lett. 481:105-108. [DOI] [PubMed] [Google Scholar]