Abstract
Rsc1 and Rsc2 are alternative bromodomain-containing subunits of the ATP-dependent RSC chromatin remodeling complex in Saccharomyces cerevisiae. Smk1 is a sporulation-specific mitogen-activated protein kinase homolog that is required for the postmeiotic events of spore formation. In this study we show that RSC1 and RSC2 are haploinsufficient for spore formation in a smk1 hypomorph. Moreover, diploids lacking Rsc1 or Rsc2 show a subset of smk1-like phenotypes. High-copy-number RSC1 plasmids do not suppress rsc2-Δ/rsc2-Δ sporulation defects, and high-copy-number RSC2 plasmids do not suppress rsc1-Δ/rsc1-Δ sporulation defects. Mid-late sporulation-specific genes, which are normally expressed while key steps in spore assembly occur and which include genes that are required for spore wall formation, are not expressed in cells lacking Rsc1 or Rsc2. We speculate that the combined action of Rsc1 and Rsc2 at mid-late promoters is specifically required for the proper expression of this uniquely timed set of genes. Our data suggest that Smk1 and Rsc1/2 define parallel pathways that converge to provide signaling information and the expression of gene products, respectively, that are required for spore morphogenesis.
Meiosis is the evolutionarily conserved pathway used by sexually reproducing organisms to generate haploid products from diploid precursors. Meiosis is coupled to species-specific differentiation programs that produce gametes that are capable of sexual fusion. In the yeast Saccharomyces cerevisiae, meiosis is coupled to spore formation. Multiple nutritional signals induce the sporulation of mitotically dividing diploid cells; nitrogen and fermentable carbon inhibit sporulation, while nonfermentable carbon sources, such as acetate, promote sporulation (13). Once they are induced, diploid cells undergo the landmark events of meiosis, including a single round of DNA replication, synapsis, and genetic recombination, followed by two rounds of chromosome segregation without an intervening S phase. Homologous pairs of chromosomes are segregated from each other in the first, meiosis-specific division, while sister chromatids are segregated in the second, mitosis-like division. Spore formation involves the biogenesis of prospore membranes which grow from the spindle pole bodies as the second meiotic division is being completed (11, 15, 17). After the haploid products of meiosis are surrounded by the prospore membranes, multiple layers are assembled within and around this structure to give rise to the mature spore wall (16).
Meiosis and spore formation are intimately controlled through the transcriptional program of sporulation (6, 21). The genes that are expressed during the program can be broadly grouped into four categories, early, middle, mid-late, and late, based on the timing of their expression (6). Early genes are involved in the initiation of the program as well as in DNA replication and recombination. Middle genes control the meiotic divisions and the initiation of spore morphogenesis. The mid-late and late genes are expressed during spore morphogenesis and maturation, respectively (6, 21).
SMK1 encodes a mitogen-activated protein (MAP) kinase homolog whose transcription is controlled by a tightly regulated middle sporulation-specific promoter (12, 20). Once Smk1 is expressed, it is activated by phosphorylation in its regulatory loop (24). The deletion of SMK1, or mutations that prevent its activation, leads to spores that display an array of aberrant spore wall defects, even within the same ascus (12, 24, 30). Experiments performed with hypomorphic alleles of SMK1 showed distinct blocks during spore formation and demonstrated that these blocks can be suppressed by increasing gene dosages of these alleles (29).
In eukaryotes, nucleosome positioning can inhibit the access of transcription factors to the promoter regions of DNA. This inhibition can be overcome through the actions of ATP-dependent chromatin remodeling enzymes (27). The yeast Swi/Snf complex, which contains a Swi2/Snf2 catalytic subunit, was the first to be identified (8, 19). Since then, five other complexes containing Swi2-like catalytic cores have been identified in yeast (4, 25, 28). Only one of these, the RSC (for “remodels the structure of chromatin”) complex, is essential for viability (4). The RSC complex is comprised of 15 subunits, including the ATP-dependent catalytic subunit, Sth1 (4). In addition to being essential for mitotic growth, the RSC complex is also required for sporulation. Temperature-sensitive mutants of STH1 have been shown to cause a decrease in transcription of the early class of sporulation-specific genes, as well as poor spore formation (32). Two other subunits of the RSC complex, RSC1 and RSC2, are homologous bromodomain-containing proteins that are mutually exclusive, thus defining two distinct forms of RSC (5). Neither RSC1 nor RSC2 is essential for viability, but they are both synthetically lethal, suggesting that they have some redundant function (5). The deletion of either RSC1 or RSC2 causes a decreased sporulation efficiency and aberrant spore formation (33).
Sensitized genetic backgrounds have been used to identify and characterize genes that function in a variety of pathways in a wide spectrum of organisms. One approach to using sensitized genetic backgrounds is based on the premise that if the activity of a key component of a pathway is reduced to a threshold level of function, other components of the pathway can be made susceptible to a reduction in gene dosage. This approach has been particularly useful for studying signal transduction pathways in diploid organisms (9). Currently, little is known about the upstream components that signal to the Smk1 MAP kinase homolog or the downstream effector molecules that are controlled by this protein kinase.
We have used a sensitized smk1 genetic background to demonstrate that this gene shows specific genetic interactions with RSC1 as well as with RSC2. Moreover, cells lacking Rsc1 or Rsc2 show a subset of smk1-like phenotypes. The overexpression of one homolog in the absence of the other does not rescue these phenotypes. We show that both RSC1 and RSC2 are specifically required for expression of the mid-late class of genes. We propose that both Rsc1 and Rsc2 are specifically required for the proper expression of mid-late sporulation-specific genes, thus coupling the expression of gene products required for spore formation with the Smk1 signaling pathway that controls this morphogenetic program.
MATERIALS AND METHODS
Yeast strains and plasmids.
All strains used for this study are in the SK1 genetic background and are listed in Table 1. The DBY14 and DBY15 deletion strains were generated by first extracting genomic DNAs from knockout strains obtained from the American Type Culture Collection (35266-rsc2-Δ and 34686-rsc1-Δ). Primers flanking the deleted open reading frame (ORF) (for RSC2, the sense sequence was TATTGGCGTTTATGGGGAAG and the antisense sequence was AAGCCCAATTCCTTCTCAGC; for RSC1, the sense sequence was GCGCGAAATCAAATAGCAT and the antisense sequence was TACGGCATGGAAAATTGTTG) were used to amplify the ORF with a >250-bp flanking sequence. This PCR fragment was transformed into LNY150. Kanamycin-positive transformants were sporulated, and tetrads were tested by PCR for the deletion of the corresponding ORFs. These haploids were then mated to generate the diploid strains. The high-copy-number RSC2 (p426.RSC2.2XHA) and RSC1 (p426.RSC1) plasmids were gifts from Bradley Cairns. These are both in the pRS426 URA3 2μm vector (26).
TABLE 1.
Yeast strains used for this study
| Strain | Genotype | Source or reference |
|---|---|---|
| LNY150 | MATa/MATα leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | Lenore Neigeborne |
| MWY43 | MATa/MATα smk1-4/smk1-4 ura3/ura3 leu2::hisG/leu2::HisG his3/HIS3 his4/HIS4 lys2/lys2 ho::LYS2/ho::LYS2 | 29 |
| MDPY10 | MATa/MATα smk1::LEU2/smk1::LEU2 leu2-hisG/leu2-hisG trp1-hisG/trp1-hisG lys2/lys2 his4-N/his4-G ura31/ura3 ho::LYS2/ho::LYS2 | 30 |
| DBY15 | MATa/MATα rsc1::KAN/rsc1::KAN leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | This study |
| DBY14 | MATa/MATα rsc2::KAN/rsc2::KAN leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | This study |
| DBY16 | MATa/MATα rsc1::KAN/rsc1::KAN smk1-4/smk1-4 leu2/leu2 ura3/ura3 trp1/trp1 his4-N/his4-G lys2/lys2 ho::LYS2/ho::LYS2 | This study |
| DBY23 | MATa/MATα rsc2::KAN/rsc2::KAN smk1-4/smk1-4 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | This study |
| DBY19 | MATa/MATα sth1::KAN/STH1 smk1-4/smk1-4 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | This study |
| DBY20 | MATa/MATα sth1::KAN/STH1 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | This study |
| DBY42 | MATa/MATα rsc2-A312::mTn3::lacZ::LEU2/RSC2 smk1-4/smk1-4 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | This study |
| DBY43 | MATa/MATα rsc2-A312::mTn3::lacZ::LEU2/rsc2-A312::mTn3::lacZ::LEU2 smk1-4/smk1-4 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 his4-N/his4-G ura3/ura3 ho::LYS2/ho::LYS2 | This study |
Growth and sporulation of cells.
Vegetative growth was maintained in YPD (1% yeast extract, 2% peptone, 2% glucose), SD (0.67% yeast nitrogen base without amino acids and 2% glucose plus nutrients essential for auxotrophic strains), or YEPA (1% yeast extract, 2% peptone, 2% potassium acetate). For synchronous sporulation, cells were grown in YEPA to an optical density at 600 nm (OD600) of 0.3 to 0.5. Prior to sporulation, cells were harvested, washed in SM (2% potassium acetate plus 10 μg of adenine, 5 μg of histidine, 30 μg of leucine, 7.5 μg of lysine, 10 μg of tryptophan, and 5 μg of uracil per ml), and resuspended in SM to 2 OD units.
Genetic screen.
MWY43 cells were transformed with a randomly integrating transposon (mTn3) library (22). Integrating transformants were selected on SD lacking leucine. These transformants were pooled, and aliquots were plated onto YPD. Colonies were subsequently replica plated onto nitrocellulose filters that were placed colony side up on sporulation plates. After incubation for 3 days at the appropriate temperature, the colonies were transferred to a nitrocellulose filter and tested by a fluorescence plate assay (see below). Isolates that failed to fluoresce at the threshold temperature (28°C) but that fluoresced at the permissive temperature (25°C) were grown in YPD, and the insertion site was identified by a rescue plasmid protocol (22).
Assays for spore wall assembly.
For the fluorescence assay, nitrocellulose filters with sporulated colonies were placed colony side up in a petri plate containing 280 μl of ascal wall lysis buffer (100 mM sodium citrate, 10 mM EDTA [pH 5.8], 750 mM β-mercaptoethanol, and 70 μl of glusulase [NEE-154, crude solution; Dupont, Wilmington, Del.]), incubated at 37°C for 4 h, briefly blotted on 3M Whatman paper, and then placed in a petri plate containing 300 μl of concentrated NH4OH. Fluorescence was observed with a 304-nm UV light source and was photographed through a blue filter (Wratten no. 98; Kodak, Rochester, N.Y.).
The spore viability after exposure to glusulase was determined by first resuspending 1 OD600 unit of sporulated cells in 800 μl of 0.1 M PIPES (pH 6.5) and 200 μl of glusulase and incubating them at 30°C for 1 or 2 h. The cells were then washed and plated on YPD. For heat shock, the cells were resuspended in water and heated at 55°C for 1 or 2 h and then plated on YPD. For both glusulase and heat shock experiments, equal amounts (OD) of untreated sporulated cells were plated and plating efficiencies were calculated. The percent survival of treated cells was calculated by dividing the number of surviving colonies by the number of survivors of untreated controls.
Northern blots.
Preparation of total RNAs and Northern blot hybridization analysis were performed as previously described (12). DNA probes for the coding regions of the indicated genes were made by isolating PCR products from preparative agarose gels and radiolabeling them with [α-32P]CTP by random priming, and the probes were used for hybridization analysis at 106 dpm/ml.
RESULTS
Generation of sensitized smk1 strain.
Members of our laboratory previously described an allelic series of temperature-sensitive smk1 loss-of-function mutants that are specifically defective in the postmeiotic program of spore formation (29). In the present study, one of these alleles (smk1-4) was used to generate a sensitized strain in the SK1 hypersporulating background that would be useful for identifying new members of the Smk1 spore formation pathway. As a first step toward this goal, a smk1-4/smk1-4 homozygote and a smk1-Δ/smk1-4 heterozygote were compared for their ability to incorporate dityrosine, a sporulation-specific component of the outer layer of the spore wall, into insoluble material at a variety of temperatures. For these experiments, a plate assay that detects dityrosine via its fluorescence properties was used. These experiments demonstrated that at 25°C, both the smk1-4/smk1-4 and smk1-Δ/smk1-4 strains formed fluorescent spore colonies. In contrast, at 28°C the smk1-4/smk1-4 strain formed fluorescent spore colonies while the smk1-Δ/smk1-4 strain did not. At a fully restrictive temperature of 34°C, neither strain formed fluorescent spore colonies (data not shown). These experiments show that at 28°C, smk1-4 is haploinsufficient for fluorescent spore colony formation. We reasoned that other genes that are haploinsufficient for fluorescent spore colony formation in this background, specifically at 28°C (referred to as the “sensitized” temperature below), might correspond to Smk1 pathway components.
RSC1, RSC2, and STH1 are haploinsufficient in the sensitized smk1 strain.
We screened 51,000 individual smk1-4/smk1-4 colonies that had been mutagenized by use of a randomly integrating transposon library (22) for isolates that were fluorescence positive at 25°C but fluorescence negative at the sensitized temperature of 28°C. Thirty-one positive isolates were selected and the insertion sites in these isolates were identified. For strains carrying the transposon inserted into nonessential genes, we tested the ability of homozygous mutants to form fluorescent spore colonies. For these studies, the heterozygous transposon-insertion diploid isolates were sporulated at the permissive temperature, and haploid transposon-containing segregants were recovered and backcrossed to generate homozygous transposon-containing strains. Using this strategy, we identified a single nonessential gene that was both haploinsufficient for fluorescent colony formation as a heterozygote at the sensitized temperature and required for spore formation as a homozygote at the fully permissive temperature in the smk1-4/smk1-4 background. This gene was RSC2, a component of the RSC chromatin remodeling complex (isolated twice) (Fig. 1A). In addition, we isolated insertions in two essential genes that correlated with the haploinsufficient phenotype in the smk1 sensitized background. However, specific defects in the homozygous mutants could not be tested due to the inviability of the alleles that were recovered. These genes were ERG2, a sterol isomerase that is essential in our genetic background (isolated three times), and RSP5, an essential ubiquitin ligase (isolated twice). In this study, we have characterized the connection between RSC2 and SMK1.
FIG. 1.
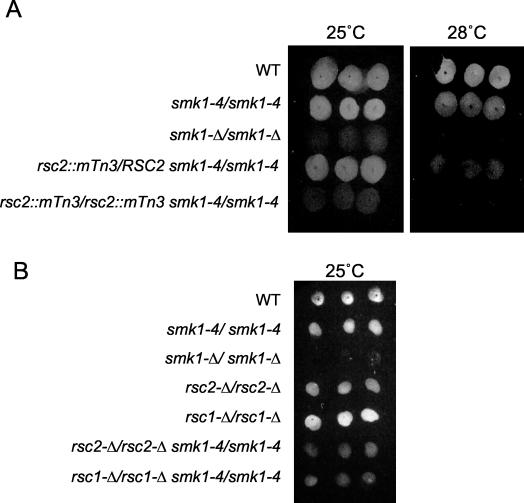
RSC1 and RSC2 genetically interact with smk1-4. (A) Fluorescence assay performed at permissive (25°C) and threshold (28°C) temperatures with wild-type (WT) and coisogenic strains harboring the indicated mutations. Three independent isolates of each strain are shown. (B) Fluorescence assay of homozygous full-length ORF deletion mutants of RSC1 and RSC2 in indicated backgrounds.
To confirm the genetic interaction between RSC2 and smk1-4, we deleted the full ORF of RSC2 from the smk1-4/smk1-4 background. This rsc2-Δ allele, like the transposon-insertion rsc2 alleles, was haploinsufficient for spore formation, as assayed by the fluorescence assay, specifically at the sensitized temperature (data not shown). We also tested the consequence of deleting both copies of RSC2 from the smk1-4/smk1-4 background and from a coisogenic SMK1/SMK1 genetic background. As shown in Fig. 1B, an rsc2-Δ/rsc2-Δ smk1-4/smk1-4 strain was unable to form fluorescent spore colonies at the permissive temperature (indistinguishable from the transposon alleles). Surprisingly, however, the rsc2-Δ/rsc2-Δ SMK1/SMK1 strain was able to form fluorescent spore colonies in the plate assay. These results confirm the genetic interaction between the SMK1 MAP kinase homolog and the RSC2 chromatin remodeling subunit genes. While the rsc2-Δ/rsc2-Δ SMK1/SMK1 strain did form spores and fluoresce in the plate assay, the kinetics of sporulation were delayed and the absolute level of spore formation was modestly reduced in this background. A more detailed phenotypic characterization of the rsc2-Δ/rsc2-Δ strain is described below.
RSC2 encodes a bromodomain-containing component of the multisubunit RSC. RSC1 encodes a homolog of RSC2. The occupancy of Rsc1 or Rsc2 has been shown to be mutually exclusive (5). The remaining subunits of RSC, including the catalytic Sth1 ATPase, appear to be encoded by unique genes. We tested whether mutations in RSC1 or STH1 showed interactions with smk1-4 which were similar to that seen with the RSC2 mutations described above. rsc1-Δ/RSC1 smk1-4/smk1-4 and sth1-Δ/STH1 smk1-4/smk1-4 strains displayed a spectrum of temperature-sensitive fluorescent phenotypes that were indistinguishable from those seen in the rsc2-Δ/RSC2 smk1-4/smk1-4 strain (haploinsufficient specifically at the sensitized temperature of 28°C) (data not shown). The rsc1-Δ/rsc1-Δ SMK1/SMK1 strain was able to form fluorescent spore colonies, while the rsc1-Δ/rsc1-Δ smk1-4/smk1-4 strain was defective in this assay (Fig. 1B). These results parallel those seen with the set of rsc2-Δ mutant strains and show that a reduced gene dosage of either of the two alternative bromodomain-containing proteins or the catalytic subunit of RSC specifically impairs the function of the SMK1 spore formation pathway in the sensitized genetic background.
rsc1-Δ and rsc2-Δ mutants display defects in meiotic progression.
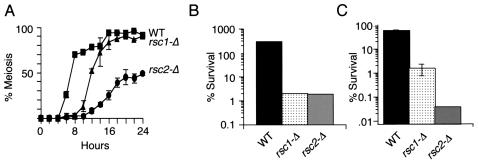
In order to gain additional insight into the roles of Rsc1 and Rsc2 in sporulation, we compared the phenotypes of cells lacking only RSC1 or RSC2 with that of the wild type. To monitor the meiotic divisions, we sporulated rsc1-Δ/rsc1-Δ cells, rsc2-Δ/rsc2-Δ cells, and wild-type cells, fixed them at various times postinduction, and stained them with 4′,6-diamidino-2-phenylindole (DAPI). Both rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants completed meiosis more slowly than wild-type cells (Fig. 2A); however, it should be noted that both of these mutant strains divide mitotically at a slower rate than the wild type. In YEPA, the medium used for presporulation growth, the generation times for the rsc1-Δ/rsc1-Δ, rsc2-Δ/rsc2-Δ, and wild-type strains were 4, 4, and 2 h, respectively. Thus, the slower meiotic kinetics in these strains could be explained in part by mitotic defects, since cells induce sporulation only during a restricted phase (G1) of the cell cycle. While the mitotic defects are likely to contribute to a lag in meiotic induction, they are unlikely to be sufficient to completely explain the differences in meiotic kinetics (see below). Moreover, rsc2-Δ/rsc2-Δ strains completed meiosis to only 50% the level seen in wild-type cells. These results suggest that Rsc1 and Rsc2 function positively to promote meiotic events that occur prior to spore formation. However, cells are still able to complete the meiotic divisions in the absence of Rsc1 or Rsc2, showing that neither of these proteins is absolutely required for these events. Interestingly, all of the rsc1-Δ/rsc1-Δ or rsc2-Δ/rsc2-Δ cells that completed meiosis in an otherwise wild-type genetic background appeared to form spore walls, as assayed by phase-contrast microscopy.
FIG. 2.
Sporulation defects of rsc1-Δ/rsc1-Δ (rsc1-Δ) and rsc2-Δ/rsc2-Δ (rsc2-Δ) strains. (A) Completion of meiosis I and II. Sporulated cells were taken at the indicated times postinduction and scored for the presence of three or four DAPI-stained foci. (B) Glusulase resistance. Cells of the indicated genotypes were sporulated for 3 days. Samples were exposed to glusulase as described in Materials and Methods. Survival is expressed as the percentage of surviving colonies from treated plates compared to untreated controls. Thus, the wild-type resistance appears to be >100% because four spore colonies are released per digested ascus. (C) Heat shock resistance. Sporulated strains were exposed to 55°C for 1 h and plated onto YPD plates. Error bars are too small to be seen (n = 3).
Strains lacking Rsc1 or Rsc2 display a subset of smk1-Δ phenotypes.
Spore formation involves the coordinated assembly of four distinct spore wall layers. Some of these layers function to protect the products of meiosis from specific environmental insults (2). Thus, the proper formation of these layers can be assayed at the phenotypic level by measuring the formation of fluorescent spore colonies as well as resistance to enzymatic (glusulase) digestion, organic solvents, and heat shock treatment. Each of these phenotypes requires increasing amounts of Smk1 activity, with fluorescent spore colony formation needing the lowest level, resistance to glusulase requiring an intermediate level, and full resistance to heat shock requiring the highest level (29). smk1-Δ/smk1-Δ mutants are defective in establishing resistance to all of these environmental insults. Although rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants form fluorescent spore colonies and spores which are viable (>85% viability for rsc1-Δ/rsc1-Δ and >45% viability for rsc2-Δ/rsc2-Δ) (see Materials and Methods), these mutants show a striking sensitivity to glusulase and heat shock treatment. Thus, the removal of Rsc1 or Rsc2 creates a spore morphogenesis phenotype that is similar to that seen in certain smk1 hypomorphs (Fig. 2B and C).
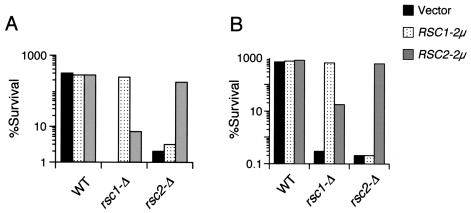
The possibility remained that the genetic interaction between the RSC complex components and SMK1 was not specific for RSC1 or RSC2, but rather was a dosage effect of lacking either protein. To test this possibility, we transformed rsc1-Δ/rsc1-Δ or rsc2-Δ/rsc2-Δ strains with high-copy-number plasmids containing RSC1, RSC2, and vector controls. These strains were then assayed for meiotic progression as well as for glusulase and heat shock resistance. As shown in Fig. 3, the overexpression of RSC1 in a rsc2-Δ/rsc2-Δ mutant did not suppress either of these phenotypes and vice versa. These data demonstrate a unique requirement for both Rsc1 and Rsc2 during spore development.
FIG. 3.
Overexpression of RSC2 does not suppress sporulation defects of rsc1-Δ/rsc1-Δ cells and vice versa. Strains transformed with the indicated plasmids were sporulated and scored for glusulase resistance (A) and heat shock resistance (B). Error bars are too small to be seen (n = 3).
Rsc1 and Rsc2 are both required for mid-late sporulation-specific gene expression.
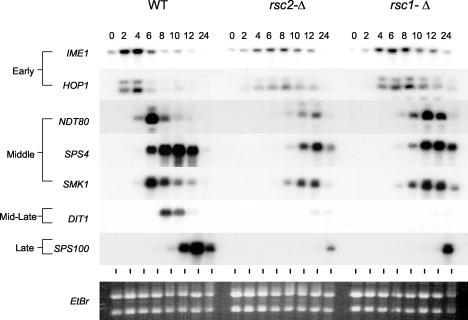
We next examined the transcriptional profile of key genes expressed during the transcriptional program of sporulation in cells lacking Rsc1 or Rsc2. For these experiments, wild-type, rsc1-Δ/rsc1-Δ, and rsc2-Δ/rsc2-Δ cultures were sporulated under synchronous conditions, and aliquots were collected at various times postinduction. RNAs from these samples were assayed with probes representing sporulation-specific genes of multiple temporal classes (early, middle, mid-late, and late) by Northern blot hybridization analysis. As shown in Fig. 4, both rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants expressed the early IME1 and HOP1 genes. However, the kinetics of early gene induction were modestly delayed in the rsc1-Δ/rsc1-Δ strain and more severely delayed in the rsc2-Δ/rsc2-Δ strain. In addition, while the levels of IME1 and HOP1 expression in the rsc1-Δ/rsc1-Δ strain approached those seen in the wild type, the magnitudes of IME1 and HOP1 expression in the rsc2-Δ/rsc2-Δ strain were reduced by about half. Thus, the duration of delays and the magnitude of early sporulation-specific gene expression correlates with the meiotic defects seen in these strains (Fig. 4). These results suggest that the delayed meiotic kinetics in cells lacking Rsc1 and Rsc2 and the reduced meiotic index seen for cells lacking Rsc2 might largely be explained by defects in IME1 expression in conjunction with the reduced mitotic growth rates seen for these strains.
FIG. 4.
Transcription of sporulation-specific genes in cells lacking RSC1 or RSC2. Total RNAs were prepared from homozygous strains of the indicated genotypes and were assayed by Northern blot analysis using the indicated probes.
In the rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ strains, the NDT80, SPS4, and SMK1 middle sporulation-specific genes were induced. In the case of the rsc1-Δ/rsc1-Δ mutant, the expression levels of all of these middle genes were comparable to those in the wild-type control strain. Moreover, the interval between the peaks of early and middle gene transcript accumulation in the rsc1-Δ/rsc1-Δ strain was comparable to that seen for the wild type. These results suggest that the rsc1-Δ/rsc1-Δ strain is largely unaffected in the expression of middle genes, despite the fact that RSC1 functions in IME1 induction earlier in the program. While middle genes are induced in the rsc2-Δ/rsc2-Δ strain, they are induced with substantially delayed kinetics and to a reduced level compared to the wild-type strain. The delay and reduction in middle gene expression seen for the rsc2-Δ/rsc2-Δ strain were comparable to the delay and reduction of early genes, making it possible that the middle gene expression defects are a downstream (dependency) consequence of the early (IME1) gene expression defects. Regardless, a key finding of this study is that middle gene induction occurs in both the rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants. In the case of the rsc1-Δ/rsc1-Δ strain, the level of SMK1 expression was comparable to that seen for the wild-type strain. These results make it unlikely that the genetic interactions observed between RSC and SMK1 can be explained through the specific regulation of the SMK1 promoter by the RSC.
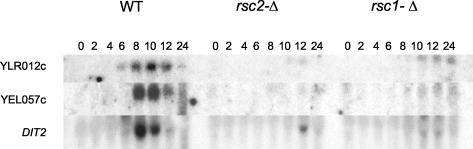
Strikingly, both rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants are specifically and severely defective in expressing DIT1. DIT1 encodes an enzyme that is required for the synthesis of the sporulation-specific dityrosine precursor that is cross-linked to the outer layer of the spore wall (2, 3). This gene is expressed in a temporal class defined as mid-late or subclass 6a (as defined by Primig et al. 21). We wanted to see if the transcription of other mid-late genes was also affected in these mutants. As seen in Fig. 5, the transcription of all mid-late genes tested was dramatically reduced in both the rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants. To our knowledge, the expression of the mid-late sporulation-specific genes is unique in its near absolute requirement for both Rsc1 and Rsc2. It was previously reported that DIT1 transcription was modestly reduced and that the late SPS100 gene was severely reduced (to 25 and 7% of wild-type levels, respectively) in smk1-Δ/smk1-Δ strains (12). The data shown here prompted us to reexamine these results. After multiple experiments comparing the wild-type and smk1-Δ/smk1-Δ strains, we were unable to see any reproducible difference in the expression of DIT1 or any other mid-late gene (data not shown). The expression of SPS100, however, was reduced to the previously reported levels. These results suggest that Smk1 does not directly regulate mid-late gene induction. The expression data support a model in which Smk1 and the RSC complex function in parallel pathways that converge on spore morphogenesis.
FIG. 5.
RSC1 and RSC2 are specifically required for the transcription of all mid-late genes tested. The filter shown in Fig. 4 was assayed with the indicated probes.
DISCUSSION
In this study, we have shown that RSC1 and RSC2 are haploinsufficient for spore formation in a strain in which Smk1 catalytic activity has been crippled (a smk1 hypomorph). In addition, we have demonstrated that diploid cells lacking RSC1 or RSC2 show a subset of spore formation phenotypes that are indistinguishable from those seen in certain hypomorphic smk1 strains. We propose that the genetic interactions between these genes reflect their shared function in the process of spore morphogenesis. Our transcriptional studies showed that there are dramatic defects in the induction of all mid-late sporulation-specific genes tested in rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ strains. Although there are transcriptional defects earlier in the program in these strains, these defects are mostly kinetic (genes are expressed later than normal) and the observed differences in magnitude are modest (less than twofold). Thus, we believe that the striking lack of mid-late gene induction in the rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ strains represents specific requirements for both Rsc1 and Rsc2 at mid-late promoters rather than a dependency of mid-late gene induction on earlier Rsc1- or Rsc2-dependent steps in the transcriptional program. Two genome-wide microarray data sets showed that there are roughly 500 and 150 examples of early and middle genes, respectively, in the yeast genome. There are roughly 60 mid-late genes, depending upon the classification scheme and the database used to compile expression parameters (21). DIT1 is the founding member of the mid-late class of genes (2). Since there is some degree of variability in assigning classes, we have focused our expression studies on genes with a similar transcriptional profile to that of DIT1. These genes are expressed as key steps in spore morphogenesis are occurring, and at least some of them have been shown to be required for spore formation. We propose that the genetic interactions between SMK1, RSC1, and RSC2 documented in this study represent an example in which the activity of a signal transduction pathway has been coupled to the availability of effector molecules through the transcriptional program (Fig. 6). In this model, the Smk1 MAP kinase homolog might represent a terminal protein kinase in the signaling pathway (itself expressed as a middle gene) whose activity is required to coordinate a process (spore morphogenesis) that requires a uniquely timed set of promoters (mid-late). This model predicts that the substrates regulated by Smk1 will include mid-late gene products or factors that coordinately function with mid-late gene products.
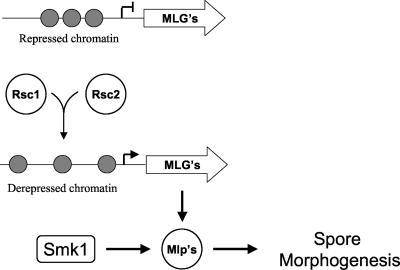
FIG. 6.
Model to explain genetic interactions between smk1, rsc1, and rsc2 mutants. Both Rsc1 and Rsc2 are required to convert an inactive chromatin structure at mid-late gene promoters to active chromatin. The products of these genes are then acted on by Smk1 to regulate spore morphogenesis.
The mid-late DIT1 and DIT2 genes encode enzymes that are required for the synthesis of dityrosine-containing precursors that are incorporated into the outer layer of the spore wall. Although rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants express dramatically reduced levels of DIT1 and DIT2 mRNAs, a sufficient amount of dityrosine precursor is still apparently produced to allow SMK1/SMK1 cells to form positive spore colonies in the fluorescence plate assay. In contrast to the SMK1/SMK1 background, the smk1-4/smk1-4 background did not form fluorescent spore colonies in the absence of Rsc1 or Rsc2. The fluorescence plate assay measures the incorporation of the dityrosine precursor into the insoluble material (soluble material is mostly released during sample preparation) (29; also see Materials and Methods). In a previous study, we showed that smk1 mutants can accumulate soluble dityrosine precursors but that they fail to incorporate these precursors into insoluble material. Taken together, these results suggest that Dit1 and Dit2 are not normally rate limiting. We speculate that Smk1 functions downstream of Dit1 and Dit2 to control the incorporation of dityrosine-containing precursors into insoluble material in the spore wall.
Yukawa et al. were the first to show that RSC plays a role in the transcriptional program of sporulation in yeast (32). Their studies showed that the essential Sth1/Nps1 ATPase subunit of RSC is required for meiosis and sporulation. Using a temperature-sensitive allele of STH1 (nps1-105), they showed specific defects in the expression of several early meiotic genes, including IME1, at the nonpermissive temperature. Our results are consistent with their demonstrated requirement for RSC during early meiosis. Yukawa et al. were also the first to analyze the role of Rsc1 and Rsc2 in sporulation (33). Their study demonstrated functional differences between Rsc1 and Rsc2 during the meiotic program which are consistent with the results of our work. There were also several differences between their study and ours that deserve comment. First, the Yukawa study showed that rsc1-Δ/rsc1-Δ, but not rsc2-Δ/rsc2-Δ, mutant cells have noticeable defects in spore formation, as assayed by light microscopy. The overexpression of IME1, the key regulator of induction, did not suppress this phenotype, suggesting a later role for Rsc1 in spore formation. In our study, rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ mutants both formed spores, as assayed by light microscopy. In addition, we observed multiple spore-specific spore wall layers that were indistinguishable from those of the wild type in a substantial fraction of the rsc1-Δ/rsc1-Δ and rsc2-Δ/rsc2-Δ cells by thin-section electron microscopy (data not shown). We speculate that differences in the strain background (our study used the hypersporulating SK1 strain while the Yukawa study used the W303 strain background) are responsible for these differences, since identical presporulation growth conditions and similar sporulation medium were used for the two studies. In the Yukawa study, the expression of the key early IME1 and IME2 genes was monitored, but genes expressed later in the program were not studied. Our results with IME1 and IME2 were mostly consistent with the results of that study. Our results showing the transcriptional defects that occur later in the program and showing that both Rsc1 and Rsc2 are specifically required for mid-late gene induction represent findings that were not documented in the Yukawa study but that are relevant for explaining the phenotypes that were described in that previous work.
In addition to RSC2, we also identified RSP5 and ERG2 as being haploinsufficient for spore formation in our sensitized background. We have not characterized these genetic interactions in detail. However, the fact that these genes were identified multiple independent times suggests that they represent bona fide genetic interactions and thus function in spore formation. RSP5 encodes a ubiquitin protein ligase that functions in membrane sorting and intracellular trafficking of membrane proteins, while ERG2 encodes a sterol isomerase that is required for ergosterol synthesis. Formation of the prospore membrane occurs through the fusion of secretory vesicles. These results are thus consistent with Erg2 and Rsp5 functioning in the postmeiotic program of spore morphogenesis, possibly by regulating membrane fusion and/or protein trafficking. Further studies will be required to test these ideas.
Our data indicate that the sensitized smk1 genetic screen as described above has not been carried out to saturation. Thus, while we expected to uncover insertions in CAK1, which is known to be required for the phosphorylation of Smk1's regulatory loop (24), and in SMK1 itself, these insertion sites were not identified in our studies. The failure to identify CAK1 and SMK1 insertions could be due to insertion site bias. However, we feel that it is more likely that relevant mutants were missed in the primary screening steps. We also noted that a high rate of false positives were uncovered in the primary screen, making a thorough analysis of all genes yielding a haploinsufficient spore phenotype an impractical goal. Further modifications to the approaches described here would be necessary to identify all of the genes that are haploinsufficient for spore morphogenesis in a weakened smk1 background.
The most striking conclusion to emerge from this study is that both Rsc1 and Rsc2 are required for mid-late gene induction. It has been shown that although neither RSC1 nor RSC2 is essential, they are synthetically lethal. This demonstrates that they do have an overlapping mitotic function and thus the potential to functionally substitute for one another. An analysis of the mitotic growth of these mutants showed only subtle phenotypic differences (5). Furthermore, genome-wide localization studies of the RSC complex have shown no discernible differences in the localization profiles of Rsc1 and Rsc2 to promoters in mitotic cells (18). These data show that Rsc1 and Rsc2 can be recruited to the same promoters and suggest that phenotypic differences observed between rsc1-Δ and rsc2-Δ mitotic cells may be a function of RSC1 and RSC2 expression levels and inherent requirements for RSC function at different promoters. Our data indicate that mid-late sporulation-specific promoters are unusual in that their induction requires both Rsc1 and Rsc2.
The divergently transcribed DIT1/DIT2 promoter is the most studied mid-late promoter. Genetic and molecular studies indicate that there are multiple regulatory regions within this promoter, including a repression site (termed the NRE), as well as additional positive and negative regulatory elements. Friesen et al. have shown that the NRE requires the global Tup1/Ssn6 repressor complex (10). Tup1/Ssn6 is thought to interact with hypo-acetylated histone tails, and these interactions are important for maintaining a transcriptionally inactive state (1, 23). In addition, a nonoverlapping site resembling a middle sporulation element (MSE) in the DIT1/DIT2 promoter has been shown to be responsive to the Ndt80 transcription factor, which induces middle sporulation-specific promoters (7). The MSE has also been shown to be responsive to the Sum1 repressor, which can compete with Ndt80 for MSE occupancy and which represses gene expression in mitotic cells (31). The Sum1 repressor can function at least in part by recruiting the Hst1 NAD-dependent histone deacetylase (14). Thus, the possibility exists that multiple chromatin-mediated repression complexes (Sum1 dependent and Tup1/Ssn6 dependent) as well as activator proteins collaborate to yield the uniquely timed pattern of mid-late gene expression. Interestingly, although some mid-late sporulation-specific promoters contain MSEs (about 36%) and others have been shown by genome-wide localization studies to bind to proteins that are thought to recruit Tup1/Ssn6 complexes, not all mid-late promoters contain both of these consensus elements and some contain neither. Thus, a common unifying feature of mid-late promoters at the DNA sequence level is not obvious. The requirement of both Rsc1 and Rsc2 at these promoters appears to be a conserved feature, however, as shown in this study. These observations, in conjunction with the diverged bromodomain-containing motifs in Rsc1 and Rsc2, lead us to speculate that both Rsc1 and Rsc2 are required for mid-late promoter induction because the repressive chromatin structure established by multiple repression complexes must be remodeled by a pathway that requires unique features of the alternative Rsc1- and Rsc2-containing RSC.
Acknowledgments
We thank Bradley Cairns for providing plasmids and for helpful discussions. We thank Andrew Boglioli and Allison Martin for technical assistance and Karen Schindler for comments on the manuscript.
This work was supported by grant GM061817 to E.W. from the National Institutes of Health.
REFERENCES
- 1.Bone, J. R., and S. Y. Roth. 2001. Recruitment of the yeast Tup1p-Ssn6p repressor is associated with localized decreases in histone acetylation. J. Biol. Chem. 276:1808-1813. [DOI] [PubMed] [Google Scholar]
- 2.Briza, P., M. Breitenbach, A. Ellinger, and J. Segall. 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4:1775-1789. [DOI] [PubMed] [Google Scholar]
- 3.Briza, P., M. Eckerstorfer, and M. Breitenbach. 1994. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble l-dityrosine-containing precursor of the yeast spore wall. Proc. Natl. Acad. Sci. USA 91:4524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 5.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 6.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 7.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 8.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 9.Daga, A., and U. Banerjee. 1994. Resolving the sevenless pathway using sensitized genetic backgrounds. Cell Mol. Biol. Res. 40:245-251. [PubMed] [Google Scholar]
- 10.Friesen, H., S. R. Hepworth, and J. Segall. 1997. An Ssn6-Tup1-dependent negative regulatory element controls sporulation-specific expression of DIT1 and DIT2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19:3657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krisak, L., R. Strich, R. S. Winters, J. P. Hall, M. J. Mallory, D. Kreitzer, R. S. Tuan, and E. Winter. 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8:2151-2161. [DOI] [PubMed] [Google Scholar]
- 13.Kupiec, M., B. Byers, R. Esposito, and A. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In E. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.McCord, R., M. Pierce, J. Xie, S. Wonkatal, C. Mickel, and A. K. Vershon. 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moens, P. B., and E. Rapport. 1971. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-Borchart, A. C., and M. Knop. 2003. Prospore membrane formation: how budding yeast gets shaped in meiosis. Microbiol. Res. 158:83-90. [DOI] [PubMed] [Google Scholar]
- 17.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 20.Pierce, M., M. Wagner, J. Xie, V. Gailus-Durner, J. Six, A. K. Vershon, and E. Winter. 1998. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 22.Ross-Macdonald, P., A. Sheehan, G. S. Roeder, and M. Snyder. 1997. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth, S. Y. 1995. Chromatin-mediated transcriptional repression in yeast. Curr. Opin. Genet. Dev. 5:168-173. [DOI] [PubMed] [Google Scholar]
- 24.Schaber, M., A. Lindgren, K. Schindler, D. Bungard, P. Kaldis, and E. Winter. 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 28.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner, M., P. Briza, M. Pierce, and E. Winter. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner, M., M. Pierce, and E. Winter. 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16:1305-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yukawa, M., S. Katoh, T. Miyakawa, and E. Tsuchiya. 1999. Nps1/Sth1p, a component of an essential chromatin-remodeling complex of Saccharomyces cerevisiae, is required for the maximal expression of early meiotic genes. Genes Cells 4:99-110. [DOI] [PubMed] [Google Scholar]
- 33.Yukawa, M., H. Koyama, K. Miyahara, and E. Tsuchiya. 2002. Functional differences between RSC1 and RSC2, components of a for growth essential chromatin-remodeling complex of Saccharomyces cerevisiae, during the sporulation process. FEMS Yeast Res. 2:87-91. [DOI] [PubMed] [Google Scholar]