Abstract
The Pkc1-mediated cell wall integrity-signaling pathway is highly conserved in fungi and is essential for fungal growth. We thus explored the potential of targeting the Pkc1 protein kinase for developing broad-spectrum fungicidal antifungal drugs through a Candida albicans Pkc1-based high-throughput screening. We discovered that cercosporamide, a broad-spectrum natural antifungal compound, but previously with an unknown mode of action, is actually a selective and highly potent fungal Pkc1 kinase inhibitor. This finding provides a molecular explanation for previous observations in which Saccharomyces cerevisiae cell wall mutants were found to be highly sensitive to cercosporamide. Indeed, S. cerevisiae mutant cells with reduced Pkc1 kinase activity become hypersensitive to cercosporamide, and this sensitivity can be suppressed under high-osmotic growth conditions. Together, the results demonstrate that cercosporamide acts selectively on Pkc1 kinase and, thus, they provide a molecular mechanism for its antifungal activity. Furthermore, cercosporamide and a β-1,3-glucan synthase inhibitor echinocandin analog, by targeting two different key components of the cell wall biosynthesis pathway, are highly synergistic in their antifungal activities. The synergistic antifungal activity between Pkc1 kinase and β-1,3-glucan synthase inhibitors points to a potential highly effective combination therapy to treat fungal infections.
Human fungal pathogens are a highly divergent group of fungal species, and Candida albicans, Aspergillus fumigatus, and Crytococcus neoformans are the three major pathogens causing severe systemic infections among the immune-compromised population (14). New emerging fungal pathogens are non-C. albicans Candida species and Fusarium species (14). As the number of immune-compromised patients increases due to human immunodeficiency virus infections, chemotherapy, organ transplant, and invasive medical procedures, the incidence of life-threatening systemic fungal infections rises accordingly. Recently, fungi have also been implicated to be causal agents for chronic rhinosinusitis, infecting tens of millions of people around the world (38). Currently, effective drug therapy to treat fungal infections is very limited and dominated by the azole class of antifungals, which selectively inhibit lanosterol demethylase activity, a late step in ergosterol biosynthesis (11, 12, 43). Although the azole antifungals have excellent safety profiles and are orally bioavailable, they are not fungicidal and thus require long therapeutic regimens. Consequently, their widespread use has led to an increasing incidence of drug resistance.
A new class of semisynthetic lipopeptide antifungal agents, commonly known as echinocandins and pneumocandins, is efficacious in the treatment of systemic Candida infections and aspergillosis (11, 12). This class of antifungals inhibits fungal cell wall biosynthesis by targeting the β-1,3-glucan synthase and, with an entirely different mode of action from the azoles, is thus effective against azole-resistant fungal strains. Because the cell wall is a unique feature of fungi and is essential for fungal cell growth (7, 44), the lipopeptide antifungal agents not only have excellent safety margins but also are highly fungicidal by causing rapid fungal cell lysis. However, despite comprehensive chemistry efforts, the lipopeptide class of antifungal agents in general has very poor oral bioavailability and requires parenteral administration, which as a result severely restricts the use of this class of effective antifungals. Furthermore, exhaustive screening of chemical and natural product libraries by Lilly and also others (35) have so far failed to identify new β-1,3-glucan synthase inhibitors with improved oral bioavailability.
In addition to β-1,3-glucan synthase, a cell wall integrity pathway mediated through a serine/threonine protein kinase, Pkc1, is also central to cell wall biosynthesis and remodeling (5, 15, 29). The cell wall, although appearing rigid, in fact undergoes constant modifications and repair, which are coordinately regulated with the cell cycle, development, and in response to environmental stresses such as heat shock and rapid osmotic changes, via the Pkc1-mediated signaling pathway (15, 17, 19, 24, 29, 30). Furthermore, genetic studies show that this Pkc1-mediated signaling pathway is highly conserved in all fungi and that Pkc1 plays a central role in this signaling pathway (5). Like inactivation of β-1,3-glucan synthase, loss of Pkc1 function also leads to rapid cell autolysis (29, 36). Therefore, Pkc1 represents a potential molecular target for developing broad-spectrum, fungicidal antifungal agents. This potential has been further bolstered by recent successful development of pharmaceutics selectively targeting protein kinases (40).
In this report, we describe further biochemical characterizations of the Pkc1 homolog of C. albicans, CaPkc1, and a high-throughput screening (HTS) based on CaPkc1 kinase activity to discover novel Pkc1 inhibitors. To illustrate the potential of Pkc1 kinase inhibitors as antifungal agents, we focus on the discovery and characterization of cercosporamide, a known antifungal natural product with a previously unknown mode of action, as a highly selective and potent Pkc1 inhibitor. In addition, we further demonstrate that the antifungal activity of cercosporamide is mediated through inhibition of Pkc1 kinase activity.
MATERIALS AND METHODS
Fungal strains, media, and general microbiological and molecular biological techniques.
C. albicans strains A26 and CAI4, A. fumigatus strain WM-1, and Saccharomyces cerevisiae diploid strain YPH501 were all obtained from the American Type Culture Collection and maintained as per their instructions. The MICs of antifungal compounds were evaluated as previously described (20). To characterize the potential synergy of combining Pkc1 kinase and β-1,3-glucan synthase inhibitors, the checkerboard microtiter plate testing method was employed and the fractional inhibitory concentrations (FICs) of each compound tested and their FIC indices were calculated based on MIC endpoints as previously described (6, 31). Standard techniques for protein analysis by gel electrophoresis, immunoprecipitation (IP), and Western blotting, for RNA analysis, and for DNA manipulation were used (4, 48). Antihemagglutinin (anti-HA) and anti-His6 antibodies were purchased from Roche (Indianapolis, Ind.). Phosphatidylserine (PS) and diacylglycerol (DAG) were obtained from Avanti Polar Lipids (Alabaster, Ala.).
Expression and purification of CaPkc1 from insect Sf9 cells.
caPKC1 was cloned into vector pCR2.1 (Invitrogen, Carlsbad, Calif.) after reverse transcription-PCR from total RNA isolated from C. albicans strain CAI4 by using primers 5′-GTAGTCGACCGTCACAACAACAAAACCCAG-3′ and 5′-TGCGGCCGCAAATCGTTGCATTGTCAGATATATGC-3′ and then subcloned into pFastBacHT (Invitrogen) as a SalI and NotI fragment. Subsequent baculovirus generation and transfection of Sf9 insect cells were as per the instructions of Invitrogen.
Transfected Sf9 cells were lysed in 30 mM Tris-HCl, pH 7.5, containing protease inhibitors (leupeptin [13 μg/ml], soybean trypsin-chymotrypsin inhibitor [13 μg/ml], aprotinin [13 μg/ml], N-tosylphenylalanine chloromethyl ketone [13 μg/ml], 3 mM Na-p-tosyl-l-arginine methyl ester, 7 mM benzamidine, and 0.3 mM phenylmethylsulfonyl fluoride) and phosphatase inhibitors (100 mM β-glycerophosphate, 20 mM sodium fluoride, 1 mM sodium vanadate, 20 mM p-nitrophenyl phosphate, 10 nM microcystin, 1 μM okadaic acid, and 0.2% [vol/vol] Nonidet P-40). Okadaic acid was obtained from Upstate Biotechnology (Lake Placid, N.Y.), microcystin was from BRL-GIBCO, and all the other reagents were obtained from Sigma (St. Louis, Mo.). The lysates were sonicated on ice and centrifuged at 35,000 rpm for 60 min at 4°C on a Beckman ultracentrifuge with a Vti50 rotor. The CaPkc1 protein was purified from the supernatant as described below.
The fast-flow chelating Sepharose column (Amersham Biosciences, Piscataway, N.J.) was charged with Ni2+ and equilibrated with buffer A (20 mM sodium phosphate, [pH 7.2], 500 mM sodium chloride, 10 mM sodium fluoride, and 20% glycerol) mixed with 2% buffer B (20 mM sodium phosphate [pH 7.2], 500 mM sodium chloride, 10 mM sodium fluoride, 20% glycerol, and 500 mM imidazole). Protein lysates were loaded onto the column at 2 ml/min. After loading, the column was first washed with six column volumes of buffer A mixed with 2% buffer B and then with 10 column volumes of buffer A mixed with 12% buffer B. The CaPkc1 protein was eluted over a gradient of 60 to 500 mM imidazole with a flow rate of 2 ml/min. The presence of the CaPkc1 protein in the fractions was followed both by a kinase activity assay and by gel electrophoresis.
Construction of Candida GAL1-10 promoter-regulated expression vector and expression of 2× HA-tagged CaPkc1 in Candida.
The CaGAL1-10 promoter sequence of about 1.2 kb was PCR amplified from the genomic DNA isolated from C. albicans strain CAI4 by using a pair of primers, 5′-GCGGCCGCGGTATAACTCTTTCTTATAAAAATCGG-3′ and 5′-GAGCTCTTCCTTGGTTTAATTCCAAACGAAACG-3′. After cloning into pCR-2.1, the GAL1-10 promoter sequence was subcloned as a NotI and SacI fragment into pBluescript II SK(+). A 2× HA tag sequence was then added at the NotI site of the above-resulting plasmid by insertional mutagenesis using the QuikChange mutagenesis kit (Stratagene, La Jolla, Calif.) as described previously (48) with the pair of primers 5′-CTGAGCAGCGTAATCTGGAACGTCATATGGATAGGAGCCCGCATAGTCAGGAACATCGTATGGGTAAAAGACCATGGTATAACTCTTTCTTATAAAAATCGGTTTG-3′ and 5′-ATGGTCTTTTACCCATACGATGTTCCTGACTATGCGGGCTCCTATCCATATGACGTTCCAGATTACGCTGCTCAGGCGGCCGCTCTAGAACTAGTGGATCCC-3′. The URA3 gene of C. albicans as a selection marker was cloned as a XhoI and SalI fragment after PCR amplification, using pMB7 as template (1) with primers 5′-CCGCTCGAGTCTAGAAGGACCACCTTTGATTGTAAA-3′ and 5′-ACGCGTCGACAGTACTAATAGGAATTGATTTGGATGGTATAAA-3′ to give rise to the regulated expression vector pGal1-1 for C. albicans. Finally C. albicans PKC1 was amplified from genomic DNA with primers 5′-AATCACTAAAAAGCGGCCGCACCGTCACAACAACAAAACCCAGAAC-3′ and 5′-GTTCTCGTCGACCAACACTAACTCGTGAATCAACAATG-3′ and cloned as a NotI and SalI fragment into pGal1-1, resulting in pGal1-Pkc1. Transformation of strain CAI4 with pGal1-Pkc1 DNA was carried out as previously described (10).
To induce GAL1-10-mediated Pkc1 expression, transformed C. albicans cells were cultured in glucose-containing synthetic medium to early log phase. Cells were collected by centrifugation and washed once in synthetic medium containing no carbon source. Then, the cells were transferred to synthetic medium containing galactose (2%, wt/vol) as sole carbon source.
Cloning and expression of CaRho1 in Escherichia coli and GTP-binding assay.
CaRHO1 was cloned into pGEX as a BamHI and EcoRI fragment and expressed as a glutathione S-transferase (GST) fusion protein in BL21 E. coli cells. The bacterial lysates were prepared in the same lysate buffer as for insect cells (described above), and the GST-CaRho1 protein was purified through a glutathione-Sepharose column as per the instructions of Pharmacia. GTP-binding and nucleotide competition assays of the purified GST-CaRho1 protein were performed as previously described (42).
CaPkc1 kinase assay development.
The filter-binding kinase assays were carried out in 96-well titer plates in a 50-μl reaction volume typically consisting of 50 mM Tris-HCl (pH 8.0), 1 mM dithiothreitol, 10 mM MgCl2, 115 μM peptide substrate (GPLGRHGSIRQKKEEV), 0.25 μg of CaPkc1, 43 μM cold ATP, and 1 μM [γ-32P]ATP. Termination of the reaction, filter binding and, subsequently, measurements were done as previously described (48). PS and DAG in chloroform were dried under a stream of nitrogen gas and then were sonicated into distilled water to produce a 2 mM suspension, immediately before addition into the kinase assay mixtures.
For automated HTS, kinase assays were done in the scintillation proximity-based assay (SPA) format in 384-well titer plates using the N-terminal biotinylated peptide. The kinase reaction was carried out in 40 μl of 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 0.01% Triton X-100, 1 mM dithiothreitol, 1 μM ATP, 0.375 μM peptide substrate, 160 ng of CaPkc1, and 0.125 μCi of [γ-33P]ATP for 4 h at room temperature and terminated by addition of 40 μl of the termination solution (40 mM EDTA and 0.2 mg of streptavidin-coated beads [Amersham] in 73.4% cesium chloride). Plates were counted with a TopCount (Perkin-Elmer, Boston, Mass.) for 30 s per well 2 h after termination of the kinase reaction.
Generation of TetR-regulated Pkc1 S. cerevisiae strain and development of hypersensitive cell-based assay for selective Pkc1 inhibitors.
A two-step transformation process was used to create the S. cerevisiae strain dependent on TetR-regulated expression of Pkc1. First, a heterozygous PKC1/pkc::HIS1 strain was constructed by gene replacement in the diploid yeast strain YPH501, with a HIS1 fragment containing 50 base-flanking sequences homologous to the 3′ and 5′ ends of PKC1. A resulting heterozygous diploid strain was then isolated and subsequently transformed with a plasmid containing the PKC1 gene under the control of a TetR-regulated promoter derived from pCM188. After sporulation, the tetrads were screened to identify the haploid strain SH91-2A containing the pkc1::HIS1 allele in the chromosome and also the TetR-regulated PKC1 plasmid.
In the agar diffusion cell-based assay, yeast cells were mixed into the melt top agar medium kept at 55°C to a final concentration of 2 × 106 cells/ml. Doxycycline was then added to the medium at 5 μg/ml and mixed thoroughly before pouring over the bottom agar plates prewarmed to 50°C. After cooling at room temperature for 3 to 4 h, various amounts of compounds in a 5-μl volume of dimethyl sulfoxide were spot applied to the agar plates to test their antifungal activities by measuring the sizes of the cleared zones on a lawn of yeast cells. In assays to stabilize yeast cell wall defects, 1 M sorbitol was incorporated into both bottom and top agar.
Fermentation, isolation, and characterization of cercosporamide.
The fermentation was carried out as described previously (22). The combined fermentation broth (26 liters) was centrifuged, and the cell mass was extracted twice with 9 liters of methanol. The combined methanolic extract was diluted with an equal volume of water and charged onto an Amberchrom CG-161 column. The column was initially washed with 4 liters of 50% aqueous methanol, followed by 4 liters of methanol. The methanol concentrate (13.2 g) was further chromatographed over a Sephadex LH-20 column to yield 4.2 g of a crude cercosporamide. Additional chromatography of a portion of this material (0.88 g) over a Waters symmetry prep column (50 by 250 mm) using a 15-to-50% acetonitrile gradient afforded 660 mg of over 95% pure cercosporamide. The structure of cercosporamide was then established by mass spectrometry (MS) and nuclear magnetic resonance (NMR) data.
RESULTS
CaPkc1 expressed in insect cells retains proper biochemical properties.
To conduct an enzymatic activity-based HTS for Pkc1 kinase inhibitors, we first needed to generate a large amount of active Pkc1 protein. As CaPkc1 has a calculated molecular mass of 126 kDa, we suspected that it would be too large to be expressed in the soluble, active form by bacteria and, furthermore, active Pkc1 is also a phosphoprotein (3, 46). Thus, we directed our effort to express CaPkc1 in insect Sf9 cells by using a baculovirus expression system. CaPkc1 was expressed as an N-terminal His-tagged protein to facilitate purification. As shown in Fig. 1A, CaPkc1 protein was expressed very well by insect cells compared to the chloramphenicol acetyltransferase (CAT) expression positive control. The presence of CaPkc1 was further confirmed by Western blotting using an anti-His tag antibody (Fig. 1A).
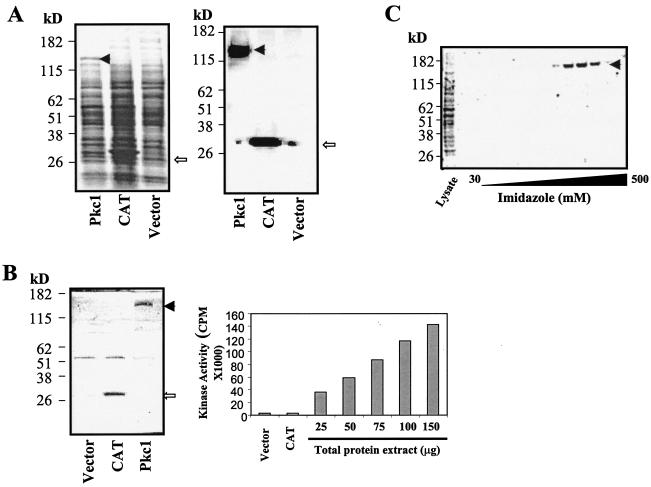
FIG. 1.
Expression of active CaPkc1 kinase in insect cells. (A) Left panel, Coomassie blue-stained gel after electrophoresis of insect cell lysates containing CaPkc1 and CAT. Right panel, Western blot analysis of insect cell lysates as shown in the left panel with an anti-His tag antibody. (B) Left panel, Coomassie blue-stained gel after electrophoresis of the IP complex with the anti-His tag antibody of the insect cell lysates as shown in panel A. Right panel, kinase activity present in the immuno-complex after IP with the anti-His tag antibody of the insect cell lysates. (C) Coomassie blue-stained gel showing purification of the N-terminal His-tagged CaPkc1 protein from insect cell lysates through a Ni2+-charged affinity column with an imidazole gradient. The solid arrowhead indicates the CaPkc1 protein, and the open arrowhead shows the CAT protein.
To determine whether the Pkc1 protein expressed in insect cells is active as a protein kinase, we carried out IP using the anti-His tag antibody and then assayed the kinase activity present in the immuno-complex. The immuno-complex contained high kinase activity and, furthermore, the level of kinase activity present in the immuno-complex was proportional to the amount of protein extract of the insect cells harboring the Pkc1 expression construct (Fig. 1B). By contrast, immuno-complex from vector control and CAT-expressing insect cells did not contain significant kinase activities (Fig. 1B). Gel electrophoresis and subsequent Coomassie blue staining showed that the IP specifically isolated a protein with an expected molecular mass of that of CaPkc1 as seen in Fig. 1A (Fig. 1B). Together, the results demonstrated that the Pkc1 protein expressed by insect cells is active as a protein kinase and, furthermore, that the kinase activity detected in the immuno-complex is specific to CaPkc1.
CaPkc1 protein was affinity purified from insect cell extracts (Fig. 1C). The corresponding kinase assay showed that the purified Pkc1 protein retained high kinase activity. Two lines of evidence showed that this kinase activity was specific to the purified CaPkc1 protein. Firstly, levels of kinase activity closely tracked with the amounts of the CaPkc1 protein present in the fractions. Secondly, the fractions collected in an identical manner from vector control insect cell extracts did not contain significant kinase activities (data not shown).
Our goal in this effort was to discover novel Pkc1 inhibitors through an enzyme-based HTS to be developed into antifungal drugs. Thus, as antifungals, Pkc1 inhibitors must be active against the endogenous Pkc1 kinase activity in vivo under conditions where Pkc1 is known to be active and required for cell viability. To maximize the potential of inhibiting the endogenous Pkc1 protein, the CaPkc1 protein used in the HTS should, therefore, have physiologically relevant biochemical properties as the endogenous Pkc1 protein. To obtain the endogenous CaPkc1 for a comparison with the insect cell-expressed CaPkc1, we engineered a regulated expression system based on the GAL1-10 promoter of C. albicans. We found that C. albicans grows well in medium containing galactose as the sole carbon source, and sequence analysis of the C. albicans genome revealed that the galactose metabolic pathway is highly conserved between C. albicans and S. cerevisiae. In particular, the GAL1 and GAL10 genes are arranged in a similar divergent fashion and share a common intergenic promoter sequence as in S. cerevisiae (21). The GAL1-10 promoter of S. cerevisiae has been used widely for regulated expression of genes in this yeast system (21). We suspected that the GAL1-10 promoter of C. albicans could be similarly used in C. albicans. To investigate if the GAL1-10 promoter is glucose repressible and galactose inducible, we did a Northern analysis of GAL1 expression in C. albicans cells at various time intervals upon transfer to galactose-containing medium and then after subsequent addition of glucose to the galactose-containing medium. Indeed, as shown in Fig. 2A, the expression of the GAL1 gene was tightly repressible by glucose and highly inducible by galactose.
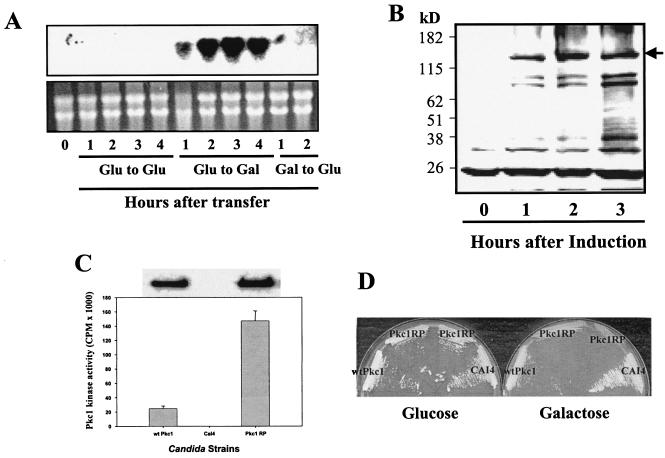
FIG. 2.
Regulated expression of CaPkc1 protein in C. albicans under control of the C. albicans GAL1-10 promoter. (A) Upper panel, Northern blot analysis of GAL1 gene expression, showing induction by galactose and repression by glucose in C. albicans. Lower panel, ethidium bromide-stained gel of total RNA used for the Northern blot analysis as shown in the upper panel. An overnight culture of C. albicans was collected by centrifugation and washed two times with a medium containing no carbon source. The collected C. albicans cells were then added into fresh medium containing either glucose or galactose as the sole carbon source. To repress GAL1 gene expression, glucose was directly added to cells growing in the galactose-containing medium. (B) Galactose-induced expression of HA-tagged CaPkc1 (solid arrow) by C. albicans cells. Induction of CaPkc1 expression was done similarly as described for panel A, and expression of the HA-tagged CaPkc1 was detected by Western blotting with an anti-HA tag antibody. (C) Kinase activity of the wt and the constitutively active mutant CaPkc1 (CaPkc1RP). The wt and the mutant CaPkc1 proteins were first isolated by IP with the HA tag antibody, and the immuno-complex was then analyzed by Western blotting (upper panel) and assayed for kinase activity (lower panel). (D) Agar plates of various C. albicans strains growing in glucose- or galactose-containing medium, showing the lethality by overexpression of the mutant CaPkc1RP protein on the galactose-containing medium.
We then constructed several vectors containing various lengths of the 1.2-kb intergenic promoter sequence between GAL1 and GAL10 and found that for maximal expression the entire 1.2-kb promoter sequence is required. To facilitate analysis and purification, a 2× HA tag sequence was introduced at the N terminal of CaPkc1. As shown in Fig. 2B, the CaPkc1 protein under the control of the Candida GAL1-10 promoter was highly expressed 1 to 3 h after transfer of the culture from glucose- to galactose-containing medium. Enzyme assays with the immuno-complex after IP showed high kinase activity (Fig. 2C). It was previously reported that an activated R398P mutant allele of S. cerevisiae Pkc1 has much higher kinase activity than the wild-type (wt) Pkc1 (34, 46). Furthermore, when overexpressed, the constitutively active mutant Pkc1 causes lethality in S. cerevisiae (46). To see whether such a mutation has a similar effect on CaPkc1 activity and function, we created a mutation at the same position (R400P) and expressed the mutant protein under the control of the GAL1-10 promoter in C. albicans. The mutant protein expressed equally well as the wt Pkc1, and a kinase assay after IP showed that indeed the mutant CaPkc1 has a much higher protein kinase activity (Fig. 2C). Furthermore, overexpression of the mutant CaPkc1, as in S. cerevisiae, caused lethality in C. albicans (Fig. 2D).
We next biochemically characterized the CaPkc1 protein expressed in insect cells and that in C. albicans and found that they had virtually identical biochemical properties. Its enzyme activity preferred alkaline conditions, with an optimal pH of 8.0 and an optimal temperature of 30 to 37°C. As previously reported for S. cerevisiae Pkc1, CaPkc1 also did not require second messengers for kinase activity. The CaPkc1 kinase has a Km of 19 μM for ATP and 2 μM for the peptide substrate and is highly sensitive to the broad-spectrum kinase inhibitor staurosporine, with a 50% inhibitory concentration (IC50) of <5 nM.
PS activation of CaPkc1.
Studies in S. cerevisiae show that Rho1 and Pkc1 both play key roles in the cell wall integrity-signaling pathway and, as a downstream target of Rho1, Pkc1 both genetically and physically interacts with Rho1 (5, 9, 15, 26, 34). It was further shown in vitro that active Rho1 (bound to GTP) conditions Pkc1 for activation by PS (25). To determine if CaRho1 in the presence of PS also similarly regulates CaPkc1 activity, we expressed CaRho1 as a GST fusion protein in bacteria and affinity purified it (Fig. 3A). A nucleotide-binding competition assay was carried out to determine if the bacterially expressed GST-CaRho1 was functional as a G-protein. As shown in Fig. 3B, the GST-CaRho1 fusion protein bound to GTP very efficiently and specifically, as CTP, ATP, and UTP failed to compete against GTP binding even at high concentrations, thus demonstrating that bacterially expressed GST-CaRho1 is indeed a functional G-protein.
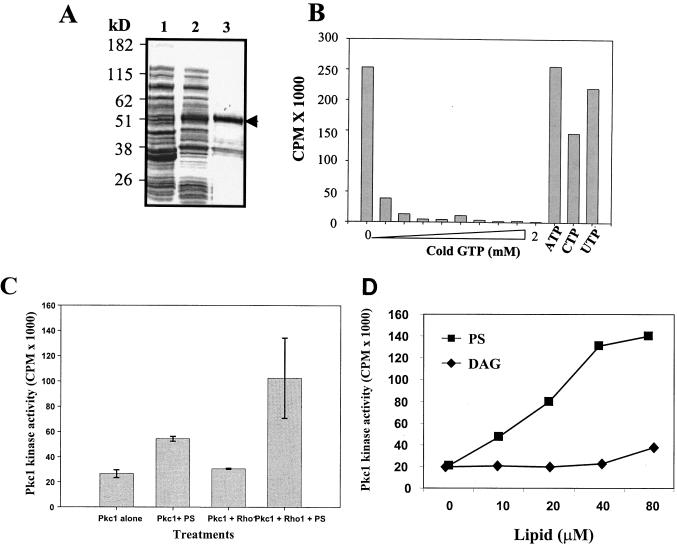
FIG. 3.
Activation of CaPkc1 kinase activity by Rho1 and PS. (A) Coomassie blue-stained gel showing expression and purification of the GST fusion CaRho1. Lane 1, soluble bacterial lysate before induction with isopropyl-β-d-thiogalactopyranoside (IPTG); lane 2, soluble bacterial lysates after IPTG induction; lane 3, affinity-purified GST-CaRho1(arrow). (B) GTP binding of the purified GST-CaRho1 protein as shown in panel A and the competition assay with cold GTP and other trisphosphate nucleotides. (C) CaPkc1 kinase activity in the presence or absence of CaRho1 and PS as indicated. (D) Dose-dependent activation of CaPkc1 kinase activity by PS. PS and DAG were added immediately after sonication into the kinase assay mixture, right before the start of the kinase reaction.
CaPkc1 kinase assay in the presence of CaRho1 and PS was carried out as described previously for S. cerevisiae Pkc1 (25). GST-CaRho1 was preloaded with the nonhydrolyzable GTP[γ]S. Addition of the CaRho1 protein alone had no effect on CaPkc1 kinase activity, whereas addition of PS alone showed a small but consistent activation of CaPkc1 kinase activity (Fig. 3C). However, when Rho1 and PS were added together to the assay mixture, CaPkc1 kinase activity was markedly increased (Fig. 3C), similarly to that previously reported for Rho1 activation of S. cerevisiae Pkc1 in the presence of PS (25).
As CaPkc1 has both a C-1 (for Ca2+ and diacylglycerol binding) and a C-2 (for PS binding) domain (33, 37), we were thus intrigued by the consistent, although small, activation of CaPkc1 by PS in the experiments shown in Fig. 3C. We thus further explored potential lipid activation of CaPkc1. To our surprise, we found that PS alone could markedly activate CaPkc1 in a concentration-dependent manner, but only when applied freshly after sonication into the kinase assay mixture (Fig. 3D). By contrast, DAG showed no activating effect on CaPkc1 activity (Fig. 3D). Additionally, we also found that Ca2+ had no effect on CaPkc1 activity either by itself or in combination with DAG or PS. To further explore the stoichiometry of activation of CaPkc1 by PS, we investigated the activation of CaPkc1 by using Triton X-100-PS-mixed micelles. Mixed micelles of Triton X-100 and lipids have been used widely to study the lipid activation of mammalian protein kinase Cs (PKCs) (16). However, we found that Triton X-100 at concentrations above its critical micelle concentration prevented CaPkc1 activation by PS. Triton X-100 by itself did not inhibit CaPkc1 activity up to the critical micelle concentration (data not shown).
Selective human PKC inhibitors have no activity on CaPkc1.
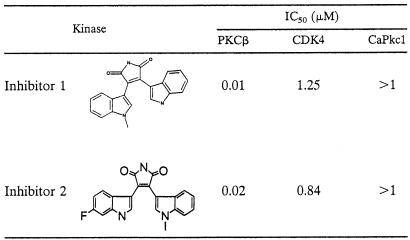
The primary sequence of fungal Pkc1 kinase domains shares high homology with mammalian PKCs. Although fungal Pkc1 exhibits different biochemical properties from mammalian PKCs, for Pkc1 as a viable antifungal target it must have sufficient structural diversity from human PKCs to allow the development of selective Pkc1 inhibitors. To demonstrate this possibility, we first tested human PKCβ selective kinase inhibitors against CaPkc1. We reasoned that if selective human PKC inhibitors lack activity against fungal Pkc1, then it should be equally possible to develop selective CaPkc1 inhibitors that lack activity against human protein kinases. Indeed, the PKCβ inhibitors showed no significant activity against CaPkc1 (Table 1). Thus, our results demonstrate that CaPkc1 not only has different biochemical properties, but it also has considerable structural differences from human PKCs. These biochemical and structural differences should, therefore, afford the potential to develop selective fungal Pkc1 inhibitors as antifungal agents.
TABLE 1.
PKCβ kinase inhibitors lack activity against CaPkc1
HTS.
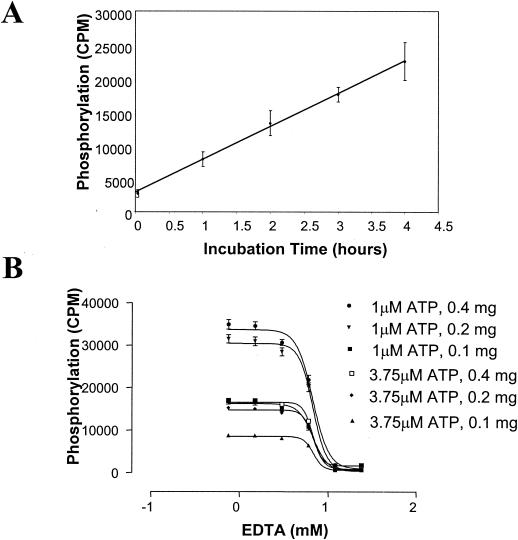
To carry out a totally automated HTS, we developed a SPA in a 384-well format using the N-terminal biotinylated peptide substrate instead of the traditional filter-binding assay in the experiments described above. The SPA allows direct readout of the kinase activity from the assay mixture. We also used the filter-binding kinase assay in a low-throughput format to confirm screen activities, to help eliminate some false positives identified by SPA due to the intrinsic fluorescence-quenching properties of some compounds in the chemical libraries. Streptavidin-coated PVT SPA beads and [33P]ATP are the two most expensive components of the SPA-based HTS and, furthermore, a large amount of 33P radioactive waste is also a potential health hazard. Therefore, we carried out a series of experiments to optimize specific assay signals with less SPA beads and [γ-33P]ATP. Because CaPkc1 kinase activity remained linear for up to 4 h (Fig. 4A), we therefore extended the reaction time to up to 3 to 4 h. In addition, we also lowered the concentration of cold ATP to 1 μM and that of peptide substrate to 0.375 μM. We found that CaPkc1 at 160 ng per reaction mixture could generate a highly robust signal, with only 0.2 mg of SPA beads and 0.125 μCi of [γ-33P]ATP (Fig. 4B). Since the CaPkc1 protein already has a highly robust kinase activity in the absence of second messengers, we omitted PS in the HTS assay in order to reduce the complexity of the HTS.
FIG. 4.
High-throughput kinase assay development. (A) Time course of the CaPkc1 kinase reaction. Phosphorylation of the biotinylated peptide substrate was determined at 1-h intervals. (B) Evaluation of signal strength of kinase activity readout with various combinations of hot ATP, cold ATP, peptide substrate concentrations, and various amounts of streptavidin-coated SPA beads.
We conducted a very successful HTS under the assay conditions shown in Fig. 4B. The HTS identified a variety of structurally diverse Pkc1 kinase inhibitors from a synthetic compound and natural product library. Here, we focused on the discovery and isolation of a previously known antifungal compound as a highly potent and selective Pkc1 kinase inhibitor, to further demonstrate the potential of targeting the Pkc1-mediated cell wall integrity pathway for developing broad-spectrum fungicidal antifungal drugs.
Cercosporamide, an antifungal natural product, is a potent and selective Pkc1 inhibitor.
Isolation and identification of active compounds from natural product extracts is always an arduous process, particularly when active molecules are minor components. In order to rapidly evaluate these natural products to identify novel and selective Pkc1 inhibitors, we developed an agar diffusion cell-based assay that rapidly discriminates selective Pkc1 kinase inhibitors from nonselective inhibitors. This assay was developed with the assumption that if a compound selectively inhibited Pkc1, then lowering the Pkc1 cellular concentration should render the cells hypersensitive to the compounds. To that end, we generated an S. cerevisiae mutant strain dependent on the regulated expression of Pkc1 from a TetR promoter in the presence or absence of doxycycline. We first used staurosporine to validate this assay, as staurosporine was previously shown to preferentially inhibit Pkc1 in S. cerevisiae (49). Indeed, as expected, addition of doxycycline to repress Pkc1 expression greatly enhanced the sensitivity of the mutant yeast strain to staurosporine and, by contrast, it had no effect on the sensitivity of the mutant strain to fluconazole (see Fig. S1 in the supplemental material).
We then rapidly evaluated all the natural products to identify those showing increased activity in the presence of doxycycline in the agar diffusion assay. We reasoned that those showing increased whole-cell activity in the presence of doxycycline would likely contain selective Pkc1 inhibitors and, thus, we focused our efforts on the isolation and identification of Pkc1 kinase inhibitors from these natural products only. One of the Pkc1 inhibitors identified, as described in detail below, is a known antifungal natural product, cercosporamide, whose mode of antifungal activity was not previously known.
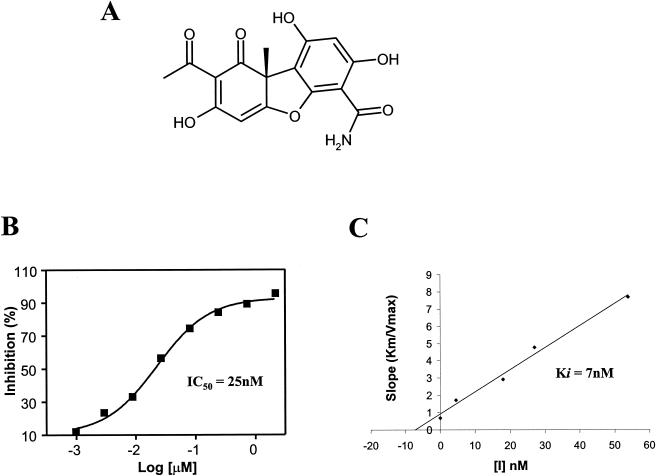
A partially purified sample derived from a fungal organism had a highly potent CaPkc1 activity, with an IC50 of <0.25 μg/ml, and further showed markedly enhanced antifungal activity against the mutant yeast strain in the presence of doxycyline, suggesting that this sample contains a selective Pkc1 inhibitor(s) (see Fig. S2 in the supplemental material). The sample was fractionated into subfractions, and the activity in subfractions was then followed by both the agar diffusion assay and enzyme activity assay. A highly potent Pkc1 inhibitor was then isolated from active fractions by chromatography and structurally determined by NMR and MS to be cercosporamide (Fig. 5A; see Fig. S3 and Table S1 in the supplemental material), a previously known antifungal natural product (45). CaPkc1 kinase assays showed that cercosporamide is a highly potent, ATP competitive inhibitor with an IC50 of <40 nM and Ki of 7 nM (Fig. 5B and C).
FIG. 5.
Isolation and determination of cercosporamide as a selective Pkc1 kinase inhibitor. (A) The active compound purified and its structure determined by NMR and MS as cercosporamide, a previously known antifungal natural product with an unknown mode of action. (B and C) Cercosporamide is a highly potent, ATP-competitive Pkc1 kinase inhibitor, with an IC50 of <50 nM and a Ki of <7 nM.
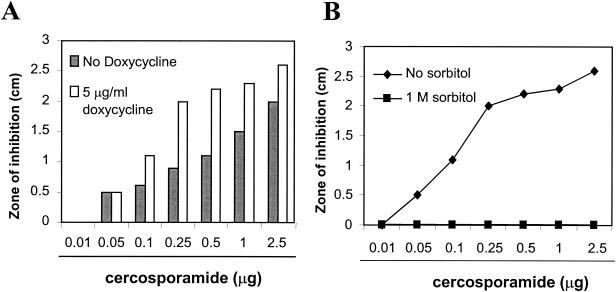
As reported previously (45), cercosporamide showed potent antifungal activity, with a MIC of 10 μg/ml against both C. albicans and A. fumigatus in the present study. Moreover, cercosporamide showed markedly enhanced antifungal activity against the mutant yeast cells in the presence of doxycyline (Fig. 6A). Genetic studies have established that growing cells in a high-osmotic medium can rescue the lethality caused by defective Pkc1 (36, 37). Thus, if cercosporamide indeed selectively inhibits Pkc1, then its antifungal activity should also be suppressible by high osmotic medium. We thus tested this hypothesis using the mutant yeast strain in the presence of doxycycline to increase assay sensitivity with or without incorporation of 1 M sorbitol. As described above, this mutant strain was highly sensitive to cercosporamide in the absence of sorbitol. However, in the presence of 1 M sorbitol, the antifungal activity of cercosporamide was completely suppressed (Fig. 6B). The results therefore demonstrate that cercosporamide is indeed a selective inhibitor of Pkc1. This selectivity was further confirmed by assays against a panel of human protein kinases (Table 2).
FIG. 6.
Antifungal activity of cercosporamide is mediated through selective inhibition of Pkc1 kinase activity. (A) Cercosporamide has markedly enhanced antifungal activity in the presence of doxycycline. (B) Suppression of the antifungal activity of cercosporamide by 1 M sorbitol.
TABLE 2.
Cercosporamide is a highly potent and selective CaPkc1 inhibitor
| Kinase | IC50 (μM) | Selectivity ratio vs CaPkc1 |
|---|---|---|
| CaPkc1 | 0.044 | 1 |
| PKCα | 1.022 | 23 |
| PKCβ | 0.349 | 8 |
| PKCγ | 5.772 | 130 |
| PKCɛ | 1.574 | 36 |
| CDK2/cyclin A | 12.548 | 283 |
| CDK1/cyclin B | 5.199 | 117 |
| CDK4/cyclin D1 | >20 | >451 |
| CDK2/cyclin E | >20 | >451 |
| GSK 3β | 1.338 | 30 |
| CAMKII | 1.505 | 34 |
| PKAa | >20 | >451 |
Synergistic antifungal activity of cercosporamide and a β-1,3-glucan synthase inhibitor.
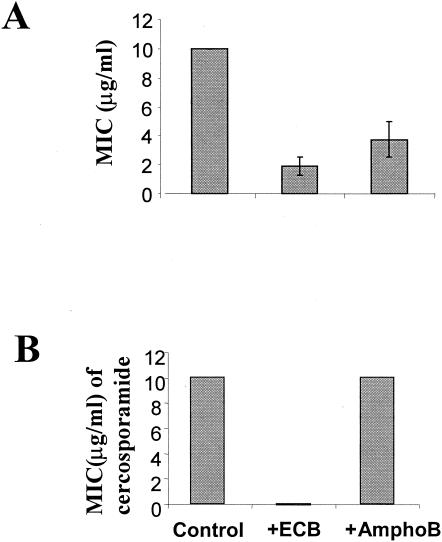
The role of Pkc1 in cell wall integrity and biosynthesis is largely mediated through expression of genes involved in cell wall biosynthesis and regulation of the β-1,3-glucan synthase activity. We thus suspected that the Pkc1 kinase inhibitor cercosporamide and a β-1,3-glucan synthase inhibitor, an analog of echinocandin (13), would show synergistic antifungal activities, as they target two key components of the cell wall biosynthetic pathway. To test this hypothesis, we first determined a concentration at which the echinocandin analog and amphotericin B by themselves did not have apparent inhibitory activity on C. albicans growth. As amphotericin B, a potent antifungal agent, has a different mode of action from the echinocandin analog, it was included in this experiment as a negative control. We then assayed the antifungal activity of cercosporamide against C. albicans in the presence of the echinocandin analog and amphotericin B at the respective concentrations. Cercosporamide, echinocandin analog, and amphotericin B have MICs of 10, 1.8, and 3 μg/ml against C. albicans, respectively (Fig. 7A). We found that the echinocandin analog at 0.16 μg/ml and amphotericin B at 1.25 μg/ml showed no effect on C. albicans growth. However, in the presence of 0.16 μg of the echinocandin analog/ml, the MIC of cercosporamide against C. albicans was dramatically reduced by more than 270-fold (Fig. 7B). By contrast, amphotericin B had no effect on the MIC of cercosporamide (Fig. 7B). To further prove a synergistic antifungal activity between cercosporamide and the echinocandin analog, the MICs of cercosporamide and the echinocandin analog were tested over increasing concentrations of each other in a checkerboard format in 96-well titer plates as described previously (31). Taking the MICs as an endpoint, the FIC indices of all the tested concentration combinations between cercosporamide and the echinocandin analog were <1, whereas the FIC indices between cercosporamide and amphotericin B equaled 1. Together, the data thus demonstrate that cercosporamide and echinocandin indeed have synergistic antifungal activities.
FIG. 7.
Cercosporamide and a β-1,3-glucan synthase inhibitor, an echinocandin analog, together have markedly enhanced antifungal activity. (A) MICs of cercosporamide (Cercos), echinocandin analog (ECB), and amphotericin B (amphoB) against C. albicans. (B) Dramatically increased antifungal activity of cercosporamide, seen as a marked decrease in its MIC in the presence of ECB at an ECB concentration that by itself has no apparent antifungal activity.
DISCUSSION
The highly conserved Pkc1 protein kinase plays a central role in the cell wall integrity-signaling pathway and is essential for cell wall biosynthesis and remodeling. We conducted a CaPkc1 kinase activity-based HTS to discover Pkc1 kinase inhibitors as potential broad-spectrum fungicidal antifungal drugs. As reported in this paper, the successful discovery of cercosporamide, a previously known antifungal compound, as a highly selective Pkc1 kinase inhibitor and subsequent establishment of its mode of antifungal activity via inactivation of Pkc1 have convincingly demonstrated the enormous potential of targeting the cell wall integrity-signaling pathway for antifungal drug discovery.
Cercosporamide, originally isolated as a phytotoxin from the plant fungal pathogen Cercosporadium henningsii, was shown to have broad-spectrum antifungal activity (45). Subsequently, it was shown that S. cerevisiae cells with cell wall defects are highly sensitive to this compound (18), suggesting that it might have an activity against the fungal cell wall. However, its mode of antifungal activity was not established. Here we present several lines of evidence demonstrating that fungal Pkc1 is its molecular target, thus both providing a molecular explanation for the previous observations and also establishing its mode of action. Firstly, in vitro enzyme assays show that cercosporamide is a highly potent, ATP-competitive Pkc1 kinase inhibitor. Moreover, this activity against Pkc1 appears to be highly selective, as it has no or very little activity against a panel of other serine/threonine protein kinases. Secondly, S. cerevisiae cells with lowered Pkc1 kinase activity are highly sensitive to cercosporamide. This is in perfect agreement with Pkc1 as its cellular target and also provides further evidence for its Pkc1 selectivity, because lowering the cellular level of its target Pkc1 would obviously require much less cercosporamide to achieve a complete inhibition of Pkc1 activity. Thirdly, and most importantly, high osmolarity in the growth medium can completely suppress the antifungal activity of cercosporamide. This is consistent with previous findings from genetic studies that high-osmotic growing conditions help stabilize cell wall defects, thus allowing apparently normal cell growth even in the presence of an inactive cell wall integrity-signaling pathway (29, 36, 37). Together, the results further demonstrate that Pkc1 indeed is the molecular target of cercosporamide. Therefore, our present studies helped elucidate the mode of action of a previously known antifungal compound.
The fungal Pkc1 protein kinases are highly conserved both structurally and functionally in fungi. Some limited biochemical studies of the Pkc1 kinases from a few fungal species have shown that they appear also to share very similar, if not identical, biochemical properties (3, 28, 46). Although the fungal Pkc1 kinase is biochemically and functionally different from mammalian PKCs, fungal Pkc1 and mammalian PKCs do share high homology in their catalytic domains (33). Most protein kinase inhibitors are ATP competitive; therefore, in the beginning we were concerned whether such high homology in the catalytic domains provides sufficient structural differences in the ATP-binding pockets to allow the development of selective Pkc1 inhibitors as antifungal drugs for human application. To test the possibility, we first assayed some highly potent and selective PKCβ inhibitors we developed at Lilly for activity against CaPkc1 and found that these kinase inhibitors had virtually no activity against CaPkc1, even at high concentrations. Lack of activity of these selective PKCβ inhibitors indicates that the ATP-binding pocket of fungal Pkc1 indeed has sufficient structural differences to allow the development of Pkc1 selective inhibitors. The fact that our investigators have been able to develop selective PKCβ inhibitors (8), even though human C family kinases share even higher overall homology in their kinase domains than that of Pkc1, further boosted our confidence in developing selective Pkc1 inhibitors. The subsequent discovery of a selective Pkc1 inhibitor in cercosporamide, as described in this paper, therefore has further validated this possibility.
In this study we also further biochemically characterized CaPkc1 both expressed in insect cells and in C. albicans. To express CaPkc1 in C. albicans, we engineered a regulated expression system based on the GAL1-10 promoter. This expression system, as in S. cerevisiae, should be broadly applicable to studying gene functions in this important human pathogen. Like a previous report for S. cerevisiae Pkc1 (25), we also found that CaRho1 markedly activates Pkc1 kinase activity in vitro in the presence of PS. However, we also found that PS alone can markedly activate Pkc1 activity when applied immediately after sonication into the assay mixture. Perhaps, binding of Rho1 to the C-1 domain normally allows easy access of PS to the C-2 domain to activate Pkc1 kinase activity. Then, when applied freshly by sonication, PS is in a physical state that is more conducive to binding to the C-2 domain, even in the absence of Rho1 protein. PS activation of Pkc1 kinase from other fungi in the absence of Rho1 has also been observed (28). However, the exact nature of this activation of Pkc1 by PS remains to be elucidated. It is puzzling though that, unlike activation of mammalian PKCs (16), PS when prepared in a mixed micelle system failed to activate Pkc1. Therefore, the physiological significance of PS activation of Pkc1 remains to be determined. In fact, a role of PS in Pkc1 regulation in vivo has not been observed. Fungal Pkc1 kinase also has a C-1 domain (33). In a recent study, Pkc1 was shown to play a role in activation of Cdc28 at START for progression through G1 in S. cerevisiae, and Pkc1-dependent activation of Cdc28 is associated with an increase in the cellular level of DAG (32). However, DAG activation of Pkc1 kinase so far has not been demonstrated in vitro. Similarly we also did not observe any activation of CaPkc1 in vitro by DAG when applied in an identical manner as PS. More in-depth future research employing combined molecular and biochemical approaches, such as domain shuffling as reported recently by Schmitz et al. (41), will help dissect the function of C-1 and C-2 domains and perhaps also shed some light on potential roles, if any, of PS and DAG in Pkc1 regulation within biological contexts.
The Pkc1-mediated cell wall integrity-signaling pathway has been best characterized in S. cerevisiae. This pathway is activated in response to cell wall perturbation and regulates the expression of a core set of genes implicated in cell wall biogenesis via activation of two transcription factors, Rlm1 and Swi4, by phosphorylation (23, 27, 47). Interestingly, we found that the Pkc1 kinase inhibitor cercosporamide and the β-1,3-glucan synthase inhibitor echinocandin analog together showed a remarkably high synergy in their antifungal activities. This finding is consistent with a recent study showing that S. cerevisiae mutants of the Pkc1-mediated cell wall integrity-signaling pathways are hypersensitive to the β-1,3-glucan synthase inhibitor caspofungin (39). Furthermore, it has been shown by genome-wide expression profiling in S. cerevisiae that the Pkc1-mediated signaling pathway is activated as a compensatory response to inhibition of β-1,3-glucan synthase activity (39). As Pkc1 and β-1,3-glucan synthase are two key components required in cell wall biosynthesis and they have nonoverlapping and essential functions, it is thus expected that together they would exert a synergy in their antifungal activities by eliminating the aforementioned feedback compensatory responses. This finding points to a potential highly powerful combination therapy to treat fungal infections. Perhaps, because of its effectiveness and targeting of two key and nonoverlapping functions of cell wall biosynthesis, the combination therapy would also minimize the emergence of resistance. In fact, because the current antifungal agents have limited effectiveness against many fungal infections, combination therapy with antifungal agents with different modes of action has been increasingly advocated and used to treat refractory infections (2).
In conclusion, we explored the potential of targeting the Pkc1-mediated cell wall integrity-signaling pathway for antifungal drug discovery by conducting an HTS based on the C. albicans Pkc1 protein kinase. Among many potent Pkc1 kinase inhibitors identified from a chemical library was a known antifungal compound, cercosporamide, previously with an unknown mode of action. We showed that cercosporamide is actually a highly selective Pkc1 kinase inhibitor, and we further established that its antifungal activity is mediated through inhibition of the Pkc1 kinase activity. The discovery of a known antifungal compound as a highly potent and selective Pkc1 inhibitor thus demonstrates the great potential of the cell wall signaling pathway for the discovery of novel antifungal drugs.
Supplementary Material
Acknowledgments
We thank Robert Dean, Shannon Brier, Eddie Angleton, Xiangdong Qu, John Locklear, and Nailya Gilyazova for technical support and Sheng-bin Peng for critically reading the manuscript.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org.
REFERENCES
- 1.Alani, A., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiple disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniadou, A., and D. P. Kontoyiannis. 2003. Status of combination therapy for refractory mycoses. Curr. Opin. Infect. Dis. 16:534-545. [DOI] [PubMed] [Google Scholar]
- 3.Antonsson, B., S. Montessuit, L. Friedli, M. A. Payton, and G. Paravicini. 1994. Protein kinase C in yeast: characteristics of the Saccharomyces cerevisiae PKC1 gene product. J. Biol. Chem. 269:16821-16828. [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seiman, J. A. Smith, and K. Struhl (ed.). 1992. Current protocols in molecular biology, 2nd ed. Wiley, New York, N.Y.
- 5.Banuett, F. 1998. Signaling in the yeasts: an information cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 62:249-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berenbaum, M. C. 1978. A method for testing for synergy with any number of agents. J. Infect. Dis. 137:122-130. [DOI] [PubMed] [Google Scholar]
- 7.Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19678-19682. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., A. M. Jack, and N. E. Cameron. 2002. Effects of the protein kinase Cβ inhibitor LY333531 on neural and vascular function in rats with streptozotocin induced diabetes. Clin. Sci. 103:311-321. [DOI] [PubMed] [Google Scholar]
- 9.Drgonova, J., T. Grgon, K. Tanaka, R. Kollar, G. C. Chen, R. A. Ford, C. S. M. Chan, Y. Takai, and E. Cabib. 1996. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272:277-279. [DOI] [PubMed] [Google Scholar]
- 10.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fostel, J. M., and P. A. Lartey. 2000. Emerging novel antifungal agents. Drug Disc. Today 5:25-32. [DOI] [PubMed] [Google Scholar]
- 12.Geogopapadakou, N. H., and T. J. Walsh. 1996. Antifungal agents: chemotherapeutic targets and immunologic strategies. Antimicrob. Agents Chemother. 40:279-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, L. J., P. Marder, L. L. Mann, L. C. Chio, and W. L. Cirrent. 1999. LY303366 exhibits rapid and potent fungicidal activity in flow cytometric assays of yeast viability. Antimicrob. Agents Chemother. 43:830-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groll, A. H., and T. J. Walsh. 2001. Uncommon opportunistic fungi: new nosocomial threats. Clin. Microbiol. Infect. Dis. 7:8-24. [DOI] [PubMed] [Google Scholar]
- 15.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannun, Y. A., C. R. Loomis, and R. M. Bell. 1985. Activation of protein kinase C by Triton X-100 mixed micelles containing diacylglycerol and phosphatidylserine. J. Biol. Chem. 260:10039-10043. [PubMed] [Google Scholar]
- 17.Harrison, J. C., E. S. Bardes, Y. Ohya, and D. J. Lew. 2001. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3:417-420. [DOI] [PubMed] [Google Scholar]
- 18.Hong, Z., P. Mann, N. H. Brown, L. E. Tran, K. J. Shaw, R. S. Hare, and B. DiDomenico. 1994. Cloning and characterization of KNR4, a yeast gene involved in (1,3)-beta-glucan synthesis. Mol. Cell. Biol. 14:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor SWI4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-50013. [PMC free article] [PubMed] [Google Scholar]
- 20.Jamison, J., S. Levy, X. Sun, D. Zeckner, W. Current, M. Zweifel, M. Rodriguez, W. Turner, and S. H. Chen. 2000. Synthesis and antifungal activity of pseudomycin side-chain analogues. Bioorg. Med. Chem. Lett. 10:2101-2105. [DOI] [PubMed] [Google Scholar]
- 21.Johnston, M. 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51:458-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julian, R. K., R. E. Higgs, J. D. Gygi, and M. D. Hilton. 1998. A method for quantitatively differentiating crude natural extracts using high-performance liquid chromatography-electrospray mass spectrometry. Anal. Chem. 70:3249-3254. [DOI] [PubMed] [Google Scholar]
- 23.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 24.Kamada, Y., U. S. Jung, J. Piotrowsky, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genet. Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 25.Kamada, Y., H. Qadota, C. P. Python, Y. Anraku, Y. Ohya, and D. E. Levin. 1996. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 271:9193-9196. [DOI] [PubMed] [Google Scholar]
- 26.Kondoh, O., Y. Tachibana, Y. Ohya, M. Arisawa, and T. Watanabe. 1997. Cloning of the RHO1 gene from Candida albicans and its regulation of β-1,3-glucan synthesis. J. Bacteriol. 179:7734-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken, J. Arroyo, J. D. Hoheisel, and J. Francois. 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278:20345-20357. [DOI] [PubMed] [Google Scholar]
- 28.Lendenfeld, T., and C. P. Kubicek. 1998. Characterization and properties of protein kinase C from the filamentous fungus Trichoderma reesei. Biochem. J. 330:689-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin, D. E., and E. Bartlett-Heubusch. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 31.Marchetti, O., P. Moreillon, M. P. Glauser, J. Bille, and D. Sanglard. 2000. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 44:2373-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marini, N. J., E. Meldrum, B. Buehrer, A. V. Hubberstey, D. E. Stone, A. Traynor-Kaplan, and S. I. Reed. 1996. A pathway in the yeast cell division cycle linking protein kinase C (Pkc1) to activation of Cdc28 at START. EMBO J. 15:3040-3052. [PMC free article] [PubMed] [Google Scholar]
- 33.Mellor, H., and P. J. Parker. 1998. The extended protein kinase C superfamily. Biochem. J. 332:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nonaka, H., K. Tanaka, H. Hirano, T. Fujiwara, H. Kohno, M. Umikawa, A. Mino, and Y. Takai. 1995. A downstream target of Rho1 small GTP-binding protein is Pkc1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14:5831-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onishi, J., M. Meinz, J. Thompson, J. Curotto, S. Dreikorn, M. Rosenbach, C. Douglas, G. Abruzzo, A. Flattery, L. Kong, A. Cabello, F. Vicente, F. Pelaez, M. T. Diez, I. Martin, G. Bills, R. Giacobbe, A. Dombrowski, R. Schwartz, S. Morris, G. Harris, A. Tsipouras, K. Wilson, and M. B. Kurtz. 2000. Discovery of new antifungal (1,3)-β-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 44:368-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paravicini, G., M. Cooper, L. Friedli, D. J. Smith, J. L. Carpentier, L. S. Klig, and M. A. Payton. 1992. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol. Cell. Biol. 12:4896-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paravicini, G., A. Mendoza, B. Antonsson, M. Cooper, C. Losberger, and M. A. Payton. 1996. The Candida albicans PKC1 gene encodes a protein kinase C homolog necessary for cellular integrity but not dimorphism. Yeast 12:741-756. [DOI] [PubMed] [Google Scholar]
- 38.Ponikau, J. U., D. A. Sherries, E. B. Kern, H. A. Homburger, E. Frigas, T. A. Gaffey, and G. D. Roberts. 1999. The diagnosis and incidence of allergic fungal sinusitis. Mayo Clin. Proc. 74:877-884. [DOI] [PubMed] [Google Scholar]
- 39.Reinoso-Martin, C., C. Schuller, M. Schuetzer-Muehlbauer, and K. Kuchler. 2003. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2:1200-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawyers, C. L. 2003. Opportunities and challenges in the development of kinase inhibitor therapy for cancer. Genet. Dev. 17:2998-3010. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz, H. P., J. Jockel, C. Block, and J. J. Heinisch. 2001. Domain shuffling as a tool for investigation of protein function: substitution of the cystein-rich region of Raf kinase and Pkcη for that of yeast Pkc1p. J. Mol. Biol. 311:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Self, A. J., and A. Hall. 1995. Measurement of intrinsic nucleotide exchange and GTP hydrolysis rates. Methods Enzymol. 256:67-76. [DOI] [PubMed] [Google Scholar]
- 43.Sheehan, D. J., C. A. Hitchcock, and C. M. Sibley. 1999. Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 12:40-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smits, G. J., J. C. Kapteyn, H. van den Ende, and F. M. Klis. 1999. Cell wall dynamics in yeast. Curr. Opin. Microbiol. 2:348-352. [DOI] [PubMed] [Google Scholar]
- 45.Sugawara, F., S. A. Strobel, G. A. Strobel, R. D. Larsen, D. Berglund, G. Gray, N. Takahashi, T. J. Stout, and J. Clardy. 1991. The structure and biological activity of cercosporamide from Cecosporidium henningsii. J. Org. Chem. 56:909-910. [Google Scholar]
- 46.Watanabe, M., C. Y. Chen, and D. E. Levin. 1994. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with substrate specificity to that of mammalian PKC. J. Biol. Chem. 269:16829-16836. [PubMed] [Google Scholar]
- 47.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye, X. S., L. Wu and S. A. Osmani. 2001. Signal transduction in filamentous fungi: regulation of protein kinases, p. 157-174. In N. Talbot (ed.), Molecular and cellular biology of filamentous fungi. Oxford Press, New York, N.Y.
- 49.Yoshida, S., E. Ikeda, I. Uno, and H. Mitsuzawa. 1992. Characterization of a staurosporine- and temperature-sensitive mutant, stt1, of Saccharomyces cerevisiae: STT1 is allelic to PKC1. Mol. Gen. Genet. 231:337-344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.