Abstract
Objective
We tested the ability of Notch pathway receptors Notch1 and Notch2 to regulate stem and epithelial cell homoeostasis in mouse and human gastric antral tissue.
Design
Mice were treated with the pan-Notch inhibitor dibenzazepine (DBZ) or inhibitory antibodies targeting Notch1 and/or Notch2. Epithelial proliferation, apoptosis and cellular differentiation were measured by histological and molecular approaches. Organoids were established from mouse and human antral glands; growth and differentiation were measured after treatment with Notch inhibitors.
Results
Notch1 and Notch2 are the predominant Notch receptors expressed in mouse and human antral tissue and organoid cultures. Combined inhibition of Notch1 and Notch2 in adult mice led to decreased epithelial cell proliferation, including reduced proliferation of LGR5 stem cells, and increased apoptosis, similar to the response to global Notch inhibition with DBZ. Less pronounced effects were observed after inhibition of individual receptors. Notch pathway inhibition with DBZ or combined inhibition of Notch1 and Notch2 led to increased differentiation of all gastric antral lineages, with remodelling of cells to express secretory products normally associated with other regions of the GI tract, including intestine. Analysis of mouse and human organoids showed that Notch signalling through Notch1 and Notch2 is intrinsic to the epithelium and required for organoid growth.
Conclusions
Notch signalling is required to maintain gastric antral stem cells. Notch1 and Notch2 are the primary Notch receptors regulating epithelial cell homoeostasis in mouse and human stomach.
INTRODUCTION
The adult gastric epithelium is constantly renewed due to a population of actively cycling stem cells located in the gastric glands. These stem cells generate daughter cells that, upon exiting the stem cell niche, differentiate into the various epithelial cell lineages of the stomach. In the distal, antral stomach, active stem cells express the R-spondin receptor LGR5, which also marks stem cells in the intestine and other tissues.1,2 Antral LGR5 stem cells give rise to all antral lineages, including surface mucous cells, endocrine cells and deep mucous cells. The signalling pathways regulating gastric stem cell proliferation and differentiation are currently poorly understood.
Significance of this study.
What is already known on this subject?
Notch signalling controls mouse gastric epithelial cell homoeostasis.
Mouse antral LGR5 stem cell function is regulated by Notch.
Constitutive Notch activation in mice leads to gastric tumours.
Expression of Notch components is increased in some human gastric cancers.
What are the new findings?
Notch1 and Notch2 are the primary receptors mediating Notch effects in the mouse antrum.
Antral LGR5 stem cells are regulated by Notch1 and Notch2.
Notch inhibition induces antral cell remodelling to express corpus and intestinal markers.
Human gastric antral organoid growth is regulated by Notch1 and Notch2.
How might it impact on clinical practice in the foreseeable future?
Activation of the Notch signalling pathway may contribute to the pathogenesis of human gastric proliferative diseases.
Targeting the Notch signalling pathway to treat human disease might disturb gastric epithelial cell homoeostasis. Thus GI side effects need to be taken into account to evaluate the effectiveness of therapeutic interventions that target Notch.
Notch signalling is well described to maintain intestinal stem cells,3–7 and recent studies suggest that gastric stem cells are similarly regulated by Notch.8,9 In the stomach, pan-Notch inhibition led to reduced gastric stem and epithelial cell proliferation and increased differentiation of mucous and endocrine cell lineages. In contrast, activation of Notch through constitutive expression of the Notch intracellular domain (NICD) induced stem cell proliferation, gland fission and ultimately hyperproliferative polyps.8,9 Furthermore, increased expression of Notch signalling components has been associated with gastric cancer, suggesting Notch pathway involvement.10,11
Four Notch receptors (Notch1–4) exist in vertebrates that are single-pass transmembrane proteins.12 Receptor signalling involves proteolytic receptor cleavage to release the intracellular signalling component NICD, which activates target gene transcription, such as those in the Hes and Hey families.13 Notch1 and Notch2 are the primary receptors involved in intestinal stem cell homoeostasis, with Notch1 having a predominant function.5,7,14,15 Global pharmacological Notch inhibition leads to intestinal toxicity,3 but inhibition of Notch1 alone revealed a partial Notch-inhibition phenotype while avoiding major toxicity.7,14,15
The specific Notch receptors regulating the stomach have not been described. In this study we examined the role of Notch receptors in epithelial and LGR5 stem cell homoeostasis in the gastric antrum of genetic mouse models. We find that Notch1 and Notch2 are key regulators of stem cell proliferation, differentiation and apoptosis. Furthermore our studies demonstrate that Notch1 and Notch2 function to regulate growth of antral organoid cultures generated from human and mouse tissue.
METHODS
Mice
Mice of both sexes aged 2–3 months were used. Lgr5-EGFP-IRES-CreERT2 (Lgr5-green fluorescent protein (GFP)),2 (Jackson Labs #008875), Notch1-CreERT2SAT (N1Cre) and Notch2-CreERT2SAT (N2Cre),16 CBF:H2B-Venus,17 ( Jackson Labs #020942) and ROSA26-CAG-LSL-tdTomato-WPRE (ROSA-Tom),18 ( Jackson Labs #007909) mice were previously described. All mice were on a C57BL/6 background except for CBF:H2B-Venus, which was on a mixed background (CD1 and FVB/N). Mice were housed under specific pathogen-free conditions in automated watered and ventilated cages on a 12-h light/dark cycle. Experimental protocols were approved by the University of Michigan Committee on the Use and Care of Animals. For lineage tracing Notch1-expressing or Notch2-expressing cells, N1Cre; ROSA-Tom and N2Cre; ROSA-Tom mice were treated with either one injection of tamoxifen (1 mg/20 g body weight) followed by a 3-day chase or five daily injections of tamoxifen followed by a 2-week chase.
Notch pathway inhibition
For in vivo Notch inhibition, the γ-secretase inhibitor (GSI) dibenzazepine (DBZ, 30 μmol/kg intraperitoneal, SYNCOM, Groningen, The Netherlands) or vehicle (0.1% Tween-80, 0.5% hydroxypropylmethylcellulose, 0.1% dimethyl sulfoxide (DMSO) in water) was administered to Lgr5-GFP mice once per day for 5 days, with tissue collected the 6th day. Humanised IgG1 neutralising monoclonal antibodies specific for the Notch1-negative or Notch2-negative regulatory region (αN1 or αN2), or an irrelevant control IgG1 antibody interacting with herpes simplex virus gD protein (αGd) were described previously.15 Antibodies were injected intraperitoneally at 5 mg/kg on day 1 and day 4, with collection of stomach tissue on day 6, day 14 or day 28.
For in vitro treatment of mouse and human organoids, the GSI N-(N-(3,5-Difluorophenacetyl-L-alanyl))-(S)-phenylglycine t-butyl ester (DAPT,1 μM; EMD4Biosciences, Gibbstown, New Jersey, USA), αGd, αN1 or αN2 (10 μg/mL) were added to culture media and renewed every other day for 5 days in established organoid lines.
Tissue collection and histological analysis
Mice were fasted overnight with free access to water before tissue collection. For some experiments, mice were injected with 5-ethynyl-2′-deoxyuridine (EdU, 25 mg/kg, Invitrogen, Grand Island, New York, USA) 1.5 h prior to tissue collection. Stomachs were processed for cryo and paraffin histology as described.9 For analysis of proliferation, EdU incorporation was visualised with the Click-iT EdU Alexa Fluor 488 Imaging Kit (Life Technologies, Carlsbad, California, USA). Tissue sections were incubated with primary antibodies (see online supplementary table S1) followed by appropriate secondary antibodies (1:400, Invitrogen) and mounted with ProLong Gold containing 4,6-diamidino-2-phenylindole dihydrochloride (4′,6-diamidino-2-phenylindole dihydrochloride (DAPI); Invitrogen) as described.19 To analyse proliferating LGR5 stem cells, sections were immunostained for GFP (chicken) and Ki67 as described.9 Antral glands were isolated and immunostained as previously described.9 Imaging by digital microscopy was done using a Leica SP5X Inverted 2-Photon FLIM Confocal Microscope or a Nikon E-800 Microscope.
For ultrastructural analysis, tissue was fixed and Epon embedded as described.6 Electron micrographs were captured using a JEOL JEM 1400 plus electron microscope.
Gene expression analysis
RNA was isolated from antral tissue or gastric organoids as described.9 RNA was isolated from human antral tissue using Trizol (Invitrogen), followed by DNase treatment and purification using the RNeasy Mini Kit (Qiagen). cDNA was prepared from 500 ng total RNA and quantitative RT-PCR was performed as described,20 using primers listed in online supplementary table S2, normalised to Gapdh (mouse) or ACTB (human).
Gastric organoid culture
Mouse gastric organoid culture was carried out as described.9,21 In brief, antral glands were resuspended in Matrigel (Corning, Tewksbury, Massachusetts, USA) and overlayed with culture media (50% L cell line expressing Wnt3a, R-spondin3, Noggin (L-WRN) conditioned media, Advanced Dulbecco’s modified eagle’s medium (DMEM)/F12, 10% fetal bovine serum (FBS), 1X Pen-Strep, 2 mM L-Glutamine), with the addition of Y-27632 (10 μM, Tocris, Bristol, UK) only upon initial plating. L-WRN conditioned media containing Wnt3a, R-spondin3 and Noggin was generated as described.21 Human antral tissue (surgical resection or Gift of Life) was obtained under Institutional Review Board approved protocols and organoids were established as described,22 with modifications. Tissue was incubated with 10 mM dithiothreitol (Invitrogen) in Dulbecco’s phosphate-buffered saline (DPBS) with Pen-Strep and Gentamycin for 15 min at room temperature, followed by incubation in 12 mM EDTA in DPBS with Pen-Strep and Gentamycin for 1 h at 4°C on a rocking platform, vigorously shaken for 1 min to release glands and pelleted at 100 g for 5 min. Glands were resuspended in Matrigel and after 30 min at 37°C, the culture media described above with the addition of Y-27632 and SB431542 (10 μM, Tocris Bioscience) was added to each well. Media was renewed every other day. Studies were performed in three independent established organoid lines that had been passaged at least three times before analysis.
Morphometrics
Morphometric analysis used ImageJ software (1.46r, Wayne Rasband, National Institutes of Health (NIH), USA). For EdU incorporation, cleaved caspase-3 cells and gastrin cells, the entire length of the antrum for each animal was imaged (n=4–6 animals per group) and cell counts were normalised to epithelial length (μm). For LGR5 stem cell proliferation, the number of GFP/Ki67 double-positive cells per gastric antral gland was counted in αGd control (n=833 glands), αN1 (n=674 glands), αN2 (n=662 glands) and αN1+αN2 (n=548 glands) (n=5–7 mice/group). For organoid measurements, the area of at least 300 organoids per treatment was measured and data are presented as fold-change compared with control (vehicle or αGd) organoids.
Statistics
GraphPad Prism software was used for statistical analysis. Quantitative data are presented as mean±SEM and analysed using Student’s t test (comparing DBZ/DAPT to vehicle) or one-way analysis of variance (ANOVA) with Dunnett’s or Tukey’s post hoc test (comparing αN1, αN2, αN1+αN2 and αGd); p<0.05 was considered statistically significant (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). qRT-PCR data are expressed as mRNA fold-change versus control (vehicle or αGd).
RESULTS
Notch signalling in antral stem cells
We used the CBF:H2B-Venus reporter mouse to identify cells undergoing Notch signalling in the gastric antrum. This transgenic mouse expresses a nuclear fluorescent protein in response to Notch receptor activation.17 Isolated antral glands showed scattered Venus-positive cells that were primarily located in the gland base (figure 1A). Costaining showed Venus expression in differentiated cell types, including chromogranin A-expressing enteroendocrine cells (figure 1B) and doublecortin like kinase 1-expressing tuft cells (figure 1C). Crossing the Notch reporter strain to Lgr5-GFP mice showed that some of the nuclear Venus-positive cells were also marked with cytoplasmic GFP, indicating Notch signalling in antral LGR5 stem cells (figure 1D).
Figure 1.
Notch signalling in gastric antral cells. (A–D) Antral glands isolated from CBF:H2B-Venus mice (green, nuclear) costained for (B) chromogranin A (CHGA) (red), (C) doublecortin like kinase 1 (DCLK1) (red), or (D) Lgr5-GFP (green, cytoplasmic) and DAPI (blue). Insets show higher-powered views for each channel. (E) Notch receptor expression was determined by qRT-PCR analysis of total RNA isolated from full-thickness antral tissue or antral glands (mean±SEM; n=3–5 mice). ND, not detected. (F) Confocal imaging of paraffin sections from N1-Cre;Tom and N2-Cre;Tom mice costained for red fluorescent protein (RFP) (red), E-cadherin (green) and DAPI (blue) 3 days or 2 weeks after tamoxifen activation of Cre recombinase. Arrows and arrowheads indicate epithelial and non-epithelial Tomato expression, respectively. (G) Frozen tissue sections from CBF: H2B-Venus mice costained for the endothelial marker Platelet/Endothelial Cell Adhesion Molecule 1 (PECAM-1) (left, red) or the smooth muscle cell marker α-smooth muscle actin (SMA) (right, red). Costained cells in boxed regions are marked (arrowheads) in higher-powered insets. Scale bar: 50 μm.
Gene expression analysis for the four Notch receptors showed that Notch1 and Notch2 were the primary Notch receptors expressed in full thickness antral tissue and isolated glands (figure 1E, see online supplementary figure S1). To test if Notch receptors are present in antral stem cells, we performed a lineage tracing experiment with mice expressing Notch receptor-CreERT2 genes crossed to ROSA-Tom mice. We examined N1Cre;ROSA-Tom and N2Cre;ROSA-Tom mice 3 days and 2 weeks post tamoxifen treatment. Tomato-marked epithelial cells were evident in the antral gland base of N1Cre,ROSA-Tom and N2Cre;ROSA-Tom mice at 3 days post induction (figure 1F, arrows). At 2 weeks post tamoxifen epithelial lineage stripes were detected in the antrum of N1Cre;ROSA-Tom mice (arrows), suggesting that Notch1 is expressed in antral stem cells. The lack of lineage stripes in N2Cre;ROSA-Tom mice suggests that this receptor is expressed in short-lived cells.
In addition to epithelial cell labelling there was extensive non-epithelial Notch lineage marking with N1Cre;ROSA-Tom and N2Cre;ROSA-Tom (figure 1F, arrowheads). Histological analysis of tissue sections from the CBF:H2B-Venus reporter mouse demonstrated that the majority of Venus-positive stromal cells are Platelet/Endothelial Cell Adhesion Molecule 1 (PECAM-1)-expressing endothelial cells and α-smooth muscle actin-expressing smooth muscle cells (figure 1G).
Notch regulation of LGR5 stem cells
To test the function of Notch1 and Notch2 to regulate antral epithelial cell homoeostasis we first examined proliferation after treatment with either inhibitory antibodies that selectively target Notch1 and Notch2 or the GSI DBZ (a pan-Notch inhibitor). Analysis of EdU incorporation showed that proliferation was reduced with combined αN1+αN2 treatment to a similar extent as DBZ (figure 2A–H). Treatment with αN1 or αN2 alone significantly reduced the number of proliferating cells, but to a lesser extent (figure 2D–H). Analysis at 2 weeks and 4 weeks after αN1 or αN2 treatment demonstrated normal proliferation after the antibody treatment was discontinued (figure 2H). However, morbidity prevented analysis of the αN1+αN2 group at these later time points.
Figure 2.
Notch1 and Notch2 regulate antral cell proliferation. (A, B and D–G) Proliferating cells were detected in (A) vehicle, (B) dibenzazepine (DBZ), (D) Gd antibody control (αGd), (E) αN1+αN2, (F) αN1 and (G) αN2 tissues by staining for EdU (green) with DAPI (red). (C and H) Morphometric quantification of EdU cells (mean±SEM; n=5–7 mice). (I and J) Proliferating LGR5 stem cells were visualised in treated Lgr5-GFP mice by GFP (green) and Ki67 (red) costaining (arrow) with DAPI (blue) and quantified by morphometrics (mean±SEM; n=5–7 mice; 40–100 glands/mouse). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 versus vehicle using Students’t test or versus αGd using one-way ANOVA. Scale bar: 50 μm (A, B and D–G) or 25 μm (I).
To directly investigate LGR5 stem cells, we treated Lgr5-GFP mice with αN1 and/or αN2 and analysed stem cell proliferation via coimmunostaining for GFP and Ki67 (figure 2I, arrow). Notch1 and/or Notch2 inhibition reduced LGR5 stem cell proliferation, with αN1+αN2 having the most substantial effect (figure 2J), which was similar to our previous analysis of pan-Notch inhibition with DBZ.9 The similarity of combined N1 and N2 receptor blockade to pan-Notch inhibition suggests that Notch1 and Notch2 are the key receptors mediating Notch effects on stem cell proliferation.
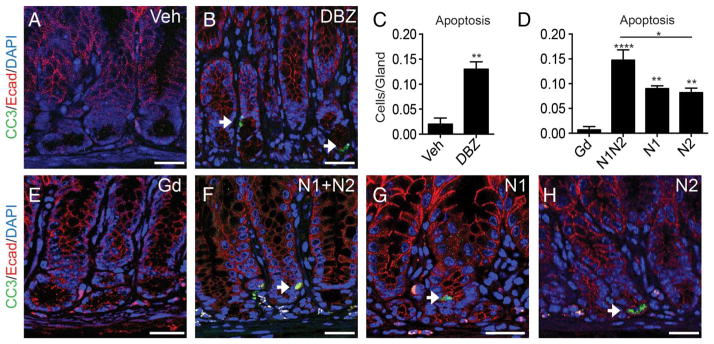
We next analysed cell death by staining for the apoptotic marker cleaved caspase 3. Apoptotic cells are normally rare in the antral epithelium. Notch inhibition with DBZ or receptor blockade was observed to increase the number of caspase-positive cells, with apoptotic cells predominantly in the gland base (figure 3). As with proliferation, αN1+αN2 treatment was comparable to DBZ, with more modest effects observed after treatment with antibodies targeting individual receptors (figure 3C, D).
Figure 3.
Enhanced apoptosis after Notch inhibition. (A, B and E–H) Apoptosis was measured by costaining for cleaved caspase-3 (CC3) (green) and E-cadherin (Ecad) (red) with DAPI (blue) in (A) vehicle, (B) dibenzazepine (DBZ), (E) αGd, (F) αN1+αN2, (G) αN1 and (H) αN2-treated mice. (C and D) Morphometric quantification of apoptotic CC3/E-cad costained cells (arrows) (mean±SEM; n=5–7 mice). Scale bar: 20 μm.
Notch regulates antral organoid growth
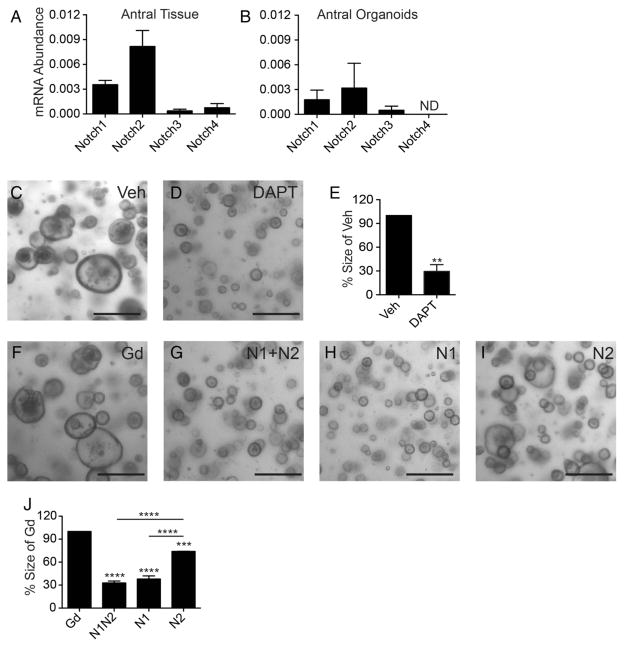
We established organoids to test whether the Notch effects are intrinsic to epithelial cells. Organoids established from the CBF: H2B-Venus reporter mouse showed extensive Venus labelling, demonstrating active Notch signalling in organoid culture (figure 4A). Gene expression analysis showed that, similar to what was observed in gastric glands, Notch1 and Notch2 were the primary Notch receptors expressed in antral organoids (figure 4B). To investigate Notch function we treated organoids with the pan-Notch inhibitor DAPT and observed reduced overall growth (figure 4C–E). Reduced growth was also observed after receptor targeting, with combined N1+N2 inhibition similar to pan-Notch inhibition, and individual receptor targeting showing a larger role for Notch1 to regulate growth (figure 4F–J). These findings demonstrate that intrinsic Notch signalling is required for antral organoid growth.
Figure 4.
Notch1 and Notch2 regulate mouse antral organoid growth. (A) Cells undergoing Notch signalling (green) were observed in antral organoids established from the CBF:H2B-Venus Notch reporter mouse strain. (B) Gene expression of Notch receptors determined by qRT-PCR analysis of antral organoids (mean±SEM n=3 independent organoid lines). ND, not detected. Morphology (C, D and F–I) and quantitative measure of organoid size (E and J) following 5 days of treatment with (C) vehicle, (D) N-(N-(3,5-Difluorophenacetyl-L-alanyl))-(S)-phenylglycine t-butyl ester (DAPT), (F) αGd, (G) αN1+αN2, (H) αN1 or (I) αN2 (mean±SEM; n=3 lines). Scale bar: 50 μm (A) or 250 μm (C, D and F–I).
Notch1 and Notch2 regulate antral epithelial cell differentiation
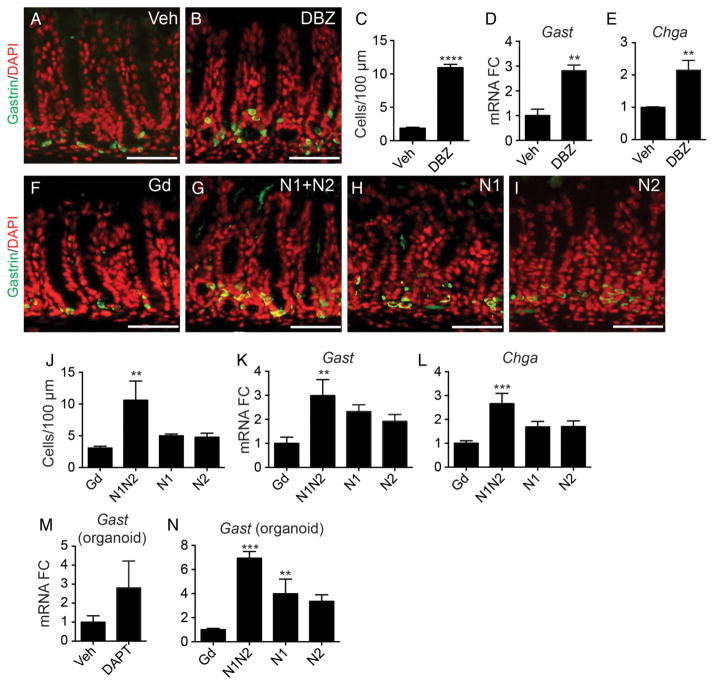
We previously showed that Notch inhibition was associated with increased antral cell differentiation.9 Thus, we analysed the role of Notch1 and Notch2 for the major antral cell lineages: enteroendocrine, surface mucous and deep mucous cells. Analysis of enteroendocrine cells showed increased numbers of gastrin-expressing cells and increased abundance of Gast and Chga mRNAs with Notch inhibition (figure 5). Similar effects were observed after treatment with DBZ (figure 5A–E) and αN1 +αN2, but not with αN1 or αN2 treatment, suggesting that these two receptors are fully redundant for this function (figure 5F–L). In accordance with the in vivo findings, increased Gast mRNA was observed in organoids after DAPT or αN1+αN2 treatment (figure 5M, N).
Figure 5.
Notch1 and Notch2 regulate antral endocrine cell differentiation. (A, B and F–I) Gastrin immunostaining (green) of paraffin sections from mice treated with (A) vehicle, (B) dibenzazepine (DBZ), (F) αGd, (G) αN1+αN2, (H) αN1, and (I) αN2 with DAPI (red). (C and J) Gastrin cells were quantitated by morphometrics (mean±SEM, n=4–5). Gene expression analysis of gastrin (Gast) and chromogranin A (Chga) in (D, E, K and L) full-thickness tissue or (M and N) organoids (mean±SEM; n=5–7 mice or 3 independent organoid lines). Scale bar: 50 μm.
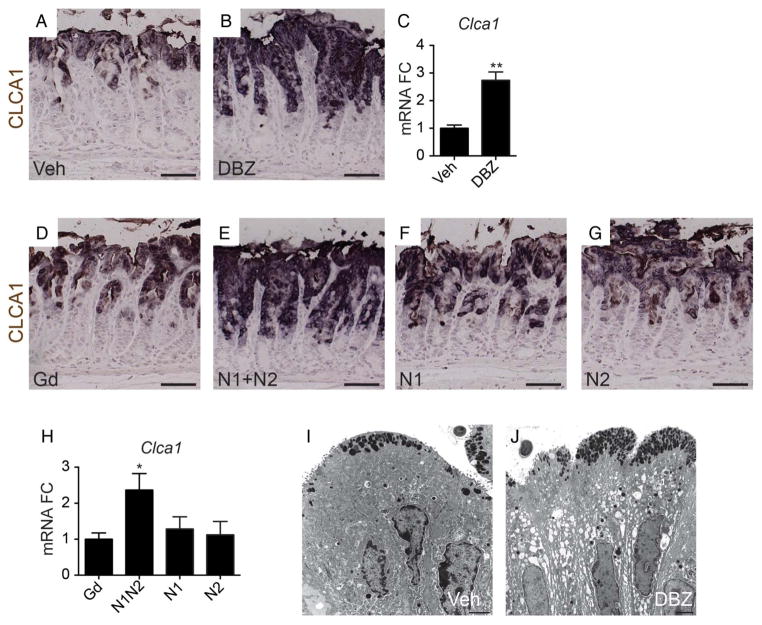
Similarly, analysis of the surface mucous cell marker CLCA1 showed expansion of this cell population after DBZ (figure 6A–C) or combined αN1+αN2 treatment, but not after individual receptor blockade (figure 6D–H). Consistent with the marker expression data, ultrastructural analysis showed a marked expansion of surface mucous cells with increased numbers of granules in these cells after DBZ treatment (figure 6I, J; see online supplementary figure S2).
Figure 6.
Notch1 and Notch2 regulate surface mucous cell differentiation. (A, B and D–G) Immunostaining for the surface mucous cell marker CLCA1 in (A) vehicle, (B) dibenzazepine (DBZ), (D) αGd, (E) αN1+αN2, (F) αN1 and (G) αN2-treated mice. (C and H) qRT-PCR analysis of Clca1 expression (mean±SEM; n=5–7 mice). (I and J) Transmission electron microscope images of surface mucous cells from (I) vehicle or ( J) DBZ-treated mice. Scale bar: 50 μm (A, B and D–G) or 2 μm (I and J).
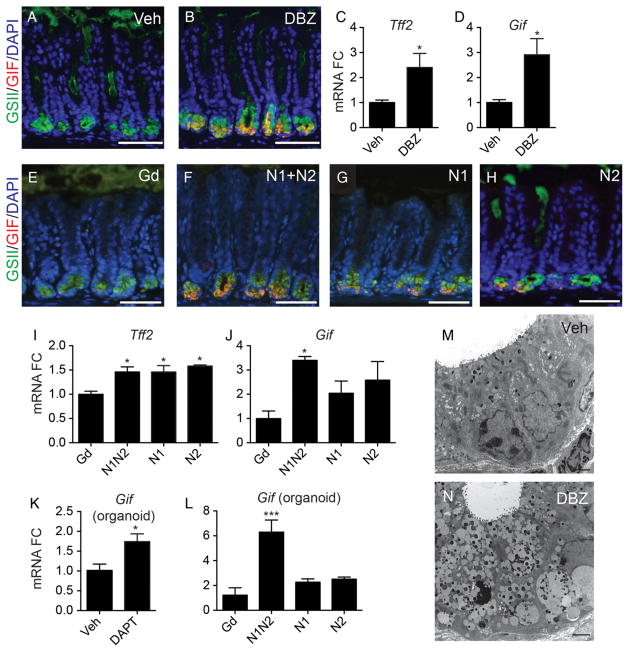
Analysis of H&E stained tissues suggested increased mucous cells at the gland base after Notch inhibition (see online supplementary figure S3). Accordingly, griffonia simplicifolia lectin II (GSII)-lectin staining showed an apparent expansion of this cell lineage after DBZ or αN1+αN2 treatment (figure 7A, B, E–H), which was confirmed by increased trefoil factor 2 (Tff2) mRNA abundance (figure 7C, I). Surprisingly, further analysis of these cells also showed increased staining for the chief cell marker gastric intrinsic factor (GIF), which was confirmed by increased Gif mRNA (figure 7D, J). High-powered confocal analysis showed GSII and GIF colabelled cells at the gland base, primarily localised in separate granules (see online supplementary figure S4). Ultrastructural analysis showed a marked increase in secretory granule number and granule size in cells at the gland base after Notch inhibition (figure 7M, N and see online supplementary figure S2). Consistent with the in vivo data, increased Gif expression was also observed in DAPT or αN1+αN2 treated antral organoids (figure 7K, L).
Figure 7.
Notch inhibition remodels cells at the antral gland base. (A, B and E–H) Costaining with GSII lectin (green) and an anti-gastric intrinsic factor (anti-GIF) antibody (red) with DAPI (blue) on (A) vehicle, (B) dibenzazepine (DBZ), (E) αGd, (F) αN1+αN2, (G) αN1 and (H) αN2-treated mice. (C, D and I–L) Gene expression analysis of trefoil factor 2 (Tff2) and Gif by qRT-PCR analysis of full-thickness tissue or organoids (mean±SEM; n=4–7 mice). (M and N) Transmission electron microscope images of gland base cells from (M) vehicle or (N) DBZ-treated mice. Scale bar: 50 μm (A, B and E–H) or 2 μm (M and N).
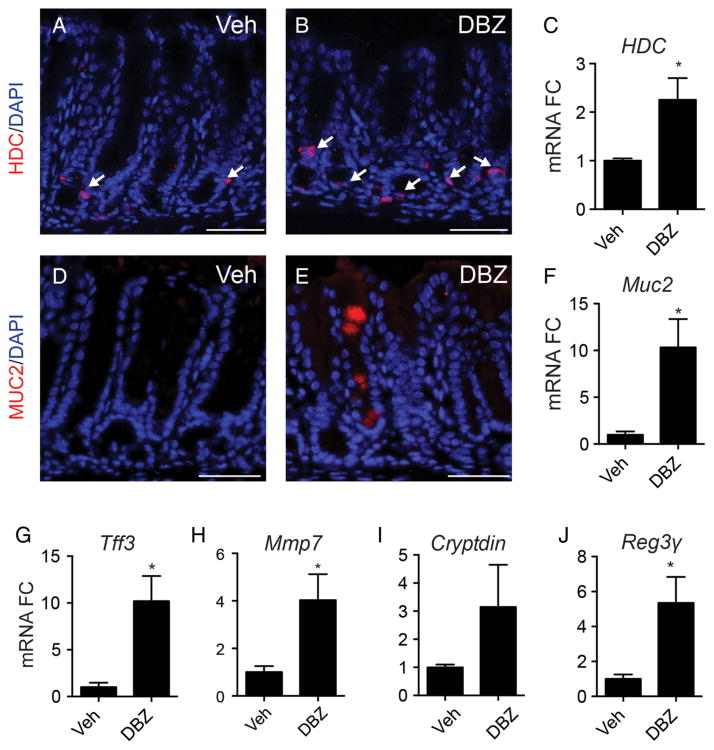
To better understand the cellular remodelling observed in the Notch-inhibited antrum we examined markers associated with the gastric corpus and intestine. Increased immunostaining and gene expression was observed for the corpus enterochromaffin-like (ECL) cell marker histidine decarboxylase (figure 8A–C); however there was no change in expression of the corpus parietal cell marker H,K-ATPase (data not shown). We also observed increased expression of markers of intestinal cells, including goblet cells (Muc2 and Tff3) and Paneth cells (Mmp7, Cryptdin, Reg3γ) in the DBZ-treated antrum (figure 8D–J). Immunostaining revealed scattered Muc2-positive cells in the distal antrum that were localised to the mid-region of the gland. Similar to the corpus markers, the increase in intestinal markers was not associated with a general intestinalisation as the DBZ-treated antrum did not increase expression of the intestinal markers Cdx2, Vil1, Cck or Lyz1 (data not shown) and continued to express gastric specific markers (Tff2 and Gif).
Figure 8.
Increased expression of gastric corpus and intestinal cell markers in dibenzazepine (DBZ)-treated antral tissue. (A–C) Immunostaining and gene expression analysis of histidine decarboxylase (HDC). (D–F) Immunostaining and gene expression analysis of Muc2. (G–J) Gene expression analysis of trefoil factor 3 (Tff3), matrix metalloproteinase-7 (Mmp7), cryptdin, and regenerating islet-derived 3 γ (Reg3γ) in antral RNA isolated from vehicle or DBZ-treated mice (mean±SEM; n=3–6 mice).
Together these findings suggest that global Notch inhibition with DBZ or combined αN1+αN2 treatment results in a generalised increase in cellular differentiation, with some cellular remodelling to express markers of other regions of the GI tract. Dual receptor blockade is required for the differentiation effect, suggesting that Notch1 and Notch2 function together during differentiation.
Notch is necessary for growth of human gastric organoids
We took advantage of the design of the inhibitory antibodies to target mouse and human Notch1 and Notch2 to test the regulation of human gastric stem cells in vitro in organoids derived from human gastric glands. We first determined which Notch receptors were expressed by qRT-PCR, showing that similar to our findings in mouse, Notch1 and Notch2 were the predominant receptors expressed in human antral tissue and organoids (figure 9A, B and see online supplementary figures S5 and S6, table S3).
Figure 9.
Human antral organoid growth is regulated by Notch1 and Notch2. (A and B) Notch receptor mRNA abundance was measured by qRT-PCR in (A) full-thickness human tissue or (B) antral organoids (mean±SEM, n=3 patients or patient-derived organoid lines). See online supplementary figures S5 and S6, table S3 for detailed information on human tissues. Morphology (C, D and F–I) and quantitative measure of organoid size (E and J) following treatment with (C) vehicle, (D) N-(N-(3,5-Difluorophenacetyl-L-alanyl))-(S)-phenylglycine t-butyl ester (DAPT), (F) αGd, (G) αN1+αN2, (H) αN1 or (I) αN2 (mean±SEM, n=3 independent patient-derived organoid lines). Scale bar: 250 μm. ND, not detected.
We confirmed that intrinsic Notch signalling was required for growth of human organoids, with reduced organoid size observed after addition of DAPT to the culture media (figure 9C–E). Treatment with αN1+αN2, αN1 or αN2 also reduced growth, with a pattern remarkably similar to what we observed in mouse organoids. These findings suggest that Notch1 and Notch2 together regulate human antral organoid growth and that the mouse serves as a valid model to study human gastric epithelial stem cell function.
DISCUSSION
Here we report that Notch signalling regulates gastric antral epithelial cell homoeostasis through the Notch1 and Notch2 receptors. Inhibition of Notch1 and Notch2 signalling mimicked effects on proliferation and differentiation observed with global Notch inhibition with DBZ treatment. Notch signalling in antral LGR5 stem cells was shown by Notch reporter expression and lineage tracing studies. We also showed that Notch signalling is crucial for proliferation of gastric LGR5 stem cells, with Notch1 and Notch2 functioning to promote stem cell proliferation. Our findings suggest that Notch1 is the primary receptor in LGR5 stem cells as we observed full lineage stripes with N1Cre; ROSA-Tom mice but only short-term epithelial cell marking in N2Cre; ROSA-Tom mice. In addition αN1 was more effective than αN2 in inhibiting organoid growth. Further studies will be needed to understand how Notch1 and Notch2 regulate transit amplifying progenitors versus stem cells.
Our finding that Notch signalling stimulates gastric stem and progenitor cell proliferation is consistent with previous reports associating Notch pathway activation with gastric tumorigenesis. Upregulation of Notch pathway components has been reported in human gastric cancer, including NOTCH1, NOTCH2, DLL4 and HES1,10,11,23 with NOTCH1 being associated with diffuse type and JAGGED1 being overexpressed in diffuse and poorly differentiated type cancers.24 Consistent with a potential role for Notch in promoting gastric tumorigenesis, genetic mouse models of Notch overexpression in the stomach induced hyper-plastic polyps.8,9
We further demonstrate that Notch1 and Notch2 regulate epithelial cell differentiation. Inhibition of Notch1 and Notch2 signalling resulted in significant increases in all differentiated lineages, with increases in endocrine, surface mucous and deep mucous cells, similar to what is seen with global Notch inhibition. Our observation that individual inhibition of Notch1 or Notch2 did not broadly change differentiated cell types suggests that these receptors generally have redundant roles in differentiation. Notch regulation of differentiation in the stomach differs from the action of Notch in the intestine, where it has been well established that pathway inhibition induces a cell fate switch leading to secretory cell hyperplasia.3,4,6 Thus, while Notch signalling in the intestine promotes enterocyte differentiation, Notch signalling in the stomach appears to generally repress differentiation of all lineages.9 It is important to note however that in contrast to the intestine, the adult stomach only contains secretory cells.
Interestingly, we observed that Notch inhibition resulted in marked cellular remodelling. Cells at the surface exhibited an increased number of secretory granules to accompany the observed increases in immunostaining and gene expression of surface mucous cell markers. Tissue remodelling in the gland base was characterised by increased density and size of secretory granules and upregulation of intrinsic factor immunostaining. We also observed an increase in expression of selected intestinal and corpus cell markers, although the expression pattern did not suggest conversion of the antrum to these GI tissues. Further studies are needed to understand the role Notch plays in maintaining antral identity.
Cells that coexpress GIF and mucous cell markers, such as TFF2, have been previously described in immature and adult stomach. In mouse, coexpressing cells are observed throughout the immature newborn stomach, including the corpus and antral regions.20 Moreover, in adult human and mouse corpus, a coex-pressing metaplastic cell lineage termed spasmolytic polypeptide-expressing metaplasia (SPEM) has been observed.25 SPEM is induced in response to loss of parietal cells and has been proposed to be a precursor to gastric cancer.26–28 How the remodelled antral cells in the Notch-inhibited antrum relate to these previously described cell types will be an interesting future question.
The development of methods to grow epithelial organoids from primary mouse and human tissue is a powerful approach to study antral stem cells.1,22,29,30 Either complete Notch blockade with GSI, or specific receptor targeting with αN1 or αN2 inhibitory antibodies had similar effects of reducing overall organoid growth, with Notch1 playing a more significant role than Notch2. This finding agrees with our data showing active Notch1 signalling in antral LGR5 stem cells. Furthermore these studies showed that Notch signalling is intrinsic to the gastric epithelium, with signalling required for organoid growth in culture. In addition, we observed that Notch regulates organoid differentiation, with upregulation of zymogenic and endocrine cell marker expression after Notch inhibition with DAPT or combined αN1+αN2 treatment, similar to what we observed in vivo.
Our finding that Notch1 and Notch2 receptor signalling regulates human stomach epithelium has therapeutic implications. Notch pathway dysregulation is associated with the development of several cancers and thus this pathway is a prime target of therapeutic significance.31 The use of pan-Notch inhibitors such as GSIs for Notch-related pathologies has been of clinical relevance for years; however GI toxicity severely limits their use.3 Thus more specific pathway targets are being explored. In particular, the development of humanised inhibitory antibodies against either Notch1 or Notch2 might allow for dose-dependent inhibition of each receptor while avoiding the side effects seen with global Notch inhibition.15 However, our findings predict that targeting Notch1 or Notch2 to treat cancer might be associated with disruption of gastric epithelial cell homoeostasis and thus gastric cellular changes should be considered in patients undergoing targeted Notch inhibition.
Acknowledgments
The authors thank Dr Thaddeus Stappenbeck for L cell line expressing Wnt3a, R-spondin3, Noggin (L-WRN) cells, Dr John Williams and Bradley Nelson for assistance with electron microscopy, Dr Spyros Artavanis-Tsakonas for use of the N1Cre and N2Cre mice and Christopher Altheim for generating L cell line expressing Wnt3a, R-spondin3, Noggin (L-WRN) conditioned media.
Funding GBG was supported by NIH T-32-GM0008322 and NIH T32-DK094775, ESD was supported by NIH F32-DK093349, and ESD and PHD were supported by NIH T32-HD007505. The research was funded by an award from Biogen to AL, by NIH P01-DK06041 and NCI P50-CA130810 project awards to LCS and Core support from the Michigan Gastrointestinal Research Center Grant NIH P30-DK34933 and the University of Michigan Cancer Center Support Grant NCI P30-CA6592.
Footnotes
Competing interests CWS is an employee of Genentech and owns shares of Roche.
Ethics approval Human tissue use was approved by University of Michigan IRBMed Institutional Review Board. Animal experiments were approved by the University of Michigan Committee on the Use and Care of Animals.
Provenance and peer review Not commissioned; externally peer reviewed.
Contributors GBG designed and performed experiments, analysed data and wrote the manuscript. ESD, TMK, AT and NLC performed experiments and analysed data. PHD, JRS, DMS, AL, IS and CWS provided reagents or collected samples. LCS and AL obtained funding, designed experiments and wrote the manuscript. All authors critically reviewed the manuscript.
References
- 1.Barker N, Huch M, Kujala P, et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 3.Van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–63. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 4.Fre S, Huyghe M, Mourikis P, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–8. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 5.Riccio O, van Gijn ME, Bezdek AC, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–83. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.VanDussen KL, Carulli AJ, Keeley TM, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–97. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carulli AJ, Keeley TM, Demitrack ES, et al. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TH, Shivdasani RA. Notch signaling in stomach epithelial stem cell homeostasis. J Exp Med. 2011;208:677–88. doi: 10.1084/jem.20101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demitrack ES, Gifford GB, Keeley TM, et al. Notch signaling regulates gastric antral LGR5 stem cell function. EMBO J. 2015;34:2522–36. doi: 10.15252/embj.201490583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh TS, Wu CN, Hsu KW, et al. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res. 2009;69:5039–48. doi: 10.1158/0008-5472.CAN-08-4021. [DOI] [PubMed] [Google Scholar]
- 11.Sun Y, Gao X, Liu J, et al. Differential Notchl and Notch2 expression and frequent activation of Notch signaling in gastric cancers. Arch Pathol Lab Med. 2011;135:451–8. doi: 10.5858/2009-0665-OA.1. [DOI] [PubMed] [Google Scholar]
- 12.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 14.Tran IT, Sandy AR, Carulli AJ, et al. Blockade of individual notch ligands and receptors controls graft-versus-host disease. J Clin Invest. 2013;123:1590–604. doi: 10.1172/JCI65477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–7. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 16.Fre S, Hannezo E, Sale S, et al. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE. 2011;6:e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowotschin S, Xenopoulos P, Schrode N, et al. A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Dev Biol. 2013;13:15. doi: 10.1186/1471-213X-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Diaz L, Hinkle KL, Jain RN, et al. Parietal cell hyperstimulation and autoimmune gastritis in cholera toxin transgenic mice. Am J Physiol Gastrointest Liver Physiol. 2006;290:G970–9. doi: 10.1152/ajpgi.00461.2005. [DOI] [PubMed] [Google Scholar]
- 20.Keeley TM, Samuelson LC. Cytodifferentiation of the postnatal mouse stomach in normal and Huntingtin-interacting protein 1-related-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1241–51. doi: 10.1152/ajpgi.00239.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–82. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanDussen KL, Marinshaw JM, Shaikh N, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64:911–20. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Leung S, Yuen S, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–15. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du X, Cheng Z, Wang YH, et al. Role of Notch signaling pathway in gastric cancer: a meta-analysis of the literature. World J Gastroenterol. 2014;20:9191–9. doi: 10.3748/wjg.v20.i27.9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt PH, Lee JR, Joshi V, et al. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest. 1999;79:639–46. [PubMed] [Google Scholar]
- 26.Halldórsdóttir AM, Sigurdardóttrir M, Jónasson JG, et al. Spasmolytic polypeptide-expressing metaplasia (SPEM) associated with gastric cancer in Iceland. Dig Dis Sci. 2003;48:431–41. doi: 10.1023/a:1022564027468. [DOI] [PubMed] [Google Scholar]
- 27.Nozaki K, Ogawa M, Williams JA, et al. A molecular signature of gastric metaplasia arising in response to acute parietal cell loss. Gastroenterology. 2008;134:511–22. doi: 10.1053/j.gastro.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weis VG, Sousa JF, LaFleur BJ, et al. Heterogeneity in mouse spasmolytic polypeptide-expressing metaplasia lineages identifies markers of metaplastic progression. Gut. 2013;62:1270–9. doi: 10.1136/gutjnl-2012-302401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartfeld S, Bayram T, van de Wetering M, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–36. e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 31.Takebe N, Miele L, Harris PJ, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445–64. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]