Abstract
In Trypanosoma brucei, two classes of transcripts are produced from two distinct mitochondrial genome components. Guide RNAs (gRNAs) are usually minicircle encoded and exist as primary transcripts, while the maxicircle-encoded rRNAs and mRNAs are processed from a polycistronic precursor. The genes for the gRNAs gMURF2-II and gCYb(560) each have uncommon kinetoplast DNA (kDNA) locations that are not typically associated with transcription initiation events. We demonstrate that the conserved maxicircle gRNA gMURF2-II has an unusual location within the ND4 gene. This is the first report of a completely intragenic gene in kDNA. In addition, the gMURF2-II and ND4 transcripts are generated by distinctly different events; the ND4 mRNA is processed from a polycistronic precursor, while transcription of the gRNA initiates downstream of the 5′ end of the ND4 gene. The gCYb(560) gene has an atypical minicircle location in that it is not flanked by the inverted repeat sequences that surround the majority of minicircle gRNA genes. Our data indicate that the mature gCYb(560) gRNA is also a primary transcript and that the 5′-end heterogeneity previously observed for this gRNA is a result of multiple transcription initiation sites and not of imprecise 5′-end processing. Together, these data indicate that gRNA genes represent individual transcription units, regardless of their genomic context, and suggest a complex mechanism for mitochondrial gene expression in T. brucei.
Trypanosoma brucei is a parasitic protozoan with an extraordinarily complex mitochondrial genome. This genome, the kinetoplast, consists of a network of thousands of 1-kb minicircles that are unique to kinetoplasts and 50 copies of a 22-kb maxicircle that is analogous to mitochondrial DNAs in other organisms (23). The mRNAs produced from most maxicircle genes must be edited by the insertion or deletion of uridylate residues to create mature transcripts. This process is mediated by small, complementary guide RNAs (gRNAs) encoded on minicircle DNA. The process of editing occurs posttranscriptionally and has been studied extensively (for recent reviews, see references 16, 24, and 25). However, the initial step in mitochondrial gene expression, transcription, has been largely ignored.
While no kinetoplast DNA (kDNA) promoter elements have been described to date, there are regions from which transcription has been shown to initiate. In T. brucei, most minicircle gRNA genes encode primary transcripts located within cassettes of imperfect inverted 18-bp repeats (13, 20). These inverted repeats have been implicated in playing a role in transcription, but there have been no direct tests of their function in transcription initiation. Although precise maxicircle transcription start sites have not been identified, transcription of the major strand has been shown to begin at least 1,200 nucleotides (nt) upstream of the 5′ end of the mature 12S rRNA and to proceed polycistronically (17). Mature transcripts are rapidly processed from this precursor, often by mutually exclusive events, as many of the coding regions overlap (15, 21). Transcripts corresponding to the maxicircle minor strand are also present (18), but the transcription start site has not been identified.
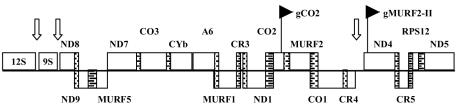
Observations for relatives of T. brucei indicate that not all maxicircle genes are transcribed as part of a polycistronic precursor. The maxicircles of both Leishmania tarantolae and Crithidia fasciculata encode numerous gRNAs located primarily within intergenic spaces (2, 27). Many of these mature gRNAs are primary transcripts (3), indicating that several transcription units exist on these maxicircle genomes. In contrast, the coding region of the T. brucei maxicircle is much more compact and is thought to encode only two gRNAs (27) (Fig. 1). Interestingly, both of these gRNAs are located within mRNA coding sequences. The COII guide RNA, gCOII, is unusual in that it is encoded within the 3′ end of the COII mRNA and is thought to function in cis, as no discrete gCOII transcript has been detected (2, 15). The other gRNA, gMURF2-II, has not been tested for expression. This gene is located within the 5′ end of the ND4 message, indicating that these two transcripts could be generated via mutually exclusive processing events from a precursor RNA. Intriguingly, this gRNA is also located downstream of one of three intergenic regions on the compact maxicircle (Fig. 1), raising the possibility that the gMURF2-II gene is a discrete transcription unit regulated by a promoter located within this region. This locus therefore represents a unique region on the T. brucei maxicircle that may be regulated by alternative processing and/or transcription initiation events.
FIG. 1.
Linearized map of coding region of T. brucei maxicircle. Major- and minor-strand genes are represented above and below the line, respectively. Overlapping regions are stippled. Intergenic spaces are indicated by white arrows. The gRNA sequences are represented by black flags. MURF, maxicircle unidentified reading frame; CR, C-rich; CO, cytochrome oxidase; A6, ATPase 6; ND, NADH dehydrogenase; CYb, apocytochrome b; RPS12, ribosomal protein subunit 12.
In order to further characterize potential transcription units in T. brucei kDNA, we sought to more closely investigate the product of the conserved maxicircle gMURF2-II gene. This gRNA is expressed exclusively from the maxicircle gMURF2-II locus and is contained entirely within the 5′ end of the ND4 gene. A characterization of the structure of the 5′ ends of these two transcripts demonstrated that the gMURF2-II RNA is a primary transcript, while the ND4 mRNA is processed, likely from the polycistronic precursor. In addition, to examine unusual minicircle transcription units, we investigated the expression of gCYb(560), which is not located between inverted repeats. This gRNA is also a primary transcript, indicating that minicircle gRNA transcription initiation signals exist outside of the inverted repeat regions. These results imply that gRNA genes operate as individual transcription units, regardless of their genomic context, and suggest a complex mechanism of transcription and processing of mitochondrial transcripts.
MATERIALS AND METHODS
Cell growth and mitochondrial isolation.
The procyclic T. brucei EATRO 164 IsTaR 1 serodeme, derived from VAT 1.7 clone A, was grown in SDM-79 medium supplemented with 5% fetal bovine serum (Sigma) at 27°C. Mitochondrial isolations were prepared from cells harvested at a density of ∼3 × 107 cells ml−1. Cells were lysed in a Dounce homogenizer under hypotonic conditions, and mitochondrial vesicles were isolated on Nycodenz step gradients as described previously (11). In order to obtain precursor transcripts for the ligation experiments, we incubated isolated mitochondria in transcription buffer (5 mM HEPES [pH 7.6], 3 mM potassium phosphate [pH 7.7], 125 mM sucrose, 6 mM KCl, 10 mM MgCl2, 1 mM EDTA, 2 mM dithiothreitol) and a 100 μM concentration of each ribonucleoside triphosphate for 30 min (10). Mitochondria were snap frozen in a dry ice ethanol bath, and stored at −80°C prior to RNA extraction as described below.
Oligonucleotide probes and primers.
All DNA oligonucleotides were synthesized by IDT. The RNA oligonucleotide was synthesized by Dharmacon. Forward primers (F) match the RNA sequence, while reverse (R) primers are complementary. The following primers were used: gMURF2-II F, 5′-GAAAGCACAAAAATAAAATTAAATTAGAG-3′; gMURF2-II R, 5′-CATTCAATTACTCTAATTTAATTTTATTTTTGTGC-3′; gA6-14sU R, 5′TAATTATCATATCACTGTCAAAATCTGATTCGTTATCGGAGTTATAGCCCTATAGTGAGTCGTATTAAATT-3′; gND7(506) R, 5′-CACTAACTATACTACAGGTTATTTACATCG-3′; gCYb-(560A/B) R, 5′-CCTCCCYATTACTCAGAAATCTACATTGTC-3′; oligo dA-XBS R, 5′-GATCTAGAGGATCCCGGGAAAAAAAAAAAAAAA-3′; XBS F, 5′-GATCTAGAGGATCCCGGG-3′; and RNA oligonucleotide, 5′-AGAUUUUGACAGUGAUAUGAUAAUUA-3′.
Nucleic acid isolation and Northern and Southern blot analysis.
Total T. brucei genomic DNA was isolated as described previously (7). Mitochondrial RNAs (mtRNAs) were isolated by the acid guanidinium-phenol-chloroform method (6). gRNAs were gel purified in an 8 M urea-6% acrylamide gel. For Northern blot analysis, mtRNAs were electrophoresed through an 8 M urea-5% (37.5:1 ACRYL/BIS) acrylamide gel and transferred to a Nytran membrane (Schleicher & Schuell) by use of a semiphor electroblotter (Hoeffer). Northern and Southern blots were hybridized with 32P end-labeled oligonucleotide probes under standard conditions.
5′ and 3′ end mapping.
Gene-specific DNA oligonucleotides were labeled with γ-32P by the use of T4 polynucleotide kinase (Invitrogen) and were gel purified in an 8 M urea-12% acrylamide gel. mtRNA (2.5 μg) was reverse transcribed as previously described (7) in the presence of 100 kcpm of primer. RNA sequencing ladders were generated by adding equal amounts of deoxy- and dideoxy-ATP or -TTP to the extension reaction. Reactions were stopped by the addition of an equal volume of 95% formamide gel-loading buffer and resolved in an 8 M urea-8% acrylamide sequencing gel. Gels were fixed in 7% acetic acid plus 7% methanol, dried, and exposed on film.
Enzymatic treatments of RNA.
mtRNA (10 μg) or 2 μg of gel-purified gRNA was dephosphorylated with alkaline phosphatase (New England Biolabs) according to the manufacturer's instructions. Both untreated and phosphatase-treated RNAs (5.0 μg of mtRNA or 1.0 μg of gRNA) were phosphorylated with T4 polynucleotide kinase (Invitrogen) supplemented with 1 mM ATP. The alkaline phosphatase was inactivated by proteinase K digestion prior to phenol-chloroform extraction. All enzymatic reactions were terminated by phenol-chloroform extraction followed by ethanol precipitation in the presence of a linear acrylamide carrier.
RNA ligations and poisoned primer extension analysis.
A 26-nt synthetic RNA oligonucleotide (Dharmacon) was used as the acceptor RNA in all ligation reactions. This RNA oligonucleotide possesses a 5′ OH and is consequently unable to serve as a donor molecule in the ligation reaction. mtRNA (5.0 μg) or gRNA (0.5 μg) was used in each ligation reaction. Untreated, alkaline phosphatase-treated, kinase-treated, or alkaline phosphatase- and kinase-treated RNA was mixed with 200 pmol of the acceptor RNA oligonucleotide (a more than sixfold molar excess) and incubated overnight at room temperature with 20 U of T4 RNA ligase (New England Biolabs) in the supplied 1× buffer supplemented with 40% polyethylene glycol 400. The ligations were phenol-chloroform extracted, ethanol precipitated, and resuspended in 1× reverse transcription buffer (50 mM KCl, 20 mM Tris [pH 8.5], 0.5 mM EDTA, 8 mM MgCl2). Poisoned primer extension reactions were performed with each population of ligated RNA (1.0 μg of mtRNA or 0.1 μg of gRNA), with 100 kcpm (∼50 fmol) of primer in the presence of a 1.6 mM concentration (each) of dATP, dCTP, dTTP, and ddGTP, as described previously (7). Reactions were resolved in 8 M urea-8% acrylamide sequencing gels. The gels were fixed in 7% acetic acid plus 7% methanol, dried, and exposed on a phosphorimager cassette (Molecular Dynamics). For each gel, the signals for the ligated and unligated products were quantified with ImageQuant software (Molecular Dynamics), and the percent ligation efficiency was represented by the following equation: amount of ligated product/(amount of ligated product + amount of unligated product) × 100.
RESULTS
gMURF2-II maxicircle localization and expression.
In T. brucei, the gMURF2-II gene is located within the ND4 coding region downstream of one of three mapped regions of intergenic space on the maxicircle (Fig. 1). While the position of the gMURF2-II gene is conserved in L. tarantolae and C. fasciculata, in these organisms transcription initiates within an intergenic region upstream of ND4. The gMURF2-II transcript has been detected in both Leishmania and Crithidia as a discrete RNA (1, 2) and has been shown to be a primary transcript in Leishmania (3). The conservation of the gMURF2-II gene in T. brucei indicates the possibility of a promoter element within the coding region of the maxicircle genome, in spite of the extensive overlap between the gRNA and mRNA genes.
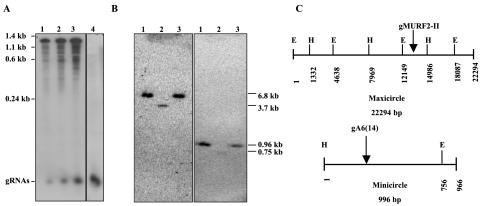
In order to confirm that the gMURF2-II RNA was indeed expressed in T. brucei, we performed a Northern blot of isolated mtRNA by using an oligonucleotide probe complementary to both the gMURF2-II gRNA and the 5′ end of the ND4 message. As shown in Fig. 2A (lanes 1 to 3), the gMURF2-II probe hybridizes to a small RNA of about 60 to 70 nt, representing the gMURF2-II RNA, as well as to the ∼1.3-kb ND4 transcript. The gRNA signal detected by the gMURF2-II probe is similar in size to the minicircle-encoded gRNA gA6(14), as seen in the blot probed with an oligonucleotide complementary to gA6(14) in lane 4.
FIG. 2.
Expression and localization of gMURF2-II gene. (A) Northern blot of 1 μg (lane 1), 2.5 μg (lane 2), 5 μg (lane 3), and 10 μg (lane 4) of mtRNA probed with an oligonucleotide complementary to gMURF2-II (lanes 1 to 3) or an oligonucleotide complementary to the minicircle gRNA gA6(14) (lane 4). The gMURF2-II probe detects both the ∼65-nt gRNA transcript and the ∼1.3-kb ND4 mRNA, while the gA6(14) probe hybridizes to a gRNA band only. (B) Restriction digestion and Southern blot analysis of T. brucei genomic DNA. Three micrograms of DNA was digested with HindIII (lanes 1), HindIII plus EcoRI (lanes 2), or EcoR1 (lanes 3), separated in a 1% agarose gel, transferred to a Nytran membrane, and probed with an end-labeled gMURF2-II oligonucleotide (left). The blot was then stripped and rehybridized with an end-labeled gA6(14) probe (right). (C) Restriction maps of T. brucei maxicircle and gA6(14) minicircle (H, HindIII; E, EcoR1).
Since the majority of the gRNAs are encoded by minicircle DNA in T. brucei, we performed a Southern blot of T. brucei DNA in order to confirm the maxicircle location of the gMURF2-II gene. As seen in the left panel in Fig. 2B, the gMURF2-II probe hybridized exclusively to the predicted maxicircle fragments and not to the ∼1-kb minicircle DNA. In contrast, a probe for a minicircle-encoded gRNA, gA6(14), hybridized exclusively to the predicted minicircle fragments (13) (Fig. 2B, right panel). These results confirm that no additional copies of the gMURF2-II gene exist in the kinetoplast genome and that this gRNA must be transcribed from this region of maxicircle DNA.
The gMURF2-II gene is located within the ND4 gene downstream of an intergenic region.
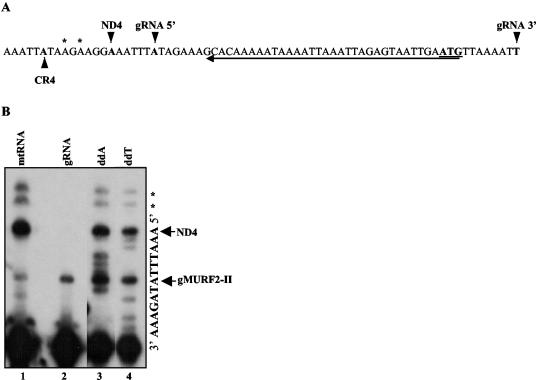
In order to determine the exact location of the gMURF2-II gene, we mapped both the 5′ and 3′ ends of the gRNA transcript. The 5′ end of gMURF2-II was mapped by a primer extension assay. The same gMURF2-II probe used for the Northern blot shown in Fig. 2A was used in the extension experiment and is underlined in Fig. 3A. Because the 5′ end of ND4 overlaps with the gMURF2-II gRNA sequence, this probe hybridized to both transcripts to produce two major extension products. As shown in Fig. 3B, the smaller of the two is located 7 nt from the 3′ end of the primer, while the larger is 13 nt from the 3′ end of the primer and corresponds to the mapped 5′ end of ND4 (8). Two additional minor products, of 17 and 19 nt, are also visible (Fig. 3B, lane 1). In order to determine which of these bands corresponded to the 5′ end of gMURF2-II, we performed primer extension analysis with gel-purified gRNAs (Fig. 3B, lane 2). For this experiment, only the 7-nt product was present, demonstrating that this band represents the 5′ end of the gMURF2-II transcript while the two larger products in lane 1 represent alternate minor mRNA 5′ ends. The maxicircle sequence within this region and the location of the mapped 5′ ends of the major- and minor-strand products are shown in Fig. 3A. These results demonstrate that the 5′ end of the gMURF2-II gene is located downstream of the 5′ end of the ND4 gene, within the 5′ untranslated region.
FIG. 3.
(A) Nucleotide sequence of gMURF2-II locus including the mapped 5′ ends of gMURF2-II and ND4 transcripts as well as the minor-strand CR4 gene (22). The 5′ end of the gRNA was determined by primer extension analysis using the gMURF2-IIR primer, indicated by the arrow underneath the sequence. The 3′ end of the gRNA indicated was obtained by sequencing RT-PCR products. The start codon for ND4 is underlined. (B) Primer extension mapping of gMURF2-II and ND4 5′ ends. An end-labeled gMURF2-IIR primer was extended in the presence of 1 μg of mtRNA (lane 1) or gel-purified gRNA (lane 2). The nucleotide sequence was obtained via primer extension of the total mtRNA in the presence of ddA or ddT (lanes 3 and 4, respectively). Extension products were run in an 8% acrylamide-urea gel. The 5′ ends of the gRNA and mRNA are indicated by arrows. Two minor mRNA products are marked with asterisks.
The 3′ end of the gMURF2-II gRNA was mapped by 3′ rapid amplification of cDNA ends. cDNA was synthesized from mtRNA by using a 5′-tagged oligo(dA) primer which hybridizes to the poly(U) tail common to the 3′ end of all gRNAs. The PCR product obtained with a gMURF2-II gene-specific primer and a tag-specific primer was sequenced to identify the 3′ end of the gRNA (data not shown). As seen for many gRNAs, the 3′ end of gMURF2-II ends in a short stretch of T's (indicated in the sequence shown in Fig. 3A) (26). Since gRNAs are posttranscriptionally modified by the addition of a short poly(U) tail (3), it is unclear whether the 3′ end of this gRNA is generated via transcription termination at this site or processing of a larger transcript. Together, the results of the 5′ and 3′ mapping of the gMURF2-II gRNA demonstrate that the gRNA is encoded completely within the ND4 gene. Although it is common for maxicircle genes to overlap in T . brucei, this is the first case in which a discrete intragenic transcript has been detected.
The mature gMURF2-II and ND4 transcripts are generated via different events.
Studies of mitochondrial gene expression in T. brucei have shown that the maxicircle is transcribed as a polycistronic precursor which is rapidly processed into mature transcripts (17). Due to the close packing and overlap of genes on the maxicircle, the generation of mature transcripts often involves mutually exclusive events (15, 21). According to this model of polycistronic transcription, the intragenic location of the gMURF2-II gRNA suggests that either gMURF2-II or the ND4 transcript could be made from a given precursor RNA, but not both. However, minicircle gRNAs in T. brucei are primary transcripts, as are both the minicircle and maxicircle gRNAs from Leishmania and Crithidia. Consequently, gMURF2-II may in fact be a primary rather than a processed transcript. In addition, if a promoter does exist within the intergenic region upstream of the gMURF2-II/ND4 region, it is possible that ND4 and/or other downstream mRNAs are primary transcripts.
In order the distinguish among these possibilities, we employed a modified version of a technique described by Bruderer et al. (4) to characterize the structure of the 5′ ends of gMURF2-II and ND4. Mitochondrial transcripts do not possess the 5′ cap structure seen in most nuclear RNAs. Consequently, primary transcripts possess triphosphate 5′ ends while processed transcripts have monophosphate 5′ ends. This technique takes advantage of the specificity of the enzyme T4 RNA ligase to ligate an RNA oligonucleotide to the 5′ end of transcripts bearing a monophosphate moiety. Transcripts with a di- or triphosphate or hydroxyl group at the 5′ end will not serve as substrates for this enzyme. Thus, a processed transcript will ligate to an exogenous RNA linker oligonucleotide while primary transcripts will not. The negative control consists of a phosphatase treatment that results in all transcripts bearing unligatable 5′ OH ends. The subsequent treatment of phosphatase-treated RNAs with kinase results in 5′-monophosphorylated molecules that serve as the positive control for ligation. It is the comparison of the abilities of the various treated samples to ligate to the RNA oligonucleotide that allows one to infer the nature of the 5′ end of the molecule. Primary transcripts, by nature of their polyphosphate ends, will remain unligated in all but the positive control, while processed transcripts will ligate in all but the negative control. Figure 4A shows the predicted ligation outcome for each treatment for a processed and a primary transcript.
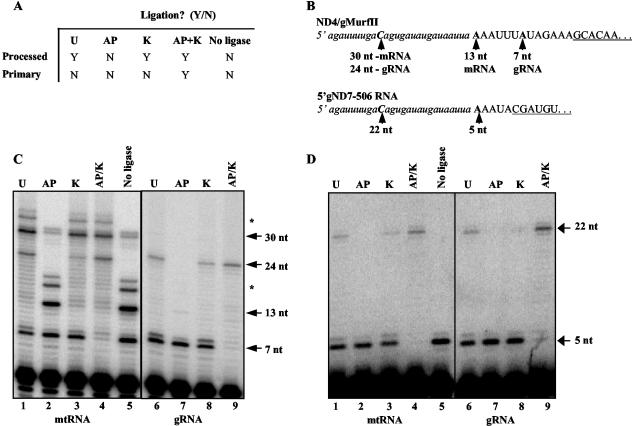
FIG. 4.
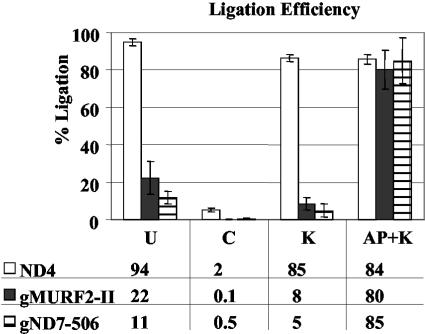
Ligation-dependent poisoned primer extension assay of total mtRNA and gel-purified gRNA. (A) Table of expected ligation outcomes for processed and primary transcripts subjected to various enzymatic treatments. Treatments: U, untreated; K, kinase; AP, alkaline phosphatase; AP/K, alkaline phosphatase followed by kinase; no ligase, untreated RNA not subjected to ligation. (B) Nucleotide sequence of the synthetic RNA oligonucleotide (lowercase italics) and the 5′ ends of ND4-gMURF2-II and gND7(506) (capital letters). The 3′ ends of the primers used in the extension reactions are underlined and the sizes of the expected products are indicated by arrows. Primer extension products were electrophoresed in 8% acrylamide-urea gels. (C) The 7-nt unligated gMURF2-II product is visible for all treatments but the positive control (AP/K), while the 13-nt unligated ND4 product is present only in the negative control lane (AP). (D) Minicircle-encoded gND7(506) remains unligated for all but the positive control lane (AP/K). Asterisks in panel C indicate the minor mRNA 5′ ends seen in Fig. 2A and the corresponding ligation products.
We subjected mtRNAs or gel-purified gRNAs to the various treatments and incubated them in the presence of T4 RNA ligase and a 26-nt synthetic RNA oligonucleotide containing a single C residue 17 nt from the 3′ end (Fig. 4B). We then performed a poisoned primer extension analysis in the presence of ddGTP to compare the abilities of various transcripts to ligate to the RNA oligonucleotide. This assay was sensitive enough to allow us to directly analyze the ligation results without the subsequent PCR step described in the original technique. A representative gel is shown in Fig. 4C. As expected, neither the gRNA nor the mRNA ligated in the negative control sample treated with alkaline phosphatase (Fig. 4C, lane 2). The predominant bands in this lane represent the 7-nt gMURF2-II 5′ end and the 13-nt ND4 5′ end. The larger bands seen at 17 and 19 nt represent the minor mRNA 5′ ends seen in Fig. 3. An identical result was obtained in the no-ligase negative control (Fig. 4C, lane 5). The faint doublet seen at ∼30 nt in the two negative control lanes is difficult to explain. These bands were not observed in the original primer extension mapping of 5′ ends (Fig. 3B) and may be a result of a variation in the mitochondrial isolation procedure. In order to optimize the detection of precursor transcripts, we incubated isolated mitochondria in transcription buffer and ribonucleoside triphosphates for 30 min before RNA isolation (10). Consequently, this band may represent a minor precursor product that is typically not detected, and this signal was subtracted from subsequent quantifications.
In contrast to the negative control lanes, both the ND4 and gMURF2-II RNAs did ligate in the samples that were subjected to kinase after phosphatase treatment (Fig. 4C, lane 4), as evidenced by the ddGTP stops seen at 24 and 30 nt for the gRNA and mRNA, respectively, as well as the minor mRNA ligated bands seen at 34 and 36 nt. Significantly, virtually no unligated gRNA or mRNA remained in the positive control lane, indicating the efficiency of the enzymatic steps required for this experiment. Likewise, the 13-nt unligated ND4 mRNA band is not visible in the untreated and kinase-only lanes (lanes 1 and 3, respectively), although the ligated 30-nt mRNA band is present in these lanes. This indicates that the majority of the ND4 mRNA possesses a 5′ monophosphate at the 5′ end, as expected for a transcript that is processed from a polycistronic precursor. In contrast, the unligated 7-nt product corresponding to the 5′ end of gMURF2-II is present in all lanes but the positive control lane, demonstrating that the majority of the gMURF2-II population possesses an unligatable triphosphate end, as expected for a primary transcript. The result for the gRNA is more clearly visible in the right half of the panel, for which the treatments, ligation, and primer extension were performed on gel-purified gRNA. Again, the 7-nt product corresponding to the 5′ end of the unligated gRNA is the predominant product in all lanes but the positive control lane (Fig. 4C, lane 9), in which only the ligated band is visible.
Although the majority of the gMURF2-II RNA remained unligated in the untreated lanes, some ligation was observed (Fig. 4C). Consequently, to better compare the ligation abilities of the gMURF2-II and ND4 transcripts, we quantified the signals corresponding to the ligated and unligated products and calculated the ligation efficiencies. A comparison of these values showed that ND4 has a ligation efficiency of >80% in all lanes but the negative control lane, while the gRNA ligated appreciably only in the positive control lane (Fig. 5). In addition, to confirm that the pattern obtained for gMURF2-II was similar to that seen for other mitochondrial primary transcripts, we performed this experiment using a primer complementary to the minicircle-encoded gRNA gND7(506) (14). Minicircle gRNAs have been shown to be primary transcripts by their ability to be capped in vitro by the enzyme guanylyl transferase (19). As seen for gMURF2-II, the gND7(506) RNA did not serve as an efficient substrate for T4 RNA ligase unless it was first treated with alkaline phosphatase followed by a kinase treatment (Fig. 4D). The 5-nt band corresponding to the mapped 5′ end of the unligated gND7(506) RNA is the predominant band in all lanes but the AP/K positive control lane (Fig. 4D, lanes 4 and 9), in which only the 22-nt ligated product is visible. Likewise, the ligation efficiency calculated for gND7(506) for each treatment was similar to that observed for gMURF2-II (Fig. 5).
FIG. 5.
Calculated RNA ligation efficiencies. Phosphorimages of replicate gels were analyzed with ImageQuant software (Molecular Dynamics). The percent ligation was calculated as the amount of ligated product/(amount of ligated plus amount of unligated product) × 100. The percent ligation for each transcript in each treatment is shown in the bar graph and the values are shown below. ND4 values were calculated by using only the major unligated and ligated products. Error bars represent the standard deviations obtained for replicate gels.
Together, these results indicate that the majority of the gRNA populations tested contain a triphosphate 5′ end. However, it is clear from the small amount of ligated gRNA seen in the untreated lanes that a minor population does contain a monophosphate 5′ end. The 5′ phosphates of in vitro-transcribed molecules have been observed via nuclear magnetic resonance to be labile under normal storage and handling conditions, resulting in a primary transcript population containing a mixture of tri-, di-, and monophosphates (Charles Hoogstraten, personal communication). However, we cannot rule out the possibility that the minor population of ligatable gRNAs in the untreated lane was a result of a 5′ monophosphate generated by an endogenous mitochondrial RNase. Despite the presence of this small population, the pattern of ligation ability for the processed mRNA is clearly distinct from that of the primary gRNA transcripts, visibly reflecting the difference in the structures of their 5′ ends.
An atypical minicircle gRNA not encoded within a cassette is also a primary transcript but shows heterogeneity in start site selection.
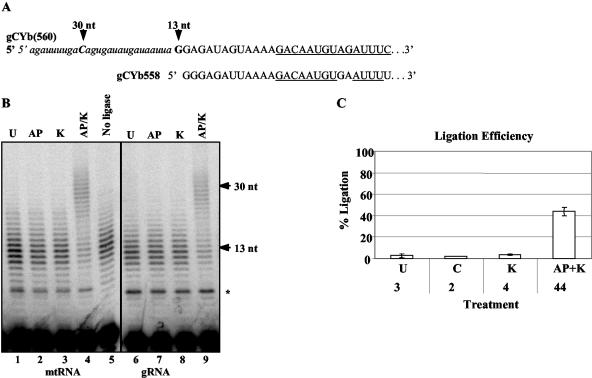
The gene for the gND7(506) gRNA is located between 18-bp imperfect inverted repeats, as are most minicircle gRNA genes. Each minicircle has, on average, three of these cassettes (20). Transcription of these minicircle gRNAs initiates a fixed distance downstream of the 3′ end of the upstream repeat, leading to speculation that the repeat sequences play a role in transcription. In addition, the stability of the resulting gRNA seems to depend upon transcription initiation at this location, as downstream gRNAs that are transcribed when the mtRNA polymerase reads through the upstream cassette are degraded (9). However, some gRNA cassettes lack functional gRNA genes while other gRNA genes are not located within cassettes (12). The gRNAs that mediate the initial editing of the CYb message are not located within inverted repeats. These gRNAs [gCYb(558) and gCYb(560A/B)] are redundant but not identical and are located on closely related minicircles that each also contain three gRNA cassettes (22). Interestingly, an analogous gRNA identified in both Leishmania and Crithidia (gCYb-I) is encoded by the maxicircle (2, 27). The T. brucei gCYb gRNAs have previously been shown to have heterogeneous 5′ ends, as determined by primer extension analysis (22). Likewise, these transcripts also have heterogeneous 3′ ends that can extend into the downstream forward repeat (22), indicating that some readthrough transcription occurs. This 5′ and 3′ heterogeneity could be due to a lack of precise transcription initiation and termination signals and/or the result of inexact processing of this RNA from a precursor generated via readthrough transcription from the upstream gRNA cassette. In order to determine whether the unusual gCYb(560) gRNA is a primary transcript, we performed a similar ligation-dependent primer extension, using a primer complementary to gCYb(560). As seen in Fig. 6, poisoned primer extension analysis did not result in a single band, but rather a series of bands, of ∼13 nt for the unligated gRNA, reflecting the 5′ heterogeneity previously observed for these gRNAs. The primer used is specific for gCYb(560), but due to the similarity in sequence among the redundant CYb gRNAs, this primer can also anneal, albeit less efficiently, to the gCYb(558) RNA (Fig. 6A). The distances from the 3′ end of the primer to the most common mapped 5′ ends of these gRNAs are very similar (difference of 1 nt) and do not account for the extent of the observed heterogeneity. From these experiments, it is clear that the RNA ligation-dependent pattern shown in Fig. 6 matches that seen for the maxicircle-encoded gMURF2-II as well as the minicircle cassette-encoded gND7(506) in that the gRNAs remained unligated in all but the positive control lane. In addition, the heterogeneity of the ligated product demonstrates that the heterogeneity seen at the 5′ end of gCYb(560) is the result of an imprecise transcription start site rather than of processing. These results, taken together with the maxicircle-encoded gMURF2-II result, indicate that mature gRNAs in T. brucei are primary transcripts, regardless of their kDNA location, and suggest important implications for potential transcription regulation of gRNA expression.
FIG. 6.
Ligation-dependent poisoned primer extension analysis of an atypical minicircle-encoded gRNA. (A) Nucleotide sequence of the synthetic RNA oligonucleotide (lowercase italics) and the 5′ ends of the redundant gCYb(560) and gCYb(558) gRNAs (capital letters). The gRNA sequence includes the most common 5′ ends (22), and the sequence corresponding to the 3′ end of the primer used for the primer extension reaction is underlined. (B) Primer extension products in an 8% acrylamide-urea gel. The arrows indicate the most common 5′ ends as shown in panel A. The asterisk indicates a 7-nt synthetic RNA oligonucleotide-dependent artifact resulting from partial hybridization of the 3′ end of the primer to the RNA oligonucleotide during the extension reaction. (C) Percent ligation efficiency for gCYb(560) as described in the legend to Fig. 5.
DISCUSSION
Although there is a core set of genes that are retained by all mitochondria, mitochondrial genomes vary dramatically in size, gene content, and organization (5). The T. brucei mitochondrial genome is unusual in that two distinct classes of transcripts are localized to two completely different genome components. The maxicircle component encodes primarily the rRNA and protein genes homologous to those encoded in the mitochondrial DNAs of other organisms. There is little room for intergenic regulatory elements within this compact genome, and the rRNA and mRNA genes are transcribed as part of a polycistronic precursor originating within the variable region (17). Interestingly, the maxicircle genomes of the related kinetoplastid organisms Leishmania and Crithidia have more potential for regulatory regions and encode additional transcription units in the form of gRNA genes (2, 27). An analysis of sequences surrounding these maxicircle gRNA genes did not reveal an obvious consensus promoter element (data not shown).
We have demonstrated that one of the two conserved maxicircle-encoded gRNA genes in T. brucei, gMURF2-II, represents an individual transcription unit located within the coding region of the maxicircle. This is the only defined maxicircle transcription initiation site reported for T. brucei. Its location is interesting for two reasons. The gRNA gene is contained entirely within the coding region of an mRNA that is processed from a polycistronic precursor. This is the first report of a discrete primary transcript generated from a completely intragenic kinetoplast gene. Additionally, the gMURF2-II locus is downstream of one of only three intergenic regions on the maxicircle genome. Given the tight packing, and in most cases, overlap of adjacent genes, the retention of this intergenic region indicates a potential regulatory function. The identification of a primary transcript within the coding region of the T. brucei maxicircle gives us a small, stable transcript to measure by in vitro transcription assays as we work to define promoter elements in T. brucei kDNA.
An analysis of the structures of the 5′ ends of the gMURF2-II and ND4 transcripts indicated that they are generated via distinctly different events. While the ND4 RNA must be precisely processed from the polycistronic precursor, the gMURF2-II RNA is initiated de novo a mere 7 nt downstream of the ND4 5′ end and is destined for termination and/or polyuridylation 50 bp downstream from the gRNA start site. Although there are two minor ND4 processing sites upstream of the major 5′ end of the mRNA, we did not detect any smaller minor products by using an ND4 mRNA-specific primer located downstream of the 3′ end of the gRNA (data not shown). This result suggests a complex model for maxicircle transcription in which distinct complexes may be involved in the specific generation of the gRNA and mRNA.
The majority of gRNAs in T. brucei are encoded on the distinct minicircle genome which is unique to these organisms and are located between inverted imperfect 18-bp repeats that have been implicated as playing a role in transcription (19, 20). Our results indicate that the unusual minicircle-encoded gCYb(560) gene also produces a mature primary transcript, demonstrating that minicircle gRNA genes located outside of these repeats also function as independent transcription units. The heterogeneity at the 5′ and 3′ ends of gCYb(560) that was previously reported and also detected in this study indicates that while the repeats are not essential for minicircle gRNA expression, they may contain signals for precise start site selection and termination. A recent analysis of minicircle genomes predicted the presence of other gRNA genes located outside of inverted repeats (12). It will be interesting to determine if these genes are indeed expressed, and if so, whether they similarly have heterogeneous ends.
Taken together, the data presented here indicate that gRNA genes in T. brucei, regardless of genome location, are individual transcription units. Although a common gRNA promoter sequence is not obvious, these results indicate that a gRNA-specific determinant of transcription exists within kDNA and may reflect an as yet unidentified gRNA transcription factor. The unique location of the intragenic transcription unit for the gMURF2-II gRNA suggests the possibility of a distinct gRNA transcription complex as well as a maxicircle polycistronic transcription complex or a single complex that is capable of both activities. In vitro and/or in organelle transcription experiments using this maxicircle locus will help us to distinguish between these possibilities and to identify such putative gRNA transcription factors.
REFERENCES
- 1.Arts, G. J., H. van der Spek, D. Speijer, J. van den Burg, H. van Steeg, P. Sloof, and R. Benne. 1993. Implications of novel guide RNA features for the mechanism of RNA editing in Crithidia fasciculata. EMBO J. 12:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: guide RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 3.Blum, B., and L. Simpson. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the pre-edited region. Cell 62:391-397. [DOI] [PubMed] [Google Scholar]
- 4.Bruderer, T., L. C. Tu, and M. G. Lee. 2003. The 5′ end structure of transcripts derived from the rRNA gene and the RNA polymerase I transcribed protein coding genes in Trypanosoma brucei. Mol. Biochem. Parasitol. 129:69-77. [DOI] [PubMed] [Google Scholar]
- 5.Burger, G., M. W. Gray, and B. F. Lang. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19:709-716. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 7.Clement, S. L., and D. J. Koslowsky. 2001. Unusual organization of a developmentally regulated mitochondrial RNA polymerase (TBMTRNAP) gene in Trypanosoma brucei. Gene 272:209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corell, R. A., P. Myler, and K. Stuart. 1994. Trypanosoma brucei mitochondrial CR4 gene encodes an extensively edited mRNA with completely edited sequence only in bloodstream forms. Mol. Biochem. Parasitol. 64:65-74. [DOI] [PubMed] [Google Scholar]
- 9.Grams, J., M. T. McManus, and S. L. Hajduk. 2000. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 19:5525-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, M. E., D. R. Moore, and S. L. Hajduk. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265:11368-11376. [PubMed] [Google Scholar]
- 11.Hauser, R., M. Pypaert, T. Hausler, E. K. Horn, and A. Schneider. 1996. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J. Cell Sci. 109:517-523. [DOI] [PubMed] [Google Scholar]
- 12.Hong, M., and L. Simpson. 2003. Genomic organization of Trypanosoma brucei kinetoplast DNA minicircles. Protist 154:265-279. [DOI] [PubMed] [Google Scholar]
- 13.Jasmer, D. P., and K. Stuart. 1986. Sequence organization in African trypanosome minicircles is defined by 18 base pair inverted repeats. Mol. Biochem. Parasitol. 18:321-331. [DOI] [PubMed] [Google Scholar]
- 14.Koslowsky, D. J., G. J. Bhat, A. L. Perrollaz, J. E. Feagin, and K. Stuart. 1990. The MURF3 gene of Trypanosoma brucei contains multiple domains of extensive editing and is homologous to a subunit of NADH dehydrogenase. Cell 62:901-911. [DOI] [PubMed] [Google Scholar]
- 15.Koslowsky, D. J., and G. Yahampath. 1997. Mitochondrial mRNA 3′ cleavage/polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol. Biochem. Parasitol. 90:81-94. [DOI] [PubMed] [Google Scholar]
- 16.Madison-Antenucci, S., J. Grams, and S. L. Hajduk. 2002. Editing machines: the complexities of trypanosome RNA editing. Cell 108:435-438. [DOI] [PubMed] [Google Scholar]
- 17.Michelotti, E. F., M. E. Harris, B. Adler, A. F. Torri, and S. L. Hajduk. 1992. Trypanosoma brucei mitochondrial ribosomal RNA synthesis, processing and developmentally regulated expression. Mol. Biochem. Parasitol. 54:31-41. [DOI] [PubMed] [Google Scholar]
- 18.Myler, P. J., D. Glick, J. E. Feagin, T. H. Morales, and K. D. Stuart. 1993. Structural organization of the maxicircle variable region of Trypanosoma brucei: identification of potential replication origins and topoisomerase II binding sites. Nucleic Acids Res. 21:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard, V. W., and S. L. Hajduk. 1991. Trypanosoma equiperdum minicircles encode three distinct primary transcripts which exhibit guide RNA characteristics. Mol. Cell. Biol. 11:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard, V. W., S. P. Rohrer, E. F. Michelotti, K. Hancock, and S. L. Hajduk. 1990. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell 63:783-790. [DOI] [PubMed] [Google Scholar]
- 21.Read, L. K., P. J. Myler, and K. Stuart. 1992. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J. Biol. Chem. 267:1123-1128. [PubMed] [Google Scholar]
- 22.Riley, G. R., R. A. Corell, and K. Stuart. 1994. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J. Biol. Chem. 269:6101-6108. [PubMed] [Google Scholar]
- 23.Shapiro, T. A., and P. T. Englund. 1995. The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 49:117-143. [DOI] [PubMed] [Google Scholar]
- 24.Simpson, L., S. Sbicego, and R. Aphasizhev. 2003. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA 9:265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart, K., and A. K. Panigrahi. 2002. RNA editing: complexity and complications. Mol. Microbiol. 45:591-596. [DOI] [PubMed] [Google Scholar]
- 26.Thiemann, O. H., and L. Simpson. 1996. Analysis of the 3′ uridylylation sites of guide RNAs from Leishmania tarentolae. Mol. Biochem. Parasitol. 79:229-234. [DOI] [PubMed] [Google Scholar]
- 27.van der Spek, H., G. J. Arts, R. R. Zwaal, J. van den Burg, P. Sloof, and R. Benne. 1991. Conserved genes encode guide RNAs in mitochondria of Crithidia fasciculata. EMBO J. 10:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]