Abstract
Background
Chronic alcohol consumption leads to a loss of white adipose tissue (WAT) but the underlying mechanisms for this lipodystrophy are not fully elucidated. The present study tested the hypothesis that the reduction in WAT mass in chronic alcohol-fed mice is associated with a decreased protein synthesis specifically related to impaired function of mTOR.
Methods
Adult male mice were provided an alcohol-containing liquid diet for 24 wk or an isonitrogenous isocaloric control diet. In vivo protein synthesis was determined at this time and thereafter epididymal WAT (eWAT) was excised for analysis of signal transduction pathways central to controlling protein synthesis and degradation.
Results
While chronic alcohol feeding decreased whole-body and eWAT mass, this was associated with a discordant increase in protein synthesis in eWAT. This increase was not associated with a change in mTOR, 4E-BP1, Akt or PRAS40 phosphorylation. Instead, a selective increase in phosphorylation of S6K1 and its downstream substrates, S6 and eIF4B was detected in alcohol-fed mice. Alcohol also increased eEF2K phosphorylation and decreased eEF2 phosphorylation consistent with increased translation elongation. Alcohol increased Atg12-5, LC3B-I and –II, and ULK1 S555 phosphorylation, suggesting increased autophagy, while markers of apoptosis (cleaved caspase-3 and -9, and PARP) were unchanged. Lipolytic enzymes (ATGL and HSL phosphorylation) were increased and lipogenic regulators (PPARγ and C/EBPα) were decreased in eWAT by alcohol. Although alcohol increased TNFα, IL-6 and IL-1β mRNA, no change in key components of the NLRP3 inflammasome (NLRP3, ACS and cleaved caspase-1) were detected suggesting alcohol did not increase pyroptosis. Plasma insulin did not differ between groups.
Conclusions
These results demonstrate that the alcohol-induced decrease in whole-body fat mass resulted in part from activation of autophagy in eWAT as protein synthesis was increased and mediated by the specific increase in the activity of S6K1.
Keywords: protein metabolism, fat, mTOR, S6K1, autophagy
INTRODUCTION
There is an increasing appreciation of the metabolic activity and endocrine functions of white adipose tissue (WAT) and its important role in the development of various metabolic diseases (Kloting and Bluher, 2014). However, considerably less information is available on adipose tissue in the setting of chronic alcohol consumption. The consensus of studies in rodents indicates a loss of WAT mass and a reduction in adipocyte size with chronic alcohol consumption (Pravdova et al., 2009, Zhang et al., 2015, Zhao et al., 2015, Kang et al., 2007a), a finding also reported in human alcoholics (Liangpunsakul et al., 2010). This alcohol-induced lipodystrophy appears to result in part from an increase in the export of fatty acids via enhanced lipolysis in WAT (Kang et al., 2007a, Wei et al., 2013).
The mammalian target of rapamycin (mTOR) is emerging as a central regulator of lipid metabolism in adipocytes and other cell types (Laplante and Sabatini, 2009). Additionally, the canonical mTOR signaling pathway regulates differentiation and maintenance of adipocytes, adipose morphogenesis and growth, and hormone secretion by this tissue (Lynch, 2001). Like other tissues, mTOR kinase activity in eWAT is increased by nutrients (e.g. leucine) and growth factors (e.g., insulin) leading to the phosphorylation of eukaryotic initiation factor (eIF)-4E binding protein-1 (4E-BP1) and ribosomal protein S6 kinase-1 (S6K1), with a coordinate rapid up-regulation of protein synthesis (Lynch et al., 2002b, Lynch et al., 2002a, Zhang et al., 2009, Chakrabarti et al., 2010). Conversely, inhibition of mTOR by rapamycin decreases eWAT mass (Blanchard et al., 2012). The effect of chronic alcohol consumption on mTOR activity and protein synthesis in eWAT has not been examined, although alcohol suppresses mTOR-dependent protein synthesis in skeletal muscle, heart, liver and intestine (Preedy and Peters, 1990, Lang et al., 2007, Lang et al., 1999). Moreover, whether any observed effect of chronic alcohol in eWAT can be reproduced acutely (either in vivo or in vitro) or directly by ethanol has not been investigated. Hence, the present study tested the hypothesis that the reduction in eWAT mass in chronic alcohol-fed mice is associated with a decreased protein synthesis specifically related to impaired function of mTOR complex 1 (mTORC1).
MATERIALS AND METHODS
Animals
Male viral antibody free C57BL/6 mice aged 11–13 wk Charles River Laboratories; Wilmington, MA) were acclimated for at least 1 wk to the vivarium prior to the start of experiments. Mice were housed two per cage in shoe-box cages with corn cob bedding under environmental controlled conditions (22 ± 1 °C and 35–75% humidity; 12:12 h light/dark cycle), and during this period were provided standard rodent chow (Teklad Global 2019; Harlan Teklad, Boston, MA) and water ad libitum. All experimental procedures were performed in accordance with the National Institutes of Health (NIH) guidelines for the use of experimental animals and were approved by the Institutional Animal Care and Use Committee of The Pennsylvania State University College of Medicine (protocol #46587).
Experimental design
Chronic study
Mice were randomized and individually housed and provided the alcohol-free control Lieber-DeCarli rodent liquid diet (product # F1259SP; Bio-Serv, Flemington, NJ) for 1 wk. After this period of acclimation, mice were randomly assigned to receive either the alcohol-containing liquid diet (product # F1258SP; Bio-Serv; n = 11) or to the control (no alcohol; n = 14) liquid diet. Control mice were pair-fed the same volume of an isocaloric and isonitrogenous liquid diet. Food consumption was assessed daily and fresh food was provided daily. The alcohol-fed group was acclimated to the alcohol-containing diet using an escalating step-wise protocol which included 1 day of 10%, and 2 days each of 16%, 22%, and 30% kcal from alcohol. Feeding was continued for a total of 24 wks. Whole-body fat mass was assessed noninvasively on conscious animals using a 1H-NMR analyzer (Bruker LF90 Proton-NMR Minispec: Bruker Optics, Woodlands, TX) prior to the introduction of the alcohol-containing diet and at 6 wk intervals.
Acute study
Although the primary focus of this study was on the effect of chronic alcohol consumption on adipose tissue protein synthesis, an additional study was performed to determine whether acute alcohol intoxication produced similar changes. Mice were randomly administered ethanol at a dose of 3 g/kg or an equal volume of 0.9% sterile saline (Control) via intraperitoneal (IP) injection (Steiner and Lang, 2014). Mice were anesthetized at 4 h (control, n = 10 and EtOH, n = 12) after alcohol/saline administration and eWAT was collected. Experiments were performed between 08:00 – 11:00 with animals allowed ab libitum access to the liquid diet. The experimental time points were specifically chosen to correspond with the peak BAC and alterations in protein synthesis in other tissues (Steiner and Lang, 2014).
Protein synthesis and plasma concentrations of alcohol, insulin and cytokines
In vivo protein synthesis was assessed using the SUnSET method and an anti-puromycin antibody (Kerafast, Boston, MA). Mice were administered an intraperitoneal injection of 0.04 µmol/g BW of puromycin dissolved in sterile saline 30 min prior to the removal of fat. Western blotting was performed as below to visualize puromycin incorporation as reported (Steiner et al., 2015). Mice were anesthetized via isoflurane inhalation (3–4% in 100% oxygen) and the entire epididymal fat depot was excised from both sides and weighed. Tissue was immediately frozen in liquid nitrogen and stored at −80°C until analysis. Blood was collected from the aorta and the plasma used to determine the concentrations of alcohol (GM7, Analox Instruments, Lunenburg, MA) and insulin (ALPCO mouse ultrasensitive ELISA; Windham, NH). Plasma cytokine concentrations were measured using V-PLEX Proinflammatory Panel (MSD, Rockville, MD) from the animals in the chronic alcohol consumption study. The cytokine analysis included interferon (IFN)-γ, interleukin (IL)-1β, IL-2, IL-4, IL-5, IL-6, KC/GRO, IL-10, IL-12p70 and tumor necrosis factor (TNF)-α.
Western blotting
Western blot analysis was performed as described (Lang et al., 2007, Steiner et al., 2015, Steiner and Lang, 2014). Briefly, eWAT was homogenized in ice-cold buffer containing (in mM): 20 HEPES (pH 7.4), 2 EGTA, 0.2 EDTA, 100 KCl, 50 β-glycerophosphate, 50 NaF, 0.5 sodium orthovanadate, 1 Benzamidine, 0.1 PMSF and 1 DTT. Samples were centrifuged at 10,000 g for 10 min and the liquid phase under the top lipid layer used in subsequent analyses. Protein concentration was quantified (Bio-Rad Protein Assay; Hercules, CA) and SDS-PAGE was performed using equal amounts of total protein per sample. Following Ponceau S (Aqua Solutions, Deer Park, TX) staining to verify equal protein loading, PVDF membranes were blocked in 5% nonfat dry milk, and then incubated overnight at 4°C with primary antibody. Antibodies included (Cell Signaling, Beverly, MA, unless otherwise noted): S6K1, S6K1 (Thr389 and Thr421/Ser424), rpS6, rpS6 (Ser240/244), 4E-BP1 (Bethyl Laboratories, Montgomery, TX), 4E-BP1 (Thr37/46 and Ser65), eEF2, eEF2 (Thr56), ERK, ERK1/2 (Thr202/Tyr204), mTOR, mTOR (Ser2448 and Ser2481), REDD1 (ProteinTech, Chicago, IL), Akt, Akt (Thr308 and Ser473), PRAS40, PRAS40 (Thr246), eIF4B, eIF4B (Ser422), programmed cell death protein 4 (PDCD4), PDCD4 (Ser67), eukaryotic elongation factor (eEF)-2 kinase (eEF2K), eEF2K (Ser366), AMP-activated protein kinase α (AMPK), AMPKα (Thr172), tuberous sclerosis complex 2 (TSC2), TSC2 (Thr1462 and Ser1387), Unc-51 like autophagy activating kinase 1 (ULK1), ULK1 (Ser757 and Ser555), p62, light-chain 3B (LC3B)-I and -II, ATG12/5 complex, ATG7, ATG3, adipose tissue triglyceride lipase (ATGL), hormone sensitive lipase (HSL), HSL (Ser660), peroxisome proliferator-activated receptor gamma (PPARγ), CCAAT/enhancer-binding protein alpha (C/EBPα), NLRP3 (NLR pyrin domain containing 3), adaptor protein containing a C-terminal caspase-recruitment domain (ACS), caspase-1 (BioVision, Inc, Milpitas, CA), β-actin and tubulin. Apoptosis was assessed by Western blotting for caspase-3, cleaved caspase-3, poly(ADP-ribose) polymerase (PARP), cleaved PARP, procaspase-9, cleaved caspase-9 and Bcl-2 (all from Cell Signaling). The appropriate secondary antibody was added in 5% nonfat milk for 1–2 h prior to washing with TBST. The fat homogenate was also incubated with monoclonal anti-human eIF4E and the immunoprecipitant isolated using IgG beads (Qiagen), as described (Lang et al., 2007). After isolation from the beads, the eIF4E immunoprecipitated was subjected to Western blotting using antibodies to 4E-BP1, eIF4E and eIF4G. Western blot images were visualized on the FluorChem M Multifluor System (ProteinSimple, San Jose, CA) after exposure to Pierce ECL Western blotting substrate (Thermo Scientific, Waltham, MA). Images were analyzed using AlphaView (ProteinSimple) and Image J software (NIH).
RNA extraction and real-time quantitative PCR
Total RNA was extracted using Tri-reagent (Molecular Research Center, Inc., Cincinnati, OH) and RNeasy mini kit (Qiagen, Valencia, CA) following manufacturers’ protocols, exactly as described previously (Lang et al., 2012). On-column DNase I treatment was used to remove residual DNA contamination. RNA was eluted from the column with RNase-free water and an aliquot was used for quantitation (NanoDrop 2000, Thermo Fisher Scientific, Waltham, MA). Quality of the RNA was analyzed on a 1% agarose gel. Total RNA (1 µg) was reversed transcribed using superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). Real-time quantitative PCR was performed using 25 ng of cDNA in a StepOnePlus system using TaqMan gene expression assays (Applied Biosystems, Foster City, CA) using primers as described previously (Lang et al., 2012). The comparative quantitation method 2−ΔΔCt was used in presenting gene expression of target genes in reference to the endogenous control.
Culture of 3T3-F442A adipocytes
Murine 3T3-F442A pre-adipocytes were kindly provided by Dr. Christopher J. Lynch, Penn State College of Medicine. Cells were seeded at 0.1 × 106 cells in 6-well plates and grown to confluence (1–2 days) in Dulbecco’s Modified Eagle’s medium (DMEM) containing high glucose (4.5 g/L) (Mediatech, Manassas, VA), supplemented with 10 % (v/v) bovine calf serum (BCS) (Gemini Bio-Products, West Sacramento, CA), penicillin 100 U/ml and streptomycin 100 µg/ml (Invitrogen Life-technologies, Carlsbad, CA) at 37 °C in a humidified atmosphere of 5% CO2/air. At confluence, cells were switched to differentiation medium [DMEM + 10% (v/v) fetal calf serum (FCS) (Gemini Bio-Products) + 5 µg/ml recombinant human insulin (Lilly, Indianapolis, IN) and penicillin/streptomycin] at designated day 0, and incubated at 37 °C in 10% CO2 /air for 12 days to obtain differentiated adipocytes. Cells were inspected daily and differentiation medium was changed every 2–3 days. On Day 12, differentiation medium was removed and replaced with DMEM but without serum or insulin. Cells were returned to the incubator (37 °C for 2 h) and treated with 80 mM ethanol for 2 h or 6 h. This ethanol dose and time course was selected based on previous studies using 3T3-L1 cells (Tang et al., 2012). Cells were washed 3 × with ice cold PBS and harvested by scraping in PBS containing 2% SDS. Extracts were sonicated on ice using the microtip of a Misonix sonicator and thereafter centrifuged at 20,000 gav for 20 min. The lipid droplet layer was aspirated from the surface of the centrifuged extracts and protein concentrations were determined on the soluble extract using the BCA assay (Pierce, ThermoScientific, Rockford, IL). Western blotting was performed as described above. Three wells of cells from both control and alcohol-treated cells were run in triplicate.
Statistics
Data were analyzed using 2-tailed Students’ t-test (GraphPad, San Diego, CA and SigmaPlot, Systat, San Jose, CA) for treatment effect. Data are presented as mean ± SEM and considered significant when P < 0.05; trends were predetermined to be 0.05 < P < 0.09.
RESULTS
Body composition, plasma alcohol, insulin and cytokines
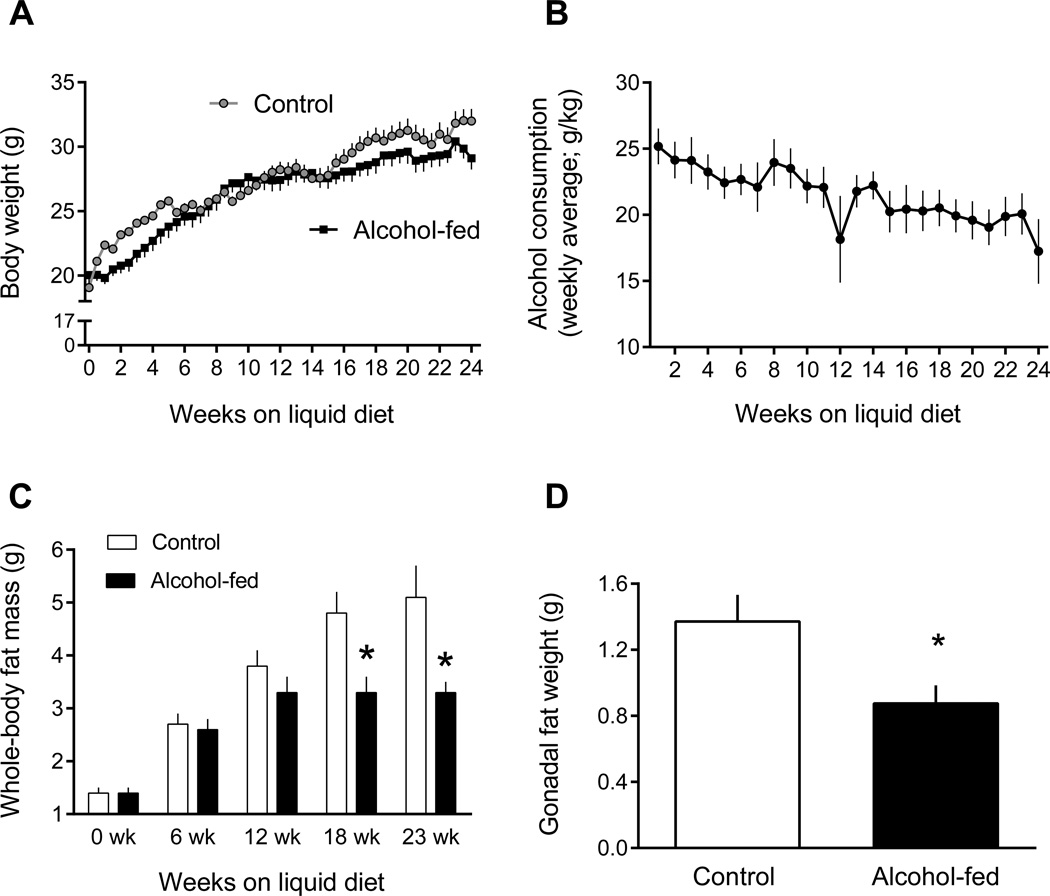
Body weight did not differ between control- and alcohol-fed mice at the start of the study (19.1 ± 0.5 g and 20.0 ± 0.6 g) and increased in both groups over the 24-wk feeding period (Figure 1A). The absolute increment in body weight was lower (P < 0.05) in the alcohol-fed group (8.3 ± 0.9 g) compared to time-match control group (11.6 ± 0.9 g). Alcohol consumption ranged from ~25 g/kg body weight at the start of the study to ~21 g/kg at the conclusion as body weight of the mice increased but they consumed a similar volume of the liquid diet (Figure 1B). At the time of sacrifice, the blood alcohol concentration was 4.6 mM. Whole-body fat mass did not differ between groups at the start of the study, but was reduced 30% and 35% after 18 and 23 wks on the alcohol-containing diet (Figure 1C). At the time of sacrifice, epididymal fat mass was reduced 36% (Figure 1D).
Figure 1.
Change in body weight, alcohol consumption, and fat mass in chronic alcohol-fed and pair-fed control mice. Values are means ± SEM; n = 11 and 14, respectively. *P < 0.05 compared to time-matched control values.
The plasma insulin concentration did not differ between control (1.62 ± 0.17 ng/ml) and alcohol-fed (1.45 ± 0.22 ng/ml) mice. Whereas alcohol feeding did not alter the plasma concentration of IL-1β, IL-4, IL-6, IL-10, or IL-12p70, compared to control values, alcohol feeding did increase TNFα, IFNγ and IL-5 (76%, 40% and 42%, respectively) and tended to increase IL-2 and KC/GRO (32% and 48%, respectively) (Table 1).
Table 1.
Plasma cytokine concentrations in control- and alcohol-fed mice
| Control-fed | Alcohol-fed | |
|---|---|---|
| TNF-α | 21.5 ± 1.7 | 37.2 ± 3.5* |
| IFN-γ | 1.5 ± 0.1 | 2.1± 0.3* |
| IL-1-β | 4.0 ± 0.5 | 4.1 ± 0.5 |
| IL-2 | 18.5 ± 1.8 | 24.5 ± 2.9 (P = 0.08) |
| IL-4 | 1.4 ± 0.1 | 1.6 ± 0.2 |
| IL-5 | 3.5 ± 0.3 | 5.0 ± 0.4* |
| IL-6 | 9.9 ± 1.5 | 12.2 ± 2.3 |
| IL-10 | 23.8 ± 2.0 | 26.8 ± 3.7 |
| IL-12p70 | 169.5 ± 15.0 | 178.6 ± 24.7 |
| KC/GRO | 256 ± 29 | 379 ± 61 (P = 0.06) |
Values are means ± SEM; n = 14 and 11, respectively.
P < 0.5 compared to time-match control-fed value. Specific P-values are presented for two cytokines that showed a trend (i.e., 0.05 < P < 0.09) to increase. All values have units of pg/ml.
Protein synthesis and signal transduction
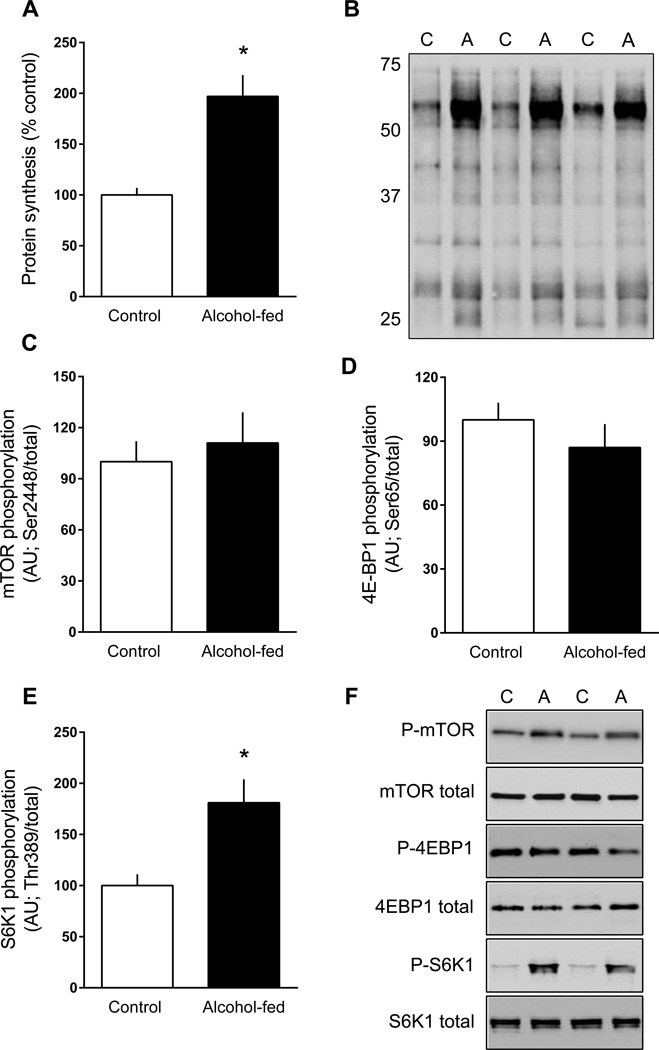
Global protein synthesis in adipose tissue was ~2-fold greater in alcohol-fed mice than pair-fed control animals (Figure 2A, 2B). To assess the potential mechanism for this change, the phosphorylation state of mTOR per se and its downstream substrates were determined (Figure 2C–2F). There was no difference in the phosphorylation of mTOR on Ser2448 (Akt-dependent (Nave et al., 1999); Figure 2C, 2F) or on Ser2481 (autophosphorylation (Soliman et al., 2010); data not shown) between groups. Similarly there was no alcohol-induced change in the phosphorylation of 4E-BP1 on Ser65 (Figure 2D, 2F) or Thr37/46 (data not shown). The phosphorylation state of 4E-BP1 can alter the interaction of eIF4E and eIF4G, and thereby the rate of mRNA translation (Haghighat and Sonenberg, 1997). Consistent with the lack of change in 4E-BP1 phosphorylation, there was no difference in the amount of eIF4G bound to eIF4E or the amount of 4E-BP1 bound to eIF4E (data not shown).
Figure 2.
Protein synthesis and mTOR signal transduction in epididymal white adipose tissue (eWAT) from chronic alcohol-fed and pair-fed control mice. Global protein synthesis was determined by assessing the incorporation of puromycin into protein and is expressed as a percentage of the time-matched control value (panels A and B). Quantitation of Western blots for mTOR, 4E-BP1 and S6K1 phosphorylation normalized to the total amount of the respective protein (panels C–E). There was no difference in the total amount of these three proteins (data not shown). Representative Western blots are presented (panel F) from eWAT isolated from two control (C) and two alcohol-fed (A) mice. Values are means ± SEM; n = 11 and 14, respectively. *P < 0.05 compared to time-matched control values.
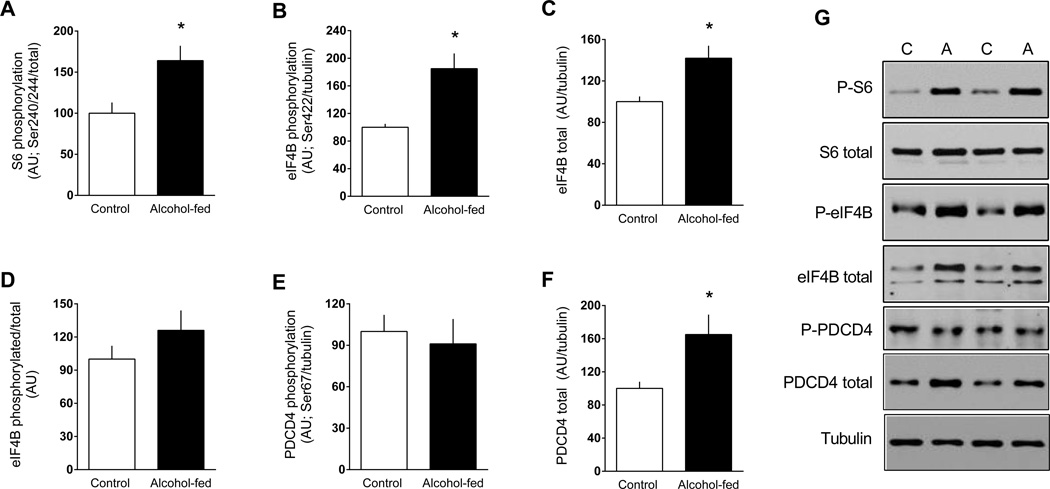
In contrast, S6K1 phosphorylation on both Thr389 (Figure 2E, 2F) and Thr421/Ser424 (data not shown) was increased in eWAT from alcohol-fed mice. To confirm activation of S6K1, phosphorylation of known downstream substrates was assessed (Dann et al., 2007). In this regard, alcohol feeding increased Ser240/244 phosphorylated S6 independent of a change in the total amount of the protein (Figure 3A and 3G). Alcohol feeding also increased Ser422 phosphorylated eIF4B but there was a coordinate increase in total eIF4B such that the phosphorylated-to-total ratio for this protein did not differ between groups (Figure 3B–D and 3G). In contrast, Ser67-phosphorylated PDCD4, another S6K1 substrate, did not differ between control and alcohol-fed mice, but total PDCD4 was increased nearly 50% (Figure 3E–G).
Figure 3.
Change in the phosphorylation state of S6K1 target proteins in eWAT from chronic alcohol-fed and pair-fed control mice. Quantitation of Western blots for the ribosomal protein S6, eIF4B (phosphorylated, total and ratio) and PDCD4 (phosphorylated and total (panels A–F). Unless, noted, values have been normalized to the total amount of the respective protein (e.g., tubulin) that did not change with alcohol (data not shown). eIF4B resolved as two bands and both bands were quantitated. Representative Western blots are presented (panel G) from eWAT isolated from two control and two alcohol-fed mice. Values are means ± SEM; n = 11 and 14, respectively. *P < 0.05 compared to time-matched control values.
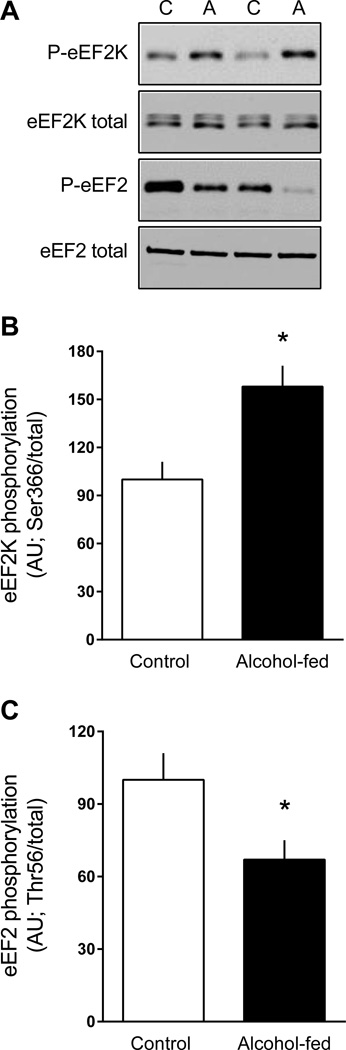
In addition, Ser366-phosphorylated eEF2K (another S6K1 substrate) was also increased in eWAT from alcohol-fed mice, compared to control values (Figure 4A, 4B). The phosphorylation of eEF2K at this site inhibits its kinase activity (Redpath et al., 1996) and this was confirmed by the concomitant reduction in Thr56-phosphorylated eEF2 (Figure 4A, 4C).
Figure 4.
Change in the phosphorylation state of proteins upstream of eEF2 kinase (eEF2K) and eEF2 in eWAT from chronic alcohol-fed and pair-fed control mice. Quantitation of Western blots for phosphorylated eEF2K and eEF2 were normalized to the total amount of the respective protein that did not change with alcohol (data not shown). Representative Western blots are presented (panel A) from eWAT isolated from two control and two alcohol-fed mice. Values are means ± SEM; n = 11 and 14, respectively. *P < 0.05 compared to time-matched control values.
Activation of the MAP kinases ERK1/ERK2 via phosphorylation also stimulates mTOR and S6 phosphorylation and ultimately protein synthesis (Rolfe et al., 2005). However, the phosphorylation of ERK1/ERK2 (Thr202/Tyr204) as well as the downstream substrate RSK1/2 (Ser380) did not differ in eWAT from control and alcohol-fed mice (data not shown).
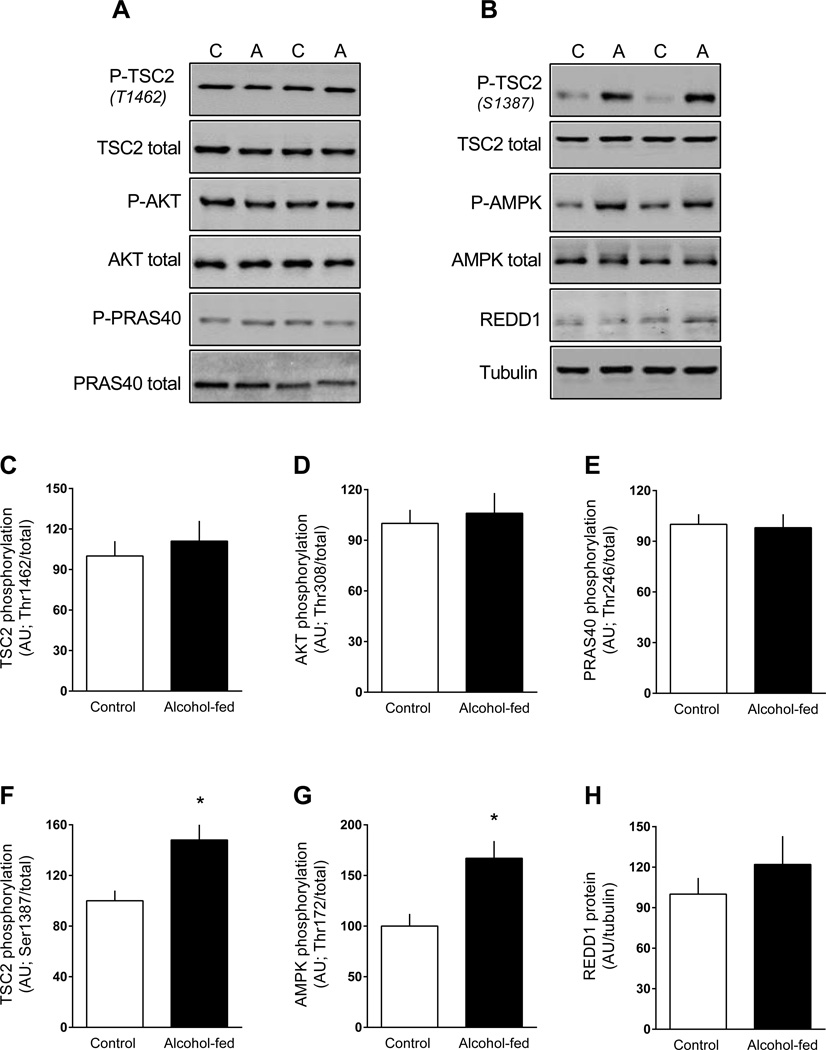
Because of the apparently discordant results noted above related to mTOR kinase activation (e.g., no change in mTOR and 4E-BP1 phosphorylation, but increased S6K1 phosphorylation), we also assessed key up-stream regulators of mTOR activity (Dibble and Cantley, 2015). TSC2 and its activity is inversely related to mTOR kinase activity (Inoki et al., 2002). TSC2 phosphorylation on Thr1462, which is Akt-dependent (Inoki et al., 2002), did not differ between control and alcohol-fed mice (Figure 5A, 5C). Consistent with this observation, there was no alcohol-induced change in Akt Thr308 phosphorylation (Figure 5A, 5D), Akt Ser473 phosphorylation (data not shown), or Thr246-phosphorylated PRAS40 (another Akt substrate) (Figure 5A, 5E). In contrast, TSC2 phosphorylation on Ser1387, regulated in part by AMPK (Huang and Manning, 2008), was increased in eWAT from alcohol-fed mice (Figure 5B, 5F), and this was consistent with the activation of AMPK as evidenced by increased AMPK phosphorylated on Thr172 (Figure 5B, 5G). Finally, AMPK can upregulate REDD1 and impair mTOR activity by potentially different mechanisms (Brugarolas et al., 2004); however, there was no alcohol effect on total REDD1 protein in eWAT (Figure 5B, 5H).
Figure 5.
Change in the phosphorylation state of proteins upstream of mTORC1 in eWAT from chronic alcohol-fed and pair-fed control mice. Quantitation of Western blots for the proteins TSC2, Akt, PRAS40, AMPK and REDD1. Unless, noted, values have been normalized to the total amount of the respective protein that did not change with alcohol (data not shown). Representative Western blots are presented (panels A and B) from eWAT isolated from two control and two alcohol-fed mice. Values are means ± SEM; n = 11 and 14, respectively. *P < 0.05 compared to time-matched control values.
Autophagy
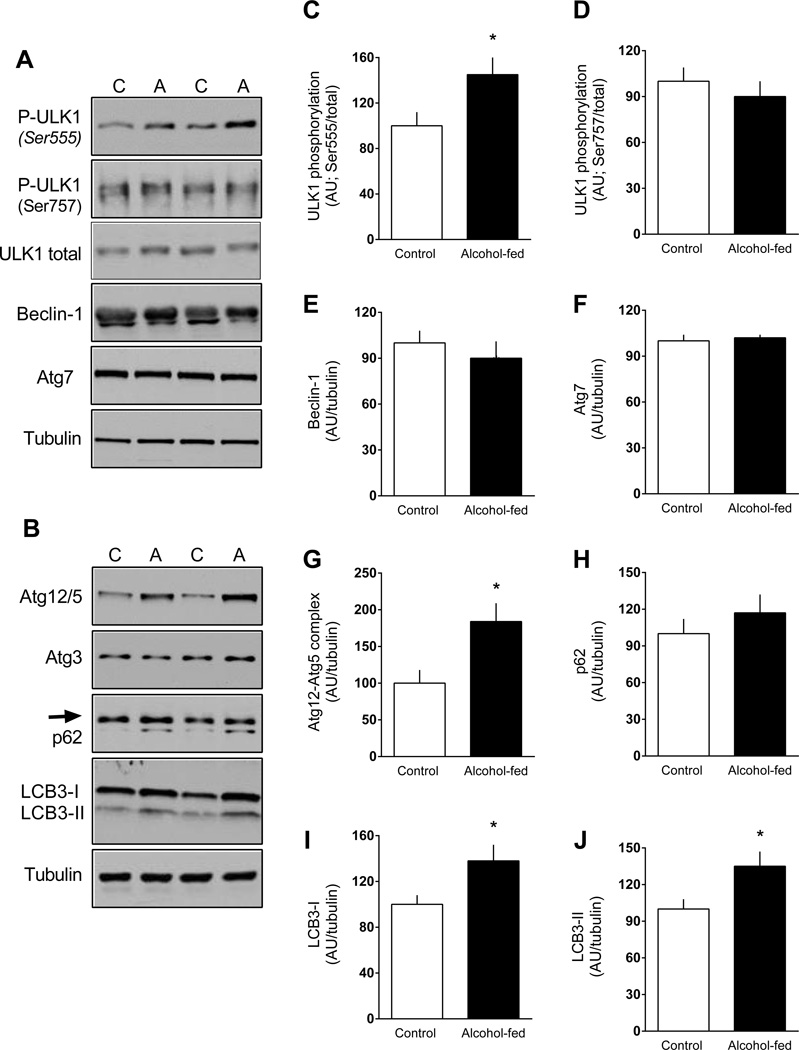
Increases in AMPK can directly phosphorylate ULK1 at multiple sites (i.e., Ser317, Ser555, and Ser777) thereby enhancing autophagy (Kim et al., 2011). Conversely, stimulation of mTORC1 inhibits autophagy via phosphorylation of ULK1 at Ser757 disrupting the interaction between ULK1 and AMPK. Ser555-phosporylated ULK1 was increased in eWAT from alcohol-fed mice, whereas ULK1 phosphorylation of Ser757 did not differ between groups (Figure 6A, 6C, 6D). Protein levels for beclin-1, Atg7, Atg3 and p62 (SQSTM1) did not differ between control and alcohol-fed mice (Figure 6A, 6B, 6E, 6H). However, there was an alcohol-induced increase in the Atg12-Atg5 complex (Figure 6B, 6G) as well as an increase in both LC3B-I and the LC3B-phosphatidylethanolamine conjugate LC3B-II (Figure 6B, 6I, 6J).
Figure 6.
Change in the phosphorylation state of proteins related to autophagy in eWAT from chronic alcohol-fed and pair-fed control mice. Quantitation of Western blots for ULK1, beclin, Atg7, Atg12/5, Atg3, p62 and LC3B-II and -I. Unless noted, values have been normalized to the total amount of the respective protein that did not change with alcohol (data not shown) or tubulin. Quantitation of Atg3 is not presented graphically as there was no difference between groups. Representative Western blots are presented (panels A and B) from eWAT isolated from two control and two alcohol-fed mice. Values are means ± SEM; n = 11 and 14, respectively. *P < 0.05 compared to time-matched control values.
Apoptosis
The protein content for a number of markers of apoptosis in eWAT was also determined. The protein content did not differ statistically (P > 0.05) between control (n = 11) and alcohol-fed (n = 14) mice for: caspase-3 (100 ± 10 AU vs 105 ± 9 AU), cleaved caspase-3 (100 ± 5 AU vs 107 ± 11 AU), total PARP (100 ± 12 AU vs 118 ± 23 AU), cleaved PARP (100 ± 13 AU vs 107 ± 9 AU), cleaved caspase-9 (below level of detection) and Bcl-2 (100 ± 11 AU vs 124 ± 16 AU). Of the endpoints assessed, only procaspase-9 (but not cleaved caspase-9) was increased in eWAT from alcohol-fed mice compared to control values (100 ± 11 vs 176 ± 22; P < 0.05).
Cytokines and lipid metabolism
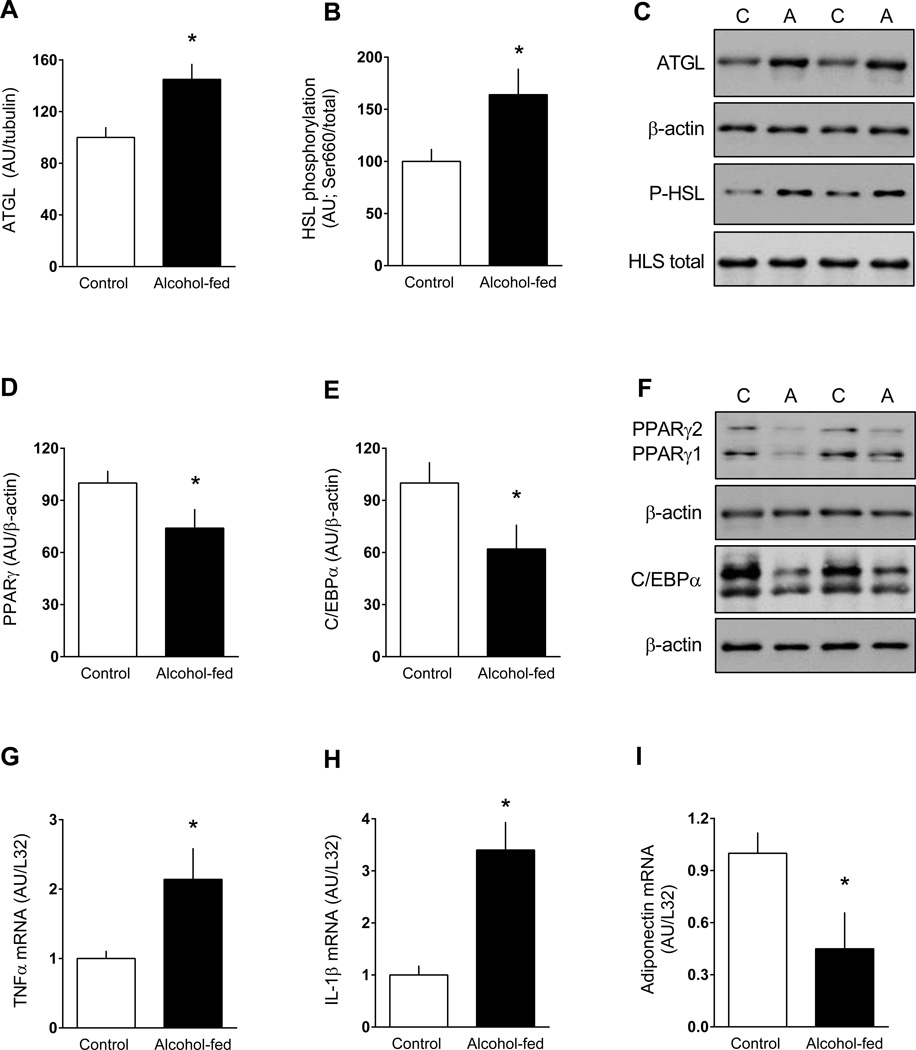
To confirm that our animal model was comparable to others in the literature regarding changes in adipose tissue metabolism produced by chronic alcohol feeding, we assessed the protein content or mRNA expression of other previously reported endpoints (Zhong et al., 2012, Sebastian et al., 2011, Kang et al., 2007b, He et al., 2015). The total amount of ATGL and the phosphorylation of HSL (e.g., lipolytic enzymes) were both increased in eWAT from alcohol-fed mice (Figure 7A–C). Conversely, the protein content for two transcription factors regulating lipogenesis, PPARγ and C/EBPα, were both decreased (Figure 7D–F). Finally, alcohol feeding increased TNFα, IL-1β and IL-6 mRNA (Figure 7G, 7H and data not shown), and decreased adiponectin mRNA content eWAT (Figure 7I).
Figure 7.
Change in the total amount or phosphorylation state of proteins related to lipolysis or lipogenesis in eWAT from chronic alcohol-fed and pair-fed control mice. Bar graphs (A, B, D and E) are quantitation of Western blots. Values have been normalized to the total amount of the respective protein that did not change with alcohol (data not shown) or β-actin. Representative Western blots are presented (panels C and F) from eWAT isolated from two control and two alcohol-fed mice. For PPARγ, the antibody recognizes both PPARγ1 and PPARγ2 which have been identified; for C/EBPα, translation from alternative start codons results in two isoforms of C/EBPα (p42 and p30), which are seen on the gel. For these proteins, both bands were quantitated and presented graphically. Panels G-I are mRNA content for TNFα, IL1β and adiponectin normalized for L32. Values are means ± SEM; n = 11 and 14, respectively. *P < 0.05 compared to time-matched control values.
Persistent low-grade inflammation in adipose tissue can activate the NLRP3 inflammasome precipitating pyroptosis (Giordano et al., 2013). However, Western blot analysis of key elements of the NLRP3 inflammasome, including NLRP3, ACS and total and cleaved caspase-1, did not differ in eWAT between control and alcohol-fed mice (data not shown).
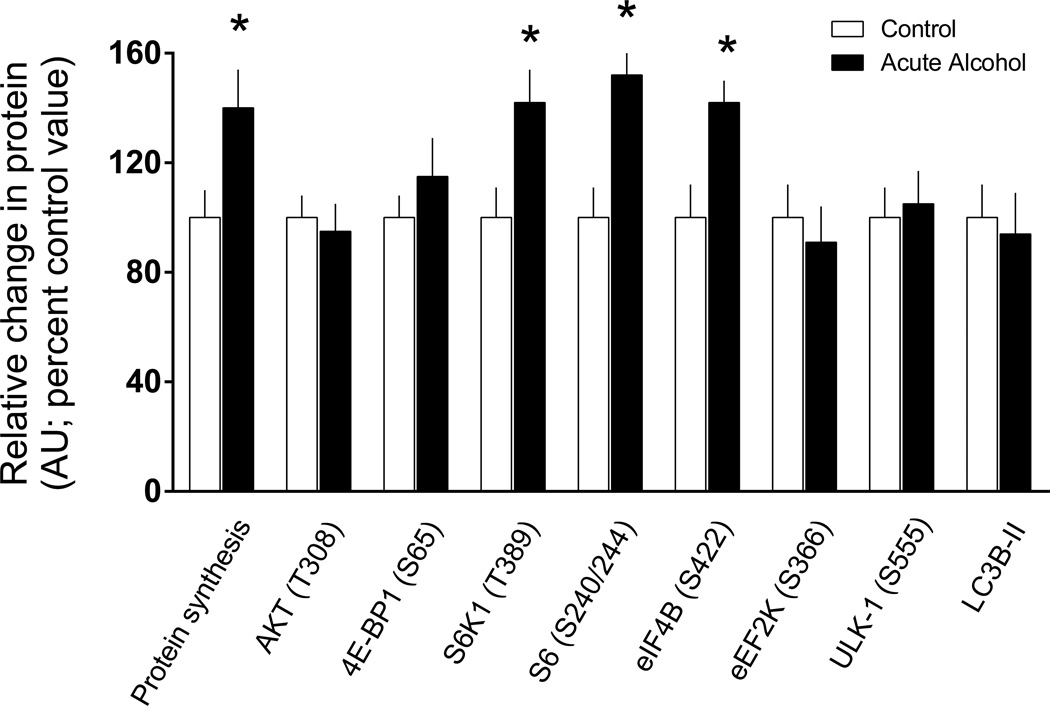
Protein synthesis and signal transduction in response to acute alcohol intoxication
Given the chronic nature of the feeding protocol used in the previous study, it was of interest to determine whether acute alcohol administration produced comparable phenotypical changes in WAT. Four hours after alcohol, we detected 30–50% increases in protein synthesis and the phosphorylation of S6K1, S6 and eIF4B (Figure 8). In contrast, acute alcohol did not alter the phosphorylation state of 4E-BP1, eEF2K or ULK1, or the amount of either LC3B-I or –II in eWAT (Figure 8 and data not shown).
Figure 8.
Protein synthesis and signal transduction in eWAT in response to acute alcohol intoxication. Mice were injected intraperitoneally with 3 mg/kg of ethanol and euthanized 4 hours thereafter. For each variable, data from control-fed mice were arbitrarily set at 100%. Values are means ± SEM; n = 10 and 12, respectively. *P < 0.05 compared to time-matched control values.
Alcohol effect on cultured adipocytes
As assessed by the addition of puromycin incorporation into protein, there was no difference in the relative rate of protein synthesis in 3T3-F442A adipocytes incubated with 80 mM alcohol for 6 hours (98 ± 4 %) compared to time-matched control values (100 ± 3%). Similarly, there was no change in the extent of Thr389-phosphorylated S6K1 between control (100 ± 10 %) and alcohol-treated (87 ± 11%) adipocytes. In contrast, incubation of adipocytes with alcohol decreased Ser65-phosphorylated 4E-BP1 30% compared to control values (control = 100 ± 7% vs alcohol = 69 ± 10%; P < 0.02). A similar trend was seen when adipocytes were cultured with alcohol for 2 h, but the difference in 4E-BP1 phosphorylation between control and alcohol-treated cells did not achieve statistical significance (data not shown). There was also no alcohol-induced change in total protein synthesis at this earlier time point. As the alcohol-induced changes in cultured adipocytes did not mimic those seen in response to either acute or chronic alcohol intake under in vivo conditions, additional analysis was not pursued.
DISCUSSION
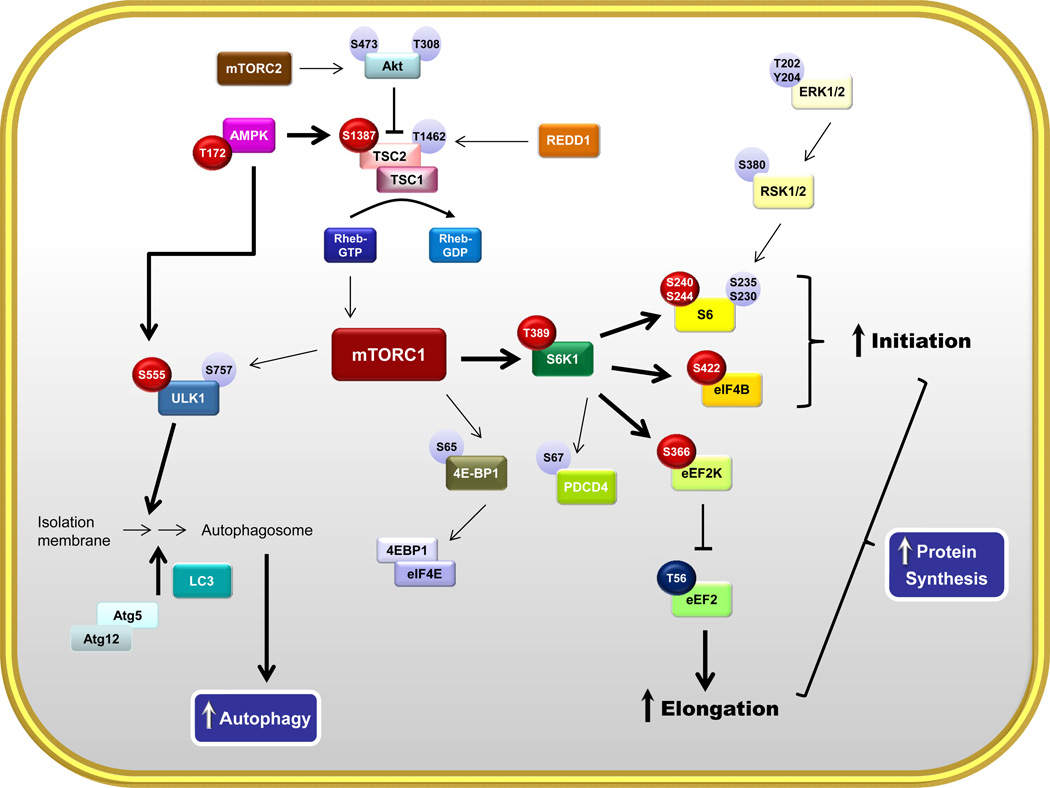
Our results demonstrate that chronic alcohol consumption in adult male mice decreases whole-body fat mass and epididymal fat weight, while unexpectedly increasing global protein synthesis this tissue. The alcohol-induced decrease in eWAT mass is consistent with the majority of rodent studies showing a loss of eWAT mass with chronic alcohol consumption (Pravdova et al., 2009, Zhang et al., 2015, Zhao et al., 2015, Kang et al., 2007a). Although adipose tissue morphology and differences in regional fat depots were not assessed in the current study, previous work has reported that chronic alcohol feeding only decreased mass and adipocyte diameter in eWAT but not subcutaneous WAT (Zhang et al., 2015, Zhong et al., 2012). Moreover, the alcohol-induced decrease in whole-body fat mass in alcohol-fed mice is consistent with results observed in human alcoholics (Liangpunsakul et al., 2010). However, in contrast to the decreased protein synthesis detected in muscle, heart, liver and intestine in chronic alcohol-fed rodents (Preedy and Peters, 1990, Lang et al., 2007, Lang et al., 1999), protein synthesis was increased in eWAT in response to alcohol. The examination of signal transduction pathways in eWAT from alcohol-fed mice provided data supporting the proposed model illustrated in Figure 9 where enhanced activation of S6K1 and its downstream substrates likely contributed to the increased protein synthesis. Furthermore, a direct effect of alcohol on fat seems unlikely as in vitro incubation of adipocytes with alcohol did not mimic changes seen in vivo.
Figure 9.
Schematic signal transduction network representing the effect of chronic alcohol on protein balance in eWAT. Abbreviations are identified within the text of the manuscript. Bold arrows indicate activation of selected pathway components; Red circles indicate an alcohol-induced increased in phosphorylation and the presumed activation of respective substrates; light colored circles indicate phosphorylation sites not altered by alcohol; dark blue circle indicates an alcohol-induced decrease in phosphorylation.
As mTORC1 is a central regulator of protein synthesis and capable of mediating dynamic changes in adipose tissue protein synthesis (Lynch et al., 2003, Lynch et al., 2002a, Laplante and Sabatini, 2009), we examined the activation of mTORC1 in eWAT from alcohol-fed mice. Stimulation of mTORC1 by growth factors is typically evidenced by increased phosphorylation of the immediate downstream substrates 4E-BP1 and S6K1. Increased phosphorylation of 4E-BP1 releases eIF4E from the 4E-BP1•eIF4E complex, thereby permitting the eIF4E•mRNA complex to bind eIF4G and then ultimately to the small ribosomal subunit; a mechanism central to initiating cap-dependent translation. However, there was no detectable change in the phosphorylation state of 4E-BP, and no change in the distribution of eIF4E between the active and inactive complex in eWAT from alcohol-fed mice.
In contrast, alcohol feeding increased the phosphorylation of S6K1 on Thr389 and Thr421/Ser424 in eWAT. An increase in S6K1 activity per se was implied by the phosphorylation of multiple known downstream target proteins. For example, phosphorylation of S6 on Ser240/244 is S6K1-dependent (Ferrari et al., 1991) and was coordinately increased by alcohol. Although an increase in S6K1 and S6 phosphorylation might be expected to increase cell growth and size (Magnuson et al., 2012), no such change in eWAT was evident based on the measurement of tissue mass. This lack of response may be due to the failure of alcohol to simultaneously increase 4E-BP1 phosphorylation that is necessary for adipogenesis (Tsukiyama-Kohara et al., 2001) or because of a correspondingly greater increase in protein degradation (discussed later). Phosphorylation of eIF4B on Ser422 is also S6K1-dependent (Raught et al., 2004) and increases its interaction with eIF3 and eIF4A, thereby increasing helicase activity of the later protein and stimulating translation (Parsyan et al., 2011). Finally, chronic alcohol consumption also increased Ser366-phosphorylated eEF2K, which is also S6K1-mediated (Wang et al., 2001). Such a phosphorylation event inhibits its kinase activity as evidenced in the current study by the concomitant decrease in Thr56-phosphorylated eEF2, that is consistent with a stimulation of peptide-chain elongation. PDCD4 is also an S6K1 substrate and can suppress translation initiation by binding to and inhibiting eIF4A helicase activity (Yang et al., 2003). However, unlike the other S6K1 substrates noted above, alcohol feeding did not increase Ser67-phosphorylated PDCD4. In contrast, chronic alcohol intake increased total PDCD4. While this increase might be anticipated to inhibit protein synthesis (Yang et al., 2003), a recent study has reported that silencing of PDCD4 can either increase or decrease protein synthesis depending upon the developmental state of the tissue (e.g., myoblasts vs myotubes, respectively) (Kakade et al., 2014). It is possible that differences between our data and those in the literature regarding the amount and phosphorylation state of PDCD4 are in part related to tissue-specific differences in regulation. Collectively, our data are largely internally consistent showing alcohol selectively increases S6K1 activity which increases protein synthesis via enhanced peptide-chain initiation and elongation.
A question implicit in these data is whether the increased S6K1 activity results from an increase in mTOR per se or whether alcohol stimulates S6K1 via an mTOR-independent mechanism. Thus, we examined signal transduction pathways upstream of mTORC1. Although Akt is central in growth factor stimulation of mTORC1 (Frost and Lang, 2007), there was no detectable increase in Akt Thr308 or Ser473 phosphorylation in eWAT from alcohol-fed mice. To confirm the lack of change in Akt activation, Thr246-phosphorylated PRAS40, an Akt substrate and negative regulator of mTOR, was assessed and shown to not differ between control and alcohol-fed mice. TSC serves as an integration point by inhibiting mTORC1 (Inoki et al., 2005). Increased Akt signaling increases Thr1462 phosphorylation of TSC2 thereby inhibiting the normal suppressive effects of this protein on mTORC1 (Inoki et al., 2005). Again, consistent with the above-mentioned changes, there was no alcohol-induced change in Thr1462 phosphorylation. Conversely, TSC2 Ser1387-phosphorylation is AMPK-dependent and Akt-independent, and increased activity of the TSC2/TSC1 complex leads to mTORC1 inhibition (Inoki et al., 2006). While this alcohol-induced increase in TSC2 Ser1387 phosphorylation was consistent with the increased Thr172 phosphorylation of AMPK, these changes were not associated with a decrease in protein synthesis. Collectively, the changes or lack thereof in phosphorylation of Akt, TSC2, PRAS40 and AMPK generally support an alcohol-induced mTORC1-independent increase in S6K1 activity. Several studies have reported S6K1 activation independent of the Akt/TSC/mTOR pathway. For example, phosphatidic acid (PA) appears to bind to S6K1 in a specific and saturable manner to increase its kinase activity, and the overexpression of phospholipase D2 which increases PA content and S6K1 activity were not inhibited by rapamycin (Lehman et al., 2007). In this regard, PA and PLD activity are increased in C2C12 myocytes exposed to alcohol (Hong-Brown et al., 2013). Also, specific atypical PKC isoforms may bind to S6K1 increasing its activity (Romanelli et al., 1999, Akimoto et al., 1998).
Autophagy was assessed as a mediator of protein turnover as this pathway is tightly regulated by alterations in both mTORC1 and AMPK (Kim et al., 2011). The phosphorylation of ULK-1 is central in controlling autophagy (Mizushima, 2010). As described above, there is no evidence that alcohol feeding regulates autophagy via an mTORC1-dependent pathway which is substantiated by the comparable extent of ULK1 Ser757 phosphorylation between control and alcohol-fed mice. In contrast, autophagy appears stimulated via AMPK-dependent mechanism which increases ULK1 Ser555 phosphorylation (Kim et al., 2011). Likewise, the Atg12-5 complex as well as the amount of both LC3B-I and –II were increased in alcohol-fed mice, suggesting an increased autophagosome formation. Furthermore, we cannot exclude the possibility of a direct effect of S6K1 on autophagy. For example, although most studies report an inverse relationship between S6K1 phosphorylation and autophagy (Kim et al., 2011), others demonstrate S6K1 is essential for a normal autophagy response under stress conditions (Hac et al., 2015). Hence, more detailed studies on autophagic flux are required that investigate the relative importance of different cellular mechanisms (AMPK vs S6K1) governing the alcohol-induced stimulation autophagy in eWAT. Regardless of the precise mechanism, we speculate the increased autophagy is sufficient to elevate the intracellular concentration of amino acids and thereby provides at least a partial explanation for the observed increase in protein synthesis in eWAT from alcohol-fed mice.
As described above, AMPK phosphorylation was increased in eWAT in alcohol-fed mice. It is noteworthy the AMPK response in fat differs from the commonly observed decrease in AMPK phosphorylation in liver (You et al., 2004) and the lack of change detected in skeletal muscle (Korzick et al., 2013) from rodents chronically consuming an alcohol-containing diet. The underlying mechanism and the functional importance of these apparent tissue-specific changes require further investigation. Activation of AMPK in adipose tissue also decreases basal and adrenergic-stimulated lipolysis (Anthony et al., 2009). Hence, the alcohol-induced increase AMPK phosphorylation in eWAT does not appear to be consistent with previous reports of increased lipolysis (Zhong et al., 2012), which is supported by our data showing increased ATGL content and increased phosphorylation of HSL in eWAT. However, an increased lipolysis is also seen after fasting and exercise, and associated with increased AMPK phosphorylation in adipose tissue (Gauthier et al., 2008). The activation of AMPK in these (and possibly in response to alcohol feeding) may represent a compensatory response mounted to minimize energy depletion and oxidative stress produced by high rates of lipolysis (Gauthier et al., 2008).
Previous work indicates chronic alcohol feeding increases lipolysis and/or decreases lipogenesis (Kang et al., 2007a, Wei et al., 2013). The increase in key lipolytic enzymes ATGL and phosphorylated HSL as well as the decrease in PPARγ and C/EBPα which are key regulators of lipogenesis demonstrate reproducibility among independent laboratories. Likewise, we also detected alcohol-induced increases in TNFα and IL-1β and decreases in adiponectin mRNA in eWAT, all of which have been reported previously (He et al., 2015, Kang et al., 2007b). Although hypoxia may produce such changes (He et al., 2015, Kloting and Bluher, 2014), the lack of an increase in REDD1 in our study does not support such a mechanism (Steiner et al., 2015). It is noteworthy, however, the alcohol-induced inflammatory response in eWAT was not associated with an increase in various NLRP3 inflammatory components suggesting that adipocytes were not dying by pyroptosis (Giordano et al., 2013). Furthermore, despite increases in TNFα in eWAT and in the circulation, that might be expected to stimulate the apoptotic pathway (Herold et al., 2013), we detected no difference in the cleavage products of caspace-9, caspace-3 or PARP between control and alcohol-fed mice.
In other tissues, acute alcohol administration reproduces many of the changes in mTORC1 and protein synthesis observed with chronic alcohol feeding (Steiner and Lang, 2015). Hence, we determined whether the observed increase in protein synthesis in eWAT required prolonged exposure to alcohol. Acute alcohol also increased eWAT protein synthesis that was associated with an elevation in S6K1, S6 and eIF4B phosphorylation, all of which were also increased in fat from chronically alcohol-fed mice. However, in contrast, we failed to detect an increase in the phosphorylation of eEF2K and ULK1, or an increase in LC3B-I and –II. These data suggest a more chronic period of exposure may be required for alcohol to alter peptide-chain elongation and increase autophagy in eWAT. Whether these represent the molecular signature for a detrimental pathological change or a compensatory change attempting to maintain tissue and protein homeostasis remains to be determined.
These data identify a novel S6K1-dependent signaling pathway in eWAT that is stimulated by alcohol, both acute administration and chronic consumption, and show alcohol produces a seemingly contradictory concurrent increase in protein synthesis with a loss of eWAT mass. Thus, the alcohol-induced increase in protein synthesis cannot explain the observed decrease in eWAT mass which likely results from enhanced lipolysis. The alcohol-induced increase in global protein synthesis in eWAT differs from the decrease observed in many other tissues under comparable situations. Also, it appears that an increase in autophagy, but not pyroptosis or apoptosis, may contribute to the decrease in eWAT mass produced by chronic alcohol intake.
Acknowledgments
We thank Maithili Navaratnarajah and Anne Pruznak for their excellent technical assistance. This work was supported by a grant from NIAAA R37 AA011290 (CHL) and by post-doctoral fellowship awards F32 AA023422 (JLS) and F32 GM112401 (KTC).
Footnotes
AUTHOR CONTRIBUTIONS
All authors conceived and designed the study; collected, analyzed, and interpreted the data; and drafted and approved the final manuscript.
None of the authors have any conflict of interests to declare.
REFERENCES
- Akimoto K, Nakaya M, Yamanaka T, Tanaka J, Matsuda S, Weng QP, Avruch J, Ohno S. Atypical protein kinase Clambda binds and regulates p70 S6 kinase. Biochem J. 1998;335(Pt 2):417–424. doi: 10.1042/bj3350417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony NM, Gaidhu MP, Ceddia RB. Regulation of visceral and subcutaneous adipocyte lipolysis by acute AICAR-induced AMPK activation. Obesity (Silver Spring) 2009;17:1312–1317. doi: 10.1038/oby.2008.645. [DOI] [PubMed] [Google Scholar]
- Blanchard PG, Festuccia WT, Houde VP, St-Pierre P, Brule S, Turcotte V, Cote M, Bellmann K, Marette A, Deshaies Y. Major involvement of mTOR in the PPARgamma-induced stimulation of adipose tissue lipid uptake and fat accretion. J Lipid Res. 2012;53:1117–1125. doi: 10.1194/jlr.M021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti P, English T, Shi J, Smas CM, Kandror KV. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Bandi HR, Hofsteenge J, Bussian BM, Thomas G. Mitogen-activated 70K S6 kinase. Identification of in vitro 40 S ribosomal S6 phosphorylation sites. J Biol Chem. 1991;266:22770–22775. [PubMed] [Google Scholar]
- Frost RA, Lang CH. Protein kinase B/Akt: a nexus of growth factor and cytokine signaling in determining muscle mass. J Appl Physiol (1985) 2007;103:378–387. doi: 10.1152/japplphysiol.00089.2007. [DOI] [PubMed] [Google Scholar]
- Gauthier MS, Miyoshi H, Souza SC, Cacicedo JM, Saha AK, Greenberg AS, Ruderman NB. AMP-activated protein kinase is activated as a consequence of lipolysis in the adipocyte: potential mechanism and physiological relevance. J Biol Chem. 2008;283:16514–16524. doi: 10.1074/jbc.M708177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano A, Murano I, Mondini E, Perugini J, Smorlesi A, Severi I, Barazzoni R, Scherer PE, Cinti S. Obese adipocytes show ultrastructural features of stressed cells and die of pyroptosis. J Lipid Res. 2013;54:2423–2436. doi: 10.1194/jlr.M038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hac A, Domachowska A, Narajczyk M, Cyske K, Pawlik A, Herman-Antosiewicz A. S6K1 controls autophagosome maturation in autophagy induced by sulforaphane or serum deprivation. Eur J Cell Biol. 2015;94:470–481. doi: 10.1016/j.ejcb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5'-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- He Z, Li M, Zheng D, Chen Q, Liu W, Feng L. Adipose tissue hypoxia and low-grade inflammation: a possible mechanism for ethanol-related glucose intolerance? Br J Nutr. 2015;113:1355–1364. doi: 10.1017/S000711451500077X. [DOI] [PubMed] [Google Scholar]
- Herold C, Rennekampff HO, Engeli S. Apoptotic pathways in adipose tissue. Apoptosis. 2013;18:911–916. doi: 10.1007/s10495-013-0848-0. [DOI] [PubMed] [Google Scholar]
- Hong-Brown LQ, Brown CR, Navaratnarajah M, Lang CH. Activation of AMPK/TSC2/PLD by alcohol regulates mTORC1 and mTORC2 assembly in C2C12 myocytes. Alcohol Clin Exp Res. 2013;37:1849–1861. doi: 10.1111/acer.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Kakade D, Islam N, Maeda N, Adegoke OA. Differential effects of PDCD4 depletion on protein synthesis in myoblast and myotubes. BMC Cell Biol. 2014;15:2. doi: 10.1186/1471-2121-15-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Chen X, Sebastian BM, Pratt BT, Bederman IR, Alexander JC, Previs SF, Nagy LE. Chronic ethanol and triglyceride turnover in white adipose tissue in rats: inhibition of the anti-lipolytic action of insulin after chronic ethanol contributes to increased triglyceride degradation. J Biol Chem. 2007a;282:28465–28473. doi: 10.1074/jbc.M705503200. [DOI] [PubMed] [Google Scholar]
- Kang L, Sebastian BM, Pritchard MT, Pratt BT, Previs SF, Nagy LE. Chronic ethanol-induced insulin resistance is associated with macrophage infiltration into adipose tissue and altered expression of adipocytokines. Alcohol Clin Exp Res. 2007b;31:1581–1588. doi: 10.1111/j.1530-0277.2007.00452.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloting N, Bluher M. Adipocyte dysfunction, inflammation and metabolic syndrome. Rev Endocr Metab Disord. 2014;15:277–287. doi: 10.1007/s11154-014-9301-0. [DOI] [PubMed] [Google Scholar]
- Korzick DH, Sharda DR, Pruznak AM, Lang CH. Aging accentuates alcohol-induced decrease in protein synthesis in gastrocnemius. Am J Physiol Regul Integr Comp Physiol. 2013;304:R887–R898. doi: 10.1152/ajpregu.00083.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang CH, Frost RA, Vary TC. Skeletal muscle protein synthesis and degradation exhibit sexual dimorphism after chronic alcohol consumption but not acute intoxication. Am J Physiol Endocrinol Metab. 2007;292:E1497–E1506. doi: 10.1152/ajpendo.00603.2006. [DOI] [PubMed] [Google Scholar]
- Lang CH, Wu D, Frost RA, Jefferson LS, Vary TC, Kimball SR. Chronic alcohol feeding impairs hepatic translation initiation by modulating eIF2 and eIF4E. Am J Physiol. 1999;277:E805–E814. doi: 10.1152/ajpendo.1999.277.5.E805. [DOI] [PubMed] [Google Scholar]
- Lang SM, Kazi AA, Hong-Brown L, Lang CH. Delayed recovery of skeletal muscle mass following hindlimb immobilization in mTOR heterozygous mice. PLoS One. 2012;7:e38910. doi: 10.1371/journal.pone.0038910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman N, Ledford B, Di Fulvio M, Frondorf K, McPhail LC, Gomez-Cambronero J. Phospholipase D2-derived phosphatidic acid binds to and activates ribosomal p70 S6 kinase independently of mTOR. FASEB J. 2007;21:1075–1087. doi: 10.1096/fj.06-6652com. [DOI] [PubMed] [Google Scholar]
- Liangpunsakul S, Crabb DW, Qi R. Relationship among alcohol intake, body fat, and physical activity: a population-based study. Ann Epidemiol. 2010;20:670–675. doi: 10.1016/j.annepidem.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CJ. Role of leucine in the regulation of mTOR by amino acids: revelations from structure-activity studies. J Nutr. 2001;131:861S–865S. doi: 10.1093/jn/131.3.861S. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Halle B, Fujii H, Vary TC, Wallin R, Damuni Z, Hutson SM. Potential role of leucine metabolism in the leucine-signaling pathway involving mTOR. Am J Physiol Endocrinol Metab. 2003;285:E854–E863. doi: 10.1152/ajpendo.00153.2003. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Hutson SM, Patson BJ, Vaval A, Vary TC. Tissue-specific effects of chronic dietary leucine and norleucine supplementation on protein synthesis in rats. Am J Physiol Endocrinol Metab. 2002a;283:E824–E835. doi: 10.1152/ajpendo.00085.2002. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Patson BJ, Anthony J, Vaval A, Jefferson LS, Vary TC. Leucine is a direct-acting nutrient signal that regulates protein synthesis in adipose tissue. Am J Physiol Endocrinol Metab. 2002b;283:E503–E513. doi: 10.1152/ajpendo.00084.2002. [DOI] [PubMed] [Google Scholar]
- Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999;344(Pt 2):427–431. [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- Pravdova E, Macho L, Fickova M. Alcohol intake modifies leptin, adiponectin and resistin serum levels and their mRNA expressions in adipose tissue of rats. Endocr Regul. 2009;43:117–125. [PubMed] [Google Scholar]
- Preedy VR, Peters TJ. Protein synthesis of muscle fractions from the small intestine in alcohol fed rats. Gut. 1990;31:305–310. doi: 10.1136/gut.31.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–1769. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath NT, Foulstone EJ, Proud CG. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. EMBO J. 1996;15:2291–2297. [PMC free article] [PubMed] [Google Scholar]
- Rolfe M, McLeod LE, Pratt PF, Proud CG. Activation of protein synthesis in cardiomyocytes by the hypertrophic agent phenylephrine requires the activation of ERK and involves phosphorylation of tuberous sclerosis complex 2 (TSC2) Biochem J. 2005;388:973–984. doi: 10.1042/BJ20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanelli A, Martin KA, Toker A, Blenis J. p70 S6 kinase is regulated by protein kinase Czeta and participates in a phosphoinositide 3-kinase-regulated signalling complex. Mol Cell Biol. 1999;19:2921–2928. doi: 10.1128/mcb.19.4.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian BM, Roychowdhury S, Tang H, Hillian AD, Feldstein AE, Stahl GL, Takahashi K, Nagy LE. Identification of a cytochrome P4502E1/Bid/C1q-dependent axis mediating inflammation in adipose tissue after chronic ethanol feeding to mice. J Biol Chem. 2011;286:35989–35997. doi: 10.1074/jbc.M111.254201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, Fingar DC. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem. 2010;285:7866–7879. doi: 10.1074/jbc.M109.096222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JL, Gordon BS, Lang CH. Moderate alcohol consumption does not impair overload-induced muscle hypertrophy and protein synthesis. Physiol Rep. 2015;3 doi: 10.14814/phy2.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JL, Lang CH. Alcohol impairs skeletal muscle protein synthesis and mTOR signaling in a time-dependent manner following electrically stimulated muscle contraction. J Appl Physiol (1985) 2014;117:1170–1179. doi: 10.1152/japplphysiol.00180.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JL, Lang CH. Dysregulation of skeletal muscle protein metabolism by alcohol. Am J Physiol Endocrinol Metab. 2015;308:E699–E712. doi: 10.1152/ajpendo.00006.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Sebastian BM, Axhemi A, Chen X, Hillian AD, Jacobsen DW, Nagy LE. Ethanol-induced oxidative stress via the CYP2E1 pathway disrupts adiponectin secretion from adipocytes. Alcohol Clin Exp Res. 2012;36:214–222. doi: 10.1111/j.1530-0277.2011.01607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, Harper ME, Tremblay ML, Sonenberg N. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shi X, Zhong W, Zhao Y, Tang Y, Sun W, Yin X, Bogdanov B, Kim S, McClain C, Zhou Z, Zhang X. Chronic alcohol exposure disturbs lipid homeostasis at the adipose tissue-liver axis in mice: analysis of triacylglycerols using high-resolution mass spectrometry in combination with in vivo metabolite deuterium labeling. PLoS One. 2013;8:e55382. doi: 10.1371/journal.pone.0055382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Matsumoto M, Pacold CM, Cho WK, Crabb DW. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 2004;127:1798–1808. doi: 10.1053/j.gastro.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Zhang HH, Huang J, Duvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One. 2009;4:e6189. doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhong W, Sun X, Sun Q, Tan X, Li Q, Sun X, Zhou Z. Visceral white adipose tissue is susceptible to alcohol-induced lipodystrophy in rats: role of acetaldehyde. Alcohol Clin Exp Res. 2015;39:416–423. doi: 10.1111/acer.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Liu Y, Xiao J, Liu L, Chen S, Mohammadi M, McClain CJ, Li X, Feng W. FGF21 mediates alcohol-induced adipose tissue lipolysis by activation of systemic release of catecholamine in mice. J Lipid Res. 2015;56:1481–1491. doi: 10.1194/jlr.M058610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Zhao Y, Tang Y, Wei X, Shi X, Sun W, Sun X, Yin X, Sun X, Kim S, McClain CJ, Zhang X, Zhou Z. Chronic alcohol exposure stimulates adipose tissue lipolysis in mice: role of reverse triglyceride transport in the pathogenesis of alcoholic steatosis. Am J Pathol. 2012;180:998–1007. doi: 10.1016/j.ajpath.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]