Abstract
G Protein Signaling Modulator-3 (GPSM3) is a leukocyte-specific regulator of G protein-coupled receptors (GPCRs), which binds inactivated Gαi·GDP subunits and precludes their reassociation with Gβγ subunits. GPSM3 deficiency protects mice from inflammatory arthritis and, in humans, GPSM3 single nucleotide polymorphisms (SNPs) are inversely associated with the risk of rheumatoid arthritis development; recently, these polymorphisms were linked to one particular SNP (rs204989) that decreases GPSM3 transcript abundance. However, the precise role of GPSM3 in leukocyte biology is unknown. Here we show that GPSM3 is induced in the human promyelocytic leukemia NB4 cell line following retinoic acid treatment, which differentiates this cell line into a model of neutrophil physiology (NB4*). Reducing GPSM3 expression in NB4* cells, akin to the effect ascribed to the rs204989 C>T transition, disrupts cellular migration toward leukotriene B4 (LTB4) and (to a lesser extent) interleukin-8 (a.k.a. IL-8 or CXCL8), but not migration toward formylated peptides (fMLP). As the chemoattractants LTB4 and CXCL8 are involved in recruitment of neutrophils to the arthritic joint, our results suggest that the arthritis-protective GPSM3 SNP rs204989 may act to decrease neutrophil chemoattractant responsiveness.
Keywords: arthritis, chemotaxis, CXCL8, fMLP, G Protein Signaling Modulator-3, leukotriene B4, neutrophils, SNP rs204989
INTRODUCTION
G Protein Signaling Modulator-3 (GPSM3) is a GoLoco motif protein (1, 2) restricted in expression to mature monocytes, lymphocytes, and neutrophils; recent expression quantitative trait locus (eQTL) analyses of stimulated primary monocytes (3) and peripheral blood CD16+ neutrophils (4) have identified human genomic sequence variations that impinge upon GPSM3 expression, including the single-nucleotide polymorphism (SNP) rs204990 (3) in a region directly upstream of the presumptive GPSM3 transcription start site. Independent, genome-wide association studies of sequence variations associated with autoimmune disease risk have highlighted additional GPSM3 SNPs as being inversely correlated (rs204989, rs204991) to the risk of developing rheumatoid arthritis (RA), ankylosing spondylitis, multiple sclerosis (refs. (5–7)), and systemic lupus erythematosus (SLE) (8). GPSM3 polymorphisms are also associated with the risk of developing type 1 diabetes (rs204991; ref. (5)), atopic dermatitis (rs176095; ref. (9)), and childhood asthma (rs176095; ref. (10)). We recently reported (11) that SNPs rs204989, rs204990, and rs204991 constitute a haplotype block within which the rs204989 C>T transition is causal to reduced GPSM3 promoter activity. The association between a promoter-weakening GPSM3 SNP and reduced risk of RA is supported by the protection afforded to Gpsm3-deficient mice in an acute model of inflammatory arthritis (12). Histologic analysis of synovial tissue from Gpsm3-deficient mice revealed a marked acellularity (12), suggesting that their protection from developing pathologic inflammation relies on a potential cellular trafficking defect.
Neutrophils are leukocytes critical to the activation of inflammation in RA and many other autoimmune-related diseases (reviewed in (13)). Neutrophils elicit this function following their initial recruitment by early-phase proinflammatory chemoattractants. In RA, this early-phase recruitment is mediated primarily by leukotriene B4 (LTB4), followed by a LTB4 positive-feedback loop in which recruited neutrophils secrete LTB4, CXCL8, and additional chemoattractants (reviewed in (14)). To date, work to identify the role of GPSM3 in disease pathophysiology has exclusively focused on circulating monocytes, which compose less than 10% of circulating leukocytes (15). However, mice made deficient in one of several monocyte chemokine receptors (i.e., CCR1, CCR2, and CCR5) do not exhibit significant protection from models of RA inflammation (16); furthermore, mice depleted of Ly6Chigh monocytes have no change in histological inflammation (17). Moreover, the initial class of leukocytes recruited to synovial tissues in human arthritis is thought to be neutrophils, which migrate in response to a sequential cascade of LTB4, IL-1β, and CXCR2 agonists (14). Further support for the primary role of neutrophils in initiating RA inflammation comes from protection afforded to mouse strains that are deficient in various proteins required for the recognition or synthesis of LTB4, including the LTB4 receptor BLT1 (18), 5-lipoxygenase activating protein (19), 5-lipoxygenase (20), and leukotriene A4 hydrolase (20).
The LTB4 receptor BLT1 and the CXCL8 receptors CXCR1 and CXCR2 are GPCRs linked to pertussis toxin-sensitive Gi-subfamily heterotrimers (21–23), and therefore sensitive to potential regulation by GoLoco motif-containing proteins (like GPSM3) that act as Gαi-selective guanine nucleotide dissociation inhibitors (1). The role of GPSM3 in neutrophil function has not previously been assessed. Here, we identify the expression of GPSM3 in an immortalized cell line that can be differentiated towards neutrophil-like function, and further identify a selective role for GPSM3 in its migration toward proinflammatory chemoattractants LTB4 and CXCL8 – a discovery that helps strengthen the association between the promoter-weakening GPSM3 SNP rs204989 and a reduced risk of RA.
RESULTS
GPSM3 expression is induced in ATRA-differentiated NB4* cells
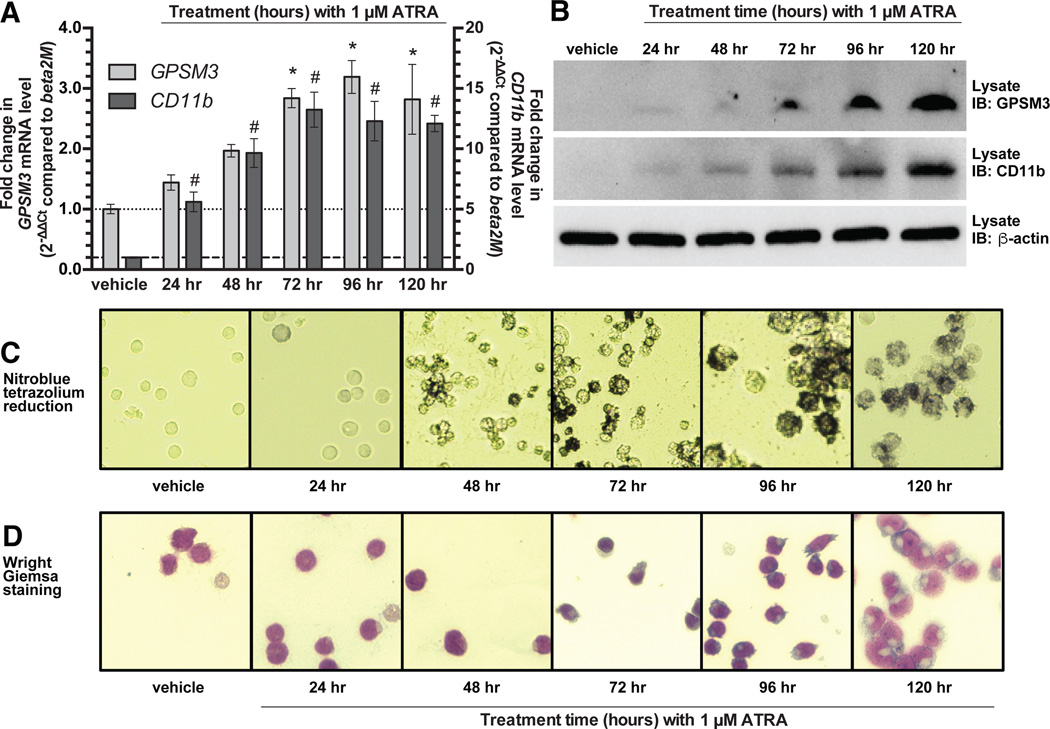
GPSM3 protein expression was undetectable within whole cell lysates of the human acute promyelocytic leukemia NB4 cell line. However, upon treatment of NB4 cell cultures with all-trans retinoic acid (ATRA), expression of GPSM3 transcript (Fig. 1A) and encoded protein (Fig. 1B) increased in a differentiation-dependent manner concurrently with the established differentiation marker CD11b. This ATRA-dependent induction of GPSM3 expression also paralleled the acquisition of other markers of differentiated neutrophil function: specifically, the activity of intracellular oxidoreductase enzymes that can reduce nitroblue tetrazolium to colored formazan dye (Fig. 1C) and morphologic changes (Fig. 1D; e.g., invagination of the nucleus), both of which have been reported previously in ATRA-differentiated NB4* cells (24).
Figure 1. GPSM3 transcript and protein expression are increased during all-trans retinoic acid (ATRA) differentiation of NB4 cells toward a neutrophil-like physiology.
(A) GPSM3 and CD11b mRNA levels in NB4 cells, as measured by qRT-PCR, both increase over treatment time with 1 µM ATRA; relative expression was normalized to cells treated with vehicle (DMSO) for 120 hours. *, significantly different to GPSM3 transcript level in vehicle control (p < 0.05); #, significantly different to CD11b transcript level in vehicle control (p < 0.05). (B) Levels of GPSM3 protein and the differentiation marker CD11b both increase with ATRA-induced differentiation. (C) ATRA-induced differentiation over time increases the neutrophil-specific ability to reduce nitroblue tetrazolium to formazan, as observed by a steady increase in formazan-positive (dark) NB4 cells over time. (D) Wright-Giemsa stained NB4* cells indicating the acquisition over time of ATRA-induced morphological changes mimicking the nuclear invagination seen in neutrophil-like precursor cells.
GPSM3 deficiency does not impair ATRA-induced differentiation of NB4 cells
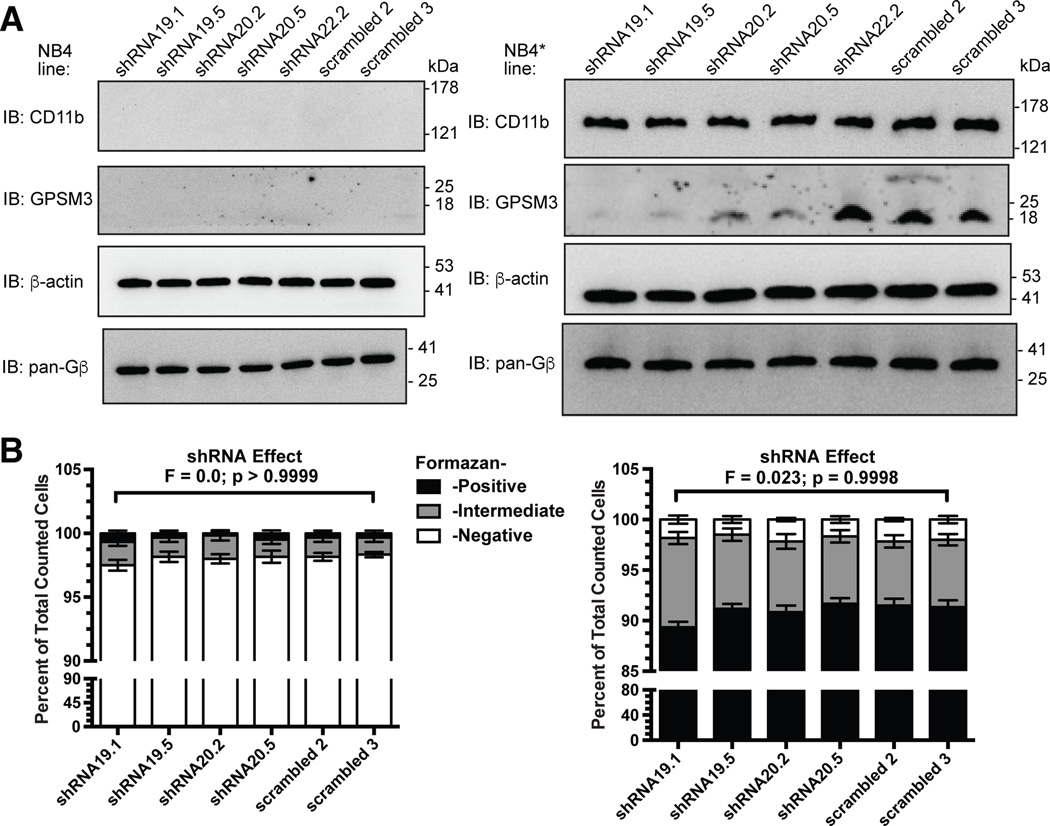
We established stable, monoclonal daughter lines from the parental NB4 cell line using previously validated short hairpin RNA (shRNA)-expressing lentiviral vectors directed against GPSM3 (25). After ATRA treatment, all daughter lines exhibited equivalent upregulation of CD11b and equivalent expression of Gβ subunits (Fig. 2A, right panel); the latter result suggests that the established interaction between GPSM3 and neo-synthesized, free Gβ subunits (25) does not affect steady-state Gβ levels in this cell line. In addition, all daughter lines exhibited equivalent induction of intracellular oxidoreductase activity upon ATRA treatment (Fig. 2B, right panel; Supplemental Table S1).
Figure 2. GPSM3 deficiency does not disrupt markers of NB4* cell differentiation.
(A) Immunoblot revealing undetectable GPSM3 and CD11b immunoreactivity within whole cell lysates from all shRNA-expressing monoclonal cell lines when these lines were mock-differentiated with vehicle only (0.01% DMSO; left panels). Immunoblot revealing decreased GPSM3 immunoreactivity after 120 hours of all-trans retinoic acid (ATRA) treatment of NB4* cells stably expressing GPSM3-targeted shRNAs shRNA19 or shRNA20 (right panels); GPSM3 knockdown was not seen to affect ATRA induction of CD11b levels nor steady-state levels of Gβ subunits. (B) Nitroblue tetrazolium reduction assay indicating that >90% of cells within each NB4-shRNA monoclonal cell line are incapable of reducing nitroblue tetrazolium after mock-differentiation with vehicle only (0.01% DMSO; left bar graph). Following differentiation with all-trans retinoic acid, >87% of differentiated NB4* cells were capable of reducing nitroblue tetrazolium, irrespective of expressed shRNA species (right bar graph).
GPSM3 deficiency impairs migration towards LTB4 and CXCL8, but not fMLP
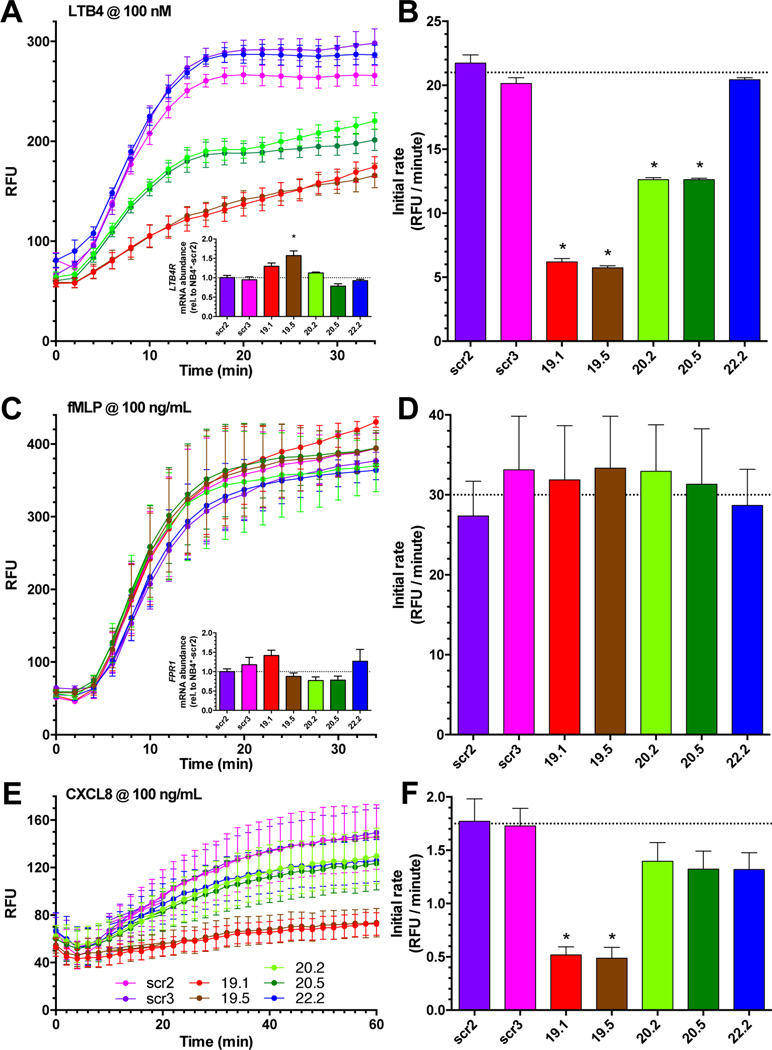
NB4* monoclonal lines with successful knockdown of GPSM3 expression were observed to have negligible spontaneous migration behaviors (Supplemental Fig. S1A) and similar overall migratory responsiveness to 10% FBS placed in the lower chamber of the Transwell apparatus (Fig. S1B). However, the NB4* lines with reduced GPSM3 expression were found to have disrupted migration towards LTB4 and (to a lesser extent) CXCL8 in Transwell migration assays (Fig. 3) when compared both to scrambled shRNA control lines and to a line expressing an ineffective shRNA (i.e., daughter line NB4*-shRNA22.2; Fig. 2A&B). In contrast to the diminished migratory responses towards LTB4 and CXCL8 elicited by GPSM3 knockdown, no significant differences were observed in migration towards 100 ng/mL of formylated peptide (fMLP; Fig. 3C,D). Additional dose-response studies failed to demonstrate any difference in fMLP-induced migration upon GPSM3 knockdown (e.g., Fig. S3 employing from 1 to 250 ng/mL fMLP concentrations). The observed reductions in migratory responses to chemoattractants LTB4 (Fig. 3A&B) and CXCR8 (Fig. 3E&F) were not correlated with any concomitant decrease in mRNA expression of the high-affinity LTB4 receptor 1 (LTB4R1; Fig. 3A inset) or the CXCL8 receptors CXCR1 and CXCR2 (Supplementary Figure S1). For example, compared to the scrambled shRNA control lines, only the NB4*-shRNA19.5 line exhibited a significant difference in LTB4R1 mRNA expression (Fig. 3A inset, Δ = +56.5%, p = 0.0134) and this difference was an increase in expression.
Figure 3. GPSM3 deficiency selectively disrupts NB4* cell migration toward LTB4 and CXCL8.
(A) Real-time Transwell migration towards 100 nM LTB4 over 30 min by indicated NB4* cell lines (after 120 hours ATRA differentiation). Significant differences in migration for all four GPSM3-deficient NB4* lines (19.1, 19.5, 20.2, 20.5; each p < 0.0001) compared to scrambled controls were determined by one-way ANOVA analyses of area under the curve with Bonferroni post-hoc. (A inset) Post-differentiated NB4* transcript abundance of the LTB4 receptor BLT1 (LTB4R); significance (p < 0.05) determined by Kruskal-Wallis test (unequal group variances, Brown-Forsythe p < 0.05) with Dunn’s post-hoc. All data plotted as mean ± S.E.M. (B) Plot of initial migration rates (RFU/min) derived from data in panel A. (C) Real-time Transwell migration towards 100 ng/mL fMLP over 30 minutes by indicated NB4* cell lines (after 120 hours of ATRA differentiation). No significant differences were observed between cell lines (F = 0.4255; p = 0.9826). (C inset) Post-differentiated NB4* transcript abundance of the fMLP receptor (FPR1); significance (p < 0.05) determined by Kruskal-Wallis test (unequal group variances, Brown-Forsythe p < 0.05) with Dunn’s post-hoc. All data plotted as mean ± S.E.M. (D) Plot of initial migration rates (RFU/min) derived from data in panel C. (E) Real-time Transwell migration towards 100 ng/mL CXCL8 (a.k.a. IL-8) over 60 minutes by indicated NB4* cell lines (after 120 hours of ATRA differentiation). Significant differences in migration for two of the GPSM3-deficient NB4* lines, namely 19.1 (F= 47.87; p < 0.0001) and 19.5 (F = 40.97; p < 0.0001), compared to scrambled controls were determined by one-way ANOVA analyses of area under the curve with Bonferroni post-hoc. Post-differentiated NB4* transcript abundance of the CXCL8 receptors CXCR1 and CXCR2 are presented in Supplemental Figure S2. (F) Plot of initial migration rates (RFU/min) derived from data in panel E.
DISCUSSION
Before its mRNA was cloned (26), GPSM3 protein was first detected in neutrophil detergent-resistant fractions by mass spectrometry (27). More recently, RNA-seq datasets indicate that human neutrophils have greater GPSM3 transcript expression than all other types of circulating leukocytes (e.g., NCBI GEO accession number GSE62408 (28)). These findings sparked our interest in determining whether GPSM3 regulates aspect(s) of neutrophil biology, especially in light of the GPSM3 SNP rs204989 being associated with decreased GPSM3 expression (11) and protection against the development of rheumatoid arthritis (5–7) – an autoimmune disease in which neutrophil function is central to its initiation. Using RNA-interference to negate the GPSM3 upregulation found to occur normally in the promyelocytic NB4 cell line upon ATRA differentiation, we have now established that GPSM3 is required for the full migratory response of this neutrophil-like cell line toward a pathophysiologically relevant concentration of LTB4 (29); in addition, GPSM3 also appears to play a role in migratory responsiveness toward CXCL8, although only observed with the highest degree of GPSM3 deficiency (i.e. the NB4*-shRNA19 monoclonal lines in which GPSM3 protein was barely detectable by immunoblotting). Migratory responses to LTB4 and CXCL8 are believed to be dependent on the relative abundance of GPSM3 protein in these cells, given that the NB4*-shRNA20 monoclonal lines 20.2 and 20.5 were seen to have an intermediate level of GPSM3 knockdown efficiency and, in parallel, an intermediate migratory phenotype toward LTB4 and no significant defect in migration toward CXCL8.
Given the primary coupling of LTB4 and CXCL8 receptors to Gαi-containing heterotrimers (21–23) and the inhibitory influence exerted by the GoLoco motifs of GPSM3 on Gαi signaling and heterotrimer recycling (26), reduced GPSM3 levels in NB4* cells likely increases the rate of inactive Gαi subunit reassociation with free Gβγ heterodimers. As signaling by the free Gβγ heterodimer is central to the migratory response (22, 23), any accelerated deactivation by increased Gαi reassociation would lead to a diminished migration phenotype. Given the diminished NB4* cell line migration toward the RA-relevant chemoattractants LTB4 and CXCL8, we surmise that the synovial acellularity seen in anti-collagen antibody challenged GPSM3-deficient mice (12) and the inverse association between RA development and the GPSM3 expression-lowering SNP rs204989 (11) could be the result of reduced neutrophil recruitment by these particular chemoattractants.
During infections, neutrophil responsiveness to bacterial-derived formylated peptides is mediated via formyl peptide receptors (30): i.e., GPCRs coupled to Gαi-containing heterotrimers (31) in a similar fashion to LTB4 and CXCL8 receptors. However, GPSM3 knockdown in NB4* cells does not appear to affect migration initiated through the formyl peptide receptor. We therefore conclude that GPSM3 knockdown does not globally affect the chemotactic machinery of NB4* cells, but instead leads to a selective abrogation of responsiveness to certain chemoattractants, like LTB4 and CXCL8 shown here. Future investigations will be directed at identifying the specific signaling mechanism(s) regulated by GPSM3 that differentially affect LTB4- and CXCL8- vs fMLP-induced migration; one such mechanism might involve direct coupling by GPSM3 of Gαi subunits to select chemokine-responsive GPCRs (i.e., in the absence of Gβγ subunits), as has been speculated on recently by Blumer and colleagues (32). The effect of GPSM3 deficiency on NB4* LTB4-induced migration, coupled with the established role that LTB4 plays in the early pathogenesis of RA, supports the therapeutic potential of GPSM3-targeted inhibitors in the treatment of RA, as we have proposed previously (11); the potential to inhibit GPSM3 pharmacologically and thereby reduce neutrophil migration to the inflammed joint while sparing fMLP responsiveness is attractive as it suggests a means of reducing RA pathology without compromising responsiveness to bacterial infections.
METHODS
Cell lines and culture conditions
The human acute promyelocytic leukemia cell line (NB4) was obtained from the Leibniz Institute German Collection of Microorganisms and Cell Cultures. Preliminary studies were performed on NB4 cells that were a generous gift from Dr. Sherman Weissman. Cells were maintained at 37°C in a humidified, 5% CO2 atmosphere in an ATCC-modification of RPMI medium supplemented with 10% heat-inactivated fetal bovine serum, 100 IU penicillin, 100 µg/ml streptomycin, and 2 µg/ml puromycin. GPSM3 expression was stably knocked-down via lentiviral transduction (WVU IBC protocol 15-04-08) as described previously (11). The shRNA hairpin sequences for shRNA19 (TRC clone ID: TRCN0000036819), shRNA20 (TRCN0000036820), and shRNA22 (TRCN0000036822) are available from The RNAi Consortium (http://www.broadinstitute.org/rnai/public) and listed in Table S2. Additional NB4 cell lines were established expressing a scrambled shRNA control (Addgene plasmid 1864), with the scrambled-hairpin sequence as reported by Addgene (https://www.addgene.org/1864/; listed in Table S2). Lentiviral ransduction was performed at a multiplicity of infection of 10 using spinoculation (ref. (33)); transduced cells were selected with 2 µg/ml puromycin and monoclonal populations established by limiting dilution. The resultant NB4 cell lines stably expressing GPSM3-targeted shRNA with the most effective knockdown of GPSM3 were selected for this study. One monoclonal cell line expressing the scrambled shRNA control (“scr1”) was lost to poor growth under selection pressure. All NB4 lines were confirmed free of Mycoplasma spp. and Acholeplasma spp. contamination using the Lookout® Mycoplasma PCR Detection kit (Sigma-Aldrich, St. Louis, MO; see Fig. S4). NB4 cell line differentiation was performed as previously reported (34) by treatment with complete media containing 1 µM all-trans retinoic acid (Acros Organics, Geel, Belgium). Cell density was maintained between 2 × 105 and 1 × 106 cells/mL.
Nitroblue Tetrazolium (NBT) reduction
Cells (5 × 105) were resuspended in PBS with 1 mg/ml NBT and 300 nM PMA, then incubated at 37°C and 5% CO2 for 30 minutes. Cells were then washed in PBS twice and mounted on slides for visualization of 500 cells per cell population per experiment. Formazan-positive (black), -intermediate (gray), and -negative (colorless) cells were scored under brightfield microscopy. Investigator was blinded to the transgene status of each experimental condition.
Wright-Giemsa staining
Cells were fixed with 100% methanol for 1 minute, then allowed to air-dry briefly. Cells were then stained using Wright-Giemsa stain for 2 minutes, after which 1.5 volumes of PBS, pH 6.8 was added, and the cells were allowed to incubate for another 2 minutes. Cells were washed and visualized on an EVOS FL Auto microscope (Thermo Scientific, Waltham, MA).
cDNA synthesis and quantitative reverse transcriptase PCR (qRT-PCR)
RNA was extracted from ATRA-differentiated NB4* cells with the RNeasy kit (Qiagen). Briefly, cells were centrifuged at 200 × g for five minutes, and the supernatant was removed. Cells were then washed with 1× PBS and centrifuged again to remove supernatant. The cell pellet was resuspended in 600 µl of the manufacturer’s lysis buffer (RLT) with 1% β-mercaptoethanol. RNA isolation was performed using an automated QIAcube workflow according to the manufacturer’s suggested protocol to minimize variability between preparations. Eluted nucleic acids were quantified by absorbance at 260 nm using a Nanodrop 2000 (Thermo Scientific) and assessed for purity using a 260/280 nm cut-off ratio ≥1.8. Immediately following isolation, a sample of eluted nucleic acids (250 ng) from each group of cells was used to synthesize complementary DNA (cDNA) with the Thermo Verso Reverse Transcriptase Kit (Thermo Scientific). cDNA was then diluted 1:5 and used immediately for qRT-PCR analyses. Samples of cDNA, derived from equal mass of input RNA from each sample, were amplified in duplicate (with an additional, independent, “no reverse transcriptase-treated” control) using the USB VeriQuest Fast SYBR Green qPCR Master Mix (Affymetrix, Cleveland, OH) and commercially-validated PCR primers (Qiagen, Hilden, Germany). Cycling conditions were established following the manufacturer’s suggested protocol. Relative quantification of gene expression was performed using the ΔΔCT method (35) with β-microglobulin chosen as the stable housekeeping gene. Primer sequences for GPSM3 and β-microglobulin were as previously described (11); all other primer pairs were procured from Qiagen.
Transwell migration assay
shRNA-expressing NB4 cell lines were serum-starved in HBSS + 1% BSA at 37°C for one hour and labeled with calcein-AM cell-permeant fluorescent dye (ex. 494 nm/em. 517 nm). Cells were washed and suspended in HBSS + 1% BSA + 10 mM HEPES, and 2.5 × 106 cells/ml were loaded into the upper well of Falcon™ HTS FluoroBlok 96-Multiwell Insert System chemotaxis chambers (Corning, NY). Cells were allowed to migrate toward vehicle or 100 nM LTB4, 100 ng/ml fMLP, 100 ng/ml CXCL8 (R&D Systems, Minneapolis, MN), or 10% FBS (ThermoFisher, Waltham, MA) in the lower chamber. Fluorescence was measured at 2-minute intervals over the course of 60 minutes at 37°C using a Flexstation 3 microplate reader (Molecular Devices, Sunnyvale, CA). All data are representative of biological triplicates with technical replicates (i.e., n = 6).
Antibodies and Western blot
Antibodies targeting GPSM3 (UNC Immunology Core; clone 35.5.1; 1:200 dilution), CD11b (Developmental Studies Hybridoma Bank; clone H5A4; 1:200 dilution), pan-Gbeta (T-20 [cat# sc-378]; Santa Cruz Biotechnology; 1:500 dilution) and β-actin (Sigma [cat# A3854]; 1:2000 dilution) were purchased from the indicated sources. Horseradish peroxidase (HRP)-conjugated anti-mouse IgG was purchased from Sigma. Cells were lysed with ice-cold RIPA buffer with protease inhibitor (ThermoScientific, Waltham, MA), sonicated on ice, then clarified by centrifugation at 20,000 × g for 5 minutes. Protein content was quantified by the Bradford protein assay (ThermoScientific, Waltham, MA) using BSA as a standard. Lysates were diluted in Laemmli buffer (BioRad, Hercules, CA) and resolved on 8–16% pre-cast SDS-polyacyrlamide gels (BioRad), transferred to nitrocellulose membranes, immunoblotted with primary (overnight, 4°C) and HRP-conjugated secondary (1:2000 dilution, 2 h at room temperature) antibodies, and visualized by chemiluminescence (ECL, ThermoScientific, Waltham, MA).
Statistical analyses
Regression analyses of Transwell migration assay data were performed using 4 parameter logistic nonlinear regression analyses over the first 30-minute period of migration with post-hoc comparison by F-test to confirm if one model adequately represents all datasets. Analyses were confirmed to accept the null hypothesis of the Shapiro-Wilk normality test (p < 0.05). Statistical analyses including initial rate determinations were performed in GraphPad Prism 6.0 using one-way ANOVA with Bonnferroni or Dunnett’s post-hoc tests where indicated.
Supplementary Material
Acknowledgments
The authors thank Dr. Sherman Weissman for his generous gift of the NB4 cell line at the onset of these studies. This work was supported in part by the National Institutes of Health under Award Number U54GM104942.
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
- 1.Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 2.Billard MJ, Gall BJ, Richards KL, Siderovski DP, Tarrant TK. G protein signaling modulator-3: a leukocyte regulator of inflammation in health and disease. Am J Clin Exp Immunol. 2014;3(2):97–106. [PMC free article] [PubMed] [Google Scholar]
- 3.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343(6175):1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naranbhai V, Fairfax BP, Makino S, Humburg P, Wong D, Ng E, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. doi: 10.1038/ncomms8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009;5(12):e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corona E, Dudley JT, Butte AJ. Extreme evolutionary disparities seen in positive selection across seven complex diseases. PLoS One. 2010;5(8):e12236. doi: 10.1371/journal.pone.0012236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357(12):1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358(9):900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 9.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, et al. Genomewide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44(11):1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Kabesch M, Bouzigon E, Demenais F, Farrall M, Moffatt MF, et al. Using eQTL weights to improve power for genome-wide association studies: a genetic study of childhood asthma. Front Genet. 2013;4:103. doi: 10.3389/fgene.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gall BJ, Wilson A, Schroer AB, Gross JD, Stoilov P, Setola V, et al. Genetic variations in GPSM3 associated with protection from rheumatoid arthritis affect its transcript abundance. Genes Immun. 2016;17(2):139–147. doi: 10.1038/gene.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giguere PM, Billard MJ, Laroche G, Buckley BK, Timoshchenko RG, McGinnis MW, et al. G-protein signaling modulator-3, a gene linked to autoimmune diseases, regulates monocyte function and its deficiency protects from inflammatory arthritis. Mol Immunol. 2013;54(2):193–198. doi: 10.1016/j.molimm.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright HL, Moots RJ, Edwards SW. The multifactorial role of neutrophils in rheumatoid arthritis. Nat Rev Rheumatol. 2014;10(10):593–601. doi: 10.1038/nrrheum.2014.80. [DOI] [PubMed] [Google Scholar]
- 14.Sadik CD, Kim ND, Luster AD. Neutrophils cascading their way to inflammation. Trends Immunol. 2011;32(10):452–460. doi: 10.1016/j.it.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parham P. The Immune System. 3rd. New York, NY: Garland Science; 2009. [Google Scholar]
- 16.Jacobs JP, Ortiz-Lopez A, Campbell JJ, Gerard CJ, Mathis D, Benoist C. Deficiency of CXCR2, but not other chemokine receptors, attenuates autoantibody-mediated arthritis in a murine model. Arthritis Rheum. 2010;62(7):1921–1932. doi: 10.1002/art.27470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seeling M, Hillenhoff U, David JP, Schett G, Tuckermann J, Lux A, et al. Inflammatory monocytes and Fcgamma receptor IV on osteoclasts are critical for bone destruction during inflammatory arthritis in mice. Proc Natl Acad Sci U S A. 2013;110(26):10729–10734. doi: 10.1073/pnas.1301001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao WH, Del Prete A, Bock CB, Haribabu B. Targeted disruption of leukotriene B4 receptors BLT1 and BLT2: a critical role for BLT1 in collagen-induced arthritis in mice. J Immunol. 2006;176(10):6254–6261. doi: 10.4049/jimmunol.176.10.6254. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths RJ, Smith MA, Roach ML, Stock JL, Stam EJ, Milici AJ, et al. Collagen-induced arthritis is reduced in 5-lipoxygenase-activating protein-deficient mice. J Exp Med. 1997;185(6):1123–1129. doi: 10.1084/jem.185.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Lam BK, Kanaoka Y, Nigrovic PA, Audoly LP, Austen KF, et al. Neutrophil-derived leukotriene B4 is required for inflammatory arthritis. J Exp Med. 2006;203(4):837–842. doi: 10.1084/jem.20052371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger M, Budhu S, Lu E, Li Y, Loike D, Silverstein SC, et al. Different G(i)-coupled chemoattractant receptors signal qualitatively different functions in human neutrophils. J Leukoc Biol. 2002;71(5):798–806. [PubMed] [Google Scholar]
- 22.Neptune ER, Iiri T, Bourne HR. Galphai is not required for chemotaxis mediated by Gi-coupled receptors. J Biol Chem. 1999;274(5):2824–2828. doi: 10.1074/jbc.274.5.2824. [DOI] [PubMed] [Google Scholar]
- 23.Neptune ER, Bourne HR. Receptors induce chemotaxis by releasing the betagamma subunit of Gi, not by activating Gq or Gs. Proc Natl Acad Sci U S A. 1997;94(26):14489–14494. doi: 10.1073/pnas.94.26.14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csomos K, Nemet I, Fesus L, Balajthy Z. Tissue transglutaminase contributes to the all-trans-retinoic acid-induced differentiation syndrome phenotype in the NB4 model of acute promyelocytic leukemia. Blood. 2010;116(19):3933–3943. doi: 10.1182/blood-2010-01-266064. [DOI] [PubMed] [Google Scholar]
- 25.Giguere PM, Laroche G, Oestreich EA, Siderovski DP. G-protein signaling modulator-3 regulates heterotrimeric G-protein dynamics through dual association with Gbeta and Galphai protein subunits. J Biol Chem. 2012;287(7):4863–4874. doi: 10.1074/jbc.M111.311712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimple RJ, Willard FS, Hains MD, Jones MB, Nweke GK, Siderovski DP. Guanine nucleotide dissociation inhibitor activity of the triple GoLoco motif protein G18: alanine-to-aspartate mutation restores function to an inactive second GoLoco motif. Biochem J. 2004;378(Pt 3):801–808. doi: 10.1042/BJ20031686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J Biol Chem. 2002;277(45):43399–43409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- 28.Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. 2014;6(10):88. doi: 10.1186/s13073-014-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klickstein LB, Shapleigh C, Goetzl EJ. Lipoxygenation of arachidonic acid as a source of polymorphonuclear leukocyte chemotactic factors in synovial fluid and tissue in rheumatoid arthritis and spondyloarthritis. J Clin Invest. 1980;66(5):1166–1170. doi: 10.1172/JCI109947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorward DA, Lucas CD, Chapman GB, Haslett C, Dhaliwal K, Rossi AG. The role of formylated peptides and formyl peptide receptor 1 in governing neutrophil function during acute inflammation. Am J Pathol. 2015;185(5):1172–1184. doi: 10.1016/j.ajpath.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullmann TJ, Cheewatrakoolpong B, Anthes JC, Siegel MI, Egan RW, Billah MM. Phospholipase C and phospholipase D are activated independently of each other in chemotactic peptide-stimulated human neutrophils. J Leukoc Biol. 1993;53(6):630–635. doi: 10.1002/jlb.53.6.630. [DOI] [PubMed] [Google Scholar]
- 32.Robichaux WG, 3rd, Oner SS, Lanier SM, Blumer JB. Direct Coupling of a Seven-Transmembrane-Span Receptor to a Galphai G-Protein Regulatory Motif Complex. Mol Pharmacol. 2015;88(2):231–237. doi: 10.1124/mol.115.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Idres N, Benoit G, Flexor MA, Lanotte M, Chabot GG. Granulocytic differentiation of human NB4 promyelocytic leukemia cells induced by all-trans retinoic acid metabolites. Cancer Res. 2001;61(2):700–705. [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.