The sudden appearance of the neural crest and neurogenic placodes in early branching vertebrates has puzzled biologists for over a century1. These embryonic tissues contribute to the development of the cranium and associated sensory organs, which were crucial for the evolution of the vertebrate “newhead”2,3. A previous study suggests that rudimentary neural crest cells existed in ancestral chordates4. However, the evolutionary origins of neurogenic placodes have remained obscure owing to a paucity of embryonic data from tunicates, the closest living relatives to those early vertebrates5. Here we show that the tunicate Ciona intestinalis exhibits a proto-placodal ectoderm (PPE) that requires inhibition of bone morphogenetic protein (BMP) and expresses the key regulatory determinant Six1/2 and its co-factor Eya, a developmental process conserved across vertebrates. The Ciona PPE is shown to produce ciliated neurons that express genes for gonadotropin-releasing hormone (GnRH), a G-protein-coupled receptor for relaxin-3 (RXFP3) and a functional cyclic nucleotide-gated channel (CNGA), which suggests dual chemosensory and neurosecretory activities. These observations provide evidence that Ciona has a neurogenic proto-placode, which forms neurons that appear to be related to those derived from the olfactory placode and hypothalamic neurons of vertebrates. We discuss the possibility that the PPE-derived GnRH neurons of Ciona resemble an ancestral cell type, a progenitor to the complex neuronal circuit that integrates sensory information and neuroendocrine functions in vertebrates.
Neurogenic placodes contribute to cranial sensory systems that mediate hearing, smell and taste2. They are considered a vertebrate innovation6 and the full repertoire, including olfactory, otic, epibranchial and trigeminal placodes, are already present in jawless hagfish and lampreys. It has been argued that the evolution of neurogenic placodes was a crucial event for facilitating the transition from filter-feeding invertebrate chordates to predatory vertebrates3.
Comparative embryological studies have failed to identify any clear evolutionary precursors of neurogenic placodes in invertebrate chordates. It has been suggested that the pre-oral organ of amphioxus, Hatschek’s pit, is related to the adenohypophyseal (anterior pituitary) placode of vertebrates; however, the fate map of this tissue remains unresolved7. In addition, the atrial primordia of Ciona have been likened to the otic and lateral-line placodes of fish, but they are not specified until tailbud stages and do not derive from the anterior neural plate border8. Also, previous studies provided evidence for a placodal-like tissue in Ciona, but it was homologized to the adenohypophyseal placode8,9, which lacks neurogenic potential. We show that the latter tissue does in fact produce ciliated sensory neurons that express GnRH and RXFP3, signatures of neuroendocrine cell types found in vertebrates10–12.
Neurogenic placodes develop from a specific domain anterior to the neural plate of vertebrate embryos2. All placode subtypes arise from this U-shaped domain, termed the preplacodal ectoderm13. This tissue expresses the transcription factor Six1, and mutations in Six1 cause craniofacial defects in humans, including deafness14. We found that the Ciona homologue of Six1, Six1/2, as well as its co-factor Eya, is initially expressed in a row of eight cells located immediately anterior to the neural plate, as marked by ZicL (Fig. 1a and Extended Data Fig. 1). This pattern is recapitulated by a Six1/2 regulatory sequence driving expression of an mCherry reporter gene (Six1/2. mCherry; Fig. 1b and Supplementary Video 1). At tailbud stages the Six1/2+ cells intercalate and become situated above the anterior brain (that is, the sensory vesicle), surrounding the future oral opening (Extended Data Fig. 2).
Figure 1.
Six1/2 expression requires BMP attenuation. a, Dorsal view of an embryo at early neurula stage electroporated with ZicL. GFP and hybridized with Six1/2 and Eya mRNA probes. b, Dorsal view of an embryo at mid-neurula stage co-electroporated with Six1/2. mCherry and Chordin. GFP after an additional division (16 Six1/2+ cells in total). c–f, Tailbud electroporated with Six1/2. mCherry and Dmrt. GFP; counterstained with phalloidin (grey). c, Tailbud co-electroporated with Dmrt. BMP2/4 (131 of 135 embryos showed no Six1/2. mCherry expression). d, Tailbud co-electroporated with Dmrt. BMP5/7 (120 of 135 embryos displayed a full expression pattern for Six1/2. mCherry). e, Tailbud co-electroporated with Dmrt. BMPR1CA (95 of 135 embryos had partial anterior Six1/2. mCherry expression). f, Tailbud co-electroporated with Dmrt. TGF-βRCA (93 of 135 embryos displayed a full expression pattern for Six1/2. mCherry and anterior neural tube defects). Arrowheads in d and e flank the derivatives of the anterior ectoderm that normally co-express Six1/2. mCherry and Chordin. GFP in control embryos (see panel b). Arrows show extrusion of sensory vesicle cells in f.
In vertebrates, the preplacodal ectoderm is specified by a two-step process during which a gradient of BMP signalling establishes preplacodal competence in the non-neural ectoderm during the blastula/early gastrula stage, followed by subsequent inhibition of BMP signalling during gastrulation13. BMP signalling may not be required for establishing proto-placodal competence in Ciona, but inhibition of BMP signalling seems to be crucial for its specification. We observe conserved expression of BMP2/4 and the Chordin (also known as Ci-Chordin) antagonist in ventral and dorsal regions, respectively, at the time when the PPE is specified during gastrulation and neurulation (Extended Data Fig. 3).
Localized Chordin expression at the lateral edges of the Ciona PPE (Fig. 1b) suggests a role in antagonizing BMPs emanating from ventral regions, similar to the situation seen in vertebrates15. We therefore explored the possibility that inhibition of BMP signalling might be required for the activation or maintenance of Six1/2 expression. We sought to overcome the presumed inhibitory effect of chordin by the targeted misexpression of BMP ligands or constitutively active forms of BMP receptors. These experiments were performed using a previously described Dmrt regulatory DNA (that is, Dmrt. GFP)16, which drives expression throughout the anterior neural plate and adjacent anterior ectoderm (Extended Data Fig. 1).
The co-electroporation of Dmrt. BMP2/4 with Dmrt. GFP and Six1/2. mCherry resulted in loss of Six1/2 expression in the anterior neural plate border, consistent with similar experiments in vertebrates17 (Fig. 1c). Conversely, misexpression of BMP5/7 had no noticeable effect on expression of the Six1/2 reporter or embryo morphology (Fig. 1d). These observations suggest that chordin may be specifically required to exclude BMP2/4 signals from the PPE.
Additional misexpression assays were performed with a constitutively active form of the BMP receptor (BMPR1CA) for cell-autonomous activation of BMP signalling. In these experiments, activation is restricted to Dmrt+ cells, which form the anterior neural plate and adjacent anterior ectoderm. Ectopic BMPR1 activity results in reduced expression of Six1/2 (Fig. 1e), similar to the results obtained upon misexpression of the BMP2/4 ligand. However, the phenotype is less severe, with persistence of Six1/2 expression in the medially derived regions of the PPE (compare Fig. 1d with Fig. 1e). Expression is lost in lateral regions, which are the source of the GnRH neurons.
To demonstrate that disrupting PPE formation is a specific result of ectopic BMP signalling, we misexpressed a constitutively active form of the transforming growth factor-β receptor (TGF-βRCA) throughout the anterior brain and adjacent ectoderm (Dmrt. TGF-βRCA). This had very little effect on the expression of Six1/2, despite severe defects in brain morphology (Fig. 1f). We therefore conclude that inhibition of BMP signalling is essential for the specification of the Ciona PPE, as observed in vertebrates15,17. However, it is possible that this inhibition indirectly regulates Six1/2 expression.
The olfactory placode is the anterior-most neurogenic placode in vertebrates and develops above the oral opening7. GnRH neurons arising from this placode migrate along the axonal tracts of olfactory neurons towards the hypothalamus11,12. They subsequently innervate the anterior pituitary and stimulate the release of gonadotropins, hormones critical for growth, sexual development and reproduction18. Previous studies have documented the expression of six different GnRH peptides encoded by two duplicated genes in Ciona19. GnRH2 (hereafter referred to as GnRH) is expressed near the oral opening (Extended Data Fig. 4) and we used the 59 regulatory sequences (GnRH. GFP) to label this lineage during development. During the tailbud stage, the posterior-most Six1/2+ cells acquire a distinctive cone-shaped morphology, permitting their direct visualization during morphogenesis (Fig. 2a and Extended Data Fig. 2). By the larval stage, these cells have differentiated into neurons that co-express GnRH. GFP and Six1/2. mCherry (Fig. 2b and Supplementary Video 2). The production of these neurons is strongly inhibited by BMP signalling (Extended Data Fig. 5).
Figure 2.
PPE-derived GnRH neurons express RXFP3. a, Tailbud electroporated with Six1/2. mCherry. Arrowhead indicates the posterior-most Six1/2+ cell that undergoes neurogenesis. b, Larva co-electroporated with GnRH. GFP and Six1/2. mCherry. Insets show Six1/2 expression (left) and GnRH expression (right) surrounding the oral opening (dotted line). The GnRH+ neuron is marked with an asterisk. c, Dorsal view of larva electroporated with SOG/Chemokine-like. mCherry. The dotted line indicates the oral opening. d, Larva electroporated with RXFP3. mCherry. e, Phylogenetic placement among human rhodopsin-class GPCRs of Ciona RXFP3 and SOG/Chemokine-like, indicated by the starred clades. Green clades comprise Ciona and human orthologues, whereas no human orthologue was found for the red clade and no known Ciona GPCR grouped within the blue clades. Support values are Bayesian posterior probability percentages. f, Juvenile co-electroporated with GnRH. GFP and Six1/2. mCherry. The dotted line indicates the oral opening. Ciona were counterstained with phalloidin (grey) in panels a–d and f. Brackets in panels b–d and f indicate PPE-derived GnRH neurons. Panels a, b, d and f are lateral anterior views.
Ciona PPE-derived GnRH neurons express two genes encoding G-protein-coupled receptors (GPCRs) implicated in neurosecretion in vertebrates, a somatostatin/opioid/galanin/chemokine-like receptor (SOG/Chemokine-like) and a relaxin-3 receptor (RXFP3) (Fig. 2c, d, Extended Data Fig. 4 and Supplementary Video 2). The relationship of these Ciona GPCRs to human rhodopsin-class GPCRs is shown in a summary tree (Fig. 2e and Extended Data Figs 6 and 7). In vertebrates, RXFP3 is expressed in hypothalamic paraventricular nuclei that provide synaptic inputs for the secretion of GnRH within the hypothalamic–pituitary–gonadal (HPG) axis20. Notably, the PPE-derived Ciona GnRH neurons survive through metamorphosis despite extensive cell death and reorganization of most of the larval nervous system (Fig. 2f). Moreover, injection of GnRH into mature Ciona causes the release of gametes and is likely to trigger spawning in wild populations21.
As both GnRH1 and chemosensory neurons arise from the olfactory placode of vertebrates, we were curious whether Ciona PPE-derived neurons might also possess chemosensory properties. Chemosensation by olfactory neurons occurs via odorant receptors in the tips of their cilia. Swimming larvae possess a group of neurons located behind the oral opening, previously described as the anterior apical trunk epidermal neurons (aATENs). These neurons possess cilia that are clearly visualized using an acetylated α-tubulin antibody (Fig. 3a). Electroporation with GnRH. mCherry followed by acetylated α-tubulin staining confirmed that the cilia project from the PPE-derived GnRH neurons (Fig. 3b). Moreover, comprehensive visualization of Six1/2+ cell morphogenesis reveals that the aATEN–GnRH neurons are derived from the a11.205 lineage (Extended Data Fig. 2). Ultrastructural analysis has shown that cilia of these neurons have a 9+ 2 microtubule arrangement22, a shared characteristic attributed to non-motile cilia of vertebrate olfactory neurons23.
Figure 3.
Ciliated GnRH neurons express functional CNGA. a–c, Larvae stained with phalloidin (grey). a, Dorsal view of a larva stained with an acetylated α-tubulin antibody; dotted line indicates the oral opening. b, Lateral view of a larva electroporated with GnRH. mCherry and stained with an acetylated α-tubulin antibody. c, Lateral view of a larva electroporated with CNGA. mCherry. Arrowheads indicate PPE-derived GnRH neurons in all panels. d, Ratiometric calcium imaging of HEK 293 cells transfected with Ciona CNGA, assayed by the intracellular Ca2+ indicator Fura-2. The graph shows the efficiency of response of CNGA to the cyclic nucleotide analogues 8-Br-cGMP and 8-Br-cAMP in seven individual cells (coloured lines) over a 120-min period. Indicated are the time points (red arrows) and durations (pink bars) of each analogue perfusion. e, Schematic that compares the PPE-derived GnRH neurons in tunicates such as Ciona (left) and olfactory-derived neurons in vertebrates (right). CNS, central nervous system; HEK, human embryonic kidney.
Conventional odorant receptors have not been identified in the Ciona genome7, but it is unlikely that a marine chordate would lack a means to detect odorants. Instead of examining divergent GPCRs, we focused on downstream effectors of chemosensory transduction. Cyclic nucleotide-gated (CNG) cation channels are critical for mediating sensory perception in photoreceptors and olfactory neurons. The binding of odorants to receptors causes an intracellular increase in cyclic nucleotides that, in turn, results in an influx of calcium, which ultimately leads to the depolarization of sensory neurons24. In rats, CNGA2 expression is detected in GnRH1 neurons but is not necessary for their activity. This expression might be due to prior activation in the shared progenitor of GnRH1 and olfactory neurons25,26.
A homologue of vertebrate CNGA1 is expressed in fully differentiated aATEN–GnRH neurons of Ciona (Fig. 3c). To determine whether Ciona CNGA forms a functional channel, we transfected a CNGA expression construct into human embryonic kidney 293 cells incubated with the intracellular calcium indicator Fura-2 AM to test for a response to cyclic nucleotides. Indeed, application of either 8-bromoguanine-39,59-cyclic monophosphate (8-Br-cGMP) or 8-bromoadenosine-39,59-cyclicmonophosphate (8-Br-cAMP) elicited an increase in intracellular calcium concentration, with a higher sensitivity to cAMP than cGMP (Fig. 3d and Extended Data Fig. 8). The occurrence of a functional CNGA in PPE-derived neurons, in addition to the presence of a 9+ 2 cilium, suggests that they are likely to possess chemosensory activities. These observations provide the first evidence, to our knowledge, that putative sensory neurons arise from a placodal-like territory in a non-vertebrate.
Our data suggest that the aATEN–GnRH neurons have dual neurosecretory and chemosensory properties. In vertebrates, neurosecretory and chemosensory cells both arise from the olfactory placode and are intimately linked. GnRH neuroblasts use the axon tracts of chemosensory neurons to guide them to their final destination in the hypothalamus. In adults, pheromones detected by chemosensory neurons cause a release of gonadotropins via hypothalamic GnRH neurons, a function crucial to sexual reproduction. Furthermore, transneuronal tracing experiments have shown that olfactory and GnRH neurons form a coherent neuronal circuit27,28, a connection that likely arose long ago.
It has been proposed that neuronal circuits evolve from the functional segregation of specialized cell types from multifunctional ancestral cells, akin to the well-known mechanism of gene duplication and subfunctionalization. A chemosensory–neurosecretory precursor is likely to have existed in ancestral chordates before the diversification of the HPG axis seen in vertebrates29,30. We have presented evidence that the PPE-derived aATEN–GnRH neurons in Ciona provide a particularly vivid example of such an ancestral cell type. Over the course of evolution within the vertebrate lineage, the proposed chemosensory and neurosecretory functions of these neurons may have become segregated into dedicated cell types that work together within a coherent circuit (Fig. 3e). Such cellular subfunctionalization might be a generally important mechanism of neuronal circuit evolution in vertebrates.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
METHODS
Embryo preparation and imaging. Ciona intestinalis adults were obtained, fertilized in vitro and electroporated for transient transgenesis as previously described16. For each electroporation, typically 70 µg of DNA was resuspended in 100 µl buffer. Embryos were fixed at the appropriate developmental stage for 15 min in 4% formaldehyde. The tissue was then cleared in a series of washes of 0.01% Triton-X in PBS. Actin was stained overnight with Alexa-647-conjugated phalloidin (Fisher, A22287) at a dilution of 1:500.
For the acetyl-α-tubulin staining, larvae were fixed for 10 min in 2% para-formaldehyde, then washed three times in 0.01% Triton-Xin PBS for 10 min each, and blocked with 1% normal goat serum for 10 min. Afterwards samples were incubated with a monoclonal anti-acetylated tubulin antibody (Sigma-Aldrich, T6793) at a dilution of 1:1,000 overnight at 4°C. Next, the tissue was washed in 0.01% Triton-X in PBS, three times for 30 min each. The samples were then incubated for 2 h at room temperature with an Alexa Fluor 488 donkey anti-mouse IgG (H+ L) antibody (Fisher, A21202) at a dilution of 1:500 in 1% normal goat serum, and finally washed in 0.01% Triton-Xin PBS (three times for 30 min each).
Samples were mounted in 50% glycerol in PBS, with 2% 1,4-diazabicyclo[2,2,2]octane (Sigma) for microscopy. Confocal images were acquired on a Zeiss LSM 700 microscope using a Plan-Apochromat 20×, 40× or 63× objective. Confocal stacks contained approximately 50 optical slices at a thickness of 1–2 µm each. Images were rendered using Volocity 6 with the 3D opacity, extended focus or XY plane visualization tools. Time-lapse images were taken on a Zeiss LSM 700 microscope at intervals of 3–4 min. Bright field in situ micrographs of larvae were acquired on a Zeiss Axio Imager microscope using an EC Plan-Neofluar 40× objective with a ProgRes C14 plus camera.
Experimental perturbations were performed in triplicate and two biological replicates were scored. No statistical methods were used to predetermine sample size. The scored replicates were totalled and these are indicated in the appropriate figure legends. Phenotypes were scored in animals exhibiting transgenesis (that is, using a reporter unaffected by a given perturbation such as Dmrt. GFP in Fig. 1). Unfertilized animals or those that failed to gastrulate were excluded from the analysis. No randomization or blinding was performed on samples.
Molecular cloning
The Genome Browser Gateway (University of California, Santa Cruz) facilitated the identification of conserved non-coding sequences between Ciona intestinalis and Ciona savignyi. Putative enhancer sequences were amplified by polymerase chain reaction (PCR) and cloned into a pCESA vector using AscI/NotI restriction sites for Chordin (KH.C6.145; 59-CGGTTACGTTAAGTTCGTGCG-39 and 59-CTTTGTTCCTGCTCGAAGTGAG-39), Six1/2 (KH.C3.553; 59-GTGACAGGAAACGTCTAGG-39 and 59-TGGCTCTGAGGCCGTGATTGTAG-39), GnRH (KH.C9.484; 59-GTGTACGATTGACAAGTTGG-39 and 59-TGTTACGTTATCTCTCTAGAAG-39), SOG/Chemokine-like (ci0100133186; 59-GGCAGATTTGACCTCAACTTG-39 and 59-GGCGTTTCC GAAAAGCCCTTT-39), RXFP3 (KH.C6.184; 59-CCGTGATTGTAATACGTATGAC-39 and 59-GCACAGTATTGTGTTATATACC-39), and CNGA (KH.C2.249; 59-GTGGAGATGCCGATTTCAACC-39 and 59-CACTTTGTTTGGCATTATTCC-39). The Dmrt and ZicL enhancers have been described previously16. A similar cloning method was used to create misexpression vectors using NotI/EcoRI sites for the BMP2/4 (KH.C4.125; 59-ATGGTGGCGCTTACGGATTGGAC-39 and 59-CTATCTACACCCACAAGCTTGC-39) and BMP5/7 (KH.C2.336; 59-ATGACTTGTCATCGCAATAAAG-39 and 59-TCAGTTGCACCCGCATGACAC-39) plasmids. The TGF-βRCA construct was made by site-directed mutagenesis to introduce a Q154D missense mutation in the regulatory glycine- and serine-rich domain of the wild type receptor. A similar strategy was used to create the previously described BMPR1CA (ref. 31). The coding sequence of SOG/Chemokine-like was amplified from cDNA (59-ATGTTGCTCGGAATCATGAAATC-39 and 59-GTATAAATATTCATACTTGTTTCTG-39) and cloned into a pCESA vector using NotI/EcoRI restriction sites.
For the calcium imaging experiments, cDNA fragments containing the full-length coding regions of C. intestinalis CNGA, CNGB and CNGC (Ghost Gene IDs: KH.C2.249,KH.L42.6, and KH.C7.605, respectively) were amplified from a cDNA pool of mid tailbud embryos (CNGA) or larvae (CNGB and CNGC) by PCR using a thermostable DNA polymerase exhibiting proofreading activity (Takara LATaq; Takara Bio) with gene-specific primers (59-AGCTAACACAGATTTTAGTGTAATG-39 and 59-GTTGCGGGTAAATTACATGTC-39 for CNGA; 59-ATGGCATTGCACATAAATGCAA-39 and 59-AGCAAAGACTTTGGTAAACATCAG-39 for CNGB; 59-TAAGGTTGATACAGGTTTCATTGG-39 and 59-GACGTAATCATGACGAAACTGTG-39 for CNGC). The PCR products were subcloned into pcDNA6 plasmid vectors and sequenced on both strands using the cycle sequencing method with an Applied Biosystems 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
In situ hybridization
The fluorescent in situ hybridizations were performed as described previously16. The colorimetric in situ hybridizations on hatched larvae were performed as previously detailed32. The in situ hybridizations for Extended Data Fig. 3 were performed as specified33. mRNA probes were synthesized using linearized cDNA clones for Six1/2 (ciad46p13), Eya (cilv29d14), Chordin (ciclo16e09), BMP2/4 (cicl060n01), GnRH2 (cilv25p08) and RXFP3 (cinc033n18) from Nori Satoh’s cDNA gene collection (OIST, Okinawa, Japan). Probe templates for CNGA (59-ATGGCATTGCACATAAATGCAA-39 and 59-AGCAAAGACTTTGGTAAACATCAG-39) and SOG/Chemokine-like (59-ATGTTGCTCGGAATCATGAAATC-39 and 59-GTATAAATATTCATACTTGTTTCTG-39) were amplified from a cDNA pool of mid tailbud embryos and subcloned into a pBSKM vector.
Phylogenetic analysis
For the initial survey tree, BLASTP34 was used to assign the two Ciona GPCR sequences of interest to the rhodopsin class. We used the MAFFT E-INS-i algorithm35 to align their seven-transmembrane domains with those of the proteins constituting the Pfam seed alignment for rhodopsin GPCRs36, as well as most Ciona rhodopsin-receptor-like proteins identified previously37. After removing ambiguously aligned amino acids with Gblocks38, the 118-sequence data set (including five outgroup sequences for Ciona Glutamate, Smoothened, Frizzled and cAMP receptor-like proteins) contained 373 positions (including gaps). For optimal tree searches and measurement of nodal support, minimum evolution (ME), maximum likelihood (ML) and Bayesian inference (BI) criteria were used39.
To classify the two Ciona GPCRs of interest with greater accuracy and support, a second set of analyses was performed using a more focused sequence sampling. Based on the initial survey tree and the identifications made by Kamesh et al.37, we selected presumed members of the specific clade containing the two Ciona GPCRs of interest, its large ‘Chemokine Cluster’ sister clade, and its two closest rootward sister clades (that is, members of the ‘SOG’ rhodopsin subclass). For these selections, GPCRDB Tools40 was used to build an alignment of human seven-transmembrane domain sequences from the Swiss-Prot database. Using Clustal Omega41, we integrated seven relevant Ciona sequences described by Kamesh et al.37 into the GPCRDB Tools alignment. After hand-editing in Jalview42, this 75-sequence data set contained 208 positions. As stated earlier, ME, ML and BI were used to analyse the final alignment.
Calcium imaging
HEK 293 cells, a gift from H. Yagisawa (Graduate School of Life Science, University of Hyogo), were grown at 5% CO2 and 100% relative humidity in DMEM medium supplemented with 10% (v/v) fetal bovine serum and 0.1 mM non-essential amino acids. HEK 293 cells do not express endogenous CNG channels43. Each Ciona CNG construct was transfected into HEK 293 cells using Targefect F-1 reagent (Targeting Systems, Santee, CA, USA) according to the manufacturer’s instructions, and the cells were incubated for 48 h.
To quantitatively visualize cyclic nucleotide-induced intracellular calcium ions, transfected HEK 293 cells were incubated for 30 min in a solution containing 140 mM NaCl, 5.4 mM KCl, 5 mM CaCl2, 5.6 mM glucose, 10 mM HEPES (pH 7.4) and 2 µM Fura-2 AM (Dojindo Laboratories, Kumamoto, Japan). After washing with PBS, cells were placed in a solution containing 140 mM NaCl, 5.4 mM KCl, 5 mM CaCl2, 5.6 mM glucose, and 10 mM HEPES (pH 7.4). 8-Br-cAMP (Sigma B7880) or 8-Br-cGMP (Sigma B1381) at various concentrations was administrated to the cells in a perfusion system. Fluorescence signals were monitored using an Argus-50/CA system (Hamamatsu photonics, Shizuoka, Japan). Each experiment was performed in at least two biological replicates. Authentication and contamination tests were not performed; however, this should not affect the reliability of the results as the untransfected negative control did not show any response to cyclic nucleotides.
Extended Data
Extended Data Figure 1.
Lineage information for Six1/2 expression in Ciona intestinalis from the gastrula to initial tailbud stage. a, Schematic of the anterior neural plate border at the mid-gastrula stage, including cell lineage nomenclature. Dmrt is initially activated in six blastomeres of 64-cell embryos (a7.9, a7.10 and a7.13). This lineage produces only the anterior neural plate (and the adjacent anterior neural plate border), which forms the PPE. The anterior-most ZicL+ cells of row IV (yellow) mark the boundary of the neural plate, which gives rise to the anterior sensory vesicle in tailbud embryos. The dotted line indicates the oral opening. b, Schematic of the anterior neural plate border during the gastrula–neurula transition, indicating the lineage-specific expression of Six1/2 (magenta). Six1/2 is initially expressed in eight cells comprising the posterior cells (row V) and the posterior lateral cells (row VI). c, Dorsal view of an initial tailbud embryo co-electroporated with Six1/2. mCherry, Dmrt. GFP and ZicL. CFP. At this stage, the cells initially expressing Six1/2 have divided once, giving rise to 16 cells in total. Brackets indicate the derivatives of the annotated lineages shown in b. Anterior is to the left.
Extended Data Figure 2.
Six1/2+ cell morphogenesis in Ciona intestinalis from the initial tailbud stage to late tailbud I. a–c, Dorsal views of tailbud embryos electroporated with Six1/2. mCherry (magenta) and counterstained with phalloidin (blue). a, At the initial tailbud stage I (according to Hotta et al.44), the Six1/2+ cells are arranged in a U shape, anterior to the neural plate. a9–c9, Underlined cell lineages are derived from the left side of the embryo. a9, Schematic indicates that the U shape is composed of 16 cells in total at the 11th generation (that is, a11.154, a11.138, etc.). There are no further divisions of these cells until after the late tailbud I stage. The green cells indicate a11.205, which are fated to become PPE-derived GnRH neurons. b, At the initial tailbud stage II, the lateral edges of the Six1/2+ cells begin to intercalate towards the midline. b9, The schematic shows a dotted circle where the future opening of the oral siphon forms. c, At the late tailbud I stage, theSix1/2+ cells have completed intercalation. The bright phalloidin signal in the centre of the pattern marks the apically localized actin of cells constricted towards the oral opening. The arrowheads indicate a PPE-derived cell fated to become a GnRH neuron. At this stage, the Six1/2+ cells are positioned on top of the anterior sensory vesicle over the ocellus. c9, The schematic shows a dotted circle where the oral opening forms. More anterior cells have undergone local cellular rearrangements. d–d0, Anterior lateral view of late tailbud I stage embryo electroporated with Six1/2. GFP and CNGA. mCherry. Asterisks indicate the ocellus. The arrowheads indicate a PPE-derived cell fated to become a GnRH neuron. d, Shows the Six1/2. GFP channel. d9, Shows the CNGA. mCherry channel (see Fig. 3c for larval expression). d0, Shows the merged image of Six1/2. GFP and CNGA. mCherry.
Extended Data Figure 3.
Dorsal–ventral BMP patterning during PPE specification in Ciona intestinalis. a, Schematic of the anterior neural plate border during the gastrula–neurula transition. Cell lineage nomenclature is used. Chordin and Six1/2 are co-expressed in the lateral posterior derivatives of rows V and VI (dark blue; also see Fig. 1b). b, Ventral view of mid-gastrula stage embryo hybridized with a BMP2/4 mRNA probe and merged with the nuclear counterstain 49,6-diamidino-2-phenylindole (DAPI). c, Lateral view of an embryo during the gastrula–neurula transition hybridized with a BMP2/4 mRNA probe. Anterior is to the left. d, Dorsal view of an embryo during the gastrula–neurula transition hybridized with a Chordin mRNA probe and merged with a DAPI nuclear counterstain.
Extended Data Figure 4.
Endogenous expression of newly described reporter genes in hatched Ciona larvae. a–d, Bright field anterior lateral views. a, Animal hybridized with a GnRH2 mRNA probe. White arrowhead indicates the position of the oral opening. b, Animal hybridized with an RXFP3 mRNA probe. c, Animal hybridized with a SOG/Chemokine-like mRNA probe. The heavily stained tunic was manually removed to reveal the expression signal. d, Animal hybridized with a CNGA mRNA probe. Red arrows mark areas of comparable expression throughout the panels in presumed PPE-derived neurons. An adjacent signal in the ocellus-associated photoreceptors makes it difficult to discriminate expression in PPE-derived neurons in panel d.
Extended Data Figure 5.
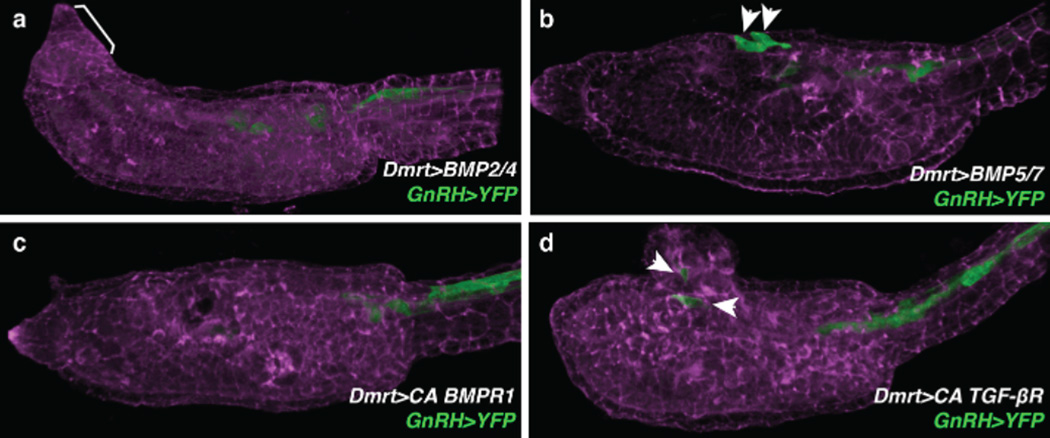
GnRH expression requires BMP attenuation. a–d, Lateral view of a larva electroporated with GnRH. YFP and counterstained with phalloidin (violet). a, Larva co-electroporated with Dmrt. BMP2/4. Of the 200 larvae, 196 had no GnRH. YFP expression in aATENs and displayed mild to severe morphogenetic defects. Bracket shows mild anterior morphological defect. b, Larva co-electroporated with Dmrt. BMP5/7. Of the 200 larvae, 124 had GnRH. YFP expression in aATENs. c, Larva co-electroporated with Dmrt. BMPR1CA. Of the 200 larvae, 197 larvae had no GnRH. YFP expression in aATENs. d, Larva co-electroporated with Dmrt. TGF-βRCA. Of the 200 larvae, 130 had GnRH. YFP expression in aATENs and most displayed severe anterior neural tube defects. Arrowheads in b and d indicate the position of GnRH expression in aATENs. Anterior is to the left in all images.
Extended Data Figure 6.
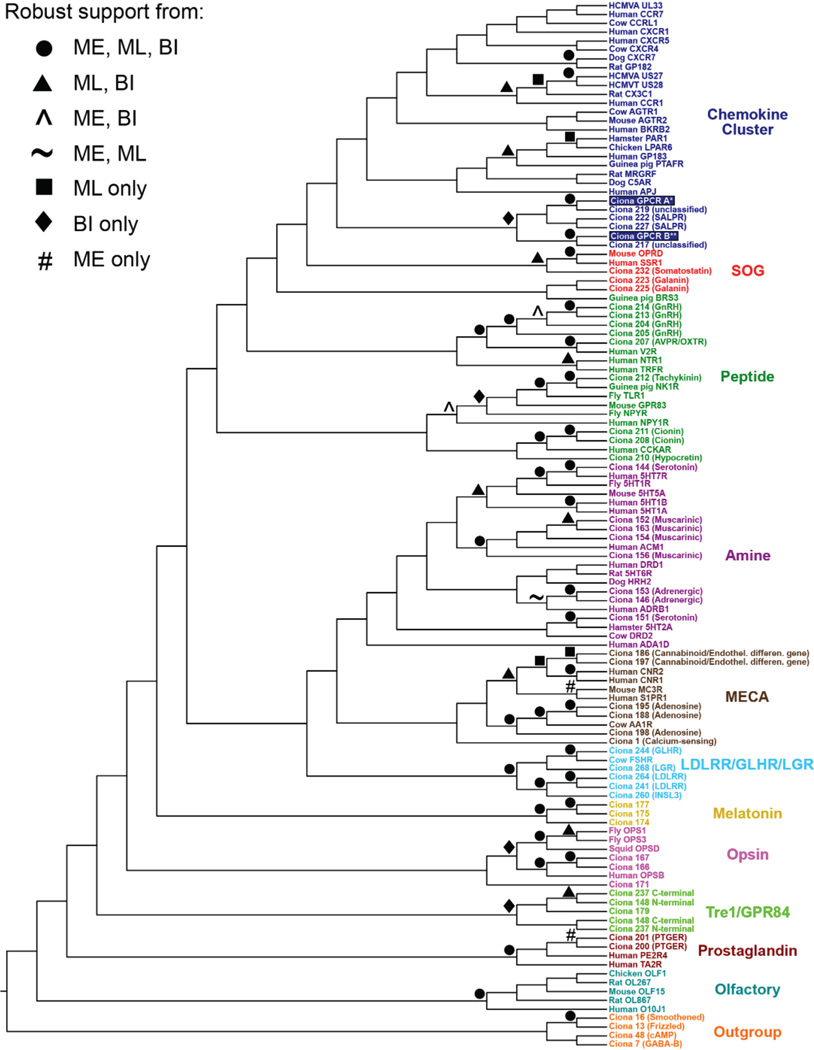
Initial phylogenetic analysis of two GPCRs expressed in the PPE-derived neurons of Ciona intestinalis. This broad survey tree, constructed according to maximum likelihood, shows the approximate placement of the two GPCRs of interest (boxed in blue) within the rhodopsin-class G-protein-coupled receptors. The numbered Ciona sequences and their receptor-type identifications in parentheses are from Kamesh et al.37. All non-Ciona sequences were downloaded in bulk from http://pfam.xfam.org; they comprise the Pfam ‘seed’ alignment of seven-transmembrane receptors for the GPCRs. The rhodopsin subclass names are given to the right of each coloured group. The asterisk and double asterisk indicate the genes labelled RXFP3 and SOG/Chemokine-like, respectively, in Fig. 2e.We judge robust nodal support as bootstrap percentages. 70 for minimum evolution (ME) and maximum likelihood (ML) and posterior probability percentages. 95 for Bayesian inference (BI).
Extended Data Figure 7.
Refined maximum likelihood phylogeny of two GPCRs expressed in the PPE-derived neurons of Ciona intestinalis. Relative to the initial survey tree (see Extended Data Fig. 6), analysis of this focused set of ‘chemokine cluster’ sequences further clarifies the phylogenetic affinities of two Ciona GPCRs expressed in the aATEN–GnRH neurons. The latter sequences are highlighted with violet boxes; all other Ciona sequences are from Kamesh et al.37. The three nodal support values are (in order): ME bootstrap percentage, ML bootstrap percentage and BI posterior probability (only values. 50 are shown). Branch lengths are proportional to molecular change (amino acid substitutions per site) between nodes; see scale bar for measurement.
Extended Data Figure 8.
Elevation of intracellular Ca2+ concentration in cells expressing Ciona CNG channels in response to cyclic nucleotides. a, Intracellular Ca2+ was visualized by Fura-2 ratiometric calcium imaging. Ca2+ influx into HEK 293 cells transfected with CNGA and CNGC was induced by 10 mM 8-Br-cGMP and 1 mM 8-Br-cAMP. In contrast, no Ca2+ influx was observed when HEK 293 cells transfected with CNGB were treated with either 10 mM 8-Br-cGMP or 1 mM 8-Br-cAMP. Coloured numbers in the ‘Before’ panels indicate cells that were subjected to a quantitative measurement of fluorescence. b, Dose-dependent response of HEK 293 cells transfected with CNGA to 8-Br-cAMP. Data were obtained from at least seven different cells in each of 16 different transfections. c, Dose-dependent response of HEK 293 cells transfected with CNGC to 8-Br-cAMP. Data were obtained from at least seven different cells in each of seven different transfections.
Supplementary Material
Acknowledgments
We thank Y. Miyamoto and M. Kotera for technical assistance and A. Stolfi for cloning Chordin. GFP. This work was supported by a grant from the National Institutes of Health (NS076542) and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (25650118, 25290067) and from the Japan Space Forum (h160179). Portions of this study were facilitated by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology in Japan. The work of C.H. in the laboratory of H. Yasuo was funded by the Agence Nationale de la Recherche (ANR-09-BLAN-0013-01). P.B.A. and A.N.K. were supported by predoctoral fellowships from the National Science Foundation and California Institute for Regenerative Medicine, respectively.
The coding sequence of SOG/Chemokine-like has been deposited in GenBank/EMBL/DDBJ under the accession KR902348.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions P.B.A. designed and performed most experiments in consultation with M.L.T.B.G. performed the Six1/2 and Eyain situ hybridizations. A.N.K. performed the larval colorimetric in situ hybridizations. C.J.W. performed the phylogenetic analysis. C.H. performed the BMP2/ 4 and Chordin in situ hybridizations. T.G.K. identified and cloned Ciona CNGs and GnRHs and analysed their expression. T.G.K., M.N., K.K. and M.T. performed functional analysis of CNGs.
The authors declare no competing financial interests.
References
- 1.Gaskell WH. The Origin of Vertebrates. Longmans, Green & Co; 1908. [Google Scholar]
- 2.Steventon B, Mayor R, Streit A. Neural crest and placode interaction during the development of the cranial sensory system. Dev. Biol. 2014;389:28–38. doi: 10.1016/j.ydbio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 4.Abitua PB, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492:104–107. doi: 10.1038/nature11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 6.Shimeld SM, Holland PW. Vertebrate innovations. Proc. Natl Acad. Sci. USA. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patthey C, Schlosser G, Shimeld SM. The evolutionary history of vertebrate cranial placodes –I: cell type evolution. Dev. Biol. 2014;389:82–97. doi: 10.1016/j.ydbio.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Mazet F, et al. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev. Biol. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Christiaen L, Bourrat F, Joly J. A modular cis-regulatory system controls isoform-specific pitx expression in ascidian stomodæum. Dev. Biol. 2005;277:557–566. doi: 10.1016/j.ydbio.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Sutton SW, et al. Distribution of G-protein-coupled receptor (GPCR) 135 binding sites and receptor mRNA in the rat brain suggests a role for relaxin-3 in neuroendocrine and sensory processing. Neuroendocrinology. 2004;80:298–307. doi: 10.1159/000083656. [DOI] [PubMed] [Google Scholar]
- 11.Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 12.Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc. Natl Acad. Sci. USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon H-J, Bhat N, Sweet EM, Cornell RA, Riley BB. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010;6:e1001133. doi: 10.1371/journal.pgen.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruf RG, et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1–SIX1-DNA complexes. Proc. Natl Acad. Sci. USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reichert S, Randall RA, Hill CS. A BMP regulatory network controls ectoderm al cell fate decisions at the neural plate border. Development. 2013;140:4435–4444. doi: 10.1242/dev.098707. [DOI] [PubMed] [Google Scholar]
- 16.Wagner E, Levine M. FGF signaling establishes the anterior border of the Ciona neural tube. Development. 2012;139:2351–2359. doi: 10.1242/dev.078485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev. Biol. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J. Neuroendocrinol. 2010;22:743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusakabe TG, et al. A conserved non-reproductive GnRH system in chordates. PLoS ONE. 2012;7:e41955. doi: 10.1371/journal.pone.0041955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGowan BM, et al. Relaxin-3 stimulates the hypothalamic-pituitary-gonadal axis. Am. J. Physiol. Endocrinol. Metab. 2008;295:E278–E286. doi: 10.1152/ajpendo.00028.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terakado K. Induction of gamete release by gonadotropin-releasing hormone in a protochordate, Ciona intestinalis. Gen. Comp. Endocrinol. 2001;124:277–284. doi: 10.1006/gcen.2001.7728. [DOI] [PubMed] [Google Scholar]
- 22.Konno A, et al. Distribution and structural diversity of cilia in tadpole larvae of the ascidian Ciona intestinalis. Dev. Biol. 2010;337:42–62. doi: 10.1016/j.ydbio.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Reese TS. Olfactory cilia in the frog. J. Cell Biol. 1965;25:209–230. doi: 10.1083/jcb.25.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaupp UB. Olfactory signalling in vertebrates and insects: differences and commonalities. Nature. Rev. Neurosci. 2010;11:188–200. doi: 10.1038/nrn2789. [DOI] [PubMed] [Google Scholar]
- 25.Constantin S, Wray S. Gonadotropin-releasing hormone-1 neuronal activity is independent of cyclic nucleotide-gated channels. Endocrinology. 2008;149:279–290. doi: 10.1210/en.2007-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Majdoubi M, Weiner RI. Localization of olfactory cyclic nucleotide-gated channels in rat gonadotropin-releasing hormone neurons. Endocrinology. 2002;143:2441–2444. doi: 10.1210/endo.143.6.8951. [DOI] [PubMed] [Google Scholar]
- 27.Boehm U, Zou Z, Buck LB. Feedback loops link odor and pheromone signaling with reproduction. Cell. 2005;123:683–695. doi: 10.1016/j.cell.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 28.Yoon H, Enquist LW, Dulac C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell. 2005;123:669–682. doi: 10.1016/j.cell.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 29.Gorbman A. Olfactory origins and evolution of the brain-pituitary endocrine system: facts and speculation. Gen. Comp. Endocrinol. 1995;97:171–178. doi: 10.1006/gcen.1995.1016. [DOI] [PubMed] [Google Scholar]
- 30.Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nature. Rev. Genet. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
References
- 31.Christiaen L, Stolfi A, Levine M. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev. Biol. 2010;340:179–187. doi: 10.1016/j.ydbio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Rehm EJ, Hannibal RL, Chaw RC, Vargas-Vila MA, Patel NH. Fixation and dissection of Parhyale hawaiensis embryos. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5127. 2009, pdb.prot5127. [DOI] [PubMed] [Google Scholar]
- 33.Hudson C, Yasuo H. Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development. 2005;132:1199–1210. doi: 10.1242/dev.01688. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamesh N, Aradhyam GK, Manoj N. The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis. BMC Evol. Biol. 2008;8:129. doi: 10.1186/1471-2148-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 39.Winchell CJ, Jacobs DK. Expression of the Lhx genes apterous and lim1 in an errant polychaete: implications for bilaterian appendage evolution, neural development, and muscle diversification. EvoDevo. 2013;4:4. doi: 10.1186/2041-9139-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isberg V, et al. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 2014;42:D422–D425. doi: 10.1093/nar/gkt1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhallan RS, Yau KW, Schrader KA, Reed RR. Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature. 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]
- 44.Hotta K, et al. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3Dreal-image embryo reconstructions: I From fertilized egg to hatching larva. Dev. Dyn. 2007;236:1790–1805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.