Abstract
Amyotrophic Lateral Sclerosis (ALS) is a severe and fatal neurodegenerative disease characterized by progressive loss of motoneurons, muscle atrophy and paralysis. Recent evidence suggests that ALS should be considered as a multi-systemic disease, in which several cell types contribute to motoneuron degeneration. In this view, mutations in ALS linked genes in other neural and non-neural cell types may exert non-cell autonomous effects on motoneuron survival and function. Induced Pluripotent Stem Cells (iPSCs) have been recently derived from several patients with ALS mutations and it has been shown that they can generate motoneurons in vitro, providing a valuable tool to study ALS. However, the potential of iPSCs could be further valorized by generating other cell types that may be relevant to the pathology. In this paper, by taking advantage of a novel inducible system for MyoD expression, we show that both control iPSCs and iPSCs carrying mutations in ALS genes can generate skeletal muscle cells. We provide evidence that both control and mutant iPSC-derived myotubes are functionally active. This in vitro system will be instrumental to dissect the molecular and cellular pathways impairing the complex motoneuron microenvironment in ALS.
Keywords: Induced Pluripotent Stem Cells, Amyotrophic Lateral Sclerosis, FUS/TLS, TDP-43, Skeletal muscle
Highlights
-
•
A novel method for inducing iPSCs differentiation into muscle is presented
-
•
Multiple inducible lines can be easily generated by a new transposable vector
-
•
Both control and iPSCs carrying ALS mutations can generate functional muscle fibers
-
•
This system will be instrumental to study non-cell autonomous contributions to ALS
1. Introduction
Amyotrophic Lateral Sclerosis (ALS) is a fatal disease that leads to death due to loss of muscle function. A subset of ALS cases has a clear genetic component and in the last years the list of genes associated with the disease has been greatly expanded (Renton et al., 2014). Among them, the gene encoding for the Cu/Zn superoxide dismutase 1 (SOD1) was the first one being associated with familial ALS (fALS) (Rosen et al., 1993). Other fALS mutations in the RNA binding proteins Fused in Sarcoma/Translocated in Liposarcoma (FUS/TLS or FUS) and Tar DNA Binding Protein 43 (TDP-43) suggest that RNA metabolism may play a relevant role in ALS pathogenesis (Lagier-Tourenne et al., 2010). Despite several pathogenic mechanisms have been proposed for the role of mutated SOD1, FUS and TDP-43 in ALS, a clear understanding of the molecular and cellular pathways leading to motoneuron degeneration and muscle atrophy is still missing. This may be partly due to the multi-systemic nature of ALS. The non-cell-autonomous effects on motoneurons of ALS mutations in other cell types have been quite extensively studied for SOD1 (Musarò, 2012). SOD1 mutant motoneurons mortality was reduced in chimaeric mice having WT nonneuronal cells and, conversely, WT motoneurons surrounded by mutant glia showed ALS hallmarks, such as ubiquitin aggregates (Clement et al., 2003, Yamanaka et al., 2008). These in vivo studies are also supported by analysis of co-culture in vitro systems, in which mutant astrocytes increased neurodegeneration of WT motoneurons derived from mouse or human pluripotent cells (Di Giorgio et al., 2007, Nagai et al., 2007, Di Giorgio et al., 2008). Non-cell autonomous effects of SOD1 mutations have been observed not only for astrocytes, but also for other non-neuronal cells. For instance, expression of mutant SOD1 in microglia affected disease progression in mice (Boillee et al., 2006). Moreover, it has been shown that mutant SOD1 expression in skeletal muscle led to muscle atrophy and functional impairment in a mouse model (Dobrowolny et al., 2008).
A new twist in the study of ALS has come by the generation of human induced Pluripotent Stem Cells (iPSCs). As iPSCs can be derived from patients carrying ALS mutations and can differentiate into a wide range of cell types, they represent a valuable opportunity for disease modeling in vitro. Several groups have reported the derivation and characterization of iPSCs derived from fALS individuals with mutations in SOD1, TDP-43 and, more recently, FUS (Dimos et al., 2008, Boulting et al., 2011, Bilican et al., 2012, Egawa et al., 2012, Lenzi et al., 2015). Relevant molecular and cellular disease phenotypes have been detected in ALS-iPSCs differentiated to motoneurons, including delocalization of mutant proteins (Bilican et al., 2012, Egawa et al., 2012, Lenzi et al., 2015), neurite degeneration (Chen et al., 2014), electrophysiological defects (Wainger et al., 2014), increased oxidative stress (Kiskinis et al., 2014) and vulnerability (Bilican et al., 2012, Egawa et al., 2012, Kiskinis et al., 2014). These reports provide the proof of principle that iPSCs can be used to model ALS disease in vitro. iPSCs are pluripotent and can generate multiple cell types, provided that appropriate differentiation protocols are used. In this work, we designed and optimized a protocol for muscle differentiation from iPSCs. As previously shown, human pluripotent cells can be converted into muscle by the ectopic expression of myogenic factors (Darabi and Perlingeiro, 2014, Tedesco et al., 2012, Tanaka et al., 2013, Abujarour et al., 2014). Our strategy is based on the inducible expression of the master gene MyoD from an integrative vector, derived from the enhanced piggyBac transposon (Lacoste et al., 2009). We show that control and ALS iPSC lines can give rise to mature myotubes endowed with functional properties. A convenient feature of our protocol is the possibility to produce stable cell lines that can be induced to muscle differentiation by doxycycline treatment.
2. Materials and methods
2.1. Generation and maintenance of human iPSCs
The iPSC lines were generated and validated as described in Lenzi et al. (2015). All iPSC lines were maintained in Nutristem-XF (Biological Industries) in plates coated with hESC-qualified Matrigel (Corning) and passaged every 4–5 days with 1 mg/ml Dispase (Gibco). Clones used in the experiments shown were: WT I clone#1; ALS I (FUSR514S/WT) clone#1; ALS II (FUSR521C/WT) clone#34 and ALS III (TDP-43A382T/A382T) clone#2.
2.2. Plasmid construction and generation of stable iPSCs lines
The epB-Puro-TT-mMyoD plasmid was generated by inserting the coding sequence of mouse MyoD (GenBank: M84918.1) in the enhanced piggyBac transposable vector epB-Puro-TT (Rosa et al., 2014). The resulting construct contains the piggyBac terminal repeats flanking a constitutive cassette driving the expression of the Puromycin resistance gene fused to the rtTA gene and, in the opposite direction, a tetracycline-responsive promoter element (TRE) driving the expression of MyoD. iPSCs were co-transfected with 4 μg epB-Puro-TT-mMyoD and 1 μg of the piggyBac transposase using the Neon Transfection System (Life Technologies) as previously described (Lenzi et al., 2015). Selection in 0.5 μg/ml puromycin gave rise to stable and inducible cell lines.
2.3. Differentiation of iPSCs into muscle cells
iPSCs were passaged (passage number around 10 to 20) in 35 mm dishes. For differentiation as colonies (Fig. 1A, Colonies), the medium was replaced with HUESM (DMEM-F12 + Glutamax, Life Technologies; 20% Knockout Serum Replacement, Life Technologies; 1X Non-Essential Aminoacids, NEAA, Life Technologies; 100 U/ml Penicillin + 100 μg/ml Streptomycin, Sigma; 0,1 mM β-mercaptoethanol, Gibco). This is considered day 0 (D0). At day 6 the medium was replaced with Growth Medium (GM; DMEM High Glucose Medium, Sigma; 20% FBS, Sigma; 25 ng/ml bFGF, Corning; 10 ng/ml EGF, Corning; 50 μg/ml Insulin, Roche; 2 mM L-Glutamine, Sigma; 100 U/ml Penicillin + 100 μg/ml Streptomycin, Sigma) in presence of 200 ng/ml doxycycline (Sigma). At day 8 the medium was replaced with Differentiation Medium (Skeletal Muscle Cell Differentiation Medium, Promocell; 100 U/ml Penicillin + 100 μg/ml Streptomycin, Sigma) in presence of 200 ng/ml doxycycline. For differentiation as single cells (Fig. 1A, Single cells), iPSCs colonies were dissociated as single cells with Trypsin-EDTA 1X (EuroClone) and 150,000 cells seeded on 35 mm dishes in Nutristem-XF supplemented with 10 μM Rock inhibitor (Y-27632, Sigma) to enhance survival upon dissociation. The next day, the medium was replaced with HUESM (D0). At day 5 the medium was replaced with GM supplemented with 200 ng/ml doxycycline. At day 7 the medium was replaced with DM supplemented with 200 ng/ml doxycycline. For the experiments shown in Fig. 2, the outline is depicted in Fig. 2A. Differentiation started from 60,000 cells seeded as single cells in each well of a 12-well plate for condition A and 1–5 and from 40,000 cells in each well of a 12-well plate for condition B. For the experiments shown in Fig. 4, 60,000 cells, dissociated as single cells, were seeded in 35 mm dishes and maintained in Nutristem-XF supplemented with 10 μM Rock inhibitor for 2 days, before the switch to HUESM (D0).
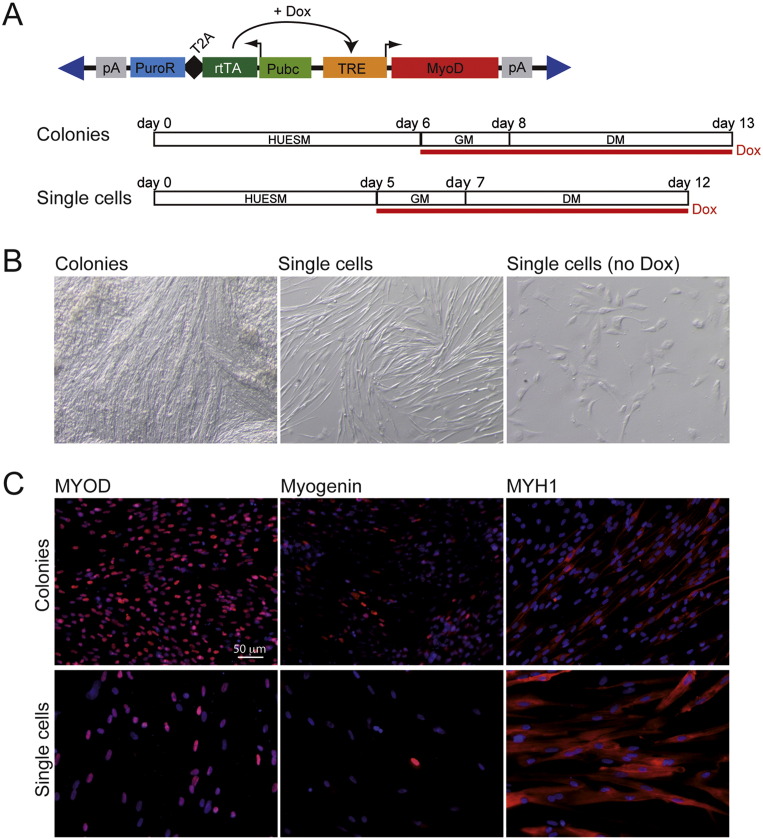
Fig. 1.
Inducible MyoD expression vector and muscle differentiation protocol.
(A) Top: schematic representation of the epB-Puro-TT-mMyoD construct. pA: polyadenylation signal; PuroR: puromycin resistance gene; T2A: self-cleavage peptide; rtTA: TET transactivator protein gene; Pubc: human Ubiquitin C constitutive promoter; TRE: TET responsive element; Dox: doxycycline. Blue triangles represent terminal repeats of the transposon. Bottom: diagrams of the differentiation protocols, starting from iPSC colonies (Colonies) or dissociated as single cells (Single cells). HUESM: iPSC differentiation medium; GM: myoblast growth medium; DM: myoblast differentiation medium. Red line: time in doxycycline (Dox). Time points of medium change are indicated above. (B) Bright field imaging of differentiated WT I-MyoD cells. “Single cells (no Dox)” indicate a control differentiation in which doxycycline had been omitted. (C) Immunostaining for the muscle markers (red): MYOD (left panels), Myogenin (middle panels) or Myosin Heavy Chain (MYH1; right panels), in WT I-MyoD cells differentiated as shown in panel (A). Nuclei are counterstained with DAPI. Scale bar for all panels: 50 μm. The percentages of positive cells were: 94.5% (Colonies; n = 1039 nuclei) versus 90.8% (Single Cells; n = 185 nuclei) for MYOD; 38.6% (Colonies; n = 690 nuclei) versus 16.1% (Single Cells; n = 31 nuclei) for Myogenin; 37.7% (Colonies; n = 2915 nuclei) versus 67.5% (Single Cells; n = 3905 nuclei) for MYH1.
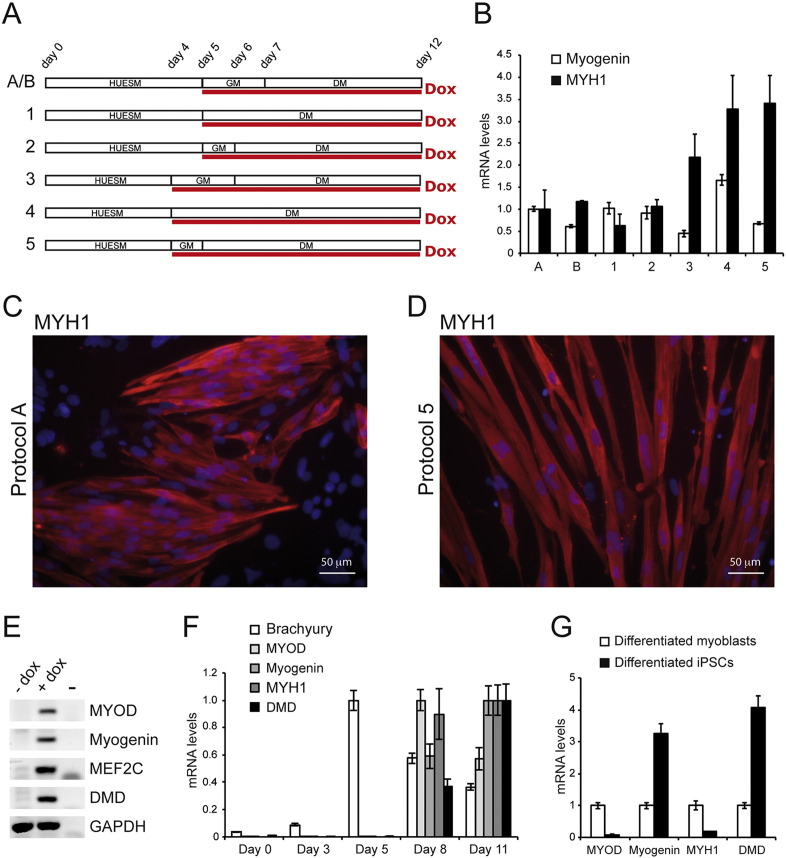
Fig. 2.
Optimization of the differentiation protocol.
(A) Diagrams of the variants of the differentiation protocol of WT I-MyoD cells. HUESM: iPSC differentiation medium; GM: myoblast growth medium; DM: myoblast differentiation medium. Red line: time in doxycycline (Dox). Time points of medium change are indicated above. See text for details. (B) Real-time qRT-PCR analysis of Myogenin (white bars) or Myosin Heavy Chain (MYH1; black bars) in cells differentiated as depicted in panel (A). Relative levels of mRNA were calculated with the delta delta Ct method and condition A is used as the calibrator sample. Error bars: standard error from a triplicate. (C–D) Immunostaining for the muscle marker MYH1 in WT I-MyoD cells differentiated with protocol A (panel C) or 5 (panel D). Nuclei are counterstained with DAPI. Scale bar for both panels: 50 μm. (E) RT-PCR analysis of muscle markers in WT I-MyoD cells differentiated with protocol 5 in absence (− Dox) or presence (+ Dox) of doxycycline. GAPDH is used as a housekeeping control. (F-G) Real-time qRT-PCR analysis of the indicated markers in a time-course experiment using protocol 5 (panel F) and in differentiated iPSCs (protocol 5, day 11) compared with differentiated human myoblasts (panel G). Relative levels of mRNA were calculated with the delta delta Ct method. In panel F, for each marker the condition with higher expression has been used as the calibrator sample. In panel G, differentiated myoblasts represent the calibrator sample. Error bars: standard error from a triplicate.
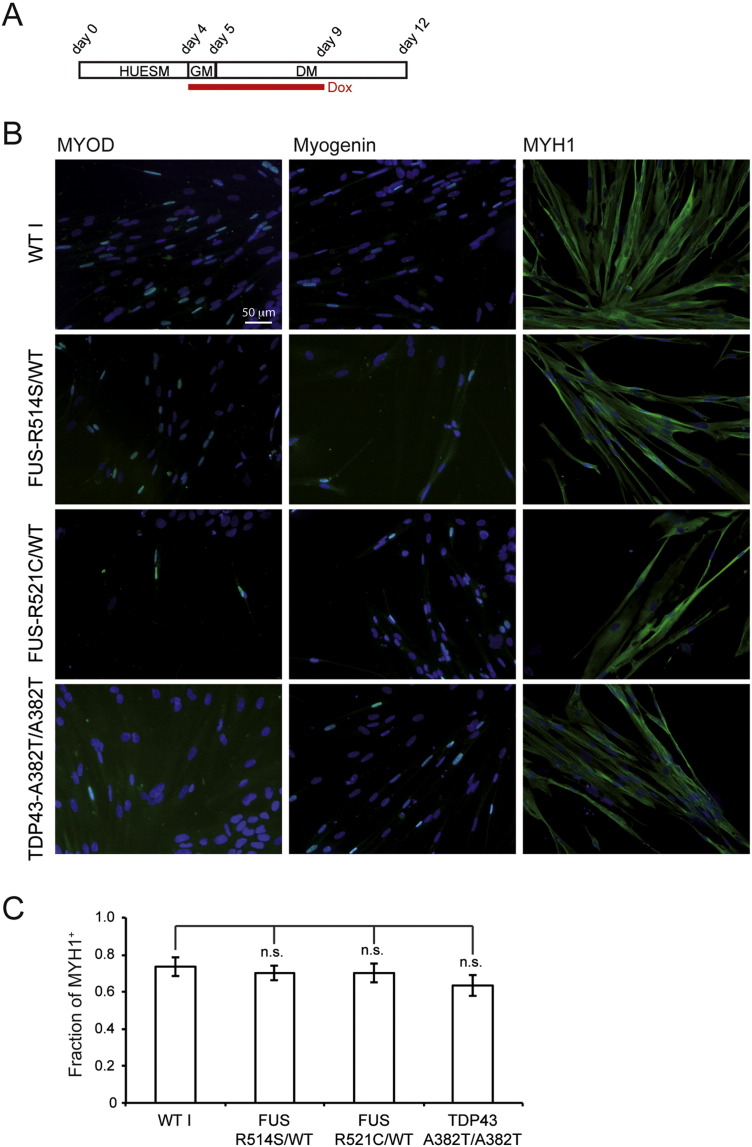
Fig. 4.
Muscle differentiation of ALS-iPSC-MyoD lines.
(A) Diagram of the differentiation protocol. HUESM: iPSC differentiation medium; GM: myoblast growth medium; DM: myoblast differentiation medium. Red line: time in doxycycline (Dox). Time points of medium change are indicated above. (B) Immunostaining for the muscle markers (green): MYOD (left panels), Myogenin (middle panels) or MYH1 (right panels), in differentiated WT I-MyoD cells or in iPSC-FUSR514S/WT-MyoD, iPSC-FUSR521C/WT-MyoD and iPSC-TDP-43A382T/A382T-MyoD differentiated ALS mutant cells. Nuclei are counterstained with DAPI. Scale bar for all panels: 50 μm. (C) Quantification of the fraction of MYH1-positive cells in control and ALS differentiated cells. Histogram bars represent averages ± S.E.M. (30 fields and at least 1000 nuclei counted per line) and n.s. indicates that the differences between mutant lines and the WT I control are not statistically significant (p > 0.1; Student's t-test).
2.4. RT-PCR and real-time qRT-PCR
Total RNA was extracted with the RNeasy kit (Qiagen). For real-time qRT-PCR and RT-PCR RNA was retrotranscribed with the SuperScriptIII kit (Invitrogen). As negative controls, minus-reverse transcriptase samples have been included in subsequent amplification reactions (not shown). For RT-PCR, cDNA was used as template with the BioTaq DNA polymerase (Bioline). Real-time qRT–PCR analysis was performed with SYBR Green QPCR Master Mix (Qiagen) in a 7500 Fast Real Time PCR System (Life Technologies) and calculations performed with the delta delta Ct method. The internal control is the housekeeping gene ATP5O (ATP synthase, H + transporting, mitochondrial F1 complex, O subunit) in real-time qRT–PCR analyses and GAPDH for RT-PCR. Primers sequences are reported in Supplemental Table 1.
2.5. Immunostaining
Cells were fixed in 4% paraformaldehyde for 20 min at room temperature and washed twice with PBS. Fixed cells were then permeabilized with PBS containing 1% BSA and 0.2% Triton X-100 and incubated with primary antibodies: monoclonal mouse anti-MyoD1, clone 5.8 A (1:50, 4 °C overnight, DAKO M351201), anti-Myosin Heavy Chain (myosin II) monoclonal mouse IgG2B from hybridoma culture (Clone #MF20) undiluted supernatant (room temperature, 1 h), anti-MyoG monoclonal mouse from hybridoma culture (Clone #F5D) undiluted supernatant (room temperature, 1 h). The secondary antibodies used are: GOAT anti-mouse Alexa Fluor 488 (1:300, room temperature 1 h, Invitrogen), GOAT anti-mouse Cy3 (1:600, room temperature 1 h, Jackson ImmunoResearch). DAPI (Sigma-Aldrich) was used to label nuclei. Cells were imaged with an Axioscope (Zeiss) microscope. For quantitative analysis, cells were imaged with an inverted Olympus iX73 microscope equipped with an X-light Nipkow spinning-disk head (Crest Optics) and Lumencor Spectra X LED illumination; images were collected using a CoolSNAP MYO CCD camera (Photometrics) and analyzed with ImageJ.
2.6. Patch-clamp recordings
Patch-clamp recordings were obtained using glass electrodes (3–5 M Ω) filled with the following intracellular solution (in mM): 140 KCl, 2 MgCl2, 10 HEPES, 2 MgATP, 5 BAPTA; pH 7.3, with KOH. During experiments, cells were continuously superfused with a normal extracellular solution (NES) containing (in mM): 140 NaCl, 2.5 KCl, 2 CaCl2, 2 MgCl2, 10 HEPES-NaOH and 10 glucose (pH 7.3), using a gravity-driven perfusion system connected to a VC-6 valve controller (Warner Instruments). All recordings were performed at 24–25 °C. ACh-evoked currents, recorded with a patch-clamp amplifier (Axopatch 200B; Molecular Devices, Foster city, CA, USA), were filtered at 1 kHz, digitized (10 kHz) and acquired with Clampex 10 software (Molecular Devices). Cells were voltage-clamped at a holding potential of − 70 mV. ACh dose–response were studied applying to each cell three different doses of the transmitter (3, 30, 300 μM) for 1 s. ACh-evoked currents were blocked treating cells with α-bungarotoxin (5 μg/ml; α-Bungarotoxin-ATTO-488, Alomone Labs) for 2 h. Data were analyzed off line with Clampfit 10 sofware; Origin 7 software was used for statistical analysis of electrophysiological data.
2.7. Intracellular calcium measurements
Fluorescence determinations were performed by using a conventional fluorescence microscopy system composed of an upright microscope (Axioscope 1, Zeiss), a digital 12-bit cooled camera (SensiCam), and a monochromator (Till Photonics). The system was driven by Metafluor (Molecular Devices). Images were acquired and stored on a HP PC, then analyzed offline. Before the experiments, cells were incubated with Fura-2 AM (3 μM) for 45 min and then extensively washed with NES. To measure Fura2 fluorescence, cells were visualized with a 40 × objective and illuminated with a Till 2 monocromator at 340 nm and 380 nm excitation wavelengths. Fluorescent images were recorded every 500 ms with a Photometrics (coolsnap ez) camera. Images were binned 2 × 2 recorded and background-subtracted ratio images produced using Metafluor software (Molecular Dynamics). Changes in intracellular calcium were determined from the changes of fluorescence ratio (F340/F380).
3. Results
3.1. Inducible MyoD expression by enhanced piggyBac converted iPSCs into muscle
The piggyBac vector is a transposable element that, in presence of the piggyBac transposase, can mediate the integration of exogenous DNA sequences into the genome of pluripotent cells. It has been previously shown that inducible expression of a MyoD transgene carried by a piggyBac vector could induce iPSC muscle differentiation (Tanaka et al., 2013, Shoji et al., 2016). The enhanced version of piggyBac (epB) carries an engineered terminal repeat, which increases transposition efficiency in human pluripotent stem cells (Lacoste et al., 2009, Rosa and Brivanlou, 2011, Rosa et al., 2014). We generated an epB construct containing the mouse MyoD coding sequence under the control of a doxycycline-inducible promoter (epB-Puro-TT-mMyoD; Fig. 1A). This vector also contains the TET transactivator (rtTA) and a puromycin resistance gene under a ubiquitous promoter. We co-transfected the epB-Puro-TT-mMyoD vector with the piggyBac transposase, expressed in trans from an independent plasmid, into iPSCs derived from a healthy donor (WT I; Lenzi et al., 2015). Upon puromycin selection we obtained a stable cell line (WT I-MyoD). When we induced MyoD expression in iPSCs maintained in pluripotency medium, we observed massive cell death in absence of myogenic differentiation (data not shown). This is in line with a recent report, showing that the absence of the epigenetic factor BAF60c in undifferentiated human pluripotent cells confers resistance to MyoD-mediated activation of skeletal myogenesis (Albini et al., 2013; Fig. S1). We bypassed this constrain by inducing multilineage differentiation before doxycycline treatment. WT I-MyoD cells were cultured in differentiation medium (HUESM) for six days, then switched to myoblast growth medium (GM) in presence of doxycycline for two days and finally cultured in myoblast differentiation medium (DM) for additional five days (Fig. 1A, Colonies). The morphology of the iPSCs become similar to myotube-like cells (Fig. 1B) and immunostaining analysis showed expression of MYOD, Myogenin (early muscle differentiation marker) and Myosin Heavy Chain (MYH1, late muscle differentiation marker) in some cells (Fig. 1C). At the end of differentiation, however, many cells were still negative for MYH1. To improve the efficiency and reproducibility of differentiation, we dissociated WT I-MyoD to single cells before differentiation (Fig. 1A, Single Cells). This step improved the morphology of differentiated cells and increased the number of MYH1 positive myotubes (Fig. 1B, C and S1; see Fig. 1 legend for quantification of positive cells).
3.2. Optimization of the differentiation protocol
With the aim to further improve the efficiency of iPSCs muscle differentiation, we repeated MyoD induction in WT I-MyoD cells under different conditions. A schematic representation of seven alternative protocols is shown in Fig. 2A. Protocol A corresponds to the “Single Cells” protocol of Fig. 1. In protocol B we started from fewer cells at day 0 (see Materials and Methods). Protocols 1–5 differ for the time of doxycycline induction and medium switch. We collected the cells at day 12 and analyzed the expression of early (Myogenin) and late (MYH1) muscle markers by real-time quantitative PCR (Fig. 2B). We observed that anticipating the switch to GM and/or doxycycline exposure resulted to an increase of MYH1 mRNA levels (protocols 3, 4 and 5). We confirmed this hypothesis by analyzing MYH1 protein levels by immunostaining (Fig. 2C and D). Compared to protocol A, protocol 5 led to an increase of the fraction of MYH1-positive cells (67.5% to 77.1%) and a slight increase in the average number of nuclei per myotube (1.19 to 1.23; n = 3905 nuclei, SD = 0.094, for condition A; n = 1995 nuclei, SD = 0.073, for condition 5; p = 0.03, t-test).
We further characterized cells differentiated with protocol 5 for the expression of other myogenic markers, such as DYSTROPHIN (DMD), MEF2C and endogenous MYOD detected with human-specific primers (Fig. 2E). A time course analysis during differentiation suggested that the expression of muscle genes might result from gradual physiological myogenic maturation. As shown in Fig. 2F, a peak of the expression of the mesoderm marker Brachyury (T) at day 5 is followed by a maximum of endogenous MYOD expression at day 8. Late markers Myogenin, MYH1 and DMD could be detected at day 8 and further increase at day 11, while MYOD levels decrease. Finally, we comparatively analyzed the expression of myogenic markers in differentiated iPSCs and in differentiated human myoblasts. Human myoblasts displayed higher levels of MYOD and MYH1, while Myogenin and DMD were more abundant in iPSC-derived muscle cells (Fig. 2G).
Taken together, these results suggested that by modifying differentiation conditions we improved the conversion of iPSCs into muscle cells, which acquire the expression of relevant markers at levels comparable to differentiated human myoblasts and in a fashion that resembles physiological myogenic maturation.
3.3. Functional analysis of WT I-MyoD iPSC-derived skeletal muscle cells
To assess the functional differentiation of WT I-MyoD cells, we first analyzed their passive electrical properties, specifically the membrane capacitance, which provides a measure of the cell surface, and the resting membrane potential. After 12 days of differentiation, membrane capacitance of WT I-MyoD cells was 35.0 ± 5.5 pF (n = 17) and their resting membrane potential was − 31.7 ± 4.4 mV (n = 16), a value similar to the one previously reported for fusion competent myoblasts and mononucleate C2C12 cells (Grassi et al., 2004).
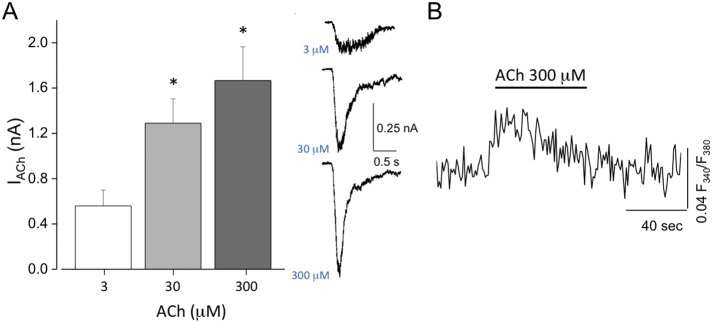
Patch-clamp recordings of differentiated WT I-MyoD cells showed the functional expression of nicotinic acetylcholine (ACh) receptors. Indeed the acute application of ACh (30 μM, 1 s) always induced inward current responses ranging between − 0.4 and − 3 nA (mean amplitude = − 1.3 ± 0.2 nA; ACh 30 μM; n = 11; HP = − 70 mV). ACh induced currents clearly showed dose dependence in a range of ACh concentrations (3, 30, 300 μM; p < 0.05, One Way ANOVA; Fig. 3A). ACh induced currents were due to the activation of nicotinic receptors, as the treatment with α-bungarotoxin completely abolished the currents (not shown).
Fig. 3.
Functional properties of WT iPSC-derived skeletal muscle cells.
(A) Left panel: dose-response relation of ACh-evoked currents in skeletal muscle cells differentiated from WT I-MyoD iPSCs with the protocol 5 described in Fig. 2 (*, p < 0.05, OneWay ANOVA). Right panel: sample traces of the three concentrations of ACh applied to a WT iPSC-derived skeletal muscle cell. (B) Average fluorescence response to ACh (300 μM), in Fura-2-loaded skeletal muscle cell differentiated from WT iPSC with the protocol 5 described in Fig. 2 (n = 22).
To further assess the functional differentiation of iPSC-derived muscle cells, we performed [Ca2 +]i imaging experiments by loading cells with Fura2 AM, a membrane-permeant and ratiometric Ca2 +-sensitive dye. In these cells, the application of ACh (3, 30, 300 μM; 1 s) induced a sustained rapid and reversible increase of intracellular calcium (n = 22 cells; Fig. 3B), indicating that ACh receptors (AChRs) activation allowed calcium influx into the cells. These results suggest that differentiated WT I-MyoD cells expressed functional AChRs and that their activation led to intracellular calcium elevation, which is necessary for muscle contraction.
3.4. ALS-iPSCs can be differentiated into functionally mature myotubes
Several patient-derived ALS iPSC lines are available in our lab (Lenzi et al., 2015). We cotransfected epB-Puro-TT-mMyoD and the piggyBac transposase in these cells generating three additional MyoD inducible lines: iPSC-FUSR514S/WT-MyoD, iPSC-FUSR521C/WT-MyoD and iPSC-TDP-43A382T/A382T-MyoD. We then induced muscle differentiation in these cells in parallel with WT I-MyoD iPSCs using an optimized protocol (depicted in Fig. 4A), based on previous results. First, we anticipated the GM switch and the addition of doxycycline to day 4. Second, as we noticed different doubling times of differentiating cells in GM in biological replicates (data not shown), we decided to keep cells in GM for only one day to increase the reproducibility. Finally, as differentiating cells expressed endogenous MYOD (Fig. 2E–G), and endogenous MYOD is expected to decrease at the end of differentiation, we removed doxycycline in the last 3 days to allow a more physiological maturation of muscle cells. Analysis of muscle markers by immunostaining showed that under these conditions both normal and ALS mutant lines could be differentiated with comparable efficiency (Fig. 4B and C).
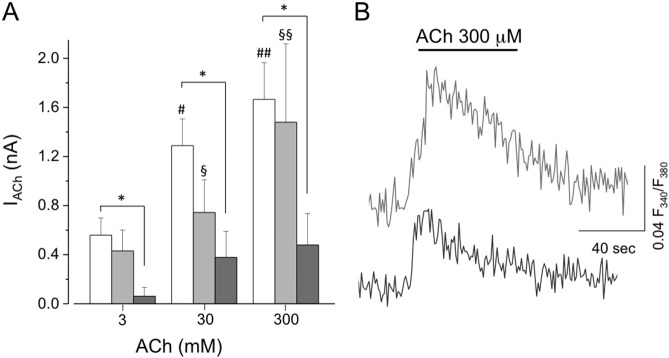
Passive electrical properties of mutant cells were similar to that measured in WT I-MyoD cells, in terms of membrane capacitance (iPSC-FUSR514S/WT-MyoD: 49.2 ± 9.6 pF, n = 8; iPSC-TDP-43A382T/A382T-MyoD: 48.8 ± 9.7 pF, n = 12; p = 0.2 vs WT I-MyoD, t-test) and resting membrane potential (iPSC-FUSR514S/WT-MyoD: − 34.8 ± 4.4 mV, n = 7; iPSC-TDP-43A382T/A382T-MyoD: − 34.8 ± 4.4 mV, n = 9; p = 0.8 vs WT I-MyoD, t-test). Patch clamp recordings of ACh-evoked currents in ALS iPSC-derived skeletal muscle cells revealed that responses evoked by ACh in iPSC-FUSR514S/WT-MyoD were similar to those recorded in WT I-MyoD cells (0.74 ± 0.27 nA, n = 7, ACh 30 μM, p = 0.1, t-test) (Fig. 5A). In the same analysis, however, the iPSC-FUSR521C/WT-MyoD mutant did not show responses to ACh (data not shown). In the case of the iPSC-TDP-43A382T/A382T-MyoD mutant, differentiated cells displayed significantly smaller currents compared to WT and FUSR514S/WT cells (0.38 ± 0.21 pA, n = 9, ACh 30 μM, p < 0.05 vs WT I-MyoD, t-test) (Fig. 5A).
Fig. 5.
Functional properties of ALS iPSC-derived skeletal muscle cells.
(A) Dose-response relation of ACh-evoked currents in skeletal muscle cells differentiated as described in Fig. 4 from FUSR514S/WT-MyoD (light gray bars, n = 7; §, p < 0.05 vs 3 μM; §§, p < 0.05 vs 3 μM and 30 μM; OneWay ANOVA) and from TDP-43A382T/A382T-MyoD (dark gray bars, n = 9; *, p < 0.05 vs WT I-MyoD cells, TwoWay ANOVA) iPSCs, compared to those recorded in WT I cells (white bars, n = 11; #, p < 0.05 vs 3 μM; #, p < 0.05 vs 3 μM and 30 μM; OneWay ANOVA). (B) Average fluorescence response to ACh (300 μM) in Fura-2-loaded iPSC-FUSR514S/WT-MyoD (light gray trace; n = 18 cells) and iPSC-TDP-43A382T/A382T-MyoD (dark gray trace; n = 13 cells) cells.
Fluorescence determination in Fura2 AM-loaded cells showed that ACh application induced transient intracellular calcium rise in FUSR514S/WT and TDP-43A382T/A382T ALS mutant cells (Fig. 5B). Notably, ACh-induced currents responses were dose-dependent in iPSC-FUSR514S/WT-MyoD cells, while iPSC-TDP-43A382T/A382T-MyoD cells displayed significantly smaller currents without dose-dependence (Fig. 5A).
Taken together these results demonstrate that the MyoD inducible system and differentiation protocol described here allowed the conversion of multiple iPSC lines, including cells carrying ALS mutations, into functionally active myotubes.
4. Discussion
The French neurologist Jean-Martin Charcot in 1874 was the first to describe anatomical lesions in the lateral spinal cord of ALS patients, thus pointing to motoneurons as the cell type primarily affected in the disease. Since then, other neural and non-neural cell types have been shown to contribute to this complex disease.
Both animal models and in vitro cell systems can contribute to the dissection of molecular pathways and cellular phenotypes underlying ALS. The SOD1 mouse model has been extensively used to study the non-cell autonomous effects of the mutations in different cell types. Muscle-restricted expression of mutant SOD1 elicited in this district toxic effects, due to increased oxidative stress, associated to signs of ALS and motoneuron degeneration in old mice (Dobrowolny et al., 2008, Wong and Martin, 2010). However, reduction of mutant SOD1 in muscles did not affect disease onset or survival of transgenic mice (Miller et al., 2006). A better understanding of the role of muscle is crucial to design effective therapeutic strategies for this multi-systemic disease. The genetic etiology of ALS also contributes to its complexity. Despite the list of genes underlying inherited ALS has been greatly expanded in the last decade, effects of non-SOD1 mutations in muscle cells, and their possible contribution to ALS onset or progression, have not been elucidated. In a recent report, Wachter et al. suggested that overexpression of FUS and TDP-43 in muscle cells affects motoneuron neurite length in an in vitro murine co-culture system (Wächter et al., 2015). This study relied on the ectopic expression of human mutant proteins in a mouse system. So far, a proper human model system in which these evidences could be confirmed and implemented is still missing. In this paper we show that both normal and ALS patient-derived human iPSCs can be converted to functionally mature myotubes in vitro. One important point of strength of our system is the expression of physiological levels of mutant proteins by human iPSC-derived cells (Lenzi et al., 2015). Notably, ectopic expression of human WT FUS in murine systems (in vivo or in vitro) produced the same detrimental effects observed when mutant proteins are used (McGoldrick et al., 2013, Mitchell et al., 2013, Wächter et al., 2015). Such dose-dependent toxicity should be taken into consideration when analyzing the results of experiments relying on transfection of exogenous ALS-related genes. Human iPSCs and their differentiated derivatives, such as those described here, are not flawed by this concern.
Our approach is based on the ectopic expression of the myogenic factor MyoD in an inducible way, in cells that had stably integrated the transgene. Transduced iPSCs represent a stable and expandable population, in which doxycycline treatment and medium switch are sufficient to induce myogenic differentiation in few days. Even if we do not provide comparative analysis, a survey in the literature suggests similar timing and efficiency of differentiation with our system compared to analogous methods (Tanaka et al., 2013, Shoji et al., 2016).
In our previous work, we have reported comparable efficiency of motoneuron differentiation between WT and FUS or TDP-43 mutant iPSCs (Lenzi et al., 2015). Here we have extended this analysis to muscle differentiation showing that ALS mutations did not impair myogenesis, as assessed by muscle markers expression. Functionally, both control and ALS mutant iPSC-derived muscles expressed AChRs and responded to ACh application with transient intracellular calcium increase, suggesting that these cells could have reached a mature phenotype. Interestingly, we detected significantly smaller currents in the TDP-43 mutant compared to the control. Moreover, one of the FUS mutants did not respond to ACh. Despite the fact that the fraction of cells expressing the muscle marker MYH1 was not significantly different among the lines, we cannot exclude that these differences were due to suboptimal maturation. It should be also taken into consideration that iPSC lines used in the present work were derived by somatic cell reprogramming and therefore are not isogenic. In order to ascribe any functional impairment to the genetic defect it would be necessary, in the future, to extend this analysis to other mutants and/or compare cell lines that differ only with respect to disease mutations, i.e. otherwise isogenic, generated by gene editing.
The ALS-MyoD iPSC lines described here can be used to reproduce in vitro the neuromuscular microenvironment affected by the pathology. Despite no data on motoneuron-muscle interactions are presented here, ALS-mutant iPSCs will allow, in the future, to establish this kind of co-cultures. Such system would be instrumental to understand whether FUS or TDP-43 mutant muscles can affect survival of WT motoneurons, or worsen the vulnerability of mutant motoneurons in human. So far, ALS phenotypes in human motoneurons have been only studied in cultures containing neural cells (i.e. motoneurons and glia, at the best). On the contrary, iPSC-derived motoneuron-muscle-glia co-cultures may provide, in the long term, a tool to screen for therapeutic compounds that target muscle-motoneuron interactions in ALS.
Funding
This work was partially supported by grants from: ERC-2013 AdG MUNCODD GA340172, AriSLA full grant 2014 ARCI, Epigen Flagship Project EPIGENOMICS, AFM Telethon GA17835, Fondazione Roma and Parent Project to I.B.
Conflict of interest
The authors declare no competing financial interests.
Author contributions
AR and JL designed the epB-Puro-TT-mMyoD construct and the differentiation protocols. JL performed differentiation experiments and analysis of muscle markers. FP and SDA performed patch clamp and calcium imaging recordings and analyzed the data. RDS performed real-time qRT-PCR. AR, IB and CL coordinated the work. AR conceived the project and wrote the paper.
Acknowledgments
We thank T. Santini (IIT-CLNS@Sapienza) and O. Sthandier (Sapienza University) for muscle immunostaining protocols and advice on muscle differentiation media, V. de Turris (IIT-CLNS@Sapienza) for confocal microscopy and advice on image analysis and I. Legnini (Sapienza University) for providing human myoblasts RNA samples. We thank the members of the IIT-CLNS@Sapienza for helpful discussion.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.scr.2016.06.003.
Appendix A. Supplementary data
Supplementary material.
References
- Abujarour R., Bennett M., Valamehr B., Lee T.T., Robinson M., Robbins D., Le T., Lai K., Flynn P. Myogenic differentiation of muscular dystrophy-specific induced pluripotent stem cells for use in drug discovery. Stem Cells Transl Med. 2014;3:149–160. doi: 10.5966/sctm.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini S., Coutinho P., Malecova B., Giordani L., Savchenko A., Forcales S.V., Puri P.L. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilican B., Serio A., Barmada S.J., Nishimura A.L., Sullivan G.J., Carrasco M., Phatnani H.P., Puddifoot C.A., Story D., Fletcher J., Park I.-H., Friedman B.A., Daley G.Q., Wyllie D.J.A., Hardingham G.E., Wilmut I., Finkbeiner S., Maniatis T., Shaw C.E., Chandran S. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- Boulting G.L., Kiskinis E., Croft G.F., Amoroso M.W., Oakley D.H., Wainger B.J., Williams D.J., Kahler D.J., Yamaki M., Davidow L., Rodolfa C.T., Dimos J.T., Mikkilineni S., MacDermott A.B., Woolf C.J., Henderson C.E., Wichterle H., Eggan K. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Qian K., Du Z., Cao J., Petersen A., Liu H., Blackbourn L.W., Huang C.-L., Errigo A., Yin Y., Lu J., Ayala M., Zhang S.-C. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement A.M., Nguyen M.D., Roberts E.A., Garcia M.L., Boillée S., Rule M., McMahon A.P., Doucette W., Siwek D., Ferrante R.J., Brown R.H., Julien J.-P., Goldstein L.S.B., Cleveland D.W. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- Darabi R., Perlingeiro R.C.R. Derivation of skeletal myogenic precursors from human pluripotent stem cells using conditional expression of PAX7. Methods Mol. Biol. 2014 doi: 10.1007/7651_2014_134. [DOI] [PubMed] [Google Scholar]
- Di Giorgio F.P., Carrasco M.A., Siao M.C., Maniatis T., Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat. Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio F.P., Boulting G.L., Bobrowicz S., Eggan K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R., Wichterle H., Henderson C.E., Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Dobrowolny G., Aucello M., Rizzuto E., Beccafico S., Mammucari C., Boncompagni S., Bonconpagni S., Belia S., Wannenes F., Nicoletti C., Del Prete Z., Rosenthal N., Molinaro M., Protasi F., Fanò G., Sandri M., Musarò A. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Egawa N., Kitaoka S., Tsukita K., Naitoh M., Takahashi K., Yamamoto T., Adachi F., Kondo T., Okita K., Asaka I., Aoi T., Watanabe A., Yamada Y., Morizane A., Takahashi J., Ayaki T., Ito H., Yoshikawa K., Yamawaki S., Suzuki S., Watanabe D., Hioki H., Kaneko T., Makioka K., Okamoto K., Takuma H., Tamaoka A., Hasegawa K., Nonaka T., Hasegawa M., Kawata A., Yoshida M., Nakahata T., Takahashi R., Marchetto M.C.N., Gage F.H., Yamanaka S., Inoue H. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004052. (145ra104) [DOI] [PubMed] [Google Scholar]
- Grassi F., Pagani F., Spinelli G., De Angelis L., Cossu G., Eusebi F. Fusion-independent expression of functional ACh receptors in mouse mesoangioblast stem cells contacting muscle cells. J. Physiol. Lond. 2004;560:479–489. doi: 10.1113/jphysiol.2004.070607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E., Sandoe J., Williams L.A., Boulting G.L., Moccia R., Wainger B.J., Han S., Peng T., Thams S., Mikkilineni S., Mellin C., Merkle F.T., Davis-Dusenbery B.N., Ziller M., Oakley D., Ichida J., Dicostanza S., Atwater N., Maeder M.L., Goodwin M.J., Nemesh J., Handsaker R.E., Paull D., Noggle S., McCarroll S.A., Joung J.K., Woolf C.J., Brown R.H., Eggan K. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste A., Berenshteyn F., Brivanlou A.H. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi J., De Santis R., de Turris V., Morlando M., Laneve P., Calvo A., Caliendo V., Chiò A., Rosa A., Bozzoni I. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cells (iPSCs) derived motoneurons. Dis Model Mech. 2015;8:755–766. doi: 10.1242/dmm.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick P., Joyce P.I., Fisher E.M.C., Greensmith L. Rodent models of amyotrophic lateral sclerosis. Biochim. Biophys. Acta. 2013;1832:1421–1436. doi: 10.1016/j.bbadis.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Miller T.M., Kim S.H., Yamanaka K., Hester M., Umapathi P., Arnson H., Rizo L., Mendell J.R., Gage F.H., Cleveland D.W., Kaspar B.K. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19546–19551. doi: 10.1073/pnas.0609411103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.C., McGoldrick P., Vance C., Hortobágyi T., Sreedharan J., Rogelj B., Tudor E.L., Smith B.N., Klasen C., Miller C.C.J., Cooper J.D., Greensmith L., Shaw C.E. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol. 2013;125:273–288. doi: 10.1007/s00401-012-1043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarò A. Understanding ALS: new therapeutic approaches. FEBS J. 2012 doi: 10.1111/febs.12087. [DOI] [PubMed] [Google Scholar]
- Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A.E., Chiò A., Traynor B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Brivanlou A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Papaioannou M.D., Krzyspiak J.E., Brivanlou A.H. miR-373 is regulated by TGFβ signaling and promotes mesendoderm differentiation in human embryonic stem cells. Dev. Biol. 2014 doi: 10.1016/j.ydbio.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J.P., Deng H.X. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Shoji E., Woltjen K., Sakurai H. Directed myogenic differentiation of human induced pluripotent stem cells. Methods Mol. Biol. 2016;1353:89–99. doi: 10.1007/7651_2015_257. [DOI] [PubMed] [Google Scholar]
- Tanaka A., Woltjen K., Miyake K., Hotta A., Ikeya M., Yamamoto T., Nishino T., Shoji E., Sehara-Fujisawa A., Manabe Y., Fujii N., Hanaoka K., Era T., Yamashita S., Isobe K.-I., Kimura E., Sakurai H. Efficient and reproducible myogenic differentiation from human iPS cells: prospects for modeling Miyoshi myopathy in vitro. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco F.S., Gerli M.F.M., Perani L., Benedetti S., Ungaro F., Cassano M., Antonini S., Tagliafico E., Artusi V., Longa E., Tonlorenzi R., Ragazzi M., Calderazzi G., Hoshiya H., Cappellari O., Mora M., Schoser B., Schneiderat P., Oshimura M., Bottinelli R., Sampaolesi M., Torrente Y., Broccoli V., Cossu G. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003541. (140ra89) [DOI] [PubMed] [Google Scholar]
- Wächter N., Storch A., Hermann A. Human TDP-43 and FUS selectively affect motor neuron maturation and survival in a murine cell model of ALS by non-cell-autonomous mechanisms. Amyotroph Lateral Scler Frontotemporal Degener. 2015:1–11. doi: 10.3109/21678421.2015.1055275. [DOI] [PubMed] [Google Scholar]
- Wainger B.J., Kiskinis E., Mellin C., Wiskow O., Han S.S.W., Sandoe J., Perez N.P., Williams L.A., Lee S., Boulting G., Berry J.D., Brown R.H., Cudkowicz M.E., Bean B.P., Eggan K., Woolf C.J. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M., Martin L.J. Skeletal muscle-restricted expression of human SOD1 causes motor neuron degeneration in transgenic mice. Hum. Mol. Genet. 2010;19:2284–2302. doi: 10.1093/hmg/ddq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Boillee S., Roberts E.A., Garcia M.L., McAlonis-Downes M., Mikse O.R., Cleveland D.W., Goldstein L.S.B. Mutant SOD1 in cell types other than motor neurons and oligodendrocytes accelerates onset of disease in ALS mice. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7594–7599. doi: 10.1073/pnas.0802556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.