Abstract
Numerous studies have indicated that the central apparatus plays a significant role in regulating flagellar motility, yet little is known about how the central pair of microtubules or their associated projections assemble. Several Chlamydomonas mutants are defective in central apparatus assembly. For example, mutant pf15 cells have paralyzed flagella that completely lack the central pair of microtubules. We have cloned the wild-type PF15 gene and confirmed its identity by rescuing the motility and ultrastructural defects in two pf15 alleles, the original pf15a mutant and a mutant generated by insertional mutagenesis. Database searches using the 798-amino-acid polypeptide predicted from the complete coding sequence indicate that the PF15 gene encodes the Chlamydomonas homologue of the katanin p80 subunit. Katanin was originally identified as a heterodimeric protein with a microtubule-severing activity. These results reveal a novel role for the katanin p80 subunit in the assembly and/or stability of the central pair of flagellar microtubules.
Cilia and flagella represent some of the most complex and highly ordered macromolecular assemblies in eukaryotic cells. The structural components of these organelles are well conserved and include the inner and outer dynein arms, radial spokes, and a central apparatus comprised of two singlet microtubules with their associated projections. While recent progress has been made toward defining the role of the central apparatus in flagellar motility (40, 43, 52, 53; for reviews, see references 56 and 66), we still know very little about the mechanism of central apparatus assembly.
The central apparatus includes two single microtubules and their associated projections, the central pair bridges linking the two tubules and the central pair caps attached to the distal (plus) ends of the microtubules (reviewed in reference 56). The two tubules, C1 and C2, are structurally and biochemically distinguishable. The two prominent projections on the C1 microtubule, the 1A and 1B projections, are longer than the two prominent projections on the C2 microtubule, termed 2A and 2B (1, 16, 22). Additional less prominent projections (1C, 1D, and 2C) have recently been described (38, 39). At least 23 polypeptides in addition to tubulin comprise the central apparatus (1, 16); 10 are unique to the C1 microtubule and 7 are unique to the C2 microtubule (16).
The central pair of microtubules assemble with their plus ends distal to the cell body (17). However, unlike the nine doublet microtubules, they are not nucleated from the triplet microtubules of the basal bodies; in fact, the proximal end of the central apparatus does not appear to attach to any flagellar structures. Using Tetrahymena, Thazhath et al. have shown that cells with lethal mutations in the polyglycylation domain of beta-tubulin assemble axonemes that lack the central pair (59). Therefore, tubulin polyglycylation may be important for central apparatus assembly or stability. Using trypanosomes, McKean et al. (34) have recently demonstrated that gamma tubulin is specifically required for central apparatus assembly, and in Chlamydomonas, gamma tubulin has been localized to the transition zone (51), where central apparatus assembly is believed to initiate. Therefore, gamma tubulin may play a role in central microtubule assembly. In addition, at the distal tips of the central tubules there are capping structures that serve as connectors between the central tubules and the flagellar membrane; several studies have indicated that the capping structures may be a site for regulating the assembly and disassembly of these microtubules (14).
To identify central apparatus components that play a role in regulating motility or that are involved in regulating the assembly and/or stability of the central microtubules, we have taken advantage of insertional mutagenesis in Chlamydomonas reinhardtii to generate central apparatus-defective mutants in which the mutant gene is tagged by a molecular marker (58). Here we report the PF15p sequence and localization. The flagella of mutant pf15 cells are paralyzed and completely lack the entire central apparatus. Using an insertional allele of pf15, we cloned a fragment of DNA flanking the plasmid integration site and obtained full-length genomic and cDNA sequences. PF15 encodes the Chlamydomonas homologue of the p80 subunit of the microtubule-severing protein katanin. PF15p localizes to the flagellar axoneme. Our results suggest a new role for katanin in the assembly and/or stability of the axonemal central pair of microtubules.
MATERIALS AND METHODS
Cell strains and media.
The B4 mutant was obtained from Lynne Quarmby (Simon Fraser University, British Columbia, Canada). The B4 mutant was generated by transforming a nit strain with the structural gene for nitrate reductase as previously described (19). Strains L5 (nit1 apm1-19 mt+) and L8 (nit1 apm1-19 mt) were provided by L. W. Tam and were used as parents in backcrosses with the B4 mutant to determine cosegregation of the nit+ phenotype with the motility defect. The central apparatus-defective mutants pf15a (CC807+) and pf20 (CC22+ and CC1030+) were provided by the Chlamydomonas Genetic Center, Duke University, Durham, N.C. The arg7 strain (32) used to construct double mutants for cotransformation experiments and the central pair mutants pf6, pf16, pf18, and pfI9 were provided by Mary Porter, University of Minnesota. All cells were grown in constant light in SGII, SGII-NO3 (27, 48, 58), or TAP medium (23). To construct a pf20 strain that expressed a hemagglutinin (HA)-tagged PF15 gene, we crossed pf20 cells with a B4 strain that had been transformed with the PF15-HA construct (see below for details of transformation and the PF15-HA construct, pPF15cHA). Meiotic progeny with paralyzed flagella were screened by PCR to determine which strains maintained the PF15-HA plasmid. Positive strains were then assessed by Western blotting (described below) to confirm the expression of the PF15-HA construct and the lack of expression of PF20.
Transformation.
High-efficiency transformation was achieved by using the glass bead procedure of Kindle (27). For the transformation of arg7 cells, cells were transformed with 1 μg of plasmid pARG7.8, which carries the wild-type Chlamydomonas arginino-succinate lyase gene (13). For transformation using a dominant selectable marker for emetine resistance, cells were transformed as described above with the plasmid pJN4, which carries a mutant Chlamydomonas gene for the ribosomal protein S14 that confers resistance to emetine upon transformation (41, 42). To test genomic lambda clones and plasmids for the rescue of mutant phenotypes, we cotransformed 1 to 3 μg of each genomic clone with plasmid DNA carrying a selectable marker gene.
Whole-cell, flagellar basal body complex, flagellar axoneme, and axonemal extract preparation.
Flagella were severed from cell bodies by the use of dibucaine (28, 65), isolated by differential centrifugation, and resuspended in HMDEdNa (10 mM HEPES, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM EDTA, 30 mM NaCl, pH 7.4). Axonemes were isolated with 0.5% NP-40 in HMDEdNa to remove flagellar membranes. For Western blots, axonemal extracts were prepared by suspending axonemes in HMDEdKI (contains 0.5 M KI) at a concentration of 6 mg/ml for 30 min on ice. The resulting extracts and extracted axonemes were isolated by differential centrifugation. Flagellar basal body complexes were prepared according to a previously described method (66). Cells (8 × 108) were treated with autolysin to remove cell walls. The cells were subsequently lysed, the flagellar basal body complexes were purified on Percoll gradients, and the final pellet was resuspended in 50 μl of HMDEdNa.
Electron microscopy.
For analyses of flagellar defects, axonemes from mutants of interest were prepared for thin-section electron microscopy. Specimens were fixed with 1% glutaraldehyde and 1% tannic acid in 0.1 M sodium cacodylate, postfixed in osmium tetroxide, dehydrated in a graded series of ethanol, and embedded in Quetal resin (EM Sciences, Fort Washington, Pa.). Uniform silver-gray sections were mounted on Formvar-coated, carbon-stabilized copper grids, stained with uranyl acetate and Reynolds lead citrate, and examined at 80 kV in a transmission electron microscope (either a JEOL model 100CX, JEOL model 1200CX, or Hitachi H-600 instrument).
Genetic analysis.
Techniques for mating and tetrad analysis were performed as described previously (25). Meiotic progeny were scored for Nit+ or Nit− phenotype by replica plating on SGII-NO3 and SGII agar plates. The motility phenotype was scored by resuspending meiotic progeny in liquid medium. Pairs of mutants were tested for complementation by mating strains containing complementing auxotrophic markers (for example, B4 arg7 × pf15 nit1) and plating them on minimal medium to select for stable diploid strains. For assessments of complementation, at least 10 diploids from each cross were scored for motility in liquid medium.
Isolation of genomic sequence flanking integrated plasmid.
Genomic DNA flanking the site of insertion of the integrating plasmid in the B4 mutant was obtained by construction of a genomic DNA library by use of the λFIX II vector (Stratagene, La Jolla, Calif.). The genomic DNA (20 μg) was partially digested with 0.075 U of MboI/μg of DNA at 37°C for 1 h. After a partial fill-in, the DNA was ligated to the λFIX II vector arms and packaged by use of Gigapack II XL packaging extract (Stratagene) according to the protocols supplied by the manufacturer. The resulting library was transfected into XL1-Blue MRA(P2) host cells (Stratagene) and screened with the pUC119 plasmid as a hybridization probe. Phage DNA from positive plaques was prepared by polyethylene glycol precipitation (8). Genomic DNA flanking the plasmid insertion site was identified by digesting the DNA from positive clones with various enzymes and identifying fragments that did not hybridize with the radiolabeled DNA from plasmid pMN56. Procedures for probe isolation, DNA blot analysis, and plaque-lift hybridizations were described previously (54).
Cloning PF15 by library screening and mutant rescue.
A genomic library constructed with a DNA from the A10 strain (55) in the lambda phage vector λFIX II (Stratagene) was screened by using a fragment of cloned sequence flanking the insertion site in the mutant B4 as a probe. Approximately 4 × 105 plaques in each of two successive rounds were screened to obtain the PF15 gene. Genomic DNA clones that could rescue the mutant phenotype upon transformation were further subcloned. Plasmid pPF15 was constructed by digesting the lambda clone with SacI and NotI, yielding a 6.5-kb fragment that was ligated into pBluescript and transformed into Escherichia coli DH10B (Gibco BRL).
RNA preparation and Northern blots.
Cells were grown in TAP medium to mid- to late-log phase and divided; one-half of the culture was deflagellated by pH shock. Total RNAs were prepared from nondeflagellated cells and from deflagellated cells 45 min after deflagellation (64). Poly(A)+ RNA was prepared by using either Magnetight oligo(dT) particles (Straight A's kit; Novagen) or an Oligotex mRNA Midi kit (Qiagen, Inc.). Approximately 10 μg of poly(A)+ RNA was fractionated in formaldehyde-agarose gels (49), transferred to Hybond N+ membranes (Amersham, Arlington Heights, Ill.), and hybridized with 32P-labeled probes according to the manufacturer's instructions.
Determining coding sequence for PF15 and sequence analysis of PF15, pf15a, and B4 mutants.
A 6.5-kb SacI-NotI fragment of genomic DNA that rescued motility upon transformation of B4 and pf15a mutants was sequenced. Putative exons were identified with the GreenGenie algorithm (http://www.cse.ucsc.edu/∼dkulp/cgi-bin/greenGenie) and by BLAST searches of sequence databases (3, 4). These combined approaches indicated that the 6.5-kb fragment contained the complete coding sequence of the p80 subunit of katanin. All exon splice junctions were confirmed by reverse transcription of RNA isolated from wild-type cells followed by PCR amplification (RT-PCR). First-strand cDNA synthesis was performed with specific 17-mer oligonucleotide primers and SuperScript RT II or ThermoScript reverse transcriptase (Gibco-BRL Life Technologies). For the amplification of double-stranded cDNAs, specific primers (17- to 25-mers, with melting temperatures [Tms] of 72 to 74°C) were used, together with purified products from the first-strand cDNA synthesis reaction, in a buffer containing Taq DNA polymerase (Fisher Scientific), buffer A from the manufacturer, and 5% formamide. Amplification conditions consisted of a hot start at 94°C for 5 min and 80°C for 10 min, followed by 35 cycles of 94°C for 1 min, 58°C for 45 s, and 72°C for 2 min. Nested primers were used for a second round of amplification. The resulting fragments were subcloned into pCRII by using an Original TA cloning kit (Invitrogen) for DNA sequencing. The 5′ and 3′ rapid amplification of cDNA ends (RACE, v. 2.0, or 5′ RACE system; Invitrogen) was used to determine the 5′ and 3′ ends of the transcript (21).
To identify the lesion in the pf15a allele, we amplified the entire coding sequence of the PF15 gene, with genomic DNA isolated from pf15a as a template and with pairs of gene-specific primers. PCR products were sequenced directly. The T→A transversion in the pf15a allele was confirmed by RT-PCR using RNA isolated from pf15a. To identify the site of plasmid insertion in the B4 mutant, we sequenced a lambda clone containing genomic DNA isolated from the B4 mutant and flanking the insertion site. The insertion site was confirmed by PCR, with B4 genomic DNA as a template and with two primers, one specific for pUC119 and one specific for the p80 gene. Transcription of the mutated gene was confirmed by performing RT-PCR using RNA isolated from the B4 mutant. RT was performed by using an oligo(dT) primer followed by two rounds of amplification using universal amplification primers and nested gene-specific primers.
For some cloned genomic DNAs and cDNAs, sequencing was carried out by using Sequenase 2.0 (United States Biochemical Corp., Cleveland, Ohio) according to the manufacturer's instructions. For some templates, sequencing was performed by the DNA Sequencing Facility, Iowa State University, Ames, or the Molecular Biology Core Facility, Dartmouth College, Hanover, N.H. The GCG programs were used for sequence assembly (15). Searches of the GenBank and SwissProt databases for sequence homologies were performed with the BLAST programs (3, 4) of the National Center for Biotechnology Information. Sequence alignments were performed with the ClustalW alignment feature of MacVector software, with an open gap penalty of 10.0 and an extend gap penalty of 0.05.
Construction of HA-tagged PF15 gene.
A plasmid encoding three copies of the nine-amino-acid HA epitope (3×HA) was obtained from Carolyn Silflow (50). For one construct, the pPF15 plasmid was digested first with XcmI and then filled in to generate blunt sites. The 3× HA plasmid was digested with ScaI and StuI to generate a blunt-ended insert. The 3× HA insert was purified and ligated to the pPF15 plasmid. When expressed, the resulting construct generates the PF15p protein with the 3× HA tag inserted 36 amino acids upstream of the carboxyl terminus (pPF15cHA). For a second construct, the pPF15 plasmid was digested with EcoRI. EcoRI cut the plasmid within the first intron (23 bp 3′ of the splice donor site) and immediately 3′ of the intron splice acceptor site. Therefore, EcoRI digestion removes the splice acceptor site. The 3× HA tag was amplified with primers that included EcoRI sites. The resulting PCR product was ligated into the pPF15 plasmid digested with EcoRI. If translated properly, the resulting construct would have eight additional amino acids (Gly, Ala, Ser, Thr, Arg, Ile, Glu, and Phe) inserted after Leu9 followed by the 3× HA tag (pPF15nHA). For both constructs, ligation of the 3× HA tag in the proper orientation and reading frame was confirmed by sequencing of the resulting plasmid.
Western blotting.
Flagella and flagellar basal body complexes were prepared from 8 × 108 cells. The final pellet of each sample was resuspended in 50 μl of HMDEdNa and equally loaded in 8% polyacrylamide gels for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. For additional Western blots, equivalent loads of flagella (6 mg/ml), axonemes, 0.5 M KI extracts, and/or extracted axonemes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 7% polyacrylamide gels. The gels were transferred for 1 h to polyvinylidene difluoride membranes (Immobilon P; Millipore). The membranes were blocked for 1 h in 5% milk in T-TBS (0.1% Tween, Tris-buffered saline, pH 7.5) and then incubated overnight at 4°C with a primary antibody. A mouse anti-HA antibody (Santa Cruz Biotechnology, Inc.) was diluted 1:500 in blocking buffer. Rabbit PF20 antibodies (55) were diluted 1:5,000. After three 5-min washes in T-TBS, the membrane was incubated for 1 h with a secondary antibody diluted in 3% milk in T-TBS. Horseradish peroxidase-conjugated anti-mouse secondary (Pierce) antibodies were diluted 1:10,000; horseradish peroxidase-conjugated anti-rabbit secondary antibodies were diluted 1:20,000 (Amersham Pharmacia Biotech). The ECL Plus Western blotting detection system was used for chemiluminescent detection (Amersham Pharmacia Biotech).
Nucleotide sequence accession number.
The Chlamydomonas PF15 sequence is available at GenBank (accession number AY597210).
RESULTS
Mutant phenotype and genetic analysis of B4 mutant with an insertional allele of pf15.
The B4 mutant was generated by transforming nit1 (nitrate reductase deficient) cells with the plasmid pMN56 containing the cloned NIT1 gene (18). B4 mutant cells have paralyzed flagella with normal lengths. A structural analysis of axonemal cross sections revealed that axonemes isolated from the B4 mutant completely lacked central microtubules (Fig. 1a), a phenotype previously described for the mutants pf15, pf18, pf19, and pf20 (45, 46, 61).
FIG. 1.
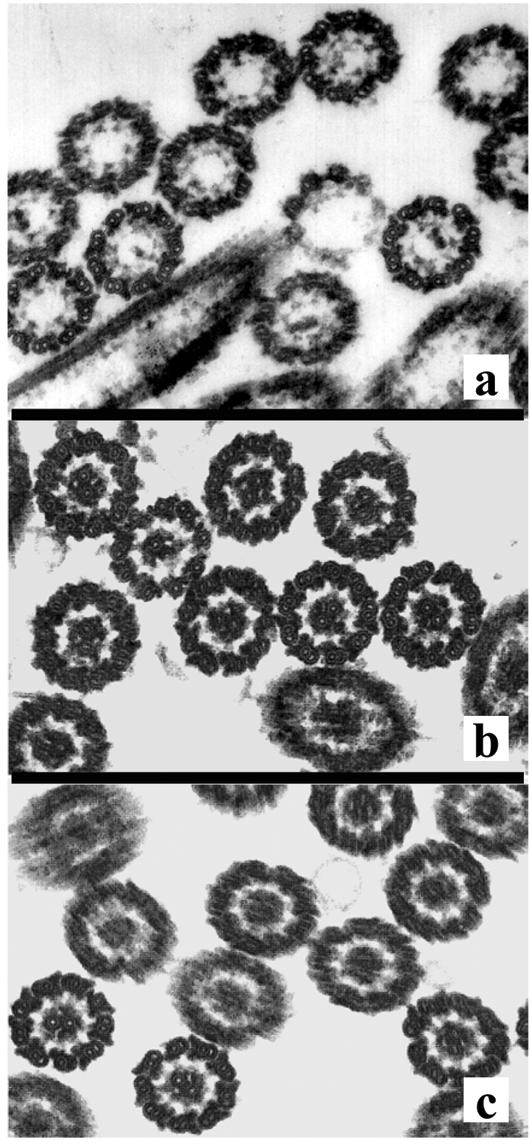
Thin-section electron microscopy of axonemes isolated from the B4 mutant (a) and from rescued mutants B4 (b) and pf15a (c) transformed with the PF15 gene. Panels are representative axonemal cross sections.
Backcrosses of the B4 mutant to a nit1 mutant strain revealed that the Nit+ phenotype cosegregated with flagellar paralysis in a minimum of 24 complete tetrads in each of two backcrosses. These results indicate that the mutant phenotype was most likely produced by gene disruption due to plasmid integration into the genome. The insertional mutant was then tested for allelism with previously identified mutants with central apparatus defects. In crosses of the B4 mutant with a pf15a mutant, no recombinants were obtained in 24 complete tetrads. Allelism was established by constructing stable diploids. Heterozygous diploids of the B4 and pf15a mutants have paralyzed flagella, indicating that these mutations are allelic.
Cloning the PF15 gene.
To clone the PF15 gene, we constructed a library with genomic DNA isolated from B4 and with lambda phage as a vector. Phage containing genomic DNA adjacent to the insertion site were identified by using the pUC119 plasmid as a probe to screen the mutant DNA library. Restriction fragments that contained only genomic DNA flanking the insertion site were identified by Southern blot analysis of the phage DNA digested with different enzymes. A 3-kb SacI fragment was identified that did not hybridize with either the nitrate reductase gene or the pUC119 vector used to generate the pf15 insertional allele (Fig. 2, probe A).
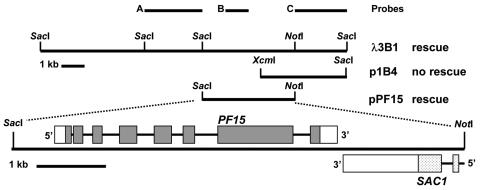
FIG. 2.
Restriction map of lambda clone and plasmids. Probe A is a 3.5-kb SacI fragment used to isolate the λ3B1 clone, and probe B is a 1.4-kb PstI fragment used as a hybridization probe for blots of DNA and RNA. Probe C is a 3-kb SacI-NotI fragment used as a hybridization probe for Southern blots. The plasmid p1B4 was generated by digesting clone λ3B1 with XcmI and SacI and ligating the indicated fragment into pBluescript. The pPF15 plasmid was created by digesting the λ3B1 clone with SacI and NotI and ligating the indicated fragment into pBluescript. The plasmid pPF15 contains coding sequences for two genes, PF15 and SAC1. Exons are indicated by dark (PF15) and light (SAC1) shading. The 5′ and 3′ untranslated regions are indicated by open squares. Introns are represented by solid lines. The pPF15 plasmid does not contain the complete coding sequence of the SAC1 gene.
To determine whether the 3-kb SacI fragment flanked the site of plasmid insertion, we used this fragment as a probe for Southern blots of genomic DNAs isolated from B4 and wild-type (A54-e18) cells digested with a variety of restriction enzymes (not shown). The restriction fragments which hybridized to this probe were of different sizes in DNAs from B4 and wild-type cells, confirming that this probe represented a piece of DNA flanking the site of plasmid insertion in the B4 mutant. The 3-kb fragment was then used as a probe to screen a wild-type genomic DNA library to obtain the complete PF15 gene (Fig. 2, probe A). A single hybridizing clone was identified (λ3B1) (Fig. 2).
Rescue of structural and motility defects in B4 and pf15 mutants.
To determine if the λ3B1 clone obtained in our screen included the complete PF15 gene, we transformed λ3B1 DNA into B4 and pf15 cells and screened the transformants for the rescue of flagellar paralysis. Successful transformants in which the genomic clone rescued the mutant phenotype were identified as swimming cells. An ultrastructural analysis of axonemes isolated from rescued cells revealed that for both the B4 and pf15 mutants, the central apparatus was restored (Fig. 1b and c). In the rescued mutants, the number of central microtubules observed in axonemal cross sections was comparable to the number of central microtubules observed in cross sections of axonemes from wild-type cells (Table 1).
TABLE 1.
Rescue of pf15 strains by transformation
| Strain | No. of central tubules present (%)a
|
||
|---|---|---|---|
| 0 | 1 | 2 | |
| Wild type | 2 | 10 | 88 |
| B4 | 100 | 0 | 0 |
| B4 transformant | 1 | 4 | 95 |
| pf15a transformant | 3 | 4 | 93 |
Percentages represent >150 transverse sections per strain.
To further delineate the PF15 gene, we used restriction fragments of λ3B1 as probes for Southern blots of genomic DNAs isolated from wild-type and B4 cells and digested with SacI (not shown). Probe A hybridized to a 3-kb SacI fragment in both wild-type and B4 cells. However, probe B hybridized to a 5.5-kb SacI fragment of B4 DNA and an 8-kb SacI fragment of wild-type DNA. Hybridizations with probe C revealed additional restriction fragment length polymorphisms. Probe C hybridized to a 2.5-kb SacI fragment of B4 DNA and an 8-kb SacI fragment of wild-type DNA. These results indicate that the plasmid insertion that produced the mutant B4 phenotype introduced an additional SacI site into the 8-kb SacI fragment. To test whether the PF15 gene was contained within the 8-kb SacI fragment, we subcloned smaller DNA fragments from this region into pBluescript (p1B4 and pPF15) (Fig. 2). A SacI-NotI restriction fragment of approximately 6.4 kb was subcloned, and it rescued motility and central apparatus assembly in both B4 and pf15a mutants upon transformation (pPF15) (Fig. 2). Therefore, the PF15 gene resides within this 6.4-kb SacI-NotI fragment.
Characterization of PF15 transcript: PF15 encodes a homologue of katanin p80.
To characterize the transcript of the PF15 gene, we prepared poly(A)+ RNA from B4, pf15a, and wild-type cells before and 45 min after deflagellation. The hybridization of probe B (Fig. 2) to blots of size-fractionated RNAs revealed a 3.5-kb transcript in wild-type cells (Fig. 3). A transcript of the same size was observed for RNA isolated from the pf15a mutant (Fig. 3). However, for RNA isolated from B4 cells, a transcript that was slightly smaller and less abundant than that seen for wild-type or pf15a cells hybridized to probe B (Fig. 3). These results indicate that the insertional mutagenesis procedure may have induced a small deletion in the corresponding gene in B4 cells. For all three strains, the hybridizing transcript was less abundant after deflagellation. This result was surprising since deflagellation in Chlamydomonas has been shown to typically induce an accumulation of transcripts from genes encoding flagellar proteins (29), including the central pair proteins PF16p and PF20p (54, 55).
FIG. 3.
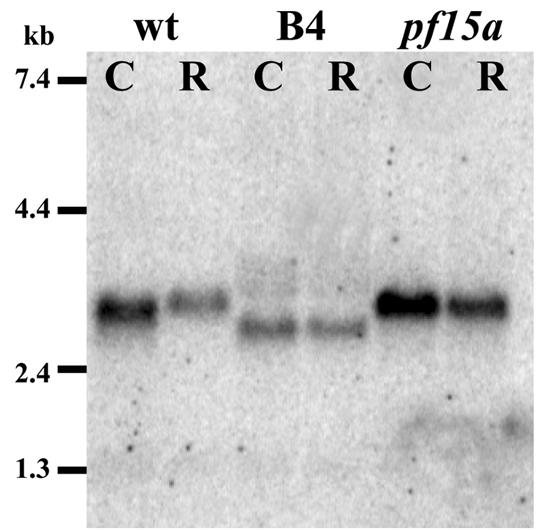
RNA blot. Ten micrograms of poly(A)+ RNA prepared from wild-type, B4, or pf15a cells before (lanes C [control]) and 45 min after deflagellation (lanes R [regenerating flagella]) was loaded in each lane. Probe B (Fig. 2) was used as a hybridization probe. The hybridization of probe B revealed a 3.5-kb transcript for RNAs isolated from wild-type cells and pf15a mutant cells. However, for RNA isolated from B4 cells, a transcript that was slightly smaller and less abundant than that seen for wild-type or pf15a cells was detected. Notably, in all cases the hybridizing transcript was slightly reduced in abundance after deflagellation.
To obtain the PF15 coding sequence, we screened several cDNA libraries, using the insert from plasmid pPF15 as a probe; however, no positive clones were identified. Therefore, the 6.4-kb fragment of genomic DNA in the pPF15 plasmid was directly sequenced. Database searches revealed that this clone contains two transcripts from divergently transcribed genes (Fig. 2). On one strand is the 3′ untranslated region and the last two exons of the SAC1 gene (11, 12). On the opposite strand is the complete coding sequence of a WD repeat-containing protein with significant amino acid similarity to the p80 subunit of katanin (24). Several results indicated that the PF15 gene encodes the p80 subunit of katanin. First, the plasmid pPF15, which rescues motility in pf15a mutants, does not contain the complete coding sequence for the SAC1 gene, and sac1 mutants are motile. Second, the plasmid p1B4 does not rescue B4 or pf15a. And finally, we did not detect any additional opening reading frames in plasmid pPF15 by BLAST searches or exon-predicting algorithms.
The predicted coding sequence of the PF15 gene was confirmed by RT-PCR using RNA isolated from wild-type cells and gene-specific primers. In addition, we also compared our sequence with the Chlamydomonas genome sequence (http://genome.jgi-psf.org/chlre2/chlre2.home.html); the coding sequence matches a portion of the sequence of scaffold 11 in the Chlamydomonas database.
The PF15 gene encodes a protein of 798 amino acids with a predicted molecular mass of 84 kDa (GenBank accession number AY597210). Database searches using the predicted amino acid sequence placed PF15p in a family of proteins containing WD repeats, first identified in the β-subunit of the heterotrimeric GTP binding protein β-transducin (20). Six contiguous WD repeats comprise most of the amino half of the protein. Many proteins with a variety of cellular functions have been identified that contain WD repeats. For example, for Chlamydomonas flagella, the WD-repeat motif has been identified in the IC69 and IC78 intermediate chains of outer arm flagellar dynein (63), the IC140 intermediate chain of inner dynein arm subform I1 (67), and the central apparatus protein PF20 (55). Yet the PF15p protein is most similar to the p80 subunit of katanin (Fig. 4).
FIG. 4.
ClustalW alignment of PF15p (CrP80, accession number AY597210) with Arabidopsis thaliana (AtP80, accession number NP_568194), sea urchin (Strongylocentrotus purpuratus SpP80, accession number AAC09329), and human (HsP80, accession number AAH01353) katanin p80 amino acid sequences. Both identical amino acids and conserved substitutions are shaded when the similarity is present in three sequences.
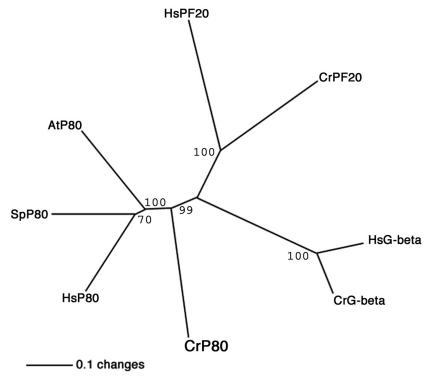
In the region of the WD repeats (amino acids 1 to 240), the protein is 41% identical (65% similar) to a p80-like protein in Arabidopsis, 44% identical (69% similar) to p80 from the sea urchin, and 41% identical (61% similar) to human p80. The midregion is largely comprised of uncharged amino acids with long stretches of glycine and alanine. The carboxyl terminus is 26% identical (52% similar) to a p80-like protein in Arabidopsis, 25% identical (40% similar) to p80 from the sea urchin, and 23% identical (39% similar) to human p80. In addition to comparing p80 sequences from other species, we compared the PF15p WD-repeat domain with those of other Chlamydomonas WD-repeat-containing proteins. PF15p is more similar to katanin p80 from other species than to the two most closely related Chlamydomonas WD-repeat-containing proteins (the G-protein beta subunit and PF20p) (Fig. 5). Based on sequence similarity and the overall organization of the WD repeats, PF15 encodes a Chlamydomonas homologue of the p80 subunit of katanin.
FIG. 5.
The ClustalW program was used to compare PF15p (CrP80) with p80 proteins from other species as well as additional WD-repeat-containing proteins. Comparisons included PF20 from Chlamydomonas (CrPF20) and humans (HsPF20) and the G-protein beta subunit from Chlamydomonas (CrG-beta) and humans (HsG-beta). The analysis was performed for the WD-repeat-containing regions only. Bootstrap values are indicated. Chlamydomonas p80 is more similar to p80 from other species than to the two most similar Chlamydomonas WD-repeat-containing proteins.
Sequence analysis of p80 in pf15a and B4 mutants.
The genetic lesion in pf15a was determined by sequencing genomic DNA corresponding to the 6.4-kb SacI-NotI fragment from the pf15a mutant. A single mutation was discovered in exon 2 of the PF15 gene (Fig. 6, asterisk) and was confirmed by RT-PCR of the PF15 gene using RNA isolated from the pf15a mutant. This missense mutation is a T-to-A transversion that results in valine 43 being replaced by aspartic acid. Valine 43 is within a highly conserved WD-repeat domain of this protein; the substitution of a charged amino acid for an uncharged amino acid in this position most likely disrupts the WD repeat. These results provide additional evidence that PF15p corresponds to the Chlamydomonas homologue of p80.
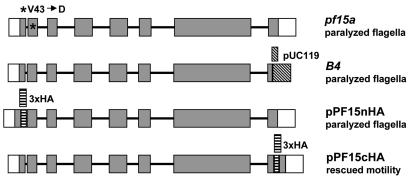
FIG. 6.
Gene structure and diagrams illustrating mutations in B4 and pf15a mutants and HA-tagged PF15 constructs. Exons are indicated by dark shading. The 5′ and 3′ untranslated regions are indicated by open squares. Introns are represented by solid lines. The pf15a mutant contains a single mutation that translates to a conversion of valine 43 to aspartic acid. In the B4 mutant, the pUC119 plasmid-integrated 3′ end of the gene deleted the nucleotides encoding the carboxyl most 29 amino acids. The plasmid pPF15nHA was generated by inserting a 3× HA tag into EcoRI sites in the first intron and exon. The plasmid pPF15cHA was generated by inserting a 3× HA tag into an XcmI site in the last exon of the coding sequence. The pPF15cHA construct rescues motility upon transformation of B4 cells, whereas pPF15nHA does not.
As noted above, Northern blots of RNA isolated from the B4 mutant revealed that a small deletion may have occurred in the PF15 gene in this mutant allele (Fig. 3). This result was somewhat surprising since insertional mutagenesis techniques generally produce large deletions in Chlamydomonas DNA. To characterize the putative PF15 transcript in the B4 mutant, we precisely determined the integration site of the transforming plasmid in B4 genomic DNA. First, we sequenced the lambda clone isolated from the library that we constructed with the B4 genomic DNA. We discovered that a copy of the pUC119 plasmid integrated into the 3′ end of the coding sequence (Fig. 6). The insertion site was confirmed by a PCR using B4 genomic DNA as a template and two primers, one specific for pUC119 and one specific for the p80 gene.
If the resulting DNA were transcribed and translated, a PF15p protein would be produced that lacks 29 amino acids at the carboxyl terminus but that includes 59 amino acids from the pUC119 sequence before reaching a stop codon. To identify any potential transcripts produced from the disrupted gene, we performed RT-PCR with RNA isolated from the B4 mutant (see Materials and Methods). We identified a transcript with the pUC119 sequence present at the 3′ end of the chimeric pUC119/PF15 transcript. The transcript extended approximately 150 nucleotides beyond the pUC119 integration site, at which point it was polyadenylated. The B4 transcript is therefore 260 nucleotides shorter than the wild-type transcript, a predicted size difference that is consistent with our results from Northern blot analysis. If the transcript were translated, the resulting polypeptide would lack the 29 most C-terminal amino acids of the wild-type PF15 gene and instead would incorporate 30 amino acids contributed from the translated pUC119 sequence.
PF15p localizes to the axoneme.
To localize the PF15 gene product, we made numerous attempts to generate polyclonal antibodies against Chlamydomonas p80, including antibodies specific for small peptides at either the carboxyl or amino terminus. In addition, we obtained antibodies against human and sea urchin p80 from Frank McNally (University of California, Davis). All of these antibodies revealed nonspecific staining on Western blots of Chlamydomonas protein. Therefore, to localize the PF15 gene product, we constructed two different PF15 genes that expressed a PF15p protein containing three copies of the HA epitope. In one construct, the 3× HA tag was inserted 33 amino acids upstream of the carboxyl terminus (pPF15cHA) (Fig. 6). In the second construct (pPF15nHA), the 3× HA tag was inserted into EcoRI sites at the 5′ end of the gene. This construct places the HA tag just upstream of the WD repeats.
For cells transformed with the 3× HA tag inserted at the 5′ end of the PF15 coding sequence (pPF15nHA), no swimmers were identified from 48 colonies that were picked and placed into liquid medium. Either the disruption of the first intron altered the expression of the 3× HA construct or the production of the PF15p protein with 3× HA at the amino terminus disrupted the protein function. In contrast, for cells transformed with the 3× HA tag inserted into the 3′ end of the PF15 coding sequence (pPF15cHA), 6 of 36 colonies that were picked and placed into liquid medium were identified as swimming cells that appeared indistinguishable from the wild type. Evidently, the placement of the 3× HA tag at the carboxyl terminus of the protein does not interfere with PF15p function.
To determine whether PF15p is localized to the flagellar basal body transition zone, the same localization as the p60 subunit of katanin (31), or if PF15p is a flagellar component, we isolated both flagellar basal body complexes and flagella from successful pPF15cHA transformants. The samples were prepared from equal numbers of cells and subjected to Western blotting using antibodies against the HA tag as well as against a known flagellar protein, PF20p (55). We predicted that if PF15p is primarily localized to the transition zone region, then we would observe little or no PF15p protein in an equivalent load of isolated flagella; flagella prepared by the dibucaine method lack this region. An immunoreactive polypeptide of the expected size was observed for both flagellar basal body complexes and flagella isolated from B4 cells transformed with the HA-tagged PF15 gene (Fig. 7A). A similar result was obtained when the same blots were probed with antibodies against PF20p. For both PF15p and PF20p, there appeared to be slightly more protein in flagella than in an equivalent amount of flagellar basal body complexes isolated from the same number of cells. This was most likely due to a slightly higher yield from the flagellar isolation preparation than from the flagellar basal body complex preparation. These results indicate that PF15p is primarily localized to flagella.
FIG. 7.
Immunoblots. (A) Flagellar basal body complexes (FBB) and flagella (FL) were prepared from equal numbers of B4 cells rescued with pPF15cHA and were equally loaded onto the gel. The resulting blot was probed with antibodies against the HA tag associated with PF15p (PF15-HA) as well as PF20p, a known axonemal component. (B) Flagella were prepared from B4 cells rescued with pPF15cHA (lane 1), pf20 cells (lane 2), and two strains of pf20 cells expressing PF15-HA protein (lanes 3 and 4). All lanes represent equal loading of isolated flagella. The resulting blot was probed with antibodies against the HA tag associated with PF15p (PF15-HA) as well as PF20p. PF15-HA assembles into flagella in the absence of the central apparatus (lanes 3 and 4). (C) Samples in lanes 1, 3, 5, and 6 were obtained from B4 cells. Samples in lanes 2, 4, 7, and 8 were obtained from B4 cells rescued with pPF15cHA. Lanes 1 and 2, isolated flagella (FL); lanes 3 and 4, isolated axonemes (AX); lanes 5 and 7, 0.5 M KI axonemal extracts (EX); lanes 6 and 8, the resulting extracted axonemes (EA). The anti-HA antibody recognizes an approximately 90-kDa polypeptide that is present in isolated axonemes and extractable with 0.5 M KI.
To determine if PF15p localizes to the central apparatus, we constructed mutant strains of pf20 (paralyzed flagella that completely lack the central apparatus) that express the PF15-HA protein (see Materials and Methods). Comparisons of flagella isolated from two pf20-PF15-HA strains with those from the B4 strain rescued with pPF15cHA revealed equivalent amounts of PF15-HA protein assembled into flagella (Fig. 7B). Immunoblots using anti-PF20p antibodies (55) confirmed that these strains lack the central apparatus; in the absence of PF20p, the central apparatus fails to assemble. These results indicate that although pf15 mutants lack the central pair, PF15p does localize to the axoneme.
To further characterize the localization of PF15p, we also probed blots of flagella, axonemes, and salt extracts isolated from successful transformants with antibodies against the HA tag (Fig. 7C). The PF15p polypeptide was present in axonemes prepared by removal of the membranes of isolated flagella by use of a detergent and was extractable in buffers containing 0.5 M KI. It should be noted that based on comparisons of exposure times for the chemiluminescent detection of other HA-tagged flagellar proteins using the same antibody and detection system, we conclude that PF15p is not very abundant. The exposure time for detecting the PF20p protein in Fig. 7A was 1 s, whereas the exposure times for the PF15-HA protein were 2 to 3 min. Unfortunately, using a variety of conditions and numerous commercially available anti-HA antibodies, we have been unable to unequivocally determine the precise localization of PF15p in axonemes by either immunofluorescence or immunogold electron microscopic techniques. This difficulty may also be due, at least in part, to the low expression levels of PF15p relative to other axonemal components.
DISCUSSION
Mutations in at least seven genes (pf6, pf15, pf16, pf18, pf19, pf20, and cpc1) result in obvious defects in central apparatus assembly and/or stability (1, 16, 39). The sequence and localization of the PF16, PF20, and PF6 gene products have been previously reported (47, 54, 55). However, in these three cases the amino acid sequence did not provide substantial clues as to why mutations in these genes result in central apparatus defects. Here we report that PF15 encodes the Chlamydomonas homologue of katanin p80. This finding reveals a novel cellular function for katanin and may suggest a possible mechanism of central apparatus assembly.
PF15 encodes Chlamydomonas homologue of katanin p80 subunit.
Although the genomic clone that rescued motility contained coding sequences from two genes, the SAC1 gene and a homologue of katanin p80, we conclude that PF15 encodes Chlamydomonas p80. Firstly, a subclone that rescued motility in pf15 mutants contained the complete coding sequence of the p80 gene but did not contain the complete coding sequence of SAC1. Secondly, sac1 mutants were defective in their response to sulfur deprivation but were wild type in motility, indicating that mutations in SAC1 do not affect flagellar function. The conclusion that PF15p is the p80 subunit was confirmed by the observation that pf15a is a missense mutation caused by a T-to-A transversion in the p80 coding sequence. The plasmid integration site in the insertional pf15 allele is located at the 3′ end of the p80 coding sequence; if a corresponding message were stable and translated, the transcript would produce a p80 protein that lacks the carboxyl 29 amino acids of the p80 gene product. Finally, a 3× HA-tagged construct in which the tag was inserted into the carboxyl terminus rescued motility upon transformation of pf15 mutants; in contrast, a construct in which the 3× HA tag was inserted into the amino terminus did not rescue motility upon transformation of pf15 mutants. These results, combined with our sequence comparisons of PF15p with other p80 proteins, indicate that the PF15 gene encodes a Chlamydomonas homologue of katanin p80.
This discovery was surprising in light of katanin's known function in cells lacking cilia or flagella. Katanin was originally identified and purified from sea urchin egg extracts as a protein with ATP-dependent microtubule-severing activity (36). The protein is a heterodimer of 60-kDa (p60) and 80-kDa (p80) subunits that localizes to the centrosome (35, 36). The p60 subunit has microtubule-severing activity, and the p80 subunit is thought to target the catalytic subunit (p60) to the centrosome and possibly to regulate its activity (24, 37). Mutational analysis of the p80 subunit has demonstrated that the amino-terminal WD-repeat domain of p80 is important for targeting katanin to the centrosome and that the C-terminal 130 amino acids are involved in dimerization with p60 (24).
Chlamydomonas p80 localization and function.
Mutant Chlamydomonas pf15 cells have paralyzed flagella of normal length that lack the central apparatus. Based on the pf15 mutant phenotype, PF15p is evidently involved in the assembly and/or stability of the central pair of microtubules. If PF15 encodes the p80 subunit of katanin, what role does katanin play in the assembly or stability of the central microtubules? One possibility is that PF15p is not a true Chlamydomonas homologue of p80 and is therefore not a subunit of Chlamydomonas katanin. A second possibility is that the microtubule-severing function of katanin is required for central apparatus assembly and/or stability. A third possibility is that in Chlamydomonas, p80 dimerizes with polypeptides other than p60 and is involved in cellular functions other than or in addition to microtubule severing.
Several lines of evidence support the conclusion that PF15p is the only Chlamydomonas p80 homologue. Southern blots of Chlamydomonas genomic DNA digested with several enzymes and probed with the PF15 gene indicate that there is a single PF15 gene. In searches of the Chlamydomonas genome (http://genome.jgi-psf.org/chlre2/chlre2.home.html) that used the human p80 sequence for comparison, the only sequence that exhibited significant similarity beyond the WD repeats was the PF15 gene. Therefore, if Chlamydomonas possesses katanin, PF15p appears to be the only candidate for a p80 homologue. Interestingly, however, the second draft of the Chlamydomonas genome sequence has revealed two p60 homologues. One of these is 57% identical to the p60 subunit in Arabidopsis (31), and one is 48% identical to the p60 subunit in Arabidopsis (http://genome.jgi-psf.org/chlre2/chlre2.home.html).
Lohret and colleagues cloned one of these Chlamydomonas p60 homologues and provided evidence suggesting that one role of katanin in Chlamydomonas is to mediate the flagellar excision or deflagellation response (30, 31). Antibodies generated against human p60 block the flagellar excision response in isolated flagellar basal body complexes and primarily localize this p60 subunit to the flagellar transition zone (31). To date, a Chlamydomonas strain has not been isolated that carries a mutation in either p60 gene; therefore, the phenotype of a mutant affecting p60 in Chlamydomonas is unknown. Based on the mutant phenotype caused by the loss of the p80 subunit in pf15, one might predict the mutant phenotype of p60 to be cells with paralyzed flagella that lack a central pair. Indeed, the p60 gene cloned by Lohret et al. (31) maps very close to pf19 (26), a central pairless mutant whose gene product is unknown. We have determined the entire coding sequence for this p60 gene in pf19 and found no mutation. However, it is still possible that in pf19 a mutation exists in an undefined regulatory element for p60 expression.
If the p80 subunit (PF15p) is required for the localization of p60, one prediction is that p60 does not assemble in pf15 mutants. We have been unable to test whether p60 assembles in either pf15 allele since Chlamydomonas p60 antibodies are not available, and the anti-p60 antibodies to either sea urchin or human p60 in our hands produce nonspecific immunoreactive bands on Western blots of Chlamydomonas flagellar basal body preparations. The pf15a allele contains a mutation in the WD-repeat domain, a domain that was previously noted as being required for the localization of katanin (24). It is possible that in the pf15a allele, p80 binds to p60 but that the complex is mistargeted. We suspect that no PF15p protein is produced in the B4 mutant based on the nature of the mutation in this allele; however, we cannot be certain that the mutant pf15 transcript is not translated. This assessment would require specific antibodies against p80. In addition, although previous studies have indicated that p80 is required for the correct localization of p60, p60 may assemble normally in the B4 allele. Regardless of whether either p80 or p60 correctly assembles in pf15 mutants, the fact remains that both alleles of pf15 (pf15a and B4) are indistinguishable from wild-type cells in their deflagellation response to pH shock (for examples, see Fig. 3 and 7).
Based on our comparisons of equivalent loads of flagella and flagellar basal body complexes on Western blots, p80 appears to primarily localize to the axoneme; the primary localization of p60 is at the basal body transition zone (31). Although Lohret et al. (30) did not include Western blot-isolated flagella probed with p60 antibodies, they did (31) observe that some p60 localizes to the axonemes by immunogold labeling. Therefore, axonemal p80 may colocalize with p60 in wild-type axonemes. In addition, our results do not rule out the possibility that p80 localizes to several different positions in the axoneme or that a small fraction of p80 localizes to the transition zone region. The observation that transcript levels of PF15p appear to be down regulated after deflagellation is unusual for a gene encoding an axonemal protein. This result may also indicate additional roles for p80 outside of the axoneme.
An alternative explanation for the differential localization of the p80 and p60 subunits is that these two polypeptides may have alternate binding partners in Chlamydomonas. As noted in Results, the carboxyl termini of PF15p and sea urchin p80 are less similar to each other than are the WD-repeat domains for these two proteins. Given that the carboxyl terminus is the p60 binding domain, the low degree of similarity at the carboxyl terminus may suggest alternative binding partners for Chlamydomonas p80. Potential katanin homologues have been found in other organisms that play roles in diverse microtubule-dependent cellular processes. For example, a potential homologue of katanin has been discovered in Arabidopsis that appears to play a role in organizing cortical microtubules; it localizes along cortical microtubules and is required for normal cell wall biosynthesis and cell elongation (5, 6, 62). Katanin homologues have been identified in Caenorhabditis elegans that play a role in spindle organization during meiosis (9, 10, 33, 57). In C. elegans, the katanin homologues MEI-1 and MEI-2 appear to localize to the meiotic spindle microtubules, polar bodies, and condensed chromatin (9, 33, 57). Despite the diverse localization of katanin in these systems, the only specific interaction partner reported for p80 is p60. A second possibility is that p80 binds to the additional p60 homologue in Chlamydomonas and that a failure to bind to this homologue results in the central pairless defect.
If Chlamydomonas p80 forms a dimer with the p60 catalytic subunit, the implication is that katanin's microtubule-severing activity may be important for central apparatus assembly. In cultured neurons, the microinjection of anti-katanin antibodies results in the accumulation of microtubules at the centrosome and the inhibition of axon outgrowth (2). Therefore, in neurons katanin appears to be required for the release of microtubules from the centrosome. In Chlamydomonas, one possibility is that katanin-mediated microtubule excision from the centrosome or basal bodies produces microtubule seeds that then assemble to form the central apparatus. Perhaps in pf15 mutants, this function has been disrupted. Interestingly, Buster et al. have proposed a model in which microtubule minus ends generated by katanin-mediated microtubule severing are capped by gamma-tubulin ring complexes (7); McKean et al. have recently shown that the abolished expression of gamma-tubulin in Trypanosoma brucei results in the production of flagella lacking a central pair of microtubules (34). In Chlamydomonas, gamma-tubulin has been localized to the transition zone (51), where central apparatus assembly is believed to initiate. These reports add support for the hypothesis that katanin-mediated microtubule severing may play a role in central apparatus assembly.
Based on the observed mutant phenotype of the Chlamydomonas p80 subunit, our results reveal a novel role for katanin in the assembly of the central pair of flagellar microtubules. The association of p80 with the axoneme suggests that p80 may play additional roles in axoneme assembly and maintenance. We are only beginning to identify the components required for intraflagellar transport (for an example, see reference 44); it is possible that p80 is involved in a form of intraflagellar transport that is required for central apparatus assembly. Currently, we do not know the precise localization of p80, although our results indicate that it is not a component of the central apparatus and that it is in very low abundance in the axoneme. Our preliminary biochemical analyses of Chlamydomonas p80 indicate that this protein is extractable and sediments at 11S on sucrose gradients; therefore, p80 most likely forms a complex with additional polypeptides. However, we have not been able to immunoprecipitate this complex with anti-HA antibodies. Determining the precise function of Chlamydomonas p80 will require the identification of p80-interacting proteins as well as a more precise localization of p80 within the axoneme.
Acknowledgments
We thank Louisa Howard in the Ripple Electron Microscope Facility at Dartmouth College for the collection of data for Fig. 1b and c. We thank Kevin Peterson (Dartmouth College) for assistance with Fig. 4 and 5.
This work was supported by NIH grant GM51379 as a consortium agreement (P.A.L.) and was supported in part by research grant 5-FY99-766 (to E.F.S.) from the March of Dimes Birth Defects Foundation. E.F.S. was also supported by NSF-RTG grant DIR-9113444 and ACS grant PF-3955.
REFERENCES
- 1.Adams, G. M., B. Huang, G. Piperno, and D. J. Luck. 1981. Central-pair microtubular complex of Chlamydomonas flagella: polypeptide composition as revealed by analysis of mutants. J. Cell Biol. 91:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, F. J., W. Yu, F. J. McNally, and P. W. Baas. 1999. An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 145:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., and D. J. Lipman. 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA 87:5509-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouquin, T., O. Mattsson, H. Naested, R. Foster, and, J. Mundy. 2003. The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116:791-801. [DOI] [PubMed] [Google Scholar]
- 6.Burk, D. H., B. Liu, R. Zhong, W. H. Morrison, and Z. H. Ye. 2001. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13:807-827. [PMC free article] [PubMed] [Google Scholar]
- 7.Buster, D., K. McNally, and F. J. McNally. 2002. Katanin inhibition prevents the redistribution of gamma-tubulin at mitosis. J. Cell Sci. 115:1083-1092. [DOI] [PubMed] [Google Scholar]
- 8.Chisholm, D. 1989. A convenient moderate-scale procedure for obtaining DNA from bacteriophage lambda. BioTechniques 7:21-23. [PubMed] [Google Scholar]
- 9.Clark-Maguire, S., and P. E. Mains. 1994. Localization of the mei-1 gene product of Caenorhabditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark-Maguire, S., and P. E. Mains. 1994. mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136:533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, J. P., F. Yildiz, and A. R. Grossman. 1994. Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell 6:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, J. P., F. H. Yildiz, and A. Grossman. 1996. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 15:2150-2159. [PMC free article] [PubMed] [Google Scholar]
- 13.Debuchy, R., S. Purton, and J. D. Rochaix. 1989. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dentler, W. L. 1980. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J. Cell Sci. 42:207-220. [DOI] [PubMed] [Google Scholar]
- 15.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutcher, S. K., B. Huang, and D. J. Luck. 1984. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Euteneuer, U., and J. R. McIntosh. 1981. Polarity of some motility-related microtubules. Proc. Natl. Acad. Sci. USA 78:372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez, E., R. Schnell, L. P. Ranum, S. C. Hussey, C. D. Silflow, and P. A. Lefebvre. 1989. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 86:6449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finst, R. J., P. J. Kim, and L. M. Quarmby. 1998. Genetics of the deflagellation pathway in Chlamydomonas. Genetics 149:927-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong, H. K., J. B. Hurley, R. S. Hopkins, R. Miake-Lye, M. S. Johnson, R. F. Doolittle, and M. I. Simon. 1986. Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. USA 83:2162-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodenough, U. W., and J. E. Heuser. 1985. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorman, D. S., and R. P. Levine. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 54:1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartman, J. J., J. Mahr, K. McNally, K. Okawa, A. Iwamatsu, S. Thomas, S. Cheesman, J. Heuser, R. D. Vale, and F. J. McNally. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell 93:277-287. [DOI] [PubMed] [Google Scholar]
- 25.James, S. W., L. P. Ranum, C. D. Silflow, and P. A. Lefebvre. 1988. Mutants resistant to anti-microtubule herbicides map to a locus on the uni linkage group in Chlamydomonas reinhardtii. Genetics 118:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kathir, P., M. LaVoie, W. J. Brazelton, N. A. Haas, P. A. Lefebvre, and C. D. Silflow. 2003. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2:362-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kindle, K. L. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87:1228-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King, S. M., T. Otter, and G. B. Witman. 1986. Purification and characterization of Chlamydomonas flagellar dyneins. Methods Enzymol. 134:291-306. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre, P. A., C. D. Silflow, E. D. Wieben, and J. L. Rosenbaum. 1980. Increased levels of mRNAs for tubulin and other flagellar proteins after amputation or shortening of Chlamydomonas flagella. Cell 20:469-477. [DOI] [PubMed] [Google Scholar]
- 30.Lohret, T. A., F. J. McNally, and L. M. Quarmby. 1998. A role for katanin-mediated axonemal severing during Chlamydomonas deflagellation. Mol. Biol. Cell 9:1195-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohret, T. A., L. Zhao, and L. M. Quarmby. 1999. Cloning of Chlamydomonas p60 katanin and localization to the site of outer doublet severing during deflagellation. Cell Motil. Cytoskeleton 43:221-231. [DOI] [PubMed] [Google Scholar]
- 32.Lux, F. G., III, and S. K. Dutcher. 1991. Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics 128:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mains, P. E., K. J. Kemphues, S. A. Sprunger, I. A. Sulston, and W. B. Wood. 1990. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 126:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKean, P. G., A. Baines, S. Vaughan, and K. Gull. 2003. Gamma-tubulin functions in the nucleation of a discrete subset of microtubules in the eukaryotic flagellum. Curr. Biol. 13:598-602. [DOI] [PubMed] [Google Scholar]
- 35.McNally, F. J., K. Okawa, A. Iwamatsu, and R. D. Vale. 1996. Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J. Cell Sci. 109:561-567. [DOI] [PubMed] [Google Scholar]
- 36.McNally, F. J., and R. D. Vale. 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75:419-429. [DOI] [PubMed] [Google Scholar]
- 37.McNally, K. P., O. A. Bazirgan, and F. J. McNally. 2000. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J. Cell Sci. 113:1623-1633. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell, D. R. 2003. Reconstruction of the projection periodicity and surface architecture of the flagellar central pair complex. Cell Motil. Cytoskeleton 55:188-199. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell, D. R., and W. S. Sale. 1999. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144:293-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano, I., T. Kobayashi, M. Yoshimura, and C. Shingyoji. 2003. Central-pair-linked regulation of microtubule sliding by calcium in flagellar axonemes. J. Cell Sci. 116:1627-1636. [DOI] [PubMed] [Google Scholar]
- 41.Nelson, J. A., and P. A. Lefebvre. 1995. Transformation of Chlamydomonas reinhardtii. Methods Cell Biol. 47:513-517. [DOI] [PubMed] [Google Scholar]
- 42.Nelson, J. A., P. B. Savereide, and P. A. Lefebvre. 1994. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14:4011-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter, M. E., and W. S. Sale. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:F37-F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin, H., D. R. Diener, S. Geimer, D. G. Cole, and J. L. Rosenbaum. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Randall, J. T., T. Cavalier-Smith, A. McVittie, J. T. Warr, and J. M. Hopkins. 1967. Developmental and control processes in the basal bodies and flagella of Chlamydomonas reinhardtii. Dev. Biol. 1(Suppl.):43-83. [Google Scholar]
- 46.Randall, J. T., J. R. Warr, J. M. Hopkins, and A. McVittie. 1964. A single gene mutation of Chlamydomonas reinhardtii affecting motility: a genetic and electron microscope study. Nature 203:912-914. [DOI] [PubMed] [Google Scholar]
- 47.Rupp, G., E. O'Toole, and M. E. Porter. 2001. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell 12:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sager, R., and S. Granick. 1953. Nutritional studies in Chlamydomonas reinhardtii. Ann. N. Y. Acad. Sci. 56:831-838. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Silflow, C. D., M. LaVoie, L. W. Tam, S. Tousey, M. Sanders, W. Wu, M. Borodovsky, and P. A. Lefebvre. 2001. The Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153:63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silflow, C. D., B. Liu, M. LaVoie, E. A. Richardson, and B. A. Palevitz. 1999. Gamma-tubulin in Chlamydomonas: characterization of the gene and localization of the gene product in cells. Cell Motil. Cytoskeleton 42:285-297. [DOI] [PubMed] [Google Scholar]
- 52.Smith, E. F. 2002. Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol. Biol. Cell 13:3303-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, E. F. 2002. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil. Cytoskeleton 52:33-42. [DOI] [PubMed] [Google Scholar]
- 54.Smith, E. F., and P. A. Lefebvre. 1996. PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J. Cell Biol. 132:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, E. F., and P. A. Lefebvre. 1997. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol. Biol. Cell 8:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, E. F., and P. A. Lefebvre. 1997. The role of central apparatus components in flagellar motility and microtubule assembly. Cell Motil. Cytoskeleton 38:1-8. [DOI] [PubMed] [Google Scholar]
- 57.Srayko, M., D. W. Buster, O. A. Bazirgan, F. J. McNally, and P. E. Mains. 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14:1072-1084. [PMC free article] [PubMed] [Google Scholar]
- 58.Tam, L. W., and P. A. Lefebvre. 1993. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135:375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thazhath, R., C. Liu, and J. Gaertig. 2002. Polyglycylation domain of beta-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 4:256-259. [DOI] [PubMed] [Google Scholar]
- 60.Wargo, M. J., and E. F. Smith. 2003. Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 100:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warr, J. R., A. McVittie, J. T. Randall, and J. M. Hopkins. 1966. Genetic control of flagellar structure in Chlamydomonas reinhardtii. Genet. Res. 7:335-351. [Google Scholar]
- 62.Webb, M., S. Jouannic, J. Foreman, P. Linstead, and L. Dolan. 2002. Cell specification in the Arabidopsis root epidermis requires the activity of Ectopic Root Hair 3—a katanin-p60 protein. Development 129:123-131. [DOI] [PubMed] [Google Scholar]
- 63.Wilkerson, C. G., S. M. King, A. Koutoulis, G. J. Pazour, and G. B. Witman. 1995. The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129:169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilkerson, C. G., S. M. King, and G. B. Witman. 1994. Molecular analysis of the gamma heavy chain of Chlamydomonas flagellar outer-arm dynein. J. Cell Sci. 107:497-506. [DOI] [PubMed] [Google Scholar]
- 65.Witman, G. B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280-290. [DOI] [PubMed] [Google Scholar]
- 66.Wright, R. L., J. Salisbury, and J. W. Jarvik. 1985. A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 101:1903-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, P., and W. S. Sale. 1998. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 9:3335-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]