Abstract
During Saccharomyces cerevisiae mating, chemotropic growth and cell fusion are critical for zygote formation. Cdc24p, the guanine nucleotide exchange factor for the Cdc42 G protein, is necessary for oriented growth along a pheromone gradient during mating. To understand the functions of this critical Cdc42p activator, we identified additional cdc24 mating mutants. Two mating-specific mutants, the cdc24-m5 and cdc24-m6 mutants, each were isolated with a mutated residue in the conserved catalytic domain. The cdc24-m6 mutant responds normally to pheromone and orients its growth towards a mating partner yet accumulates prezygotes during mating. cdc24-m6 prezygotes have two apposed intact cell walls and do not correctly localize proteins required for cell fusion, despite normal exocytosis. Our results indicate that the exchange factor Cdc24p is necessary for maintaining or restricting specific proteins required for cell fusion to the cell contact region during mating.
The fusion of two cells is a crucial biological process and is required for fertilization (sperm-egg fusion), myotube formation (myoblast fusion), mammalian placenta formation (trophoblast fusion), and Saccharomyces cerevisiae zygote formation (haploid cell fusion during mating) (for a review, see reference 48). During this process, intervening material, such as the extracellular matrix, needs to be degraded or removed so that the plasma membranes can become tightly apposed. In Drosophila melanogaster, the small G protein Rac1 and its activator myoblast city (a DOCK-180 homolog) are necessary for myoblast fusion (for a review, see reference 17). Whether small G proteins and their activators play a central role in the cell fusion process remains unclear.
During mating, yeast cells recognize and attach to each other, cell wall material is degraded, and the two haploid cells fuse, resulting in a diploid zygote. Cells respond to peptide pheromones (a- and α-factor) secreted by cells of the opposite mating type (for a review, see references 14, 28, and 51). These mating pheromones bind to specific G protein-coupled receptors on each cell type, and receptor activation results in cell cycle arrest, transcriptional induction of mating-specific genes, morphological changes leading to a pear-shaped shmoo, and polarized growth towards a partner cell. The guanine nucleotide exchange factor (Cdc24p) for the highly conserved Cdc42 G protein and a cyclin-dependent kinase inhibitor (Far1p) are required for oriented growth along a mating pheromone gradient (chemotropism) (37, 57). During oriented growth, the actin cytoskeleton and secretory apparatus polarize towards the tip of the mating projection (3, 42, 49), and therefore new cell wall and plasma membrane material is deposited at a unique location, which becomes the site of cell contact and ultimately fusion (30, 53). Polarized growth is also likely to be important for cell attachment via agglutinins and correct positioning of the cell fusion apparatus. Following cell attachment, vesicles appear to cluster at the regions of cell contact (5, 9, 22). Subsequently, the cell wall between two attached cells must be degraded so that plasma and nuclear membrane fusions can occur, resulting in a diploid zygote.
Various screens with S. cerevisiae have identified a number of mutants defective in cell fusion (4, 19, 20, 24, 27, 32, 40, 41). These mutants all make contact with a mating partner, becoming tightly attached via their cell walls. However, as the cell wall between the mating partners is present in most fusion mutants, the plasma membranes, cytoplasms, and nuclei fail to fuse. Such unfused mating pairs are referred to as prezygotes and these intermediates are observed during wild-type matings. Three classes of proteins are important for cell fusion. The first class includes Fus1p, Fus2p, Prm1p, and Fig1p, all of which are highly induced by mating pheromone and appear to function predominantly in cell fusion (20, 24, 54). It is likely that some of these proteins make up the fusion machinery. The second class includes Ram1p, Axl1p, and Ste6p; mutations in any of the corresponding genes result in a-cell-specific fusion defects (5, 18, 19). As these proteins are all important in a-factor biogenesis and production, it is likely that high levels of mating pheromone are important for cell fusion. It also appears that Axl1p and Ste6p play a role in cell fusion that is independent of their function in a-factor biogenesis and secretion, respectively (18, 19). The last class of proteins includes Rvs161p, Fps1p, Spa2p, Pea2p, Bni1p, Chs5p, Kel1p, Bud1p (Rsr1p), and Bud3p, which are also important in processes such as morphogenesis, polarity, actin organization, bud site selection, osmotic balance, or cell wall integrity (6, 15, 18, 22, 40, 41, 45, 56). It is unclear whether these proteins have direct roles in cell fusion; furthermore, the role of polarity establishment proteins, such as the guanine nucleotide exchange factor Cdc24p, in the cell fusion process remains to be elucidated.
Here we show that the GDP-GTP exchange factor Cdc24p for the highly conserved Rho G protein Cdc42p is necessary for cell fusion. We identified two novel cdc24 mating-specific mutants, the cdc24-m5 and cdc24-m6 mutants (where m indicates mating defect), which each have a mutated residue in the conserved catalytic Dbl homology (DH) domain. These mutants do not appropriately polarize two proteins required for cell fusion and hence accumulate prezygotes during mating.
MATERIALS AND METHODS
General techniques.
Standard techniques and media were used for yeast growth and genetic manipulation (43), and unless otherwise indicated, yeast strains were grown at 30°C.
Strains and plasmids.
The strains used in this study are described in Table 1. cdc24 bud1Δ double mutants were obtained by transformation and plasmid shuffling of RAY1474.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| JY426 | MATa leu2-3,112 ura3-52 his4-34 fus1-Δ1 fus2-Δ3 | Cold Spring Harbor Laboratory |
| JY429 | MATα trp1Δ1 ura3-52 cyh2 fus1-Δ1 fus2-Δ3 | Cold Spring Harbor Laboratory |
| PJ69-4A | MATa trp1-Δ901 leu2-3,112 ura3-52 his3-Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | 25 |
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | S. Emr |
| SEY6211 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 ade2 suc2-Δ9 | S. Emr |
| RAY563 | Same as SEY6210 with sph1-Δ1::HIS3 | 2 |
| RAY567 | Same as SEY6211 with sph1-Δ1::HIS3 | 2 |
| RAY914 | Same as RAY931 with pRS414CDC24 instead of pEG(KT)CDC24 | 37 |
| RAY916 | Same as RAY931 with pRS414cdc24-m1 instead of pEG(KT)CDC24 | 37 |
| RAY918 | Same as RAY928 with pRS414CDC24 instead of pEG(KT)CDC24 | 37 |
| RAY928 | Same as SEY6210 with cdc24-Δ1::loxP HIS5Spc loxP and pEG(KT)CDC24 | 37 |
| RAY931 | Same as SEY6211 with cdc24-Δ1::loxP HIS5Spc loxP and pEG(KT)CDC24 | 37 |
| RAY950 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 ade2 cdc24::LEU2 with pRS416GalHis6CDC24 | 37 |
| RAY1042 | Same as RAY950 with pRS414CDC24 instead of pRS416GalHis6CDC24 | 37 |
| RAY1044 | Same as RAY950 with pRS414cdc24-m1 instead of pRS416GalHis6CDC24 | 37 |
| RAY1046 | Same as RAY950 with pRS414cdc24-m2 instead of pRS416GalHis6CDC24 | 37 |
| RAY1052 | Same as RAY931 but cdc24-Δ1::loxP | 36 |
| RAY1474 | Same as RAY1052 with bud1Δ::loxP HIS5SpcloxP | 36 |
| RAY1487 | Same as SEY6211 but cdc24::TRP1 CDC24 and URA3::GFPBUD1 | 36 |
| RAY1657 | Same as RAY950 with pRS414cdc24-1 instead of pRS416GalHis6CDC24 | 34 |
| RAY1660 | Same as RAY950 with pRS414cdc24-111-1 instead of pRS416GalHis6CDC24 | 34 |
| RAY1681a | Same as RAY950 with pRS414cdc24-m5 instead of pRS416GalHis6CDC24 | This study |
| RAY1683a | Same as RAY950 with pRS414cdc24-m6 instead of pRS416GalHis6CDC24 | This study |
| RAY1685b | Same as RAY950 with pRS414cdc24[R416G] instead of pRS416GalHis6CDC24 | This study |
| RAY1699 | Same as RAY950 with pRS414cdc24[E423K/N446I] instead of pRS416GalHis6CDC24 | This study |
| RAY1705b | Same as RAY950 with pRS414cdc24[E423K] instead of pRS416GalHis6CDC24 | This study |
| RAY1716 | Same as RAY950 with pRS414cdc24[R416G/N551K] instead of pRS416GalHis6CDC24 | This study |
| RAY1718 | Same as RAY950 with pRS414cdc24[R416G/E423K] instead of pRS416GalHis6CDC24 | This study |
| RAY1724 | Same as RAY950 with pRS414cdc24[R416G/E423K/N446I/N551K] instead of pRS416GalHis6CDC24 | This study |
| RAY1740b | Same as RAY928 with pRS414cdc24[R416G] instead of pEG(KT)CDC24 | This study |
| RAY1747b | Same as RAY931 with pRS414cdc24[R416G] instead of pEG(KT)CDC24 | This study |
Original isolated mutants.
The cdc24-m6 and cdc24-m5 mutants were recreated by the R416G and E423K mutations, respectively, and were used for all analyses.
HIS5Sp, HIS5 from S. pombe.
pRS414CDC24 or pRS416CDC24 contains the CDC24 open reading frame, including 258 bp upstream of the ATG and 10 unique new restriction sites in CDC24 (37). Point mutations were generated by site-directed mutagenesis with Pfu polymerase (Promega) by the DpnI method (59) and identified by the addition or removal of a silent restriction site. All mutations generated were confirmed by sequencing (ABI PRISM Big-Dye terminator cycle sequencing kit). Green fluorescent protein (GFP) fusions of Cdc24p (35-37), Fus1p and Fig1p (46, 54), Spa2p (2), and Sec3p (21) were used and a GFP fusion of Sec2p was constructed by PCR-mediated replacement of the chromosomal copy of SEC2. pAS1CDC24, pGAD424BEM1, and pGAD424FAR1 plasmids (35, 37) were used for two-hybrid analyses as previously described.
Isolation of mating mutants.
Three different regions of CDC24 (bp 681 to 1221, 1221 to 1785, and 1785 to 2565) were independently amplified using mutagenic PCR conditions with pRS414CDC24 as a template and cloned, respectively, into AatII-PstI-, PstI-XhoI-, and XhoI-NotI-digested pRS414CDC24. Each library was transformed into RAY950, and mating mutants were isolated and recreated as previously described (37).
Mating assays, pheromone response assays, and phenotypic analyses.
Patch and quantitative matings were carried out as previously described (35-37), and unless indicated, mating was with a wild-type partner. Bud scars and the actin cytoskeleton were visualized as previously described (37), except that Alexa-568 phalloidin (Molecular Probes) was used for actin. For determination of the position of the bud scar relative to the mating projection, MATα cells were stained with 10 μg of calcofluor white (Sigma)/ml as previously described (37). Pheromone-induced cell cycle arrest (halo assays), induction of a FUS1-lacZ reporter, and cell shape changes were assayed as previously described (35, 37). For pheromone confusion assays, approximately 5 × 106 log-phase cells of each mating type were incubated in the presence or absence of 20 μM α-factor (Peptide Products, Ltd., Oldham, United Kingdom) for 5 h as previously described (37).
For quantitative cell fusion assays, matings were performed with a MATa GFP-Bud1 partner (RAY1487) (36). Cells were washed from the filters, sonicated, and viewed by differential interference contrast (DIC) and epifluorescence microscopy. The percentage of prezygotes is the number of prezygotes divided by the total number of mating pairs. Agglutination and cell viability assays were carried out as previously described (52, 61). For mannoprotein labeling, mating reaction mixtures were incubated with 25 μg of Alexa-Fluor 594 concanavalin A (Alexa-ConA; Molecular Probes)/ml (36).
Microscopy.
Actin cytoskeleton and time-lapse matings were imaged with a Deltavision deconvolution microscopy system (Applied Precision) on an Olympus IX-70 microscope with a numerical-aperture (NA) 1.4 × 60 objective. Actin images were deconvolved with softWoRX, and maximum-intensity projections of Z stacks were calculated. For time-lapse analyses, cells were mixed and spotted on yeast extract-peptone-dextrose agarose pads as previously described (2), and an environmental-temperature-controlled chamber (Solent Scientific) was used. In each time lapse, five mating pairs were monitored by DIC and fluorescence microscopy. Confocal microscopy was carried out with a Leica SP1 microscope with an NA 1.3 × 63 objective and 488-nm LASER excitation. Either phase-contrast images were taken using the confocal or DIC images were captured with a Leica DC-200 charge-coupled device camera. Single optical sections or arithmetic average projections of Z stacks were calculated with ImageJ, and the distribution of GFP signals was analyzed with a 10-pixel-wide line and the plot profile function. Cells were examined with a Leica DMR epifluorescence microscope with an NA 1.35 × 63 objective. Images were recorded with a Princeton Instruments Micromax charge-coupled device (Roper Scientific), using IPLab (Scanalytics) software. For electron microscopy, mating pairs from 20 mating reaction mixtures were enriched with 10-μm-pore-size polycarbonate filters (Millipore) and prepared as previously described (26), except that cells were incubated with 2% uranyl acetate prior to dehydration. Silver sections were stained with lead citrate, and micrographs were taken with a Hitachi H-600 electron microscope.
Immunoblot analyses.
Total yeast protein extracts were prepared (35) from budding cells, pheromone-treated cells, or mating mixtures using 1× 106 to 5 × 106 cells. Extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane (Schleicher and Schuell), and probed with an affinity-purified rabbit polyclonal serum against Cdc24p (1:500 dilution) or a rabbit polyclonal serum against GFP (1:1,000 dilution) (36). Rabbits were immunized with glutathione S-transferase (GST)-Cdc24 purified from Escherichia coli (pGEX4TCDC24) (pGEX4TCDC24 contains the 382 carboxyl-terminal amino acids of Cdc24p) with glutathione agarose (Sigma) as previously described (10). Antibodies were purified with GST coupled to Affigel 10 resin (Bio-Rad), followed by GST-Cdc24 coupled to Affigel 10 resin. Antibodies were eluted with Immunopure Gentle Antigen/Antibody Elution buffer (Pierce), dialyzed into 50 mM HEPES-50% glycerol, and stored at −20°C. Mouse monoclonal antibody (MAb) 22C5 (1:2,000 dilution; Molecular Probes) is against phosphoglycerate kinase (PGK). Immunoblots were visualized by enhanced chemiluminescence (Amersham).
RESULTS
Identification of novel cdc24 mating mutants.
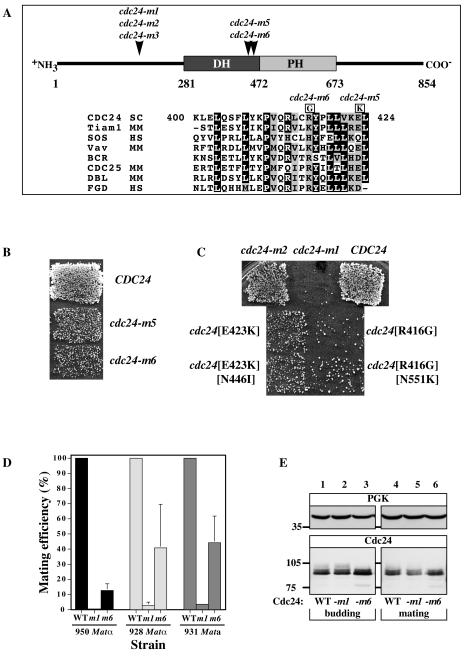
To identify additional functions of the Cdc42p guanine nucleotide exchange factor Cdc24p in the yeast mating process, we screened three libraries of cdc24 which were randomly mutagenized in regions encoding amino acids 227 to 404, 404 to 595, or 595 to 854 for mutants specifically defective in mating as previously described (37). Over 30,000 yeast colonies were tested for mating-specific defects; two mutants, the cdc24-m5 and cdc24-m6 mutants, were identified from the second library. Each of the cdc24 mutants had two amino acid alterations, E423K and N446I for cdc24-m5 and R416G and N551K for cdc24-m6. In each mutant, the first altered residue is similar in S. cerevisiae, Kluyveromyces lactis, Candida albicans, and Schizosaccharomyces pombe Cdc24p sequences. Furthermore, both residues Glu 423 and Arg 416 fall in the conserved region 3 of the catalytic domain and similarly charged amino acids are found in a number of mammalian exchange factors (Fig. 1A). cdc24-m5 and cdc24-m6 cells showed mating defects with a wild-type tester, cdc24-m6 cells having the stronger defect (Fig. 1B). The recreation of each individual mutation revealed that E423K and R416G were responsible for the mating defects of the cdc24-m5 and cdc24-m6 mutants, respectively (Fig. 1C). As the R416G mutation resulted in the strongest mating defect, we thereafter concentrated on this mutant, referred to as cdc24-m6. The cdc24-m6 mutation behaved recessively; both addition of a wild-type CDC24 copy to the mutant and addition of a cdc24-m6 copy to a wild-type strain resulted in a wild-type mating efficiency (data not shown). Quantitative matings (Fig. 1D) demonstrated that the cdc24-m6 defect is independent of cell mating type and is weaker than that of the previously characterized cdc24-m1 mutant, which is defective in chemotropism (37). When cdc24-m6 cells were mated with either the enfeebled mating partner fus1Δ fus2Δ or cdc24-m6 cells, a further decrease (less than fivefold) in mating efficiency was observed (data not shown). These results indicate that the cdc24-m6 mutant does not show a strong bilateral mating defect, in contrast to the cdc24-m1 mutant, which shows a dramatic increase (greater than 1,000-fold) in mating defect when mated with cdc24-m1 cells (37). The expression levels of Cdc24p, Cdc24-m1p, and Cdc24-m6p in budding and mating cells are similar (Fig. 1E), indicating that the cdc24-m6 mating defect is not due to an altered expression level. These results show that the cdc24-m6 mating defect is recessive, cannot be compensated by a mating partner, and is not due to an inappropriate level of Cdc24p.
FIG. 1.
Characterization of cdc24-m5 and cdc24-m6 mating mutants. (A) Locations of cdc24 mating mutations; schematic diagram of Cdc24p showing positions of cdc24-m1, cdc24-m2, cdc24-m3, cdc24-m5, and cdc24-m6 mutations. The DH catalytic domain and the pleckstrin homology domain (PH) are indicated. Below, BLAST alignments (1) of S. cerevisiae Cdc24p and mammalian exchange factor conserved region 3 sequences are shown, with residues that are similar (boxed in grey) or identical (boxed in black) in 80% or more of the sequences. Amino acid changes responsible for the cdc24-m5 (E423K) and cdc24-m6 (R416G) mutants are indicated. (B) Patch mating of the isolated cdc24-m5 and cdc24-m6 mutants. Matings using a strain derived from RAY950 with the indicated CDC24 or cdc24 gene (as the sole copy) are shown. (C) Identification of amino acid changes responsible for the cdc24-m5 and cdc24-m6 mating defect. CDC24 or cdc24 mutants were created by site-directed mutagenesis (with a sole copy of CDC24 or cdc24) in a RAY950-derived strain. Matings were carried with the enfeebled tester JY426. Combinations of R416G and E423K or all four changes did not result in a stronger defect. (D) The cdc24-m6 mating defect is independent of the mating type. Strains in which CDC24, cdc24-m1, or cdc24-m6 is the sole copy (derived from RAY950, RAY928, or RAY931) were used for quantitative matings. Values shown are the means of three to four determinations with standard error of the mean (SEM). RAY1042, RAY918, and RAY914 mating efficiencies (24.5, 9.5, and 11.1%, respectively) were set at 100%. (E) The expression level of the Cdc24-m6p is similar to that of wild-type strains during budding and mating. Cell extracts of budding (RAY914, RAY916, and RAY1747) and mating (RAY914 × RAY1042, RAY916 × RAY1044, and RAY1747 × RAY1685) cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting and probing with either anti-PGK MAb or anti-Cdc24 polyclonal sera. WT, wild type.
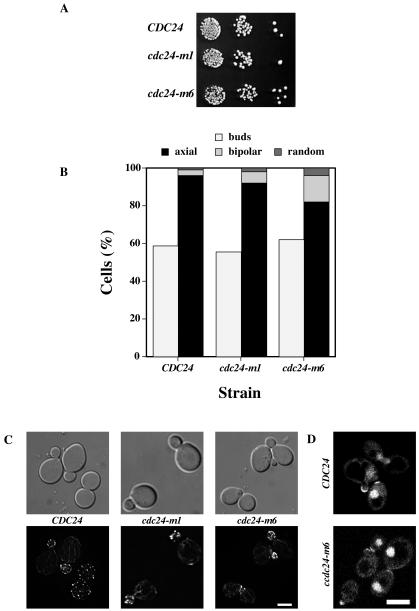
The cdc24-m6 mating mutant does not affect vegetative growth.
As the amino acid residue altered in the cdc24-m6 mutant is in the catalytic domain, we investigated whether this mutant grew normally and polarized its actin cytoskeleton. Figure 2A shows that cdc24-m6 cells grew similar to wild-type cells and that the percentage of CDC24, cdc24-m1, and cdc24-m6 cells with buds was indistinguishable (Fig. 2B). Examination of bud site selection patterns revealed a slight increase in bipolar budding with cdc24-m6 cells (Fig. 2B). We examined the morphology and actin cytoskeleton in cdc24-m6 cells. This mutant forms buds similar to those of wild-type and cdc24-m1 cells (Fig. 2C). In all three strains, both actin patches and cables were highly polarized in cells with small buds (Fig. 2C). Furthermore, both Cdc24-GFP and Cdc24-m6-GFP localized to the tips of small buds and nuclei in budding cdc24Δ cells (Fig. 2D). Together, these results indicate that cdc24-m6 cells are not defective for vegetative growth, and hence this mutant is mating specific.
FIG. 2.
cdc24-m6 cells are normal for vegetative growth. (A) cdc24-m6 cells grow normally. Serial 10-fold dilutions of the indicated cultures were spotted onto yeast extract-peptone-dextrose plates and incubated for 2 days. (B) cdc24-m6 cells bud normally. Percentages of budding cells (n = 160) and bud site selection patterns (n = 100) are shown. (C) cdc24-m6 cells polarize their actin cytoskeleton. DIC and fluorescence images of budding cells stained with Alexa-568 phalloidin are shown. Fluorescence images are maximum intensity projections of Z sections (10 × 0.1 to 20 × 0.1 μm). Bar, 5 μm. (D) Cdc24-m6p localizes to sites of growth and nuclei. Confocal microscopy images of cdc24Δ cells expressing Cdc24-GFP or Cdc24-m6-GFP are shown. Bar, 5 μm.
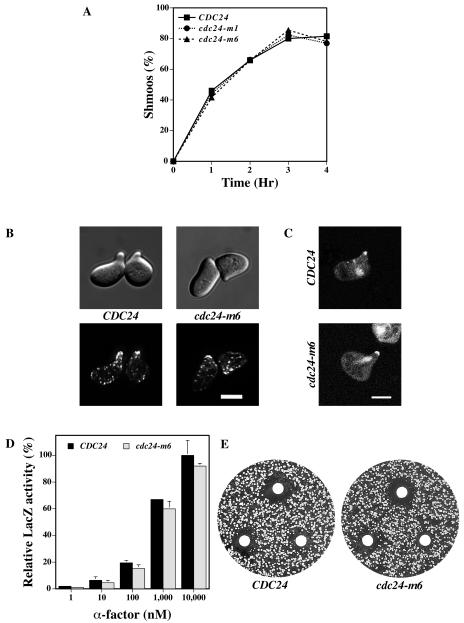
cdc24-m6 cells respond normally to mating pheromone.
We next investigated mating pheromone responses of cdc24-m6 cells. First, we examined shmoo formation as a measure of cell cycle arrest and cell shape change. The percentage of shmoos formed by CDC24, cdc24-m1, and cdc24-m6 cells was indistinguishable over a 4-h incubation period with pheromone (Fig. 3A). Thus, cdc24-m6 cells form shmoos to the same extent and with the same kinetics as the wild type. We next examined the morphology of the cdc24-m6 shmoos, their actin cytoskeleton distribution, and Cdc24p localization. In response to α-factor, cdc24-m6 cells formed pear-shaped shmoos with a polarized actin cytoskeleton similar to that of wild-type cells (Fig. 3B). Wild-type and mutant Cdc24-GFP were observed at the tips of mating projections and in the nuclei of pheromone-treated cdc24Δ cells (Fig. 3C). Pheromone-dependent gene induction was investigated with FUS1-lacZ, and Fig. 3D shows that induction levels of this gene were similar in CDC24 and cdc24-m6 cells over a range of α-factor concentrations. FUS1-lacZ induction was also measured during mating after 1, 2, and 3 h of incubation with a partner. No difference in FUS1 induction was seen between wild-type and cdc24-m6 cells, despite the observed mating defects (data not shown). These results indicate that pheromone-dependent mitogen-activated protein kinase signaling is unaffected in cdc24-m6 cells. Mating pheromone halo assays were carried out to analyze the pheromone concentration dependence of growth arrest. Figure 3E shows that the growth arrest halos with CDC24 and cdc24-m6 cells were identical. Collectively, these results demonstrate that cdc24-m6 cells respond normally to mating pheromone.
FIG. 3.
cdc24-m6 cells respond to mating pheromone. (A) cdc24-m6 cells form shmoos at the same rate and to the same extent as wild-type cells. Indicated strains were incubated with α-factor (12 μM) and shmoos were counted at the times shown (n = 125). The timing of the appearance of additional mating projections was the same in both strains. (B) The actin cytoskeleton in cdc24-m6 shmoos is polarized. DIC and fluorescence images of cells treated with 12 μM α-factor for 2 h as previously described in the legend to Fig. 2C are shown. Bar, 5 μm. (C) Cdc24-m6p localizes to mating projection tips and nuclei in shmoos. Confocal microscopy images of cdc24Δ cells expressing Cdc24-GFP or Cdc24-m6-GFP were treated with α-factor as described above. Bar, 5μm. (D) cdc24-m6 cells induce the mating-specific FUS1 gene in a pheromone-dependent fashion. Cells containing the FUS1-lacZ plasmid pSG231 were incubated with the indicated α-factor concentration for 1 h and LacZ activity was determined. The means of two independent experiments are shown with error bars indicating values. LacZ activity for CDC24 cells treated with 10,000 nM α-factor (40.8 Miller units) was set at 100%. (E) cdc24-m6 cells arrest growth in the presence of mating pheromone similar to wild-type cells. α-factor (1, 0.5, and 0.2 μg) was spotted on filters placed on a lawn of the indicated strain. Plates were incubated for 2 days. Measurements of the halo diameter indicated ≤5% difference between CDC24 and cdc24-m6 halos.
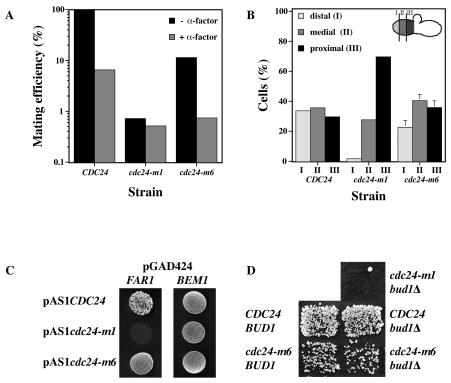
cdc24-m6 cells are not chemotropism defective.
cdc24-m1 cells respond normally to mating pheromone yet are unable to orient growth towards a mating partner (37). In contrast to wild-type cells, chemotropism mutants are unaffected by the dissipation of the pheromone gradient during mating (16, 57). We determined whether cdc24-m6 cells were similarly defective in oriented growth by the addition of a saturating level of α-factor to the mating mixture. Figure 4A shows that the addition of 20 μM α-factor to a mating mixture containing CDC24 or cdc24-m6 cells resulted in a substantial decrease in mating efficiency. In contrast, a mating mixture containing cdc24-m1 cells was barely affected by pheromone addition. Furthermore, in both cdc24 and far1 chemotropism mutants, mating projections typically form adjacent to the previous bud scar, irrespective of the location of a mating partner (37, 57). Therefore, we analyzed the position of the mating projection relative to the previous bud scar in cdc24-m6 zygotes. Mating projections of cdc24-m6 cells were randomly positioned relative to their previous bud scar, similar to what occurs in wild-type cells and in contrast to what occurs in cdc24-m1 cells (Fig. 4B) (37). Consistent with these results, Cdc24-m6p interacted normally with Far1p and Bem1p by a two-hybrid assay (Fig. 4C). In addition, cdc24-m6 bud1Δ double mutants exhibited similar mating efficiencies as the cdc24-m6 mutant alone, in contrast to chemotropism mutants such as cdc24-m1, which have a synthetic mating defect when they are combined with bud1Δ mutants (Fig. 4D) (36). Together, these four different approaches indicate that cdc24-m6 cells are not defective in chemotropic growth.
FIG. 4.
cdc24-m6 mutants are not chemotropism defective. (A) A pheromone gradient is necessary for the efficient mating of cdc24-m6 cells. Strains (∼5 × 106 RAY914, RAY916, or RAY1747 cells) were mated in the presence or absence of 20 μM α-factor as previously described (37). Values are the means of two independent matings. (B) The direction of growth in cdc24-m6 cells during mating is independent of the previous bud scar position. Strains (RAY914, RAY916, and RAY1747) were stained with calcofluor white and then mated with a wild-type tester. The position of the mating projection relative to the bud scar on the stained half of the zygote was determined in two independent experiments. Values represent the means of two or three determinations (n = 50) with the SEM shown. (C) Cdc24-m6p interacts with Far1p and Bem1p. The two-hybrid strain PJ69-4A (25) carrying the indicated GAL4 activation and DNA binding domain fusions was spotted on medium lacking Leu, Trp, and His. Growth indicates an interaction. Identical results were obtained with four transformants. (D) cdc24-m6 bud1Δ double mutants do not have an increased mating defect. Indicated strains derived from RAY1474 (cdc24Δ bud1Δ) and RAY931 (cdc24Δ BUD1) were patch mated with the enfeebled tester, JY429.
cdc24-m6 mating mixtures accumulate prezygotes with intact cell walls.
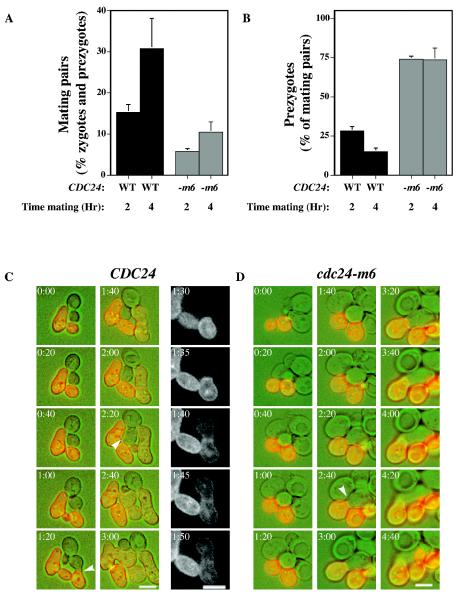
We next determined the number of mating pairs (zygotes and prezygotes) in mating mixtures of cdc24-m6 and wild-type cells. After 2 and 4 h of mating, cdc24-m6 mutant mating mixtures had threefold-fewer mating pairs than a wild-type control (Fig. 5A). When cdc24-m6 cells were mated with cdc24-m6 cells, we observed a defect in cell agglutination (agglutination index of 0.18 compared to 0.32 for wild-type pairs), suggesting that the decrease in mating pairs observed with the cdc24-m6 mutant could be due in part to an agglutination problem. In order to determine if cdc24-m6 cells were defective in cell fusion, we quantitated the percentage of mating pairs that were prezygotes after 2 and 4 h of mating with a wild-type tester whose plasma membrane was labeled with GFP-Bud1. Prezygotes were identified as mating pairs in which GFP fluorescence was observed only in one cell. In wild-type mating mixtures, prezygotes accounted for 28 and 15% of mating pairs after 2 and 4 h of incubation, respectively, whereas at both times 74% of cdc24-m6 mating pairs were prezygotes (Fig. 5B). Methylene blue dye exclusion confirmed that cdc24-m6 cells (including mating pairs) were viable (data not shown), indicating that prezygote accumulation was not due to lysis. Matings with cdc24-m5 cells also accumulated more prezygotes than controls (33% compared to 24% for control cells), suggesting that these two mutants affect the fusion process. To address the possibility that a slight reduction in Cdc24p activity resulted in prezygote accumulation during mating, we carried out matings with two different cdc24 temperature-sensitive mutants (the cdc24-111-1 and cdc24-1 mutants). The cdc24-111-1 mutant did not grow at 34°C (34), whereas the cdc24-1 mutant grew poorly. There was no substantial difference in the percentages of mating pairs that were prezygotes at 34°C: CDC24, 27% ± 1%; cdc24-111-1, 28% ± 3%; and cdc24-1, 29% ± 1%. These results indicate that a general decrease in Cdc24p function does not lead to prezygote accumulation.
FIG. 5.
cdc24-m6 cells form fewer mating pairs and accumulate prezygotes. (A) cdc24-m6 cell mating mixtures are defective in mating pair formation. RAY1042 and RAY1685 were mated with RAY1487 (GFP-Bud1). The percentage of mating pairs formed after the times indicated was determined as the number of prezygotes (fluorescence in only one cell) and zygotes (fluorescence in both cells) divided by the total number of cells, with values representing the means of four determinations (n = 150) from two independent matings with SEM. WT, wild type. (B) cdc24-m6 mating mixtures accumulate prezygotes. The same experiment as that described above was analyzed for the percentage of prezygotes. Values represent the means of two determinations (n = 125), with bars indicating actual values. WT, wild type. (C) Time-lapse images of CDC24 cells mating with a GFP-Bud1-labeled mating partner. Images taken at the indicated times with DIC (green) and fluorescence (red) are shown. The right-hand column illustrates the GFP signal. Arrowheads indicate the mating pairs just prior to fusion. Bar. 5 μm. (D) Time-lapse images of cdc24-m6 cells mating with a GFP-Bud1-labeled mating partner. The images were acquired as described above. Arrowhead indicates the prezygote. Bar, 5 μm.
To examine the dynamics of cell fusion, we used time-lapse microscopy to follow matings with a wild-type partner expressing GFP-Bud1. In each experiment, five cell pairs were imaged every 5 min at 30°C for more than 5 h. Figures 5C and D illustrate a typical CDC24 and cdc24-m6 mating time course in which the wild-type and mutant cells grew similarly during mating. CDC24 mating pairs fused after 100 and 160 min, whereas the cdc24-m6 mating pairs did not fuse even after 280 min. Single-color fluorescence images show the redistribution of the plasma membrane during fusion (Fig. 5C). In the cdc24-m6 mating, despite cell attachment which occurred at ∼140 min (2:20 image), the mutant cell continued to grow in a polarized fashion (elongated) for an additional 2 h.
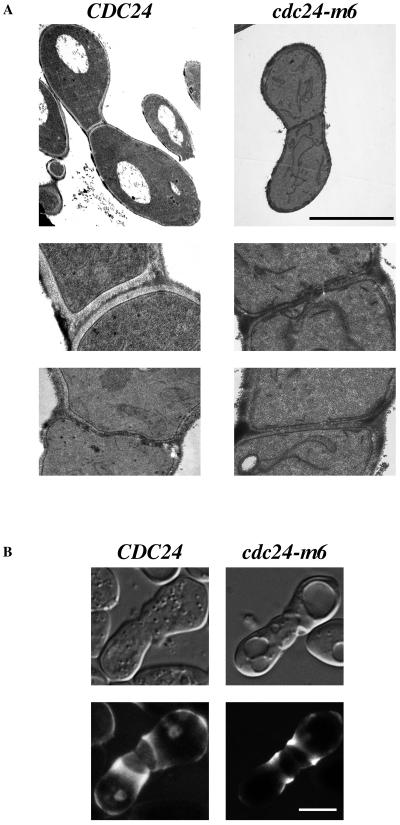
Electron microscopy and ConA labeling were used to investigate the ultrastructural features and cell wall mannoprotein distribution in cdc24-m6 prezygotes. To address whether the cdc24-m6 prezygotes had degraded their cell walls at the site of cell contact, we examined mating mixtures by electron microscopy. Figure 6A shows wild-type and mutant prezygotes in which two distinct cell walls are apparent at the region of cell contact. Examination of nuclei, endoplasmic reticula, and vesicles did not reveal any striking differences between CDC24 and cdc24-m6 prezygotes. ConA labeling indicates the location of cell wall mannoproteins, which are induced in response to mating pheromone and pass through the secretory pathway (30, 53). CDC24 or cdc24-m6 cells were mated with a wild-type tester in the presence of Alexa-ConA; prezygotes from both mating mixtures had a ring of ConA staining adjacent to the cell contact region (Fig. 6B). This distribution of cell wall mannoproteins was indistinguishable between CDC24 and cdc24-m6 prezygotes. In both wild-type and mutant prezygotes, the cell wall at the site of contact was typically less fluorescent than this ring, which we attribute to the inaccessibility of the newly synthesized mannoproteins to ConA in the mating mixture. Together, light and electron microscopy indicates that cdc24-m6 mating mixtures accumulate prezygotes whose cell walls are attached but not degraded.
FIG. 6.
Cell wall structure is unaffected in cdc24-m6 prezygotes. (A) Ultrastructural organization of CDC24 and cdc24-m6 prezygotes is indistinguishable. The indicated strains (RAY1042 × RAY914 and RAY1685 × RAY1747) were mated as previously described, and electron microscopy images are shown at magnifications of ×4,500 (top panels) and ×18,000 (bottom two panels). Prezygotes from mating mixtures of cdc24-m6 cells and a wild-type partner similarly had two distinct cell walls. Bar, 5 μm. (B) Cell wall mannoproteins are similarly distributed in CDC24 and cdc24-m6 prezygotes. The indicated strains (RAY1042 and RAY1685) were mated as previously described for quantitative matings in liquid containing 25 μg of Alexa-ConA/ml. DIC images (top panels) are taken through the central focal plane and images in the bottom panels are average projections of confocal Z sections (25 × 0.12 to 30 × 0.12 μm). Bar, 5 μm.
cdc24-m6 cells are unable to polarize proteins required for cell fusion.
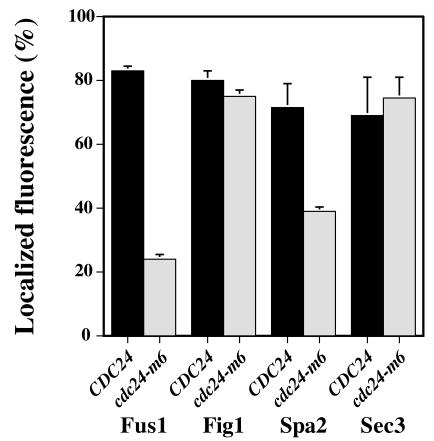
Since cdc24-m6 prezygotes appeared indistinguishable from wild-type prezygotes, we speculated that this mutant was unable to correctly localize proteins required for cell fusion. We examined the distribution of Fus1p, Fig1p, Spa2p, Sec3p, and Sec2p in prezygotes using GFP fusions. Both Fus1p and Fig1p are integral membrane proteins that are strongly induced during mating (20, 54). Figure 7A shows the distribution of Fus1-GFP in CDC24 or cdc24-m6 prezygotes. In wild-type prezygotes, Fus1p is localized predominantly at the plasma membrane, enriched at the region of cell contact and along the sides of the mating projection (46, 54). Strikingly, Fus1p was significantly delocalized in cdc24-m6 prezygotes with a concomitant increase in intracellular signal, in particular in the vacuole. Quantitation of Fus1-GFP distribution revealed a substantial decrease in the enrichment of this protein at the zone of cell contact in cdc24-m6 prezygotes (∼40% greater than the cytosolic signal) compared to that in CDC24 prezygotes (∼240% greater than the cytosolic signal). More than 80% of CDC24 prezygotes had Fus1-GFP fluorescence localized preferentially to the cell contact zone, in contrast to approximately 20% of cdc24-m6 prezygotes (Fig. 8). We further investigated Fus1-GFP distribution and levels in wild-type and cdc24-m6 cells incubated with mating pheromone. Fus1p is substantially delocalized in cdc24-m6 shmoos despite identical Fus1-GFP levels in these strains (Fig. 7B), consistent with the indistinguishable FUS1-lacZ levels in cdc24-m6 and CDC24 pheromone-treated cells and mating mixtures. In addition, there was no increase in an unglycosylated form of Fus1-GFP, as was previously observed with chs5Δ mutants (46). These results show that cdc24-m6 cells are unable to appropriately localize the cell fusion protein Fus1p and suggest that this defect is not due to an accumulation of this protein in the secretory pathway.
FIG.7.
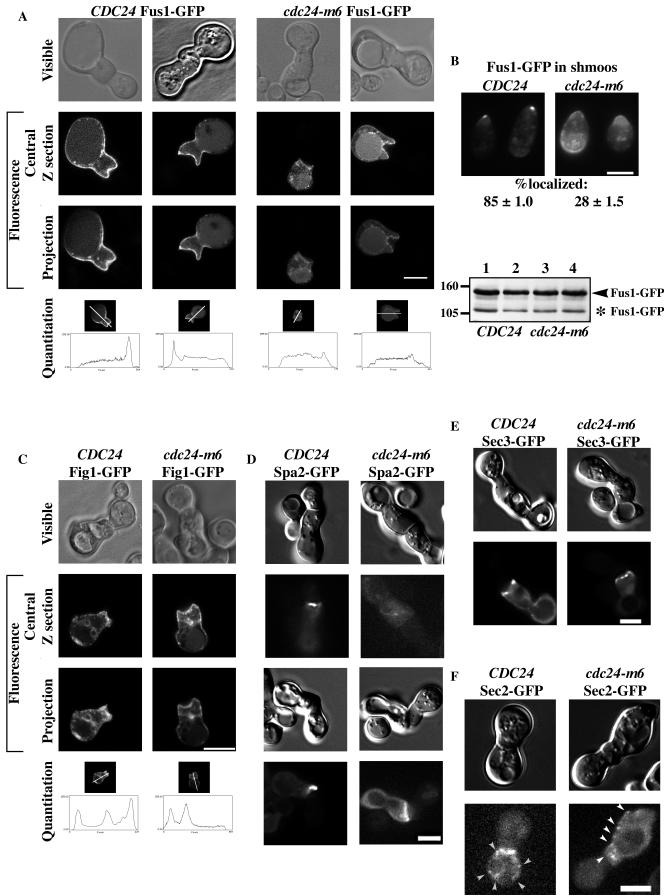
Cdc24p is required for correct localization of two cell fusion proteins. (A) Fus1p is not enriched at the cell contact zone in cdc24-m6 prezygotes. Strains RAY1042 and RAY1685 containing Fus1-GFP were mated as previously described for quantitative matings and imaged by confocal microscopy. Visible (phase contrast), central focal plane, and average projections of Z sections (10 × 0.12 to 20 × 0.12 μm) are shown. Projections were used for the quantitation of signal distribution, with a 10-pixel-wide line used for intensity determination (reading left to right in image and graph). (B) Fus1p is delocalized in cdc24-m6 shmoos. Strains RAY914 and RAY1747 carrying Fus1-GFP were incubated with α-factor as described in the legend to Fig. 3A, and fluorescence images were taken. The mean percentage of cells with an observable GFP signal polarized to the shmoo tip was determined from two independent experiments (n = 110), with ± indicating actual values. Immunoblots of cell extracts from the same experiment with unglycosylated Fus1-GFP indicated (*) are shown. Probing with anti-PGK MAb indicated that equal amounts were loaded in each lane. (C) Fig1p localization is unaffected in cdc24-m6 prezygotes. The indicated strains (RAY1042 and RAY1685) containing Fig1-GFP were mated and imaged as described for panel A. (D) Spa2p is not correctly localized in cdc24-m6 prezygotes. Strains RAY1042 and RAY1685 containing Spa2-GFP were mated as described for panel A and imaged as described for panel B. (E) Sec3p is localized to the cell contact region in cdc24-m6 prezygotes. Strains RAY1042 and RAY1685 containing Sec3-GFP were mated and imaged as described for panel D. (F) Secretory vesicle number and distribution are unaffected in cdc24-m6 prezygotes. Strains RAY1042 and RAY1685 containing Sec2-GFP were mated as described for panel A. Prezygotes expressing Sec2-GFP were imaged at 25°C every second for 20 s, and a representative image is shown. Secretory vesicles are indicated by arrowheads. Bar, 5 μm (panels A to F).
FIG. 8.
Quantitation of protein localization in cdc24-m6 prezygotes. Strains carrying the indicated GFP fusion were mated as described in the legend to Fig. 7A, and the mean percentage of prezygotes with an observable fluorescence signal that was enriched to the cell contact zone was determined from two independent experiments (n = 100), with bars indicating actual values.
We subsequently investigated the localization of an additional pheromone-induced membrane protein, Fig1p (20). Fig1-GFP is localized predominantly to the plasma membrane in CDC24 and cdc24-m6 prezygotes; furthermore, we observed Fig1-GFP at bud and birth scars (Fig. 7C). Nonetheless, Fig1p was similarly enriched at the plasma membrane region of cell contact in both strains; similar percentages of CDC24 and cdc24-m6 prezygotes showed this polarized distribution (Fig. 8), indicating that there is not a general defect in the distribution of pheromone-inducible membrane proteins in cdc24-m6 prezygotes.
We examined whether cdc24-m6 cells were defective in localizing other proteins required for cell fusion, such as the scaffold protein Spa2p (15, 22). In both wild-type and cdc24-m6 budding cells, Spa2-GFP localized as a tight spot restricted to the site of growth (data not shown) as previously described (2, 50). In wild-type prezygotes, Spa2-GFP was localized to a focused spot at the cell contact site. In contrast, in cdc24-m6 prezygotes, Spa2-GFP displayed a diffuse distribution throughout the cell or along the cell contact region (Fig. 7D). Quantitation of the number of prezygotes with polarized Spa2-GFP distribution revealed a considerable decrease in cdc24-m6 cells (Fig. 8). In addition, Spa2-GFP was similarly delocalized in cdc24-m6 shmoos (data not shown).
We next examined two markers for secretion: Sec3p is a spatial landmark for exocytosis (21), and Sec2p is a component of secretory vesicles (39, 58). Sec3-GFP localizes to the plasma membrane site where exocytosis occurs; in CDC24 and cdc24-m6 prezygotes, this protein is distributed similarly in several tight clusters at the region of cell contact (Fig. 7E). Similar percentages of CDC24 and cdc24-m6 prezygotes showed this polarized distribution (Fig. 8). In contrast, Sec2p localizes to secretory vesicles that are dynamic in both budding and mating cells. CDC24 or cdc24-m6 prezygotes expressing Sec2-GFP were imaged every second for 20 s. Figure 7F shows an image of wild-type and mutant prezygotes at one time point. Vesicles (indicated by arrowheads) were observed in prezygotes from both strains adjacent to the cell contact zone as well as throughout the cell. There was no significant difference in the average numbers of vesicles observed in prezygotes over 10 s (CDC24, 6.4 ± 2.6 vesicles, n = 5 cells; cdc24-m6 mutant, 8.2 ± 1.8 vesicles, n = 6 cells). Together the localization of Fus1p, Fig1p, Spa2p, Sec3p, and Sec2p in cdc24-m6 prezygotes indicates that this mating mutant is specifically defective in restricting two proteins required for cell fusion to the cell contact zone, yet it is normal for SEC-dependent exocytosis. These results suggest that Cdc24p is necessary for restricting or maintaining the polarized distribution of specific proteins required for cell fusion during mating.
DISCUSSION
Polarized growth is essential for yeast mating and occurs prior to, during, and subsequent to cell fusion. A complex of Cdc24p, Far1p, and Gβγ is required for directional growth of a yeast cell towards its mating partner (8, 35, 37, 57). Here we show that in addition to this role, Cdc24p is required for cell fusion during mating. We have isolated and characterized novel cdc24 mutants that are normal for vegetative growth yet specifically defective for yeast mating. Both of these mutants have a single amino acid change in the conserved DH domain. These mutants, cdc24-m5 and cdc24-m6, respond normally to mating pheromone and orient growth towards a mating partner; however, they also accumulate prezygotes during mating. This cell fusion defect is not the result of altered Cdc24p levels, distribution, or a general reduction in exchange factor function. cdc24-m6 prezygotes have two apposed intact cell walls, indicating that the fusion defect occurs prior to cell wall degradation. Strikingly, two proteins required for cell fusion do not localize correctly in cdc24-m6 prezygotes and shmoos, despite normal SEC-dependent secretion. Thus, Cdc24p is necessary for maintaining or restricting particular proteins required for cell fusion to the cell contact region during mating.
We screened for mating-specific cdc24 mutants to identify additional roles of this exchange factor. cdc24-m6 cells exhibit two temporally distinct mating defects, one prior to mating pair formation at the cell agglutination step and another at the prezygote-to-zygote transition. The agglutination defect, while important for mating in solution, is unlikely to affect mating on solid media (29). The cdc24-m6 mutant also accumulates a substantial percentage of prezygotes. Different cell polarity defects result in a cell fusion defect via different mechanisms. For example, chemotropism mutants are unable to orient growth towards a mating partner, and instead mating projection growth occurs adjacent to the previous bud site. In such mutants, the cell fusion machinery is correctly localized to the shmoo tip; however, as the mating pairs do not grow towards one another, a cell fusion defect is observed because the fusion machinery is not at the region of cell contact (47). In contrast, cdc24-m5 and cdc24-m6 mutants do not accumulate prezygotes as an indirect result of a chemotropism defect but rather due to an inability to correctly localize fusion proteins to the shmoo tip. With these mutants, cells grow towards one another; however, the proteins required for cell fusion are not appropriately restricted to the region of cell contact. These two classes of mutants highlight the fundamental differences between chemotropism (directional growth) and correct polarization of the fusion machinery.
Superficially, the fusion defects of chs5 mutants resemble those of cdc24-m6 cells (15, 45). However, Chs5p is required for the polarized distribution of Fus1p, but not Spa2p, to the shmoo and prezygote tip (46). The accumulation of Fus1p in an internal compartment in chs5Δ mutants indicates that Chs5p is necessary for polarized targeting of Fus1p to the plasma membrane. In contrast, our results indicate that cdc24-m6 cells are unaffected for SEC-dependent secretion and Fus1p membrane targeting during mating yet do not maintain the asymmetric distribution of Fus1p and Spa2p. Consistent with the notion that Cdc24p and Chs5p have distinct functions in cell fusion, overexpression of Chs5p did not alter the mating defect of cdc24-m6 cells (S. Barale and R. A. Arkowitz, unpublished data). In addition, overexpression of Fus1p and Fus2p did not suppress the mating defect of cdc24-m6 cells (Barale and Arkowitz, unpublished) as was observed with chs5Δ cells (46). Together our results suggest that Cdc24p is required for maintaining the polarized distribution of proteins necessary for cell fusion to the region of contact during mating.
How might Cdc24p regulate the asymmetric distribution of proteins required for cell fusion? One possibility is that this exchange factor is important for localized endocytosis, which has been proposed as a mechanism to maintain yeast cell polarity (55). Endocytosis is required for the polarized distribution of Fus1p in shmoos (55). As the distribution of another membrane protein (Fig1p) is unaffected in cdc24-m6 shmoos and prezygotes, we consider it unlikely that this mating-specific mutant affects overall endocytosis. Furthermore, endocytosis mutants, such as end4, are not defective for cell fusion (6). Vesicle accumulation proximal to the zone of cell fusion has been observed in prezygotes by electron microscopy (5, 9, 18, 22), yet the origin (exocytic or endocytic) of these vesicles is unknown. Another possibility is that Cdc24p regulates the mobility of cell fusion proteins either directly or via Cdc42p. The redistribution of GFP-Bud1 from the plasma membrane of one cell in a prezygote to both cells in the zygote occurs on a time scale of 5 min (Fig. 5C), similar to the time estimated for diffusion of Snc1p across a yeast cell by fluorescence recovery after photobleaching (FRAP) analyses (55), suggesting the absence of large alterations in plasma membrane mobility upon cell fusion. FRAP experiments also showed that Fus1p on the plasma membrane diffuses much more slowly than SNAREs Snc1p and Sso1p, leading to the suggestion that this cell fusion protein is immobilized via interactions with the cell wall and/or cortex. As the polarized distribution of Spa2p is also affected in cdc24-m6 shmoos and prezygotes, we favor the proposition that Cdc24p restricts the distribution of proteins required for cell fusion via interactions with the cell cortex.
The amino acid residues altered in the cdc24-m5 and cdc24-m6 mutants localize to its conserved catalytic exchange factor domain, suggesting that perhaps the G protein Cdc42p is also necessary for cell fusion. For example, in the Tiam1-Rac1 crystal structure (44, 60) the equivalent residue of Arg 416 of Cdc24p makes contacts to the G protein switch 1 and 2 residues. Indeed, a previously isolated mating-specific cdc42 mutant also accumulates prezygotes during mating (S. Barale, D. McCusker, and R. A. Arkowitz, unpublished data). Furthermore, Nelson et al. recently showed that Cdc42p-GTP interacts with Fus1p by two-hybrid (33). These results suggest that Fus1p is inappropriately localized in cdc24-m6 cells due to an altered level or location of Cdc42p-GTP.
The processes of myoblast fusion and yeast fusion during mating have many similar requirements (13, 22). Drosophila myoblast plasma membrane fusion requires Rac but not Cdc42 (13, 23, 31), myoblast city (38), the Drosophila DOCK180 homolog, and loner, an ARF6 guanine nucleotide exchange factor (11). myoblast city is part of a large family of guanine nucleotide exchange factors that can activate Rac or Cdc42 (7, 12, 38), whereas loner is required for the proper membrane localization of Rac (11). The absence of an S. cerevisiae Rac homolog suggests that Cdc42p and its guanine nucleotide exchange factor, Cdc24p, can carry out similar functions as Rac and DOCK180. Therefore, the requirement for activated G protein during cell fusion appears to be evolutionarily conserved between yeast and flies and perhaps is central to all cell fusion processes.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, and La Ligue Contre le Cancer. S.B. was supported by a Fellowship from La Ligue Contre le Cancer.
We thank J. Cherfils and M. Bassilana for constructive criticism, P. Chang for aid with electron microscopy, C. Matthews for aid with microscopy, S. Clement for technical assistance, and M. Snyder and M. Knop for strains and plasmids.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arkowitz, R. A., and N. Lowe. 1997. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J. Cell Biol. 138:17-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, M., N. Baba, Y. Ohsumi, K. Kanaya, and M. Osumi. 1989. Three-dimensional analysis of morphogenesis induced by mating pheromone alpha factor in Saccharomyces cerevisiae. J. Cell Sci. 94:207-216. [DOI] [PubMed] [Google Scholar]
- 4.Berlin, V., J. A. Brill, J. Trueheart, J. D. Boeke, and G. R. Fink. 1991. Genetic screens and selections for cell and nuclear-fusion mutants. Methods Enzymol. 194:774-792. [DOI] [PubMed] [Google Scholar]
- 5.Brizzio, V., A. E. Gammie, G. Nijbroek, S. Michaelis, and M. D. Rose. 1996. Cell fusion during yeast mating requires high levels of a-factor mating pheromone. J. Cell Biol. 135:1727-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brizzio, V., A. E. Gammie, and M. D. Rose. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141:567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brugnera, E., L. Haney, C. Grimsley, M. Lu, S. F. Walk, A. C. Tosello-Trampont, I. G. Macara, H. Madhani, G. R. Fink, and K. S. Ravichandran. 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4:574-582. [DOI] [PubMed] [Google Scholar]
- 8.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 9.Byers, B., and L. Goetsch. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, C. W., R. Altman, D. Schieltz, J. R. Yates, and D. Kellogg. 1998. The septins are required for the mitosis-specific activation of the Gin4 kinase. J. Cell Biol. 143:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, E. H., B. A. Pryce, J. A. Tzeng, G. A. Gonzalez, and E. N. Olson. 2003. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell 114:751-762. [DOI] [PubMed] [Google Scholar]
- 12.Cote, J. F., and K. Vuori. 2002. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115:4901-4913. [DOI] [PubMed] [Google Scholar]
- 13.Doberstein, S. K., R. D. Fetter, A. Y. Mehta, and C. S. Goodman. 1997. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 136:1249-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 15.Dorer, R., C. Boone, T. Kimbrough, J. Kim, and L. H. Hartwell. 1997. Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics 146:39-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorer, R., P. M. Pryciak, and L. H. Hartwell. 1995. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J. Cell Biol. 131:845-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dworak, H. A., and H. Sink. 2002. Myoblast fusion in Drosophila. Bioessays 24:591-601. [DOI] [PubMed] [Google Scholar]
- 18.Elia, L., and L. Marsh. 1998. A role for a protease in morphogenic responses during yeast cell fusion. J. Cell Biol. 142:1473-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elia, L., and L. Marsh. 1996. Role of the abc transporter Ste6 in cell-fusion during yeast conjugation. J. Cell Biol. 135:741-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erdman, S., L. Lin, M. Malczynski, and M. Snyder. 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140:461-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finger, F. P., and P. Novick. 1998. Spatial regulation of exocytosis: lessons from yeast. J. Cell Biol. 142:609-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gammie, A. E., V. Brizzio, and M. D. Rose. 1998. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9:1395-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakeda-Suzuki, S., J. Ng, J. Tzu, G. Dietzl, Y. Sun, M. Harms, T. Nardine, L. Luo, and B. J. Dickson. 2002. Rac function and regulation during Drosophila development. Nature 416:438-442. [DOI] [PubMed] [Google Scholar]
- 24.Heiman, M. G., and P. Walter. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151:719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser, C. A., and R. Schekman. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61:723-733. [DOI] [PubMed] [Google Scholar]
- 27.Kurihara, L. J., C. T. Beh, M. Latterich, R. Schekman, and M. D. Rose. 1994. Nuclear congression and membrane fusion—two distinct events in the yeast karyogamy pathway. J. Cell Biol. 126:911-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leberer, E., D. Y. Thomas, and M. Whiteway. 1997. Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 7:59-66. [DOI] [PubMed] [Google Scholar]
- 29.Lipke, P. N., and J. Kurjan. 1992. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 56:180-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipke, P. N., A. Taylor, and C. E. Ballou. 1976. Morphogenic effects of alpha-factor on Saccharomyces cerevisiae a cells. J. Bacteriol. 127:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, L., Y. J. Liao, L. Y. Jan, and Y. N. Jan. 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8:1787-1802. [DOI] [PubMed] [Google Scholar]
- 32.McCaffrey, G., F. J. Clay, K. Kelsay, and G. F. Sprague. 1987. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2680-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson, B., A. B. Parsons, M. Evangelista, K. Schaefer, K. Kennedy, S. Ritchie, T. L. Petryshen, and C. Boone. 2004. Fus1p interacts with components of the Hog1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics 166:66-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nern, A. 2000. The role of the GTP/GDP exchange factor Cdc24p in the control of cell polarity during yeast cell mating. Johann Wolfgang Goethe-Universität, Frankfurt, Germany.
- 35.Nern, A., and R. A. Arkowitz. 1999. A Cdc24p, Far1p, Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nern, A., and R. A. Arkowitz. 2000. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell 5:853-864. [DOI] [PubMed] [Google Scholar]
- 37.Nern, A., and R. A. Arkowitz. 1998. A GTP-exchange factor required for cell orientation. Nature 391:195-198. [DOI] [PubMed] [Google Scholar]
- 38.Nolan, K. M., K. Barrett, Y. Lu, K. Q. Hu, S. Vincent, and J. Settleman. 1998. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes Dev. 12:3337-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz, D., M. Medkova, C. Walch-Solimena, and P. Novick. 2002. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 157:1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philips, J., and I. Herskowitz. 1998. Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J. Cell Biol. 143:375-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philips, J., and I. Herskowitz. 1997. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 138:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Read, E. B., H. H. Okamura, and D. G. Drubin. 1992. Actin- and tubulin-dependent functions during Saccharomyces cerevisiae mating projection formation. Mol. Biol. Cell 3:429-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose, M. D., F. Winston, and P. Hieter. 1991. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Rossman, K. L., D. K. Worthylake, J. T. Snyder, D. P. Siderovski, S. L. Campbell, and J. Sondek. 2002. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 21:1315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos, B., A. Duran, and M. H. Valdivieso. 1997. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:2485-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos, B., and M. Snyder. 2003. Specific protein targeting during cell differentiation: polarized localization of Fus1p during mating depends on Chs5p in Saccharomyces cerevisiae. Eukaryot. Cell 2:821-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrick, K., B. Garvik, and L. H. Hartwell. 1997. Mating in Saccharomyces cerevisiae: the role of the pheromone signal transduction pathway in the chemotropic response to pheromone. Genetics 147:19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shemer, G., and B. Podbilewicz. 2003. The story of cell fusion: big lessons from little worms. Bioessays 25:672-682. [DOI] [PubMed] [Google Scholar]
- 49.Smith, M. G., S. R. Swamy, and L. A. Pon. 2001. The life cycle of actin patches in mating yeast. J. Cell Sci. 114:1505-1513. [DOI] [PubMed] [Google Scholar]
- 50.Snyder, M. 1989. The SPA2 protein of yeast localizes to sites of cell growth. J. Cell Biol. 108:1419-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sprague, G. F. J., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, . Cold Spring Harbor Laboratory Pressvol. 2, Cold Spring Harbor, N.Y. [Google Scholar]
- 52.Terrance, K., and P. N. Lipke. 1981. Sexual agglutination in Saccharomyces cerevisiae. J. Bacteriol. 148:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tkacz, J. S., and V. L. MacKay. 1979. Sexual conjugation in yeast. Cell surface changes in response to the action of mating hormones. J. Cell Biol. 80:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valdez-Taubas, J., and H. R. Pelham. 2003. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 13:1636-1640. [DOI] [PubMed] [Google Scholar]
- 56.Valtz, N., and I. Herskowitz. 1996. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J. Cell Biol. 135:725-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valtz, N., M. Peter, and I. Herskowitz. 1995. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 131:863-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner, W., P. Bielli, S. Wacha, and A. Ragnini-Wilson. 2002. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 21:6397-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiner, M. P., G. L. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. C. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151:119-123. [DOI] [PubMed] [Google Scholar]
- 60.Worthylake, D. K., K. L. Rossman, and J. Sondek. 2000. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408:682-688. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, M., D. Bennett, and S. E. Erdman. 2002. Maintenance of mating cell integrity requires the adhesin Fig2p. Eukaryot. Cell 1:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]