Abstract
Ribozymes and riboswitches are RNA motifs that accelerate biological reactions and regulate gene expression in response to metabolite recognition, respectively. These RNA molecules gain functionality via complex folding that cannot be predicted a priori, and thus requires high-resolution three-dimensional structure determination to locate key functional attributes. Herein, we present an overview of the methods used to determine small RNA structures with an emphasis on RNA preparation, crystallization, and structure refinement. We draw upon examples from our own research in the analysis of the leadzyme ribozyme, the hairpin ribozyme, a class I preQ1 riboswitch, and variants of a larger class II preQ1 riboswitch. The methods presented provide a guide for comparable investigations of noncoding RNA molecules including a 48-solution, “first choice” RNA crystal screen compiled from our prior successes with commercially available screens.
Keywords: RNA, Riboswitches, Ribozymes, RNA synthesis, Purification, X-ray crystallography, Crystal screen
1. Introduction
Fast sequencing technology has produced myriad new genomes that expand the number of novel noncoding RNA sequences including those of ribozymes and riboswitches (1). To keep pace with ascribing a functional role to these sequences, few approaches are as powerful as X-ray crystallography when it comes to producing an all-atom model of an RNA molecule that serves as framework to support rational analyses. In this chapter, we describe a series of protocols for the preparation, crystallization, and X-ray refinement of small ribozymes and riboswitches. Our experience is drawn from: (1) the leadzyme ribozyme—a 24-mer comprising 11-mer and 13-mer synthetic strands (2), (2) the hairpin ribozyme, a 61-mer comprising 12-mer, 13-mer, 17-mer, and 19-mer synthetic strands (3); and (3) a synthetic 33-mer riboswitch that binds the metabolite preQ1 (4). In each of these investigations, the resulting crystal structures were used to gain insight into function. In addition, we present progress on the analysis of an 80-mer preQ1 riboswitch, and its 77-mer variant, produced by in vitro transcription and crystallized from several commercial screens. These results have been compiled with other successful RNA crystallization conditions into a 48-solution “first choice” RNA crystal screen (see Table 1).
Table 1.
Summary of “first choice” RNA crystal screening conditions
| Number | Precipitant | Salt | Additive 1 | Additive 2a | Bufferb | pH |
|---|---|---|---|---|---|---|
| 1 | 5% PEG 2K | 0.10 M KCl | 0.005 M MgCl2 | Spermine | MES | 5.6 |
| 2 | 10% PEG 2K | 0.10 M NH4Cl | 0.010 M MgCl2 | Spermine | Na-cacodylate | 6.0 |
| 3 | 15% PEG 2K | 0.10 M Li2SO4 | 0.020 M MgSO4 | Spermine | Na-cacodylate | 6.5 |
| 4c | 20% PEG 2K | 0.20 M NH4Cl | 0.010 M Mg(Acetate)2 | Spermine | HEPES | 7.0 |
| 5 | 25% PEG 2K | 0.10 M NaCl | 0.001 M Co(NH3)6 | Spermine | Tris | 7.5 |
| 6 | 5% PEG 8K | 0.20 M (NH4)2SO4 | 0.010 M MgSO4 | Spermine | MES | 5.6 |
| 7d | 10% PEG 8K | 0.10 KCl | 0.020 M MgCl2 | Spermine | Na-cacodylate | 6.5 |
| 8 | 15% mmePEG 2K | 0.20 NH4Acetate | 0.010 M Mg(Acetate)2 | Spermine | Na-cacodylate | 6.5 |
| 9 | 20% mmePEG 2K | 0.20 NH4Cl | 0.010 M MgCl2 | Spermine | HEPES | 7.0 |
| 10e | 25% mmePEG 2K | 0.20 Li2SO4 | 0.001 M Co(NH3)6 | Spermine | Tris | 7.5 |
| 11 | 10% mmePEG 550 | 0.05 M NH4C2H3O2 | 0.001 M Co(NH3)6 | Spermine | Na-cacodylate | 6.0 |
| 12 | 15% mmePEG 550 | 0.05 M NH4Cl | 0.010 M MgCl2 | Spermine | Na-cacodylate | 6.5 |
| 13 | 20% mmePEG 550 | 0.05 M KCl | 0.020 M MgCl2 | Spermine | HEPES | 7.0 |
| 14 | 25% mmePEG 550 | 0.05 M NH3C2H3O2 | 0.005 M MgCl2 | Spermine | Tris | 7.5 |
| 15f | 12% mmePEG 8K 12% PEG 200 |
0.10 M KCl | 0.010 M Mg(Acetate)2 | Spermine | Na-cacodylate | 6.0 |
| 16f | 10% mmePEG 8K 12% PEG 200 |
0.10 M KCl | 0.010 M Mg(Acetate)2 | Spermine | Na-cacodylate | 6.0 |
| 17f | 11% mmePEG 4K 10% PEG 400 |
0.050 M KCl | 0.015 M Mg(Acetate)2 | Spermine | Na-cacodylate | 6.0 |
| 18f | 10% mmePEG 8K 15% MPD |
0.10 M KCl | 0.005 M MgCl2 | Spermine | Na-cacodylate | 6.0 |
| 19 | 10% PEG 400 | 0.10 M KCl | 0.001 M CoCl2 | Spermidine | Na-cacodylate | 6.0 |
| 20 | 15% PEG 400 | 0.10 M NaCl | 0.010 M MgCl2 | Spermidine | Na-cacodylate | 6.5 |
| 21 | 20% PEG 400 | 0.10 M NH4Cl | 0.020 M MgCl2 | Spermidine | HEPES | 7.0 |
| 22 | 30% PEG 400 | 0.10 M Mg(Acetate)2 | Tris | 8.0 | ||
| 23 | 10% MPD | 0.050 M LiSO4 | 0.010 M MgSO4 | Spermidine | Na-cacodylate | 6.0 |
| 24 | 15% MPD | 0.050 M KCl | 0.005 M MgCl2 | Spermidine | Na-cacodylate | 6.5 |
| 25 | 20% MPD | 0.050 M NH4Cl | 0.005 M CaCl2 | Spermidine | HEPES | 7.0 |
| 26 | 25% MPD | 0.050 M LiAcetate | 0.010 M MgCl2 | Spermidine | Tris | 7.5 |
| 27 | 30% MPD | 0.050 M Mg(Acetate)2 | Spermidine | Na-cacodylate | 6.0 | |
| 28 | 15% MPD | 0.10 M NaCl | 0.010 M MgSO4 | Spermine | Na-cacodylate | 6.5 |
| 29c | 20% MPD | 0.10 M KCl | 0.010 M MgCl2 | Spermine | HEPES | 7.0 |
| 30 | 25% MPD | 0.10 M NaAcetate | 0.010 M MgCl2 | Spermine | Tris | 7.5 |
| 31g | 30% MPD | 0.10 M NH4Cl | 0.005 M MgSO4 | Spermine | Na-cacodylate | 6.0 |
| 32 | 1.2 M (NH4)2SO4 | 0.050 M LiCl | 0.001 M Co(NH3)6 | Spermine | MES | 5.6 |
| 33 | 1.5 M (NH4)2SO4 | 0.050 M NaCl | 0.005 M MgSO4 | Spermine | Na-cacodylate | 6.0 |
| 34e | 1.8 M (NH4)2SO4 | 0.010 M MgSO4 | Spermidine | Na-cacodylate | 6.5 | |
| 35 | 2.4 M (NH4)2SO4 | 0.050 M MgSO4 | 0.005 M MgSO4 | Spermine | HEPES | 7.0 |
| 36 | 2.8 M (NH4)2SO4 | 0.050 M Mg(Acetate)2 | Spermine | Tris | 7.5 | |
| 37 | 1.5 M (NH4)2SO4 | 0.005 M MgSO4 | Spermidine | Tris | 7.5 | |
| 38 | 2.0 M (NH4)2SO4 | 2% (v/v) PEG 400 | 0.001 M CoCl2 | Spermidine | HEPES | 7.0 |
| 39 | 2.4 M (NH4)2SO4 | 0.010 M MgSO4 | Spermidine | Na-cacodylate | 6.5 | |
| 40 | 2.8 M (NH4)2SO4 | 4% (v/v) PEG 200 | 0.005 M CaCl2 | Spermidine | Na-cacodylate | 6.0 |
| 41e | 1.3 M Li2SO4 | 0.001 M Co(NH3)6 | Spermidine | Na-cacodylate | 6.0 | |
| 42h | 1.5 M Li2SO4 | 2% (v/v) PEG 1K | 0.050 MgSO4 | Spermine | HEPES | 7.5 |
| 43c | 1.8 M Li2SO4 | 0.050 NaCl | 0.020 M MgSO4 | Spermine | Na-cacodylate | 6.0 |
| 44 | 45% Tacsimate™ | 0.020 M Mg(Acetate)2 | 0.050 M KCl | Spermine | MES | 5.6 |
| 45h | 55% Tacsimate™ | 0.020 M MgCl2 | Spermine | Na-cacodylate | 6.5 | |
| 46f | 65% Tacsimate™ | 0.001 M Co(NH3)6 | Spermidine | HEPES | 7.0 | |
| 47 | 1.5 M 1.6-Hexanediol | 0.040 M NH4Cl | 0.010 M MgCl2 | Spermine | Na-cacodylate | 6.5 |
| 48 | 2.4 M 1,6-Hexanediol | 0.040 M MgSO4 | 0.050 M KCl | Spermidine | Tris | 7.5 |
All concentrations of additive 2 are 0.001 M
All buffer concentrations are 0.050 M
Conditions for diffracting crystals are noted as follows:
a 33-mer preQ1 class I riboswitch;
an 80-mer class II preQ1 riboswitch without U1A;
the minimal hairpin ribozyme;
a 77-mer class II preQ1 riboswitch without U1A;
a 77-mer class II preQ1 riboswitch without U1A;
an 80-mer class II preQ1 riboswitch with U1A
Prior to RNA synthesis, we will assume that the target RNA has been validated in terms of enzymatic or ligand-binding activity to define what is necessary and sufficient for function. It has been our experience that the best sequences for crystallization possess conserved core features, e.g., identifiable in RFAM (5), and that these sequences exhibit unique secondary and tertiary folding distinct from competing, nonproductive conformations, e.g., as indicated by HOTKNOTS (6). As a caveat, such programs do not account for the stabilizing effects of transition-state or metabolite binding, but they are a good starting point for the design of crystallization constructs. Such effects can be monitored experimentally by dynamic light scattering (DLS), which has proven useful to assess the compactness and monodispersity of RNA constructs prior to crystallization trials. In addition, we assume herein that RNA phases have been obtained already. Many authoritative reviews have been written about the RNA phase problem. For general approaches on heavy-atom phasing see refs. 7, 8; for molecular replacement see ref. 9. Keel et al. (10) provide an innovative approach for the inclusion of site-specific osmium- or iridium-hexammine binding sites that can be beneficial in experimental phasing. As such, this aspect of the discussion is confined to refinement of the model against X-ray data. Overall, this chapter should be a practical guide to RNA production, crystallization, and refinement with reference to complementary resources to fill in knowledge gaps as necessary.
2. Materials
All water is derived from a NANOpure™ UV/UF system (Thermo Scientific, Asheville, NC) with a resistivity reading of ≥18.1 MΩ. In general all solutions should be 0.22- or 0.45-µm sterile filtered and autoclaved if possible. Specialized reagents and stocks are described below. All steps should use appropriate safety measures to avoid exposure to hazardous chemicals such as ethidium bromide or acrylamide, as well as damaging radiation from UV and X-ray sources. Consult material safety data sheets for all chemicals and the manufacturer’s documentation for proper instrument operation.
2.1. DNA Template Production with Klenow Fragment
10 mM dNTP solution, cat. #N0447S (New England Biolabs, Ipswich, MA).
Klenow fragment, 5,000 U/mL, with 10× reaction buffer, cat. #M0210S (New England Biolabs, Ipswich, MA). 10× Klenow reaction buffer comprises 0.50 M NaCl, 0.10 M Tris–HCl pH 7.9, 0.10 M MgCl2, 0.010 M DTT (New England Biolabs, Ipswich, MA).
SeaKem® LE Agarose, cat. #50000 (Lonza, Allendale, NJ).
100% Formamide, cat. #BP228-100 (Fisher Scientific, Pittsburgh, PA).
10 mg/mL Bromophenol blue, cat. #343 (Allied Chemical, Morristown, NJ).
40 mg/mL Ethidium bromide, cat. #BP102-1 (Fisher Scientific, Pittsburgh, PA).
1.0 M NaCl stock solution, cat. #SX0425-3 (EMD Biosciences, San Diego, CA).
Agarose gel extraction kit, cat. #28704 (Qiagen Inc., Valencia, CA).
Prechilled neat ethanol (100%), cat. #200CSGP (Ultra Pure, LLC, CT).
OmniPur® Phenol:Chloroform:Isoamyl Alcohol, 25:24:1, cat. #6810 (EMD Biosciences, San Diego, CA).
Required apparatus: a thermostatically controlled water bath, a short/long-wavelength UV transilluminator, a microcentrifuge, and Horizontal Midi-Gel Kit CE 14×10 cm (cat. #MGU-502T, CBS Scientific Comp., Inc., Del Mar, CA).
2.2. RNA Synthesis by T 7 Polymerase
1.0 M Tris pH 7.5, Tris base, Molecular Biology Grade, cat. #648310 (CalBioChem, San Diego, CA).
1.0 M DTT, cat. #D9163 (Sigma-Aldrich, St. Louis, MO).
0.5 M Spermidine tetrahydrochloride, cat. #100472 (ICN Biomedical, Irvine, CA).
10% (w/v) Triton X-100, cat. #21568-2500 (Acros, Geel, Belgium).
50% (w/v) PEG 8000, cat. #81268 (Fluka, St. Louis, MO).
1 mg/mL Fraction V, Omnipur BSA solution, cat. #2930 (EMD Biosciences, San Diego, CA).
1.0 M MgCl2 ·(H2O)6, ACS Grade, cat. #MX0045-1 (EMD Biosciences, San Diego, CA).
0.040 M Ribonucleotide triphosphates (rNTPs) (MP Biomedicals, Solon, OH).
Recombinant T7 RNA polymerase, cat. #2085 (Ambion, Inc., Austin, TX) or equivalent.
2.3. Polyacrylamide Gel Electrophoresis Purification of RNA for Crystallography
Vertical polyacrylamide gel electrophoresis (PAGE) apparatus, cat. #LSG-400-20-NA (CBS Scientific Company, Inc., Del Mar, CA).
TBE running buffer 5× stock is: 54 g of Tris base (final 5× conc. 0.445 M), 27.5 g boric acid (final 5× conc. 0.445 M), 20 mL EDTA from a 0.50 M pH 8.0 stock (final 5× conc. 0.01 M), and sufficient water to bring the volume to 1.0 L (The final pH is 8.3; do not pH the stock).
Freshly prepared 8–15% (w/v) acrylamide:bisacrylamide (19:1) with 7.0 M urea to denature RNA. A 0.10 L 15% stock requires the following: 14.25 g acrylamide, 0.75 g bisacrylamide, 42.04 g urea, and 20 mL 5× TBE buffer. Add all solids to a flask and bring the volume to 70 mL. Place the flask on a magnetic stir plate with a heater and mix the solution at 37°C for 1 h. Add 20 mL of 5× TBE and bring the total volume to 0.10 L.
Fresh 10% (w/v) ammonium persulfate, cat. #BP179-100 (Fisher Scientific, Pittsburgh, PA).
A fresh solution of TEMED (N,N,N′,N′-Tetramethylethylenediamine) stock, cat. #T9281-100mL (Sigma-Aldrich).
Loading buffer (2×) comprising: 95% formamide, 0.025% (w/v) xylene cyanol, 0.025% (w/v) bromophenol blue, 18 mM EDTA, and 0.025% (w/v) SDS, cat. #AM8547 (Ambion, Austin, TX).
Crush-Soak Solution: 20 mM Tris pH 7.5, 2 mM EDTA, and 0.10 M ammonium acetate.
Tube Top Filter 50 mL, 0.45 µ m cellulose acetate, cat. #430314 (Corning, Inc., Corning, NY).
Chromatography Materials include: Toyopearl DEAE-650S, cat. #07472 (Tosoh Bioscience Corp., San Francisco, CA); DEAE wash buffer (20 mM Tris pH 7.5 and 2 mM EDTA); DEAE elution buffer (same as the wash buffer but containing 1.0 M ammonium acetate); desalting chromatography resin is Sephadex G-50 (DNA Grade), cat. #S5897-25G (Sigma-Aldrich, St. Louis, MO); this resin is swollen and run in water.
Various ACE Glass, Inc. columns to accommodate ion exchange (20 mm×10 cm) and desalting (15 mm×50 cm) resins. These should be wrapped in aluminum foil and autoclaved for 30 min followed by a dry cycle. Tygon tubing for gravity flow should be treated similarly, but first wet the inside of the tube to prevent damage from heating.
Fluor-coated TLC plate, cat. #AM10110 (Applied Biosystems, Carlsbad, CA).
Sterile scalpel blade and metal spatula.
A lyophilizer is used for the final desalting step although this can be substituted with a speedvac operated without heating.
2.4. Sparse Matrix Screening by the Hanging Drop Method
With RNA in hand the search for crystallization conditions begins by screening against commercial kits designed for nucleic acids such as: the Natrix and Nucleic Acid Mini Screen (Hampton Research, Aliso Viejo, CA), as well as the JBScreen Nuc-Pro (MiTeGen, Ithaca, NY). For liquid handling, we prefer a robotic system that operates in 96-well format. If lead crystallization conditions are identified, we then proceed to optimization using larger, manual setups (see Subheading 2.5). Materials for robotic crystal screening include:
A Mosquito™ robot (TTP Labtech, Melbourn, UK) or equivalent (see Note 1).
96-Well plates such as SpectraPlate-96HB, cat. #P12-106 (PerkinElmer Life and Analytical Sciences, Waltham, MA).
Clear seal film, cat. #HR4-523 (Hampton Research Inc., Aliso Viejo, CA).
Seal and sample aluminum foil lids, cat. #538619 (Beckman Coulter, Brea, CA).
ViewDrop 96-well plate seals for hanging drop, 25 per pack, cat. #4150-05100 (TTP Labtech).
Crystallization screen such as Natrix HT, cat. #HR2-131 (Hampton Research Inc.).
8-Position 2- and 5-µL micro-reservoir strips for 9-mm pitch, 50 per pack, cat. #4150-03110 (TTP Labtech).
8-Channel pipettor, P-200, cat. #89079-944 (VWR, Radnor, PA).
2.5. Materials for Manual Optimization of RNA Crystals
VDX 24-well hanging drop plate, cat. #HR3-140 (Hampton Research Inc.).
Siliconized glass circular cover slides, 22 mm diameter, cat. #HR3-231 (Hampton Research Inc.).
White petrolatum jelly cat. #S80117 (Sigma-Aldrich), which is heated to liquification on a hot plate, poured inside a 10-cc syringe with the narrow end parafilmed, and chilled on ice. The plunger is restored to extrude a column of jelly for sealing the VDX plate.
2.6. Materials for the Refinement of RNA Crystal Structures
An Intel-based computer running Mac OSX 10.5.8 or higher. Representative systems include: a MacBook Pro laptop with a 2.6 GHz Intel Core 2 Duo processor, 4 GB 667 MHz DDR2 SDRAM, and a 0.5Tb HD connected to a Zalman Trimon ZM-M220W stereo monitor; and a Quad Mac tower running OSX 10.6.5 with a 2.66 GHz Quad-Core Intel Xeon processor, 13 GB 1,066 MHz DDR3 RAM, an NVidea Quadro FX 4800 graphics card (for quad buffered stereo), and 3× 1.5Tb HDs.
3. Methods
3.1. Introduction to the Synthesis of RNA by Chemical and Enzymatic Approaches
Chemically synthesized oligonucleotides are appropriate for the production of short RNA sequences. We have had success crystallizing synthetic sequences ranging from 11 to 33 nt. Such strands are produced by solid-phase chemical synthesis (11). Major advantages include rapidity of production, accommodation of nonstandard modifications (3, 12, 13), and attainment of relatively high yields. The feasibility of synthesis can be evaluated by the heuristic: yields(%)=coupling efficiency(chain size −1)×10, where coupling efficiency depends on the nature of the nucleotide monomer (e.g., DNA, RNA, or chemical variants thereof) as well as the chemistry employed in coupling. Traditional RNA phosphoramidites were developed for DNA synthesis, and employ 5′-DMT and 3′ β-cyanoethyl (CE) protecting groups (14). This approach can couple with 98% efficiency at each step, but leaves a relatively labor-intensive workup (7, 15). More recently, the combination of a 5′-silyl ether and an acid labile 2′-OH orthoester provides superior protection chemistry identifiable as 2′-ACE phosphoramidites that exhibit >99% coupling efficiency and a relatively facile workup (16). It has been our experience that the former approach offers more choices of nonstandard nucleotide modifications (e.g., Glen Research, Sterling VA), and there are more opportunities to partner with a company that will incorporate “homemade” phosphoramidites into an RNA strand as part of a custom synthesis (12). By contrast, the latter approach is commercially more costly but the yields of longer oligonucleotides are superior. In the end the user must choose what is best for his or her application.
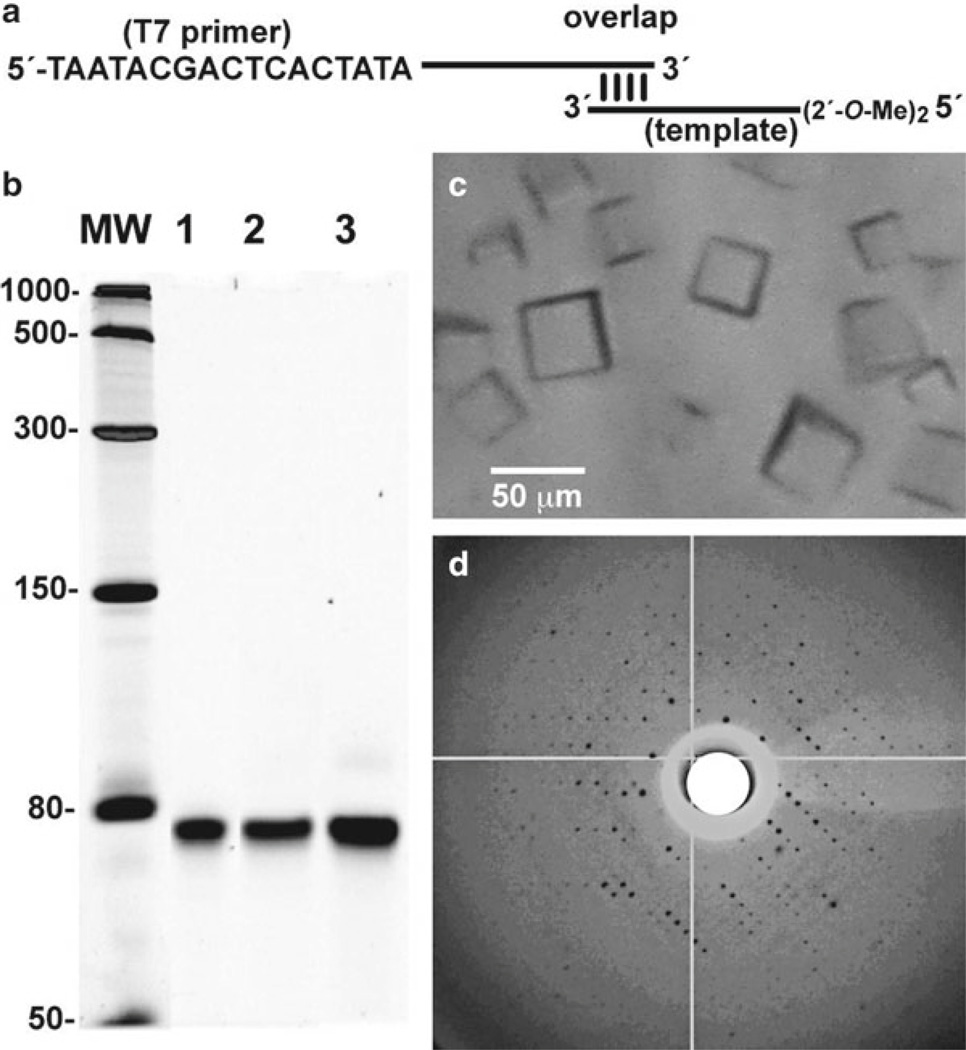
When longer RNA strands are needed in milligram quantities, chemical synthesis becomes impractical due to poor yields and high costs. In vitro transcription using short, double-stranded DNA templates derived from Klenow fragment fill-in offers an alternative to plasmid-based approaches that require cloning and DNA isolation from bacteria. Klenow allows rapid and routine production of RNA transcripts (limited to ~140 bases in part by poor yields of DNA primer and template strands), which is ideal for screening numerous different small RNA constructs for crystallization. In this method, two DNA strands are ordered in which one encodes the desired “template” for the RNA sequence to be transcribed, whereas the complementary strand harbors a 5′ T7 RNA polymerase promoter site in a “primer” strand. Due to yield limitations associated with the synthesis of very large DNA oligonucleotides, the approach utilizes strands that are relatively short (<70 nt). By design the primer and template strands exhibit complementary base pairing with a TM of 50–60°C in a central region of the duplex (see Fig. 1a). Klenow fragment is used to fill-in the primer-template pair yielding double-stranded DNA that can be used by T7 polymerase to synthesize RNA. Herein we describe this process, which adheres to the concept that the RNA construct itself is a variable in crystal screening (17). As a further consideration, the template strand is synthesized to include tandem 2′-O-methyl ribose nucleotides at the 5′-end of the DNA (18). This modification reduces untemplated transcription whereby extra nucleotides are added at the 3′-end of the RNA product, thus lowering purity and potentially confounding crystallization (18). The reader is directed to papers by Uhlenbeck for template and primer design, as well as the factors that influence a strong start by the RNA polymerase, such as use of the initial sequence 5′-GGC (see ref. 19 and references therein).
Fig. 1.
Representative steps in the preparation, purification, crystallization, and X-ray diffraction of a riboswitch aptamer domain. (a) Schematic diagram of the starting T7 polymerase primer and template that are used for Klenow fragment extension into double-stranded DNA. (b) Representative purity of an 80-mer RNA riboswitch that binds preQ1. The transcribed product was purified by denaturing PAGE and analyzed here on an 8% gel with 7.0 M urea. The sample was suspended in 2× loading buffer. RNA in lanes 1–3 were loaded in 0.055, 0.110, and 0.500 mg quantities, respectively. (c) Cubic crystals of the RNA from (b) generated by robotic screening using condition 42 of Table 1. (d) X-ray diffraction from a representative 80-mer crystal from (c) using the 22-micrometer microfocus beam at station A1 of the Cornell High Energy Synchrotron Source. The image edge is 5 Å resolution. The crystal was cryoprotected from a 1:1 mixture of paratone-N and silicone oil.
3.2. DNA Template Production with Klenow Fragment
Primer and template DNAs are dissolved in water to 100 pM/µL and used as needed or stored at −20°C.
To 460 µL of water, add 60 µL of 10× Klenow reaction buffer, 40 µL of primer, and 40 µL of template with mixing. The solution is heated to 95°C and cooled quickly to 4°C less than the TM of the overlapping sequence. The solution is held here for 20 min, then cooled quickly to 37°C prior to the addition of the Klenow reaction mixture.
The Klenow reaction mixture is prepared by adding 20 µL of 10× Klenow buffer and 36 µL of 10 mM dNTPs to 134 µL of water. After mixing, 10 µL of Klenow enzyme is added and the mixture is warmed to 37°C immediately before addition to the DNA solution.
The Klenow reaction mixture from step 3 is added to the DNA primer-template mixture with gentle mixing and then incubated at 37°C for 1 h.
The dsDNA product is purified via agarose gels. A volume of 0.70 mL of formamide is added to the synthesized DNA, heated to 70°C for 10 min, and then cooled quickly on ice. A trace amount of bromophenol blue is added, and the sample is loaded onto a 10 cm long, 2.5–3% (w/v) agarose gel containing sufficient ethidium bromide to attain 2 µg/mL. DNA mass markers can be loaded if desired. The running buffer is also made 2 µg/mL in ethidium bromide. The power supply is set to 6 V/cm and electrophoresis continues until the bromophenol blue travels 80% of the gel length.
The DNA is visualized under long-wavelength UV light and the correct DNA band is excised. The DNA is extracted and purified using an agarose extraction kit.
The DNA is phenol/chloroform extracted as follows. A 0.10 mL mixture comprising phenol/chloroform (1:1) is added per 0.10 mL of DNA eluted from the gel extraction kit. The extraction solution is shaken vigorously, and then microcentrifuged at 9,000×g for 2 min. The upper layer contains DNA and is removed, followed by addition of 0.30 mL of chloroform alone. This solution is shaken vigorously and microcentrifuged. The upper layer is removed and the chloroform extraction step is repeated again. After centrifugation, the upper layer of DNA is removed.
Ethanol precipitation of the DNA proceeds by adding 15 µL of 1.0 M NaCl and 0.30 mL of prechilled, 100% ethanol and holding the solution at −70°C for 1 h or −20°C overnight. The solution is then microcentrifuged at 15,000×g for 10 min. The supernatant is removed carefully and the pellet is washed with 70% ethanol and dried on the bench top or by a speedvac without heating.
The DNA concentration is estimated by dissolving the pellet in 0.10 mL water. The optical density is measured at 260 nm where 1.0 OD unit represents 50 µg/mL DNA.
3.3. RNA Synthesis by Bacteriophage T7 RNA Polymerase
Initial in vitro transcription reactions on a trial scale (50–100 µL) are conducted in 1.5-mL microcentrifuge tubes to determine optimal conditions.
Each reaction is prepared at 22°C to avoid precipitation of solution components such as DTT or spermidine hydrochloride.
Each trial reaction contains the following components: 8.3 µg DNA template per 1 mL reaction, 0.075 M Tris pH 7.5, 0.010 M DTT, 0.002 M spermidine, 0.01% Triton X-100, 4% PEG 8K, 50 µg/mL BSA, 0.030 M MgCl2, 4 mM rNTP, and 30 µg/mL of T7 RNA polymerase (see Note 2).
The mixture should be incubated at 37°C for no more than 24 h.
After incubation, the reaction is centrifuged at 14,000×g for 10 min before removing the soluble phase for gel purification.
The products can be analyzed by denaturing PAGE electrophoresis on a small scale. If necessary, the reaction can be optimized by changing the amount of input MgCl2, T7 RNA polymerase, or rNTPs (see Note 3).
3.4. Purification of RNA for Crystallization
Once a satisfactory trial-scale RNA reaction has been established, the conditions should be scaled up to a 1–10 mL size. The goal is to produce ≥1 mg of pure RNA for crystallization trials. Robotic screening methods (see Subheading 3.9) make it feasible to conduct 96 crystallization trials with as little as 150 µg of pure RNA, but we aspire to set up multiple 96-well plates with concentrated material. The purity and homogeneity of the RNA is a key factor in crystallization, and the primary limitation of chemical or enzymatic synthesis is incomplete polymerization (i.e., failure sequences). A secondary problem unique to in vitro transcription is the presence of n + 1 or n + 2 untemplated nucleotides that are added to the 3′-end. For purification, we prefer reverse phase HPLC for RNA strands ≤20 nt; procedures for this are described elsewhere (7, 15). For longer strands—or sequences with significant secondary structure—it is necessary to use denaturing PAGE. The electric field separates the various polymer lengths by charge, and the appropriate chain is excised from the gel. We describe this method as applied to RNA strands to be used in crystallization (see Fig. 1b, c).
3.5. First Anion Exchange Purification (DEAE)
The day before PAGE purification, prepare an Ace Glass column by pouring a 3 mL slurry of resin.
Flow the wash buffer over the resin (ten column volumes). The wash packs the resin while bringing it to the correct ionic strength.
Add the reaction(s) from Subheading 3.3 directly to the resin. Save the flow through.
Add wash buffer (five bed volumes). Save the wash volume (which may contain unbound RNA, but more likely contains mononucleotides).
Add elution buffer (ten column volumes). Collect eluate in small aliquots (0.5–1.0 mL), testing the absorption of each fraction at 260 nm for the presence of RNA.
Plot the OD vs. fraction number to determine the appropriate pooling of the RNA.
Add three volumes of 100% ethanol to the pooled RNA and place at −20°C for ~15 h.
Spin sample at 14,000 × g for 25 min to harvest the RNA as a precipitated pellet.
Gently decant the supernatant into a separate tube.
Wash the remaining RNA pellet with 70% ethanol to remove trace salts.
Allow the pellet to dry on the bench for 30 min. The pellet can be stored at −20°C.
Suspend the RNA by gentle pipetting or vortexing in water.
3.6. Denaturing Polyacrylamide Gel Electrophoresis
The polyacrylamide gel is semi-preparative with 0.75–3.0-mm Teflon spacers depending on the amount of RNA to load and the separation requirements. This will use about 0.150 L of acrylamide for the thickest spacer. Pour 0.150 L of acrylamide stock into a 0.50-L Erlenmeyer flask. Add 0.90 mL of 10% ammonium persulfate. Add 0.150 mL of TEMED to the gel solution. A comb with a single lane is preferred to evenly distribute the sample. The gel can be allowed to polymerize for 16 h, but precautions must be taken to avoid excessive drying (such as wrapping the exposed acrylamide in plastic with a damp paper towel).
Pre-run the gel at 200 V (with a starting power of 15 W) and proceed until the wattage level becomes nearly constant (2–3 h depending on the size of the gel).
Mix the RNA with an equal amount of 2× loading buffer and heat 70°C for 5 min before plunging on ice. Load the dissolved RNA onto the gel distributing evenly along its length. Usually there will be 100–200 OD 260 nm units loaded per gel.
Run the gel at 200 V until the sample completely enters the gel, then increase to 400 V. The longer the gel runs the better the resolution and it will be easier to isolate the desired RNA sequence, which should be the most prominent band.
Remove the correctly sized band by UV shadowing. First place the gel between two sheets of plastic wrap and then position the gel atop a fluor-coated TLC plate that fluoresces under UV light. In a darkened room, expose the gel to UV light. The major product will appear as a dark band on a green background. Use a Sharpie® pen to mark the boundaries of the band to excise—work quickly to avoid UV damage to the RNA.
With the lights on, carefully excise the band with a sterile scalpel (removing the plastic wrap) and cut the band into short strips (1–2 cm in length). Place the acrylamide fragments into a pre-weighted 50-mL tube. The gel should be pulverized further with a sterile metal spatula.
Add crush-soak solution to the 50-mL tube; soak volume = 3× the gel mass.
Place the sealed tube in a shaker at 50–100 rpm at 22°C for no more than 16 h.
Filter the solution using a 50-mL Tube Top Filter to remove the acrylamide. The recovered acrylamide can be soaked again ≤8 h.
3.7. Second Anion Exchange Purification (DEAE)
A fresh column is prepared as in Subheading 3.5.
Load the combined, filtered solutions from Subheading 3.6, step 9 directly to the resin making sure to collect the flow through.
Repeat steps 4–11 of Subheading 3.5.
3.8. Gel Filtration Chromatography (Desalting)
Water is added to the G50 resin, which can be swollen and degassed by autoclaving. The room temperature slurry is poured into a 15 mm×50 cm sterile column to fill 95% of the column length. The resin is allowed to pack by flowing water through by gravity for 6 h.
The RNA from Subheading 3.7 in a volume ≤1 mL is layered gently onto the G50 resin.
The column should be flowing while loading and 1.0 mL fractions can be collected.
When the RNA is done loading, gently layer the G50 column with 0.5 mL volumes of water until the RNA has entered more than 3 cm into the column. 5 mL of water is added and the column is sealed for a gravity feed.
Measure the OD 260 nm of each fraction and plot the outcome. The graph should be a single, sharp peak. Shoulders or doublets may be signs of overloading or improper loading by disruption of the sample plate during the addition of water.
Pool the appropriate fractions in a 20-mL tube and measure the OD 260 nm.
Freeze the solution in N2 (l) and make two holes in the cap with a hot needle.
Place sample on a lyophilizer until the RNA is completely dry. The sample should be a fluffy, white powder with no signs of oil, crystallinity, or discoloration, which are indications of incomplete desalting that necessitate repeating the G50 step. Lyophilized RNA can be stored at −20°C.
3.9. Robotic Crystal Screening Using the Mosquito™
Use a multichannel pipette to dispense 75 µL of each screening condition into the well of a 96-well plate.
Position the plate below the pipette assembly of the robot.
Choose a 2- or 5-µL sample strip based on the size of the desired hanging drop. We prefer 250–350 nL drops, which require a 5-µL strip whereas smaller drops use a 2-µL strip.
Pipette 5 µL for 350 nL drops into each of the receptacles of the sample strip. In general, dispense 30% more RNA solution per well than actually needed to allow for evaporation.
After pipetting the RNA, start the robot immediately. Runs can be programmed according to the manufacturer’s documentation.
When complete, invert the 96-condition hanging drop appliqué, align with the guide arrow, and press firmly with the aluminum block to seal the plate.
Peel off the protective anti-scratch layer of the appliqué for viewing.
Store the plate at 22 or 4°C for 3–5 days before checking the drops.
Record your observations for each condition using a consistent scoring system.
Promising crystals may take a few days to months to appear. Although it is possible to recover crystals from the 96-well plate by cutting the plastic appliqué encompassing the desired drop, it is often necessary to produce larger crystals under optimized conditions.
3.10. Manual Optimization of RNA Crystals Through Grid Screening
Initial screens may yield light to heavy precipitate, phase separation, microcrystals, showers of crystals, needles, or a few large crystals suited for diffraction tests. Under most circumstances, there is room for improvement. After all, high-throughput screens are designed to cover a broad number of conditions, but these often require a follow-up “grid” screen in which a single variable is changed per experiment to identify factors that best optimize crystal growth and diffraction. In general, a suitable crystal should be single with sharp edges and free of defects (see Fig. 1c); dimensions should be >100 µ m on each edge. In this section, we describe the general approaches for establishing manual screens, which are often restricted to incremental changes in pH (±0.25 U), salt (±20%), or precipitant (±20% of hit) based on initial hits from robotic screening (see Note 4). Temperature, RNA concentration, and the RNA sequence itself should not be overlooked as optimization variables (see Note 5). Methods are as follows:
Grease the circular opening of each well on a 24-well plate with a petrolatum-filled 10-cc syringe leaving a small gap to release air when the cover slide is pressed in place.
Pipette each precipitating agent into its respective reservoir. The total volume per reservoir is 1.0 mL and the final buffer concentration is 50 mM.
Pipette 1.7 µL of RNA solution to the center of a glass cover slide. Gently add 1.7 µL of reservoir solution to the RNA drop; do not mix the drop contents.
Invert the cover slide and place it onto the corresponding well solution using pressure to seal the well. Use a pipet tip to press down firmly on the cover slip edges to remove air gaps in the grease.
Store the crystallization plate in a temperature-controlled environment (see Note 6). Check the plates 3–5 days later and in 1-week intervals thereafter. Record the observations carefully every time the drops are inspected.
Single crystals can be harvested and cryoprotected as described (20, 21).
For suggestions on how to improve crystallization constructs to produce better crystals, see Note 7.
3.11. Refinement of RNA Crystal Structures
The ideal diffraction pattern should be from a single lattice, and each reflection should be nearly circular, and free of splitting and diffuse scattering; combinations of reflections should comprise distinct elliptical lunes (see Fig. 1d). For background on X-ray data collection, processing, and reduction see refs. 22, 23. To facilitate model building and refinement, the resolution of the diffraction pattern should exceed 3.2 Å resolution. Previously we noted there are many authoritative descriptions of RNA phasing and X-ray structure determination, and we will not describe this process here. Instead we will assume that the experimental phase problem has been addressed, and that the problem is one of refinement whereby the model |Fcalc| is manipulated to optimize agreement with the observed data |Fobs| while maintaining agreement with stereochemical restraints. A suitable program for RNA refinement is the PHENIX (Python-based Hierarchal ENvironment for Xtallography) software suite (24). Here we describe aspects of refinement that are pertinent to RNA crystallography. We will assume that the user is running PHENIX from a UNIX-like operating system X-window, but we point out where the GUI offers a distinct advantage.
3.12. Importing Data and Adding a Test Set
Importing structure factor amplitudes (hkl, Iobs, Iσ). There are a variety of X-ray data reduction programs. PHENIX can import or write data in various formats including: SCALEPACK (.sca), CCP4 (.mtz), CNS (.cns), and SHELX (.hkl).
- Convert intensities to amplitudes using the following command line arguments:
- % phenix.reflection_file_converter mystructure.sca--write-mtz-amplitudes--mtz-root-label=FOBS--mtz=mystructure
- To check your structure factors before starting refinement, use Phenix Xtriage. This will perform a quick analysis of your data and determine a probable solvent content, twinning analysis, Wilson scaling, and anomalous signal:
- % phenix.xtriage mystructure.mtz n_bases=61 log=mystructure_xtriage.log
To generate a new test set add “--generate-r-free-flags=True” to the argument above.
To preserve Rfree flags from a previous structure, use the “Reflection file editor” under the Reflection tools tab in the PHENIX GUI as follows: (1) if unit cell dimensions for both structures are not identical, one file must be converted into a format that does not include the unit cell dimensions, e.g., XPLOR or CNS; (2) use the GUI to import the reflection file to be refined against (i.e., “mystructure.mtz”) as well as the file that contains the desired Rfree flags (i.e., “previous_structure. mtz”); (3) highlight mystructure.mtz in the top window then click “use symmetry from selected file”; (4) in the “Input arrays” window highlight the “Data type” to be moved to the “Output arrays window” by using the “+” icon below the window; this may include: (a) FOBS, SIGFOBS, etc. from mystructure. mtz and (b) the Rfree flag from previous_structure.mtz; (5) click the “R-free flag generation …” button and in the window that opens deselect “Generate R-free flags if not already present”; then click “OK”; (6) verify that the “Extend existing R-free arrays(s) to full resolution range” option is selected and type the high and low-resolution values for your my_structure.mtz in the “high and low resolution values” boxes; (7) check that “Unit cell” and “Space group” values are correct; (8) in the “Output file” box, enter the correct path and file name for the new data file with Rfree flags, e.g., “mystructure-Rfree.mtz”; and (9) click the “Run” button.
3.13. Preparation of a Protein Data Bank File with Ligands for PHENIX Refinement
PHENIX will be unable to perform refinement if any atom, metal, or ligand names in your protein data bank (PDB) file are unrecognized. Such nonstandard atoms or residues must be identified before refinement.
- To view the coordinates and a cif (crystallographic information file) file for a particular ligand that is part of a larger structure (e.g., found in the PDB’s Ligand Expo or a downloaded cif file) use the following commands:
- % phenix.reel--chemical-components=ATP
- % phenix.reel ATP.cif
- To optimize and examine every atom in the file:
- % phenix.reel starting_model.pdb--do-all
-
The PHENIX routine “ready_set” adds hydrogens to nucleic acids and ligands in a PDB file in preparation for refinement. It also checks all the atom names and three-letter identifier codes against the PHENIX monomer library and the PDB’s ligand database, which can result in new cif files or “.edits” file (also called “edits” file) for unknown identifiers. To run this command:
- % phenix.ready_set mypdb.pdb cif_file_name=myligand.cif
Default behavior is to add hydrogens to ligands that have less than one quarter of their possible hydrogens but not to optimize_ligand_geometry. Novel linkages should also be found by ready_set and written to a custom edits file for bond restraint. This file can be written by the user to specify any custom link, bond angle, or bond length.
The format for the edits file for a 2′ –5′ phosphodiester linkage (12, 25) is:refinement.geometry_restraints.edits { bond { action = *add atom_selection_1 = name O2´ and chain A and resname 3DA and resseq 5 atom_selection_2 = name P and chain A and resname Gr and resseq 6 symmetry_operation = x,y,z distance_ideal = 1.6 sigma = 0.020 slack = None } } refinement.geometry_restraints.edits { angle { action = *add atom_selection_1 = name O2´ and chain A and resname 3DA and resseq 5 atom_selection_2 = name P and chain A and resname Gr and resseq 6 atom_selection_3 = name OP2 and chain A and resname Gr and resseq 6 angle_ideal = 109 sigma = 4 } } - Metal coordination spheres can be written to an edits file using:
- % phenix.metal_coordination mystructure.pdb
- Phenix.eLBOW (electronic Ligand Builder and Optimization Workbench) can be used to produce geometry restraints files for novel ligands. The program accepts various file input file formats (PDB, SMILES, Mol) and writes out a PDB and a cif file. Default behavior is to do a simple force field optimization of the input coordinates but alternative force fields such as AM1 are available:
- % phenix.elbow ligand.pdb
- To use eLBOW to process all unknown ligands in a pdb file:
- % phenix.elbow starting_model.pdb--do-all
- The user has the option to not optimize and keep input geometry:
- % phenix.elbow--final_geometry ligand.pdb
We recommend that all ligands output by phenix.elbow be checked visually as quality control.
Initial coordinates should be assigned a global B-factor before refinement (see Note 8).
3.14. Rigid-Body Refinement
When solving a Fourier problem in which the starting model (i.e., the phase source) and Fobs are significantly different in terms of subtle rotations and translations of the contents in the respective asymmetric units, it is possible to reposition the starting model by treating the entire model as a rigid body or to divide various known domains into groups that can be refined as independent (rigid) bodies. Rigid-body refinement can commence as follows:
-
Execute the first round of rigid-body refinement treating each whole molecule in the asymmetric unit as a separate rigid body:
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb strategy=rigid_body output.prefix=model-rigid1
This creates a rigid-body refined PDB file called modelrigid1_ 001.pdb. If there are any ligands or unusual nucleotides, then include the correct cif file after the input PDB file on the command line. Progress can be checked by examining the Rwork, Rfree, rmsd bond lengths, and rmsd bond angles located in the header of the output PDB file.
-
Multiple rounds of rigid-body refinement can be performed for RNA structures with multiple domains or when there is more than one molecule in the asymmetric unit. The method is fast and should be restricted to low-resolution data, which is sufficient to rotate and translate each rigid-body starting model to match the unknown structure to be refined. Various statements for parsing the structure into independently refined rigid-body domains may appear as follows:
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb strategy=rigid_body sites.rigid_body=“chain A or chain B” sites.rigid_body=“chain C or chain D.”
Here chains A and B are treated as one group, and C and D are treated as a separate group.
- The different chains can be treated as independent rigid bodies by the statement:
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb strategy=rigid_body sites.rigid_body=“chain A” sites. rigid_body=“chain B” sites.rigid_body=“chain C” sites. rigid_body=“chain D.”
The net effect should be a lowering of Rfree and Rwork by global repositioning of the starting model, which supplies the initial phases.
3.15. Refinement of Individual Atomic Coordinates and B-Factors
-
To perform the default three cycles of bulk solvent correction and scaling, as well as refinement of coordinates and ADP (atomic displacement parameters), use the following command:
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb
When complete PHENIX writes several files including: (1) an updated coordinate file that includes the refined structure with a header containing a summary of the refinement statistics; (2) a new MTZ file that includes electron density map coefficients compatible with interactive graphics programs such as COOT (26); (3) the log file of the refinement run; (4) a refinement parameters file (.eff); (5) a geometry restraints file (.geo); (6) a defaults file (.def) that includes all the previous commands, geometry edits, cif file locations, and modifications. This file can start the next round of refinement using the command:- % phenix.refine mystructure_002.def
- To perform different combinations of refinement steps, one can string commands together using “strategy”:
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb strategy=rigid_body+individual_sites+group_adp
- To set the number of macrocycles performed:
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb main. number_of_macro_cycles=6
Improvements in the radius of convergence are also achievable by simulated annealing (see Note 9).
3.16. TLS Refinement
- Translation, libration, screw-axis (TLS) refinement allows correction for anisotropic motion of atomic groups or domains, which can improve the model agreement with experimental data. To invoke TLS refinement type:
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb refine.adp.tls=“chain A” refine.adp.tls=“chain B” refine.adp. tls=“chain C” refine.adp.tls=“chain D”
Alternatively a “tls_group_selections.params” file can be used that specifies the TLS groups. This option will add ANISOU records to the refined pdb for atoms in the TLS groups. As such, the total B-factor includes the sum contribution of B-factor_tls and B-factor_individual values.
3.17. Optimizing Refinement Target Weights
-
As a starting point, it is best to determine a weighting scheme that gives the lowest Rfree:
- % phenix.refine Data-Rfree.mtz model.pdb optimize_wxc=true optimize_wxu=true
The command wxc=true will activate the optimization of X-ray and stereochemistry weighting, and wxu true will activate optimization of X-ray data and ADP weighting. Issuing this command will generate the lowest possible R-values, but may result in unacceptably high bond length and angle rmsd values, so examine the log file and the refined model output.
- Once an initial set of weights is calculated from step 1, it is possible to manually set the weight of the stereochemical and ADP restraint terms during refinement if Rwork and Rfree values are not satisfactory. To target a specific weight value enter:
- % phenix.refine Data-Rfree.mtz model.pdb target_weights. wxc_scale=X target_weights.wxu_scale=Y, where X determines the contribution of X-ray/stereochemical restraints and Y determines the X-ray/ADP contribution. The default values for wxc_scale and wxu_scale are X = 0.5 and Y = 1.0, respectively. Increasing either value will loosen restraints.
-
If the stereochemical restraints appear too tight, then the weighting of stereochemistry in refinement can be adjusted as follows:
- % phenix.refine Data-Rfree.mtz model.pdb wxc_scale=X
The same course of action can be taken for the ADP weight (wxu) if desired.
3.18. Use of Coot for Interactive Model Building
COOT is a program for interactive refinement of macromolecular coordinates based primarily on manual fitting of electron density maps. Here we assume the user is reading in a PDB file from PHENIX-based refinement and its corresponding MTZ map coefficients described in Subheading 3.15. Although RNA autobuilding is possible in PHENIX, cycles of PHENIX-refinement are often punctuated by manual intervention, which can improve the radius of convergence in which a model is trapped in a local energy minimum.
Start by opening an. mtz file in COOT. This can be done by choosing the “Auto Open MTZ …” under the “File” tab of the Black window GUI. This option creates both 2m Fo − D Fc and m Fo − D Fc electron density maps.
A full review of Coot capabilities is provided elsewhere (26). However, the following commands are worth reviewing since they are invoked frequently during RNA model building: (1) “Real Space Refine Zone”—fits model to electron density map selected; (2) Regularize Zone—normalize geometry to ideal values; (3) “LSQ Superpose …” located under the “Calculate” tab—allows pair-wise atomic superpositions between structures; (4) “Check/Delete Waters …”—under the “Validate” tab; (5) also under the “Validate” tab, the commands “Density fit analysis,” “ADP variance,” and “MOLPROBITY Analysis” (27); (6) “Get PDB and Map using EDS …” under the “File” tab—retrieves the requested PDB coordinates and electron density maps from the Electron Density Sever (28) if they are available.
Acknowledgments
We thank Profs. Harold C. Smith and Clara L. Kielkopf for sharing their expertise on RNA. We thank Jason Salter for assistance with diffraction analysis, as well as the staff of MacCHESS and SSRL for help with X-ray data collection. This work was supported in part by NIH grants GM063162 and RR026501 to J.E.W. MacCHESS is supported by NSF award DMR-0225180 and NIH/NCRR award RR01646SSRL. SSRL is operated by Stanford on behalf of the U.S. DOE. The SSRL Structural Molecular Biology Program is supported by the DOE, and by NIH/NCRR and NIGMS.
Footnotes
A robotic screening system is not essential but has the advantage of improving throughput with a relatively small amount of sample. Several fee-for-service screening facilities are available such as the University of Rochester Structural Biology and Biophysics Facility (http://www.urmc.rochester.edu/Structural-Biology-Biophysics/), or the batch-under-oil 1536-well screen at the Hauptman-Woodward Medical Research Institute, Inc. (Buffalo, NY) (http://www.hwi.buffalo.edu/faculty_research/crystallization.html). As an alternative, manual screening by use of the reagents in Table 1 represents a starting point.
Two factors are worth immediate consideration if difficulties arise in transcription. These include the concentration of MgCl2 and activity of T7 polymerase. The amount of MgCl2 will determine the extent of the reaction since it is depleted during the polymerization as an insoluble complex with inorganic pyrophosphate. T7 polymerase should be fresh or stored at −20°C in 20% glycerol under reducing conditions with DTT.
If it becomes apparent that the transcription products are a mixture of truncated and full-length species, it is possible that the polymerase is failing at segments of identical bases. One possible solution is to add 10% more of the nucleotide present in the repeat region.
The following variables should be taken into account when choosing or designing crystallization screens for RNA: (1) pH range: 5.6–8.0 (suggested buffers: Na-cacodylate, MES, HEPES, Tris); (2) monovalent salts of chloride: Li+, Na+, K+, ; acetate: Li+, Na+, K+, ; sulfate: Li+, ; (3) divalent cations (5–20 mM): Mg2+, Ca2+, Mn2+, and Co2+; (4) trivalents: 0.5–5.0 mM Co(NH3)6Cl3; (5) organic solvents: 5–40% (v/v) 2-methyl-2,4-pentanediol (MPD); and 1.5–2.4 M 1,6-hexanediol; (6) poly(ethylene) glycol (PEG): 5–30% (w/v) PEG 400, monomethyl ether (MME) PEG 550, MME PEG 2000, MME PEG 5000, PEG 3350, PEG 6000, and PEG 8000; (7) polyamine additives: 1–2 mM spermine and spermidine; (8) chelating agents: 0.1–0.5 mM EDTA; and (9) organic acid salts: 20–60% Tacsimate™.
The higher concentration of RNA, the more likely it is to crystallize. Ideally, the RNA will be concentrated to its solubility limit prior to initiating crystallization trials. Thus far, most of our ribozymes and riboswitches have crystallized in a range between 0.25 and 0.70 mM. Establishing the desired concentration of RNA may be as simple as resuspending the lyophilized RNA in a buffer of choice (e.g., 10 mM Na-cacodylate pH 6.0), or concentrating by centrifugation. For example, the preQ0 metabolite was added to our riboswitch aptamers under dilute conditions, then concentrated (4). For the crystals in Fig. 1c, dilute U1A was added dropwise to semi-concentrated RNA to attain a molar ratio of 1:1.05 before concentrating.
Like protein crystallization, RNA crystal growth can be sensitive to vibration and temperature. Screening is conducted in a low-vibration environment with a constant temperature (±1°C) in the range of 4–35°C. Plates are checked for crystal growth using a microscope at the temperature of screening. We typically choose 20 and 4°C to store plates during crystallization trials.
There are several documented approaches to improving RNA crystals if initial crystallization trials are unsuccessful or crystals exhibit poor diffraction. These include: adding U1A (29, 30), changing helix length (31), trying a different species (4), adding blunt or sticky ends (7), replacing nonconserved stemloops with GNRA tetraloops, or using a transition-state analog for ribozymes (12, 25).
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb set_b_iso=25
- % phenix.refine mystructure-Rfree.mtz starting_model.pdb simulated_annealing=true
References
- 1.Wedekind JE. In: Met. Ions Life Sci.: Structural and Catalytic Roles of Metal Ions in RNA. Sigel A, Sigel H, Sigel R, editors. London: Royal Society of Chemistry; 2011. pp. 299–345. [PubMed] [Google Scholar]
- 2.Wedekind JE, McKay DB. Crystal structure of the leadzyme at 1.8Å resolution: metal ion binding and the implications for catalytic mechanism and allo site ion regulation. Biochemistry. 2003;42:9554–9563. doi: 10.1021/bi0300783. [DOI] [PubMed] [Google Scholar]
- 3.Alam S, Grum-Tokars V, Krucinska J, Kundracik ML, Wedekind JE. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- 4.Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren J, Rastegari B, Condon A, Hoos HH. HotKnots: heuristic prediction of RNA secondary structures including pseudoknots. RNA. 2005;11:1494–1504. doi: 10.1261/rna.7284905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wedekind JE, McKay DB. Purification, crystallization, and X-ray diffraction analysis of small ribozymes. Methods Enzymol. 2000;317:149–168. doi: 10.1016/s0076-6879(00)17013-2. [DOI] [PubMed] [Google Scholar]
- 8.Golden BL, Gooding AR, Podell ER, Cech TR. X-ray crystallography of large RNAs: heavy-atom derivatives by RNA engineering. RNA. 1996;2:1295–1305. [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson MP, Scott WG. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr D Biol Crystallogr. 2008;D64:738–744. doi: 10.1107/S0907444908011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keel AY, Rambo RP, Batey RT, Kieft JS. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaucage SL, Reese CB. Recent advances in the chemical synthesis of RNA. Curr Protoc Nucleic Acid Chem. 2009;Chapter 2(Unit 2 16):11–31. doi: 10.1002/0471142700.nc0216s38. [DOI] [PubMed] [Google Scholar]
- 12.Torelli AT, Spitale RC, Krucinska J, Wedekind JE. Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme. Biochem Biophys Res Commun. 2008;371:154–158. doi: 10.1016/j.bbrc.2008.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spitale RC, Volpini R, Mungillo MV, Krucinska J, Cristalli G, Wedekind JE. Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core. Biochemistry. 2009;48:7777–7779. doi: 10.1021/bi9011622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha ND, Biernat J, McManus J, Koster H. Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984;12:4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spitale RC, Wedekind JE. Exploring ribozyme conformational changes with X-ray crystallography. Methods. 2009;49:87–100. doi: 10.1016/j.ymeth.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartsel SA, Kitchen DE, Scaringe SA, Marshall WS. RNA oligonucleotide synthesis via 5’-silyl-2’-orthoester chemistry. Methods Mol Biol. 2005;288:33–50. doi: 10.1385/1-59259-823-4:033. [DOI] [PubMed] [Google Scholar]
- 17.Ferre-D’Amare AR, Zhou K, Doudna JA. A general module for RNA crystallization. J Mol Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- 18.Sherlin LD, Bullock TL, Nissan TA, Perona JJ, Lariviere FJ, Uhlenbeck OC, Scaringe SA. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA. 2001;7:1671–1678. [PMC free article] [PubMed] [Google Scholar]
- 19.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 20.Garman EF, Doublie S. Cryocooling of macromolecular crystals: optimization methods. Methods Enzymol. 2003;368:188–216. doi: 10.1016/S0076-6879(03)68011-0. [DOI] [PubMed] [Google Scholar]
- 21.Garman EF, Owen RL. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2006;62:32–47. doi: 10.1107/S0907444905034207. [DOI] [PubMed] [Google Scholar]
- 22.Arndt UW, Wonacott AJ. The Rotation method in crystallography: Data collection from macromolecular crystals. New York: Elsevier/North-Holland; 1977. [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torelli AT, Krucinska J, Wedekind JE. A comparison of vanadate to a 2’–5’ linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: allatom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA. The Uppsala Electron-Density Server. Acta Crystallogr D Biol Crystallogr. 2004;60:2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 29.Ferre-D’Amare AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 30.Ferre-D’Amare AR. Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs. Methods. 2010;52:159–167. doi: 10.1016/j.ymeth.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacElrevey C, Spitale RC, Krucinska J, Wedekind JE. A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme. Acta Crystallogr. D Biol. Crystallogr. 2007;63:812–825. doi: 10.1107/S090744490702464X. [DOI] [PMC free article] [PubMed] [Google Scholar]