Abstract
In photosynthetic organisms like cyanobacteria and plants, the main engines of oxygenic photosynthesis are the pigment-protein complexes photosystem I (PSI) and photosystem II (PSII) located in the thylakoid membrane. In the cyanobacterium Synechocystis sp. PCC 6803, the slr1796 gene encodes a single cysteine thioredoxin-like protein, orthologs of which are found in multiple cyanobacterial strains as well as chloroplasts of higher plants. Targeted inactivation of slr1796 in Synechocystis 6803 resulted in compromised photoautotrophic growth. The mutant displayed decreased chlorophyll a content. These changes correlated with a decrease in the PSI titer of the mutant cells, whereas the PSII content was unaffected. In the mutant, the transcript levels of genes for PSI structural and accessory proteins remained unaffected, whereas the levels of PSI structural proteins were severely diminished, indicating that Slr1796 acts at a posttranscriptional level. Biochemical analysis indicated that Slr1796 is an integral thylakoid membrane protein. We conclude that Slr1796 is a novel regulatory factor that modulates PSI titer.
Keywords: cyanobacteria, photosynthesis, photosystem II, protein complex, redox regulation, photosystem I biogenesis
Introduction
Oxygenic photosynthesis is performed by pigment-protein complexes located in the thylakoid membranes of cyanobacteria and chloroplasts. This process generates atmospheric oxygen and chemical energy in the form of carbohydrates and is thus the ultimate source of food, feed, and fuel on the planet. Photosystem II (PSII)2 performs light-driven oxidation of water (generating a strong oxidant, P680+) and initiates the electron flow to photosystem I (PSI) via the cytochrome b6f complex during linear electron flow, whereas PSI catalyzes the last step, the oxidation of plastocyanin in the thylakoid lumen and reduction of ferredoxin to generate ATP and NADPH (a strong reductant) for CO2 fixation. Thus, oxygenic photosynthesis operates in both highly oxidizing and reducing redox environments, conditions that inevitably lead to the production of reactive oxygen species.
In photosynthetic organisms, a number of proteins are present that are involved in redox activity (1, 2). These include thioredoxins (Trxs), small proteins that can regulate enzyme activity by acting as reversible redox switches. Trxs belong to the thioredoxin superfamily of proteins, which contain a common thioredoxin fold with a characteristic two-cysteine CXXC active site motif (where X is any amino acid). The CXXC motifs have different variants, including Trx folds with only a single cysteine, which are comparatively less well studied (3). Redox regulation by Trx fold proteins via their oxidation, reduction, or disulfide exchange activities, depending on their redox environments, is an extensive system in almost all life forms, including oxygenic photosynthetic organisms.
PSI is among the largest and most complex multisubunit pigment-protein structures in nature (4). Most cyanobacterial PSI functions as a trimer with each monomer containing 12 subunits (PsaA–F, PsaI–M, and PsaX) and 127 cofactors, including 96 chlorophylls, 22 β-carotene pigments, four lipids, three Fe-S clusters, and two phylloquinone molecules (5). Although PSI structure and function are well understood, how the subunits and cofactors are incorporated into functional PSI complex remains largely unexplored (6).
To date, a few accessory protein factors involved in the stable accumulation of PSI have been identified in cyanobacteria, higher plants, and algae (6, 7). These accessory factors can be divided into four functional classes: (i) subunit assembly factors, including Alb3 (8, 9), Ycf3 (10, 11), Ycf4 (12, 13), Ycf37/Pyg7 (14, 15), Y3IP1 (16), and two lumenal proteins, PPD1 (17) and Psa2 (18); (ii) proteins involved in cofactor biogenesis, including RubA (19, 20), Hcf101 (21, 22), and CnfU/Nfu2 (23, 24); (iii) PSI stability maintenance factors such as BtpA (25, 26); and (iv) regulators for PSI gene expression (27). Although our knowledge of PSI biogenesis has advanced with these discoveries, the exact roles of many of the above factors and the details of PSI assembly remain to be investigated (6, 7, 28).
A set of protein factors involved in PSI assembly or stability has been identified in cyanobacteria (15, 19, 20, 25). Here, we report a strain of Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) lacking the protein Slr1796, which contains a Trx fold with only a single cysteine, and demonstrate that this strain has a higher PSII/PSI ratio. We show that the change in PS stoichiometry is a result of a severe reduction of PSI and that Slr1796 acts at a post-transcriptional step during PSI biogenesis. We localized Slr1796 in the cell and examined the ultrastructural changes resulting from the deletion of slr1796. Our data suggest that Slr1796 is a novel thylakoid protein required for the normal accumulation of PSI.
Results
Disruption of slr1796 Leads to Higher PSII/PSI Ratio
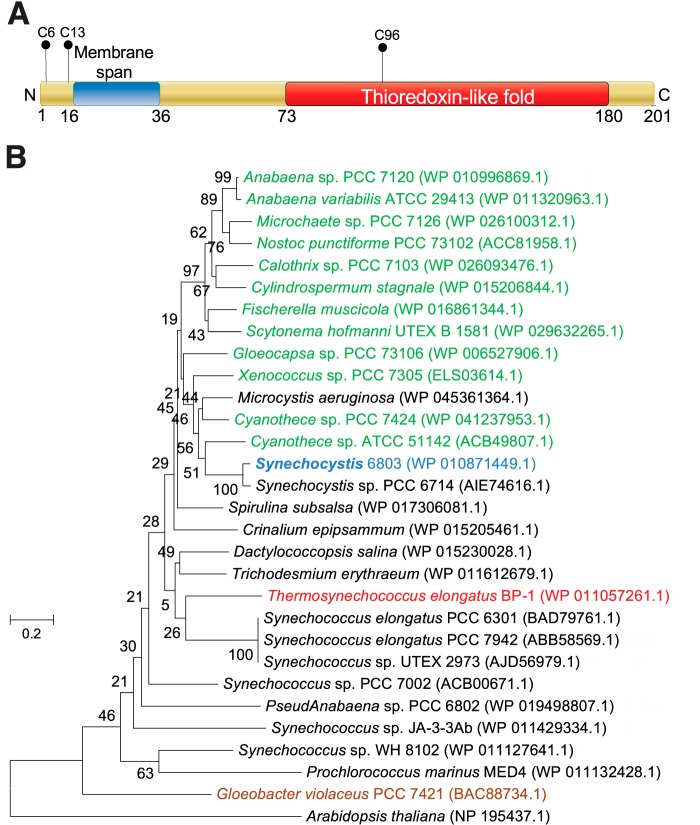
The protein Slr1796 was identified as a thylakoid membrane component by proteomics studies (29, 30), but the role of the protein remained unknown. The Slr1796 protein has three cysteine residues (Fig. 1A). Cys-6 and Cys-13 are located near the N terminus, share poor sequence similarity with other homologs, and are followed by a transmembrane segment. A single cysteine residue, Cys-96, is present in a thioredoxin fold and is highly conserved among homologs of Slr1796 (data not shown).
FIGURE 1.
Slr1796 is a single cysteine thioredoxin-like protein in thylakoid membrane. A, schematic diagram showing the domains of the Slr1796 protein in Synechocystis 6803. Transmembrane sequence prediction was performed using the TMHMM program. The single cysteine thioredoxin-like fold was predicted by UniProt. The positions of cysteine residues are labeled. B, phylogenetic analysis of the evolutionary relationship between Slr1796 and its orthologs in selected cyanobacterial species. Red, thermophile; green, nitrogen-fixing; brown, strain lacking thylakoids. The land plant A. thaliana was used as an out-group. The scale bar indicates the number of amino acid substitutions per site.
The phylogenetic relationships of orthologs of Slr1796 in a number of cyanobacteria, including many well studied strains, are shown in Fig. 1B. Phylogenetic analysis showed that close orthologs of Slr1796 are present in most (if not all) cyanobacterial strains with completely sequenced genomes, including both diazotrophic and non-diazotrophic strains (Fig. 1B). In addition, similar orthologs are predicted to be present in land plants such as Arabidopsis thaliana. It is noteworthy that an ortholog is present in Gloeobacter violaceus, a cyanobacterium without thylakoid membranes. Therefore, the slr1796 gene might have evolved before the development of the thylakoid membrane system.
To investigate the function of Slr1796, the corresponding gene was deleted in Synechocystis 6803 (Fig. 2A). The resulting Δslr1796 mutant was segregated in the presence of glucose and 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) as verified by PCR (Fig. 2B). A complemented (Comp) strain was also constructed in which the slr1796 gene was inserted in the genome of the Δslr1796 mutant at the chromosomal neutral site NS1, which has been shown to be a region of nonexpressed DNA (31) (Fig. 2C).
FIGURE 2.
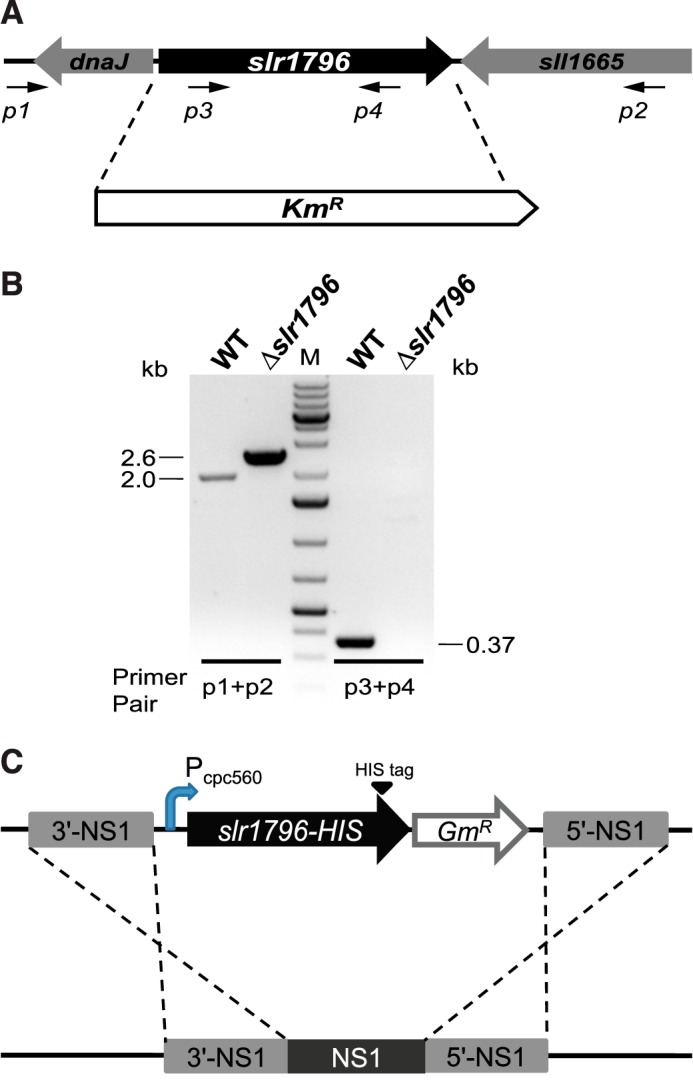
Genetic deletion and complementation of the slr1796 gene of Synechocystis 6803. A, scheme of slr1796 replacement by a KmR cassette. B, PCR confirmation of complete segregation of the Δslr1796 mutant. Primer positions are indicated. Primer pair p3 + p4 is specific to the slr1796 gene (0.37-kb product) but not to KmR. The expected size of the PCR products from primers p1 + p2 is 2.0 kb in WT and 2.6 kb in the mutant. M, DNA size standard. C, construction of the Comp strain. Slr1796 was fused at the C terminus with a His6 tag and expressed under the Pcpc560 promoter in the Δslr1796 background.
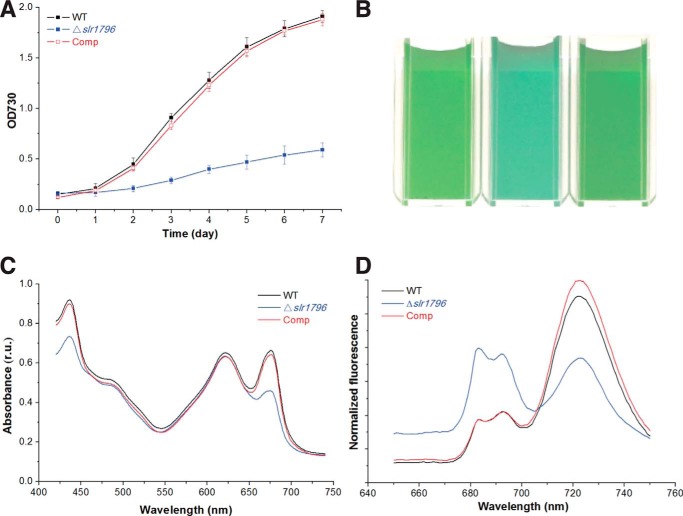
The segregated Δslr1796 mutant is stable when grown photoautotrophically and grows with a doubling time that is about 3 times slower than wild type (WT) (Fig. 3A). The visual phenotype of the Δslr1796 culture showed that the deletion strain appeared bluer in color compared with the WT culture (Fig. 3B), which could indicate an increase in the pigment ratio of phycobilin to chlorophyll. For a quantitative analysis, absorption spectra of intact cells were measured on a per cell basis (Fig. 3C). The mutant showed a large reduction in chlorophyll absorption at 436 and 676 nm compared with WT. The phycocyanin (PC) peak at 625 nm remained virtually unchanged. In contrast, the chlorophyll content of the mutant cells was nearly 75% decreased compared with WT, whereas PC levels were similar (Table 1). Thus, the 3.6-fold higher PC/Chl ratio in the Δslr1796 mutant was mainly due to the significant decrease of chlorophyll content.
FIGURE 3.
Phenotypic characterization of WT, the Δslr1796 mutant, and the complemented strain of Synechocystis 6803. A, growth curves of three strains grown under 40 μmol of photons m−2·s−1. Error bars represent the standard deviation of three biological replicates. B, strains grown in liquid culture to the same OD730 nm showing differences in the culture color. C, whole cell absorption spectra of three strains adjusted to equal cell numbers. Absorption peaks for chlorophyll a are at ∼440 and 680 nm and for phycobilins at ∼620 nm. D, low temperature (77 K) chlorophyll fluorescence emission spectra. The excitation wavelength was 435 nm. Peaks at 685 and 695 nm arise from PSII; the peak at 721 nm arises from PSI. For C and D, three or more independent replicates were performed, and representative data are presented. r.u., relative units.
TABLE 1.
Cellular pigment and membrane content and photosynthetic activity of WT and Δslr1796 mutant cells
All measurements are shown as the result for 1 × 108 cells. Errors represent the standard deviation of at least three independent biological samples. MV, methyl viologen; Asc, sodium ascorbate; DAD, diaminodurene; DCBQ, 2,6-dichloro-p-benzoquinone.
| Chl | PC | PC/Chl | Membrane protein | Photosynthetic activitya |
|||
|---|---|---|---|---|---|---|---|
| Net (H2O→CO2) | PSII (H2O→DCBQ/FeCN) | PSI (Asc/DAD→MV) | |||||
| μg | μg | μg | μmol of O2·108 cells·h−1 | ||||
| WT | 2.6 ± 0.2 | 22.3 ± 1.6 | 8.6 ± 0.6 | 58.9 ± 0.9 | 0.9 ± 0.1 | 1.1 ± 0.1 | −0.62 ± 0.04 |
| Δslr1796 | 0.7 ± 0.1 | 21.7 ± 0.8 | 31.0 ± 0.9 | 35.3 ± 0.7 | 0.6 ± 0.1 | 1.3 ± 0.2 | −0.40 ± 0.03 |
a Determined with independent biological cell samples from those used for pigment and protein quantification.
To determine the effects of the deletion of the slr1796 gene on the photosynthetic pigment-protein complexes, 77 K low temperature fluorescence emission spectra were measured (Fig. 3D). Upon excitation of chlorophyll, the Δslr1796 strain yielded three peaks as in WT; however, in the mutant, the PSI peak at 721 nm was much smaller, and the two peaks at 685 and 695 nm from PSII increased markedly, demonstrating that the mutant had a much higher PSII/PSI ratio as compared with WT. Furthermore, WT-like growth, culture color, whole cell absorption, and 77 K Chl fluorescence emission characteristics were fully restored by expression of the Slr1796 protein in the Comp strain (Fig. 3). These results indicated that the Slr1796 protein has a direct role in modulating the photosystem stoichiometry in Synechocystis 6803. We grew WT, the slr1796 mutant, and the complemented strains under light-activated heterotrophic growth conditions (32) to test the light sensitivity of the strains. Fluorescence emission spectra at 77 K showed that each of the three strains yielded peaks similar to those of cells grown under normal conditions as described above (data not shown).
PSI Content and Activity Is Specifically Reduced in the ΔSlr1796 Mutant
Because the phycobilisome light-harvesting antenna remained unaffected, we further specifically examined PSI and PSII activity in the Δslr1796 mutant. Based on equal cell numbers, the mutant showed a slightly higher PSII activity compared with WT (which corresponds to about 423 μmol of O2·mg of Chl−1·h−1 for WT) (Table 1). Net activity of photosynthesis in the Δslr1796 mutant cells was reduced to about 67% of the WT level (Table 1). We also measured PSI-mediated oxygen consumption rates and found that PSI activity in the Δslr1796 mutant cells was about 64% of WT (Table 1). Note that because the Δslr1796 strain contains much less chlorophyll per cell compared with WT we chose to present most data on a per cell basis.
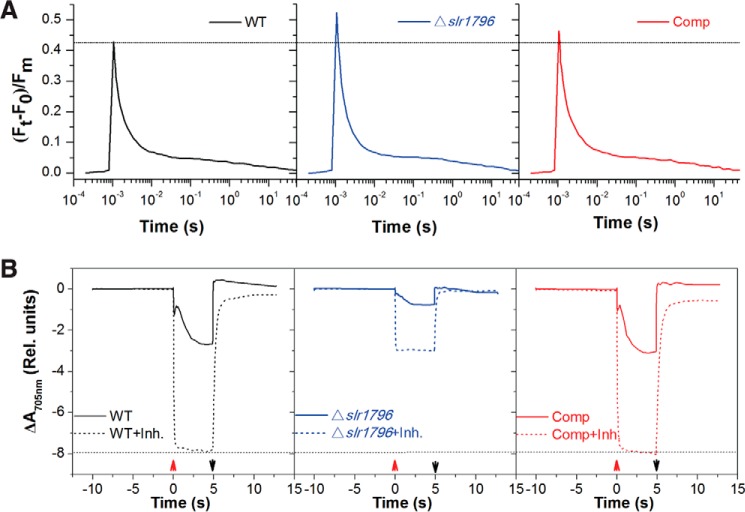
Chl a fluorescence monitoring of PSII capacity in the mutant showed results consistent with the above rates (Fig. 4A). The ratio of variable fluorescence yield (Fv) to maximum fluorescence (Fm) (33) indicates the maximum photochemical efficiency of PSII when all reaction centers are open. The mutant had a 1.2-fold higher Fv/Fm ratio than WT, indicating that PSII content and function are not compromised in the Δslr1796 mutant. The Comp strain had almost the same level of variable fluorescence as WT.
FIGURE 4.
Kinetic spectroscopic characterization of WT (black line), the Δslr1796 mutant (blue), and the complemented strain (red). Cells were suspended in BG11 to equal cell numbers at which WT cells were at a chlorophyll concentration of 5 μg/ml. A dotted line was drawn to facilitate comparisons between strains. Data are presented as the average of three biological replicates. A, reoxidation kinetics of the reduced PSII primary electron acceptor QA− in three strains. B, P700 redox kinetics in WT, Δslr1796 mutant, and Comp cells grown under the same conditions. After 10-s dark adaption, actinic light was turned on (up arrow) for 5 s and then off (down arrow) to allow the re-reduction of P700+ to P700. Inhibitors (Inh.; dotted lines), 10 μm DCMU and 10 μm 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone, were added to inhibit linear and cyclic electron flow. A negative slope indicates P700 oxidation. Rel., relative.
In contrast, the PSI content was severely reduced in the mutant as revealed by the kinetics of P700 oxidation of whole cells (Fig. 4B). The maximum oxidation of P700 in the mutant under standard light was significantly less than that of WT and the complemented strain. When electron flow from PSII and the plastoquinone pool was completely blocked by adding the inhibitors DCMU and 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone (DBMIB), ΔA705 nm (indicating the pool size of photooxidizable P700) was dramatically smaller in the mutant cells. In the complemented strain, P700 redox kinetics was fully restored to WT levels. These results led us to conclude that this mutant has a significantly reduced PSI content. This is also consistent with the observation of PSI peak reduction in the low temperature Chl fluorescence emission spectra (Fig. 3D). Taken together, the significant increase in the PSII/PSI ratio in the mutant was principally attributed to the decrease in PSI titer, rather than the increase in PSII content, with a simultaneous loss of PSI activity.
Slr1796 Regulates PSI Accumulation at a Posttranscriptional Level
The transcript levels of PSI and PSII genes in WT and the deletion mutant strains were examined by reverse transcription (RT)-PCR. Interestingly, the transcript abundances of 11 PSI subunits, four PSI assembly factors, and two PSII subunits genes were almost unchanged in the Δslr1796 strain (Fig. 5), suggesting that Slr1796 does not act at the transcriptional level or at the level of mRNA maturation or stability.
FIGURE 5.
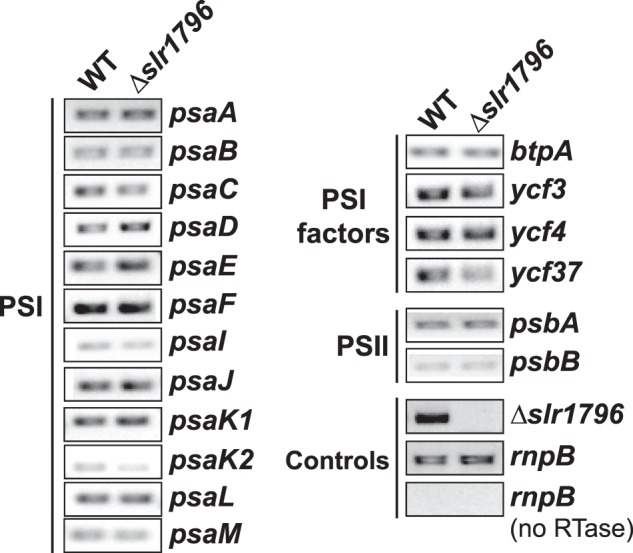
Transcript levels of PSI and PSII genes in WT and Δslr1796 mutant cells. The constitutively expressed rnpB gene was used as a reference and to show that samples do not contain contamination from genomic DNA when no reverse transcriptase (RTase) was added. The primer pair used for the detection of slr1796 gene was p3 + p4 as in Fig. 2A. Optimal PCR cycles were determined for each primer pair to ensure that the amplifications were in the linear range. Three or more independent replicates were performed, and representative results are shown.
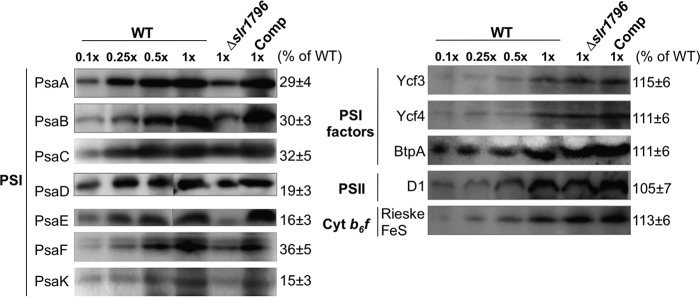
To verify the stoichiometric alteration of PSI in the Δslr1796 mutant and investigate how Slr1796 is involved in modifying the photosystems, levels of protein components in major complexes in the photosynthetic electron transfer chain were examined by immunoblotting. Total membranes were prepared from WT, mutant, and Comp cells and resolved by SDS-PAGE with equal protein amounts loaded. Western blot signals of proteins from the mutant strain were quantified relative to those in WT. For the WT samples, a dilution series was included to properly estimate the relative protein subunit levels in the mutant. Notably, the amounts of PSI subunits in the mutant strain were dramatically decreased and returned to WT levels in the Comp strain (Fig. 6). In particular, the levels of the reaction center dimer proteins PsaA and PsaB, which contain the majority of Chl a, were sharply reduced in the mutant cells. In contrast, the levels of factors involved in PSI assembly or stability (BtpA (25, 26), Ycf3 (10), and Ycf4 (33)) as well as PSII and cytochrome b6f subunits were slightly increased in the mutant.
FIGURE 6.
Representative membrane protein profiles in WT, the Δslr1796 mutant, and the complemented strain. Immunoblotting analyses of photosynthetic membrane proteins are shown. Equal amounts of membrane proteins were loaded for the detection of each protein. A dilution series of WT protein (10, 25, 50, and 100%) were loaded. Antibodies were directed against PSI subunits (PsaA, PsaB, PsaC, PsaD, PsaE, PsaF, and PsaK), PSI assembly factors (BtpA, Ycf3, and Ycf4), PSII core subunit D1, and a subunit of the cytochrome (Cyt) b6f complex (Rieske FeS). The number at the right of each band is the percentage of protein in the Δslr1796 mutant relative to that of WT (set as 100%). Values represent the standard deviation of at least 10 measurements for each protein.
We measured total membrane protein concentrations in WT and mutant samples containing equal cell numbers and found that the mutant had about 40% less total membrane protein content compared with WT (Table 1). It is notable that the loss of significant amounts of PSI, a major membrane protein component, resulted in considerable loss of overall membrane protein content. This explains why the PSII core subunit examined here, D1, showed a slightly higher content in the mutant when compared with WT in samples that were loaded on an equal protein basis (Fig. 6). The protein profiles of PSI and PSII complexes were consistent with the above evidence indicating that the deletion of Slr1796 specifically reduced the PSI content and activity in Synechocystis 6803.
Blue native PAGE showed that the levels of both PSI trimer and monomer were lower in the Δslr1796 mutant compared with WT (Fig. 7). The loss of Slr1796 appears to not affect the formation of the PSI trimer, and no new chlorophyll-protein complexes were observed in the Δslr1796 membrane complexes.
FIGURE 7.
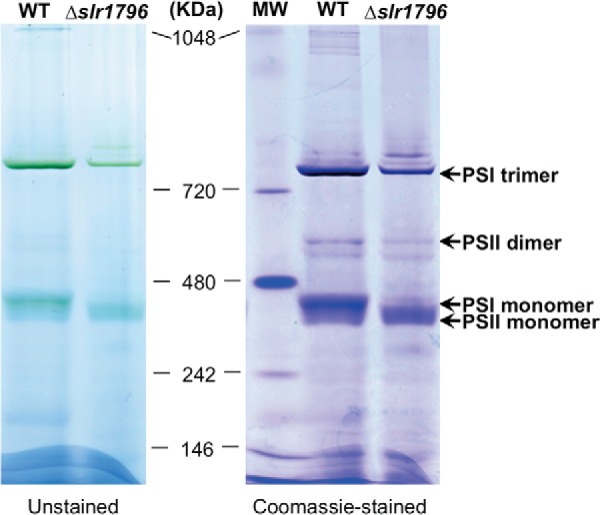
Blue native PAGE of membrane protein complexes of WT and the Δslr1796 mutant. An equal protein amount of about 35 μg was loaded to each lane. The gel is shown as unstained (left) and stained with Coomassie Brilliant Blue (right). Molecular mass markers are indicated, and the assignments of PSI and PSII complexes are given.
Slr1796 Is a Thylakoid Membrane Protein
Thylakoid membranes were prepared by aqueous polymer two-phase partitioning from cells broken with glass beads (34). Cellular subfractionation revealed that Slr1796 is present in the thylakoid membrane fraction and not in plasma membrane (Fig. 8A). Slr1796 has been detected as a component of the thylakoid membrane in proteomics studies of isolated membrane fractions (29, 30), and our result is consistent with these studies. Considering the presence of a transmembrane fragment in Slr1796, as predicted by TMHMM, it is likely an integral thylakoid protein. To examine the strength of the association of Slr1796 with the thylakoid membrane, isolated membranes were treated with detergent or salts (solubilizing agents of different ability to release membrane proteins) and then separated into pellet and soluble fractions by centrifugation. As seen in Fig. 8B, the strongest treatment of the membranes with 0.25% Triton X-100 could only partially release the Slr1796 protein, but treatment with 1 m NaCl or 0.1 m Na2CO3 did not, whereas the latter could fully dissociate the peripheral subunit PsaE from the thylakoid membrane. The overall pattern was similar to that of the integral membrane protein D1. Thus, we conclude that Slr1796 is an integral thylakoid membrane protein.
FIGURE 8.
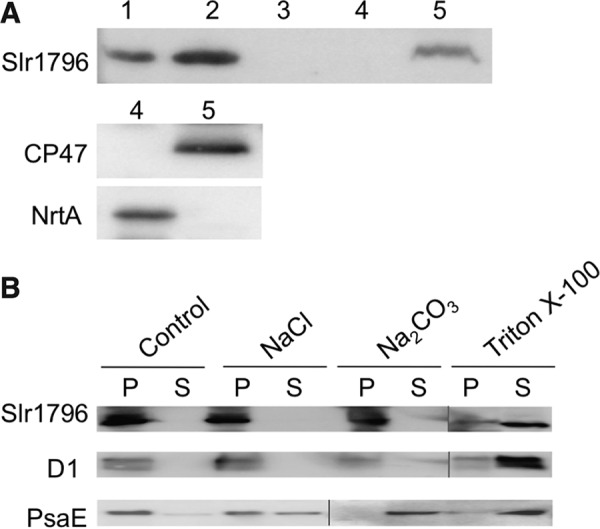
Subcellular localization of the Slr1796 protein. Protein fractions were prepared from the Comp strain. A, immunoblotting analysis of the cellular localization of the Slr1796-His protein using His tag antibodies. Lane 1, total cellular protein; lanes 2 and 3, pellet and soluble fractions after centrifugation; lanes 4 and 5, plasma and thylakoid membranes isolated by two-phase partitioning, respectively, that were validated by the detection of specific marker proteins NrtA and CP47, respectively. B, immunoblotting detection of Slr1796-His in pellet (P) and soluble (S) fractions of thylakoid membrane treated with the indicated solubilizing agents. D1 was used as a control integral membrane protein, and PsaE was used as a control extrinsic protein.
As mentioned above, we determined that Δslr1796 cells have ∼40% less membrane protein content (Table 1). We reasoned that this large difference in protein content in the membranes, concomitant with the large decrease in PSI, would likely result in altered cellular morphology in the Δslr1796 strain. We examined the ultrastructure of the mutant cells by transmission electron microscopy (TEM) and specifically investigated any changes in thylakoid membrane morphology (Fig. 9). The general cellular arrangement of the mutant strain was similar to WT with thylakoid membranes located in the cell periphery and other cellular components, including carboxysomes and polyphosphate bodies, occurring in typical quantities. The mutant strain appeared to have a less extensive thylakoid membrane system with shortened membrane segments when compared with WT (Fig. 9, C and D). In WT cells, thylakoid membranes often formed long bands of concentric layers of membranes that occupied much of the cell periphery (Fig. 9A). In contrast, in the mutant cells, thylakoid membranes were typically found in segments ∼400–700 nm in length. The thylakoid membrane spacing was also altered: in the mutant, membrane layers were separated by an interthylakoidal distance of 68 ± 9 nm (Fig. 9D) compared with a distance of 40 ± 8 nm in WT cells (Fig. 9B). In the complemented strain, these distances were closer to WT levels at 51 ± 6 nm (Fig. 9F). Thus, the deletion of Δslr1796 and the resulting changes in photosystem content and stoichiometry also resulted in profound modifications to thylakoid membrane architecture.
FIGURE 9.
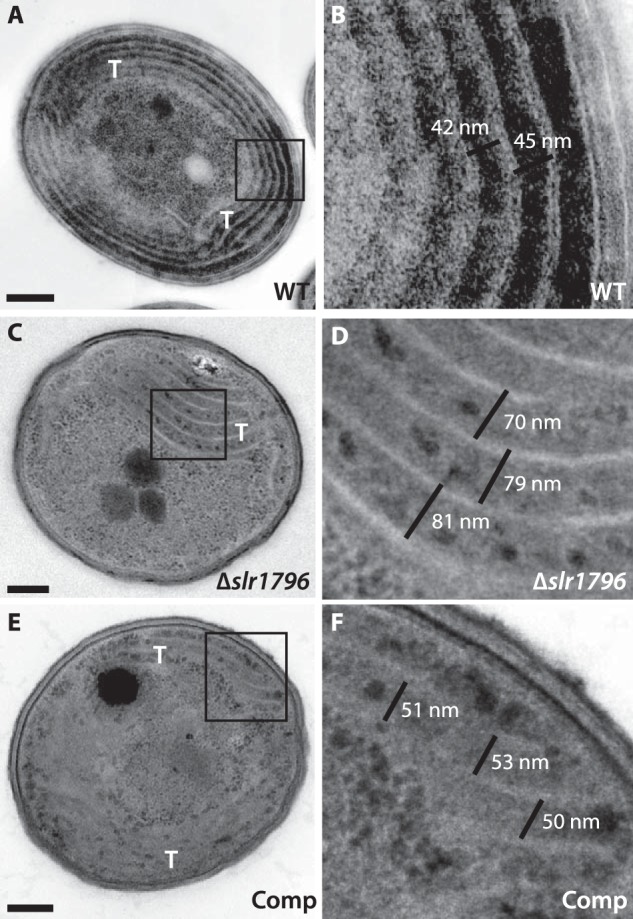
Transmission electron micrographs of WT, the Δslr1796 mutant, and the complemented Synechocystis 6803 strains. A and B, WT; C and D, Δslr1796 mutant; E and F, Comp strain. Scale bars in A, C, and E represent 200 nm. The boxed region is enlarged in B, D, and F, respectively. The black lines in B, D, and F depict the measurements of the interthylakoidal space in each strain. Thylakoid membranes (T) are also labeled.
Discussion
In this work, we have described a novel thylakoid membrane protein involved in PSI assembly or stability. Deletion of the slr1796 gene caused a specific reduction of PSI activity and an increase in the PSII/PSI ratio; however, the mutant could grow photoautotrophically under high light conditions (200 μmol of photons m−2·s−1). The complementation of the slr1796 mutation restored the WT phenotype, including growth rate and cell culture color (Fig. 3). Thus, the thioredoxin-like Slr1796 protein appears to be a critical factor for the accumulation of PSI.
Slr1796 Is Specifically Required for PSI Accumulation
The observed partial loss of PSI activity in the Δslr1796 strain results from the reduced levels of PSI rather than defective PSI complexes. Low temperature Chl fluorescence emission spectra indicated an increase in the PSII/PSI ratio (Fig. 3D), which was in parallel with the increase in the PC/Chl ratio (Table 1). Because in Synechocystis 6803 cells most Chl is associated with PSI and the light-harvesting phycobilisomes are mostly associated with PSII, it is likely that the increase in the above two ratios is due to the reduction of PSI. This is supported by evidence from kinetic spectroscopic characterizations (Fig. 4) and PSI protein profiles (Fig. 6) as well as activity measurements (Table 1). Moreover, the reduction of PSI content due to the Slr1796 mutation had only a minor effect on PSII function as evidenced from the rates of O2 evolution (Table 1) and Chl fluorescence (Fig. 4A). These data suggest that Slr1796 is specifically involved in modulation of PSI content rather than that of other photosystem protein complexes.
In our examination of the levels of PSI proteins in the Δslr1796 mutant, we observed decreases in all PSI protein subunits examined (Fig. 6). However, there were some differences in the levels detected among subunits, ranging from ∼15 to ∼36% of the levels in the WT sample. The PsaE protein showed a large decrease with ∼16% of the protein amount compared with WT. The PsaE protein is localized on the reducing side of the PSI complex and has been shown to be involved in the binding of ferredoxin or flavodoxin to PSI (35). We also found that the PSI activity level in the mutant (∼64% of WT) and the PsaA/B protein levels (∼30% of WT) are not in strict agreement. It is possible that this differential loss of PSI proteins contributes to the measured PSI activity. In particular, the lack of PsaE might result in increased access for the electron acceptor methyl viologen, leading to higher PSI activity than expected based on the PSI protein profiles.
Slr1796 May Function as a Novel Factor for PSI Accumulation
Of all known factors of PSI biogenesis, the ycf37 mutant of Synechocystis 6803 has the most similar phenotype (15) in that this mutant also has a higher PSII/PSI ratio and PC/Chl ratio with PSII amount unaffected. However, there are differences between these two mutants. The reduction in PSI content in Δslr1796 (about 20% of the WT level) is much more severe than in the ycf37 mutant (about 80% of the WT level). Slr1796 is predicted to possess a thioredoxin-like fold, whereas Ycf37 contains three tetratricopeptide repeat units resembling the structural organization of Ycf3, which is an essential factor of PSI assembly, and its loss results in complete and specific loss of PSI (10, 11). Deletion of the higher plant homolog of Ycf37, Pyg7, also shows a more severe phenotype than the ycf37 mutant with a complete loss of PSI and an inability to grow photoautotrophically (6). Moreover, the ycf4 mutant of Synechocystis 6803 resembles Δslr1796 and ycf37 except that it increases the PSII/PSI ratio, resulting from both a reduction of PSI and an increase of PSII (33). Later studies in tobacco and cyanobacteria showed that Ycf4 may serve as a non-essential factor (12). Additionally, two recently identified factors of PSI assembly localized in the thylakoid lumen, PPD1 (17) and Psa2 (18), are not found in prokaryotes, including cyanobacteria (6, 7). Psa2 may have a functional link with Ycf3 in PSI assembly, and disruption of Psa2 may affect Ycf3 abundance; however, in our work, none of the PSI biogenesis factors (Ycf3, Ycf4, and BtpA) in Synechocystis 6803 were affected upon deletion of Slr1796 (Fig. 6). Thus, Slr1796 differs from other known factors that regulate the biogenesis and/or stability of PSI.
A variety of processes, from gene expression (transcription, mRNA maturation, and translation) to protein complex assembly, can be the target point of the assembly factors (36). To investigate at what level the Slr1796 protein acts on PSI accumulation, we examined the transcript levels of PSI and PSII genes. All of these genes in the mutant cells accumulated to the same levels as in WT (Fig. 5), which implied that Slr1796 is involved in PSI accumulation at a posttranscriptional level. Another PSI assembly factor, Alb3 (8, 9), acts on early assembly steps (6, 7) and affects the insertion of the complex in the thylakoid membranes. In contrast, functional PSI complexes are inserted in the thylakoids in the Δslr1796 mutant (Figs. 4, 6, and 7), implying that the mechanism of action of Slr1796 is different from that of Alb3.
Furthermore, the deletion of Slr1796 did not affect the level of PSI assembly chaperones Ycf3 and Ycf4 but led to the simultaneous reduction of the PSI proteins examined (Figs. 5 and 6), suggesting that it is unlikely that Slr1796 acts in the early stage of PSI biogenesis. Our blue native PAGE result further showed that both the PSI monomer and trimer were present but at decreased levels in the mutant (Fig. 7). Thus, PSI could be correctly assembled and functioning, but the stability of mature PSI complexes and/or the synthesis rate of some steps may be negatively affected due to the lack of Slr1796. Furthermore, PSII content and activity in the mutant were found to be at almost the same levels as in WT as shown by the levels of D1 protein (Fig. 6), PSII fluorescence (Fig. 4A), and oxygen evolution rates (Table 1). Taken together, the Slr1796 protein may represent a novel factor that regulates PSI levels.
Absence of Slr1796 Results in Ultrastructural Changes
We have shown that the levels of PSII and phycobilisomes are essentially unchanged in the Δslr1796 strain but that PSI levels are significantly decreased. Another strain in which PSI levels are decreased is the PSI-less strain of Synechocystis 6803, which lacks the PsaA and PsaB proteins (32). Analysis of this mutant by TEM showed that the lack of PSI did not lead to changes in thylakoid distribution; however, specific measurements of interthyakoidal distances were not performed (37). Our analysis of the Δslr1796 mutant by TEM revealed a large increase in the interthylakoidal distance compared with WT cells (Fig. 9). Furthermore, the thylakoid membranes appeared to form shorter segments as opposed to longer concentric layers found in WT. Both of these phenotypes were restored to near WT levels in the complemented strain. Although the mechanism of these changes is unknown, the deletion of Slr1796 appears to effect significant changes in thylakoid membrane organization in addition to its involvement in photosystem stoichiometry.
Our results showed a specific effect of Slr1796 on PSI accumulation and activity with minimal effect on the levels of other thylakoid membrane components, in particular PSII. Based on these data, we propose a scenario in which the lack of Slr1796 results in the loss of PSI complexes, and in turn this decreases the total content of the thylakoid membrane system in the cell, resulting in the less extensive membrane system observed in the mutant. We think this is more likely than direct involvement of Slr1796 in thylakoid membrane biogenesis that results in the specific decrease of PSI in the cells. In this context, a vipp1 mutant in Synechococcus sp. PCC 7002 was found to be defective in PSI formation with a secondary effect on the thylakoid membrane system (38).
In summary, we have described Slr1796, a novel redoxin in the thylakoid membrane that has a critical role in the regulation of PSI content. Because PSI is among the largest and most complex membrane-bound macromolecular assemblies in nature and its biogenesis is rapid, the identification of relevant assembly factors is essential to understand the mechanisms underlying the biogenesis/stability of this photosystem and the biogenesis of thylakoid membranes in chloroplasts and cyanobacteria. Future studies applying methods of protein-protein interactions such as cross-linking (39) will be useful to identify the PSI subunits or their assembly chaperones interacting with the Slr1796 protein and will advance the knowledge of how and when PSI biogenesis happens in oxygenic organisms and the involvement of redox-regulating proteins in this process.
Experimental Procedures
Cyanobacterial Growth Conditions
Synechocystis 6803 WT, mutant, and complemented strains were grown in BG11 medium (40) at 30 °C under a light intensity of ∼40 μmol of photons m−2·s−1 unless otherwise indicated. Antibiotics (20 μg/ml kanamycin or 20 μg/ml gentamicin) were added where required. Growth was measured at OD730 nm on a Multi-cultivator MC 1000-OD (Photon Systems Instruments, Drasov, Czech Republic).
Strain Constructions
The gene deletion mutant was generated by replacing the slr1796 open reading frame with a kanamycin resistance (KmR) cassette. Upstream and downstream DNA fragments flanking slr1796 were amplified using the primers listed in Table 2. The KmR gene was amplified using primers with overlapping regions to the two flanking fragments, and a fusion PCR product from the three fragments was generated and inserted into the pUC118 vector backbone using the circular polymerase extension cloning method (41), resulting in plasmid pDel1796::KmR. The Synechocystis 6803 WT strain was transformed with this plasmid to generate Δslr1796::KmR. Complete segregation of the mutant was achieved in the presence of 10 mm glucose and 10 μm DCMU under low light (10 μmol of photons m−2·s−1). PCR was performed to check segregation as shown in Fig. 2.
TABLE 2.
Oligonucleotide primers used in this study
| Product | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| slr1796 left flank | cagctatgaccatgattacgaattcccaaacagccccccagg | ttcttcacgacgtggaaaaaactcagtctaaaataggg |
| Kanamycin gene | tttttccacgtcgtgaagaaggtgttgctgactca | aaagaatgagggaaagccacgttgtgtct |
| slr1796 right flank | gtggctttccctcattctttccgacaacaaaaaaacgga | atccccgggtaccgagctcgaattcggatggggacgctgaagccg |
| slr1796 deletion detection (p1 + p2) | ccaagcttgttgatgcggcc | cctccagtggaagtggcgagt |
| slr1796 detection (p3 + p4) | atcgccgctttggaagatga | atcgccgctttggaagatga |
| slr1796-His6 | cataaagtcaagtaggagattaattcaatgattatgtctgtg | catgagaccggtctcagtgatgatgatggtggtgatcgactaattcac |
| pcpc560-Slr179His6 vector backbone | tgagaccggtctcatgataaccagg | tgaattaatctcctacttgactttatgagttgg |
| psaA | gctaaggccaaagtctcggt | tgggcattggcatggagatt |
| psaB | tgtactccctgccctcctac | ggaagcccaagaagagggac |
| psbA | cccccggtggatattgatgg | accccagctacccctaacat |
| psbB | tgcccaggaatggattggtc | acaccggtttgttccacact |
| psaC | gcagccaacacaatcctcgg | ggttgtacccaatgcgtccg |
| PsaD | accagcgcctctgaacaagt | cgtagggggctttaccggag |
| psaE | ggtgattgtgcgcttcgacc | tattttgccgccgcttgcac |
| psaF | gccaagggccaaaggaaacc | ccccgaaggttatccccacc |
| psaI | acggttcctacgctgcatct | agcaaccccatggtcacagc |
| psaJ | taggggtggaaaagaagatcggg | cctttttgtcaactgctccggt |
| psaK1 | gccctaaagttgctggggtg | ccagcacagcccctaaacca |
| psaK2 | aactggggcagagccaagtc | gcgctcaaagagggggattg |
| psaL | gggcatctttccacccccat | gccatgcccacttccaaacc |
| psaM | atggcattatccgacacccaaat | cggaaagccaggaaagcagg |
| btpA | gaccaccgctggatctaccc | gagcgagcggaacaagaagc |
| ycf3 | agaggcgttacggttggagg | gcccggtgattttgagccag |
| ycf4 | tggtcacttgcccggacttt | agaagtattgggagcccgcc |
| ycf37 | tgggctatgcctgtttcgct | gaattggcccgccgttgag |
| rnpB | agttagggagggagttgc | taagccgggttctgttcc |
A complemented strain was also generated by placing the slr1796 ORF with a C-terminal His6 tag under the control of the Pcpc560 promoter to generate the plasmid pcpc560-Slr179His6::GmR. This plasmid targets genomic neutral site 1 (31) and was used for transformation of the Δslr1796 mutant to create the Comp strain Δslr1796/pcpc560-Slr179His6.
Absorption and 77 K Fluorescence Emission Spectroscopy
Synechocystis 6803 strains were adjusted to equal cell numbers, and whole cell absorption spectra were collected on a DW2000 spectrophotometer (SLM-Aminco, Urbana, IL). Phycobilin concentration was estimated using absorption data (42), and chlorophyll concentration was determined using methanol extracts as described (43).
77 K low temperature fluorescence spectra of samples adjusted to equal cell number and corresponding to 5 μg/ml Chl in WT were recorded on a Fluoromax-2 fluorometer (Jobin Yvon, Cedex, France). The excitation wavelength for chlorophyll was set at 435 nm.
Kinetic Spectroscopic Measurements
To estimate PSI and PSII activity, cells were normalized to equal numbers. The kinetics of PSI photooxidation was determined on a Joliot-type spectrometer (JTS-10, Bio-Logic, France). For P700 absorption changes, samples were illuminated for 5 s with far-red actinic light (1,400 μmol of photons m−2·s−1) and detected at 705 nm. Electron transport inhibitors DCMU and 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone (both at 10 μm) were added where indicated. Absorbance without actinic illumination was used as baseline and subtracted.
The reoxidation kinetics of the reduced PSII primary electron acceptor QA− was measured on an FL-200 dual modulation kinetic fluorometer (Photon Systems Instruments) using samples of equal cell numbers that were dark-adapted for at least 3 min before measurement.
Photosynthetic Activity Measurements
Rates of oxygen evolution and oxygen consumption in cell samples adjusted to equal cell densities were measured using a Clark-type electrode. Samples were incubated for 1 h in the dark at 30 °C before the assay under heat-filtered light of 8,250 μmol of photons m−2·s−1. Whole chain electron transport in intact cells was measured by adding 10 mm NaHCO3. PSI-mediated electron transport was measured in cells by adding 1.0 mm sodium ascorbate and 1.0 mm diaminodurene as electron donors, 2.0 mm methyl viologen as electron acceptor, and 20 μm DCMU before measurements (44, 45). For PSII-mediated electron transfer, 1 mm K3Fe(CN)6 and 0.5 mm 2,6-dichloro-p-benzoquinone were added as electron acceptors.
RT-PCR
The relative transcript levels of various PSI and PSII genes were evaluated by RT-PCR. Total RNA from Synechocystis 6803 cells at an OD730 nm of 0.4 was extracted using RNAwiz (Ambion Inc., Austin, TX), treated with RNase-free DNase I (Invitrogen), and then purified by phenol-chloroform extraction and ethanol precipitation. The RT reaction was performed using Superscript II reverse transcriptase (Invitrogen) and random primers. The RNaseP gene (rnpB) was used as a positive control with constitutive expression levels. The RT products were amplified by PCR with empirically determined cycles to ensure the PCR products represent a linear amplification range. Primer sequences are listed in Table 2.
Membrane Protein Preparations, Electrophoresis, and Western Blotting
Total membrane, thylakoid membrane, and plasma membrane were isolated as described (34) with modifications. Cells were disrupted with glass beads (0.1 mm) in a Bead Beater (Biospec Products) for 10 cycles of 30-s beating and 30 s on ice. Unbroken cells and debris were removed by centrifugation at 5,000 × g for 5 min, the total protein fraction was collected, membranes were then pelleted at 16,000 × g for 30 min and homogenized in two-phase buffer (0.25 m sucrose, 5 mm potassium phosphate buffer, pH 7.8) or other buffer where indicated, and the supernatant was collected as the soluble protein fraction. Two or three rounds of two-phase partitioning with 6.0% aqueous polymer systems (polyethylene glycol and dextran) were conducted to produce thylakoid membrane from the greener bottom phase and plasma membrane from the brown top phase.
Protein concentrations were quantified using a Pierce BCA Protein Assay kit (Thermo Scientific). Samples with equal protein amount were separated by 15% SDS-PAGE (46). After transferring the fractionated proteins onto PVDF membrane (Millipore, Billerica, MA), various proteins were detected by using specific antisera, and the signals were visualized using enhanced chemiluminescent HRP reagents (Millipore) on an ImageQuant LAS-4000 imager (GE Healthcare). Two to three independent replicates for each protein blot were performed, and image quantification was done using ImageJ software.
For blue native PAGE analysis, Synechocystis 6803 membranes were prepared and separated essentially as in Dühring et al. (47). The membranes were solubilized with 1% (w/v) n-dodecyl β-d-maltoside, and equal protein amounts were loaded onto a 1.0-mm-thick NativePAGE 3–12% Bis-Tris gel along with NativeMark unstained protein standard (Life Technologies). Electrophoresis was performed at 4 °C by increasing voltage from 70 up to 120 V during the 8-h run.
Chemical Wash of Membrane Vesicles
Ionic and non-ionic chemical wash treatments of thylakoid membranes were performed as described (48). In brief, aliquots of 50 μg of membrane proteins were washed twice and resuspended in 50 μl of HS buffer (10 mm HEPES, pH 7.6, 0.1 m sucrose) followed by adding an equal volume of HS buffer (control), 2 m NaCl, 0.2 m Na2CO3, or 0.5% Triton X-100, respectively. The mixture was incubated on ice for 30 min and centrifuged at 20,000 × g (4 °C) for 10 min, the supernatants were collected, and the pellets were washed twice with HS buffer. Samples were separated by SDS-PAGE and analyzed by Western blotting using specific antibodies as above.
Phylogenetic Analysis
The search for Slr1796 orthologs was performed using the NCBI BlastP program. A list of sequences with E value <10−6 was generated and manually curated to include entries that displayed similar sequence features as in Slr1796 of Synechocystis 6803. Protein sequences were aligned in MEGA 6.06 (49) using ClustalW, and a phylogenetic tree was constructed using the neighbor-joining method (50). Bootstrap values were calculated with 1,000 replications, and cutoff values of 50% were chosen for visualization on the tree.
Sample Preparation for Electron Microscopy
The sample preparation method was essentially as described (51). Culture aliquots were frozen in a Bal-Tec high pressure freezer. Samples were freeze-substituted in 2% osmium, acetone (3 days at −80 °C, 15 h at −60 °C, and slow thaw to room temperature) and embedded in Spurr's resin. Thin sections (∼80 nm) were cut and stained with uranyl acetate and lead citrate. Digital images were viewed and collected using a LEO 912 transmission electron microscope operating at 120 kV and a ProScan digital camera. Measurements were made from TEM images using iTEM software (Soft Imaging System) or Fiji (52). Results are given as the standard deviation of 25–35 measurements per strain.
Author Contributions
H. B. P. supervised the study. H. B. P. and Y. Z. designed the study. M. L. carried out electron microscopy experiments. Y. Z. conducted all other experiments unless indicated otherwise. Y. Z. and M. L. wrote the manuscript. All authors approved the final version.
Acknowledgments
We thank Drs. D. A. Bryant and G. Z. Shen at Penn State University for PsaD and PsaE antibodies. We also thank members of the Pakrasi laboratory for collegial discussions and Howard Berg at the Integrated Microscopy Facility at the Donald Danforth Plant Science Center for TEM assistance.
This work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, United States Department of Energy under Grant DE-FG02-99ER20350 (to H. B. P.). The authors declare that they have no conflicts of interest with the contents of this article.
- PS
- photosystem
- Trx
- thioredoxin
- DCMU
- 3-(3,4-dichlorophenyl)-1,1-dimethylurea
- Comp
- complemented
- PC
- phycocyanin
- Chl
- chlorophyll
- TEM
- transmission electron microscopy
- KmR
- kanamycin resistance
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Buchanan B. B., and Balmer Y. (2005) Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56, 187–220 [DOI] [PubMed] [Google Scholar]
- 2. Foyer C. H., and Noctor G. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905 [DOI] [PubMed] [Google Scholar]
- 3. Atkinson H. J., and Babbitt P. C. (2009) An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput. Biol. 5, e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amunts A., and Nelson N. (2009) Plant photosystem I design in the light of evolution. Structure 17, 637–650 [DOI] [PubMed] [Google Scholar]
- 5. Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., and Krauss N. (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909–917 [DOI] [PubMed] [Google Scholar]
- 6. Schöttler M. A., Albus C. A., and Bock R. (2011) Photosystem I: its biogenesis and function in higher plants. J. Plant Physiol. 168, 1452–1461 [DOI] [PubMed] [Google Scholar]
- 7. Yang H., Liu J., Wen X., and Lu C. (2015) Molecular mechanism of photosystem I assembly in oxygenic organisms. Biochim. Biophys. Acta 1847, 838–848 [DOI] [PubMed] [Google Scholar]
- 8. Göhre V., Ossenbühl F., Crèvecoeur M., Eichacker L. A., and Rochaix J.-D. (2006) One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18, 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ossenbühl F., Inaba-Sulpice M., Meurer J., Soll J., and Eichacker L. A. (2006) The Synechocystis sp PCC 6803 oxa1 homolog is essential for membrane integration of reaction center precursor protein pD1. Plant Cell 18, 2236–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boudreau E., Takahashi Y., Lemieux C., Turmel M., and Rochaix J. D. (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16, 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruf S., Kössel H., and Bock R. (1997) Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krech K., Ruf S., Masduki F. F., Thiele W., Bednarczyk D., Albus C. A., Tiller N., Hasse C., Schöttler M. A., and Bock R. (2012) The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ozawa S., Nield J., Terao A., Stauber E. J., Hippler M., Koike H., Rochaix J.-D., and Takahashi Y. (2009) Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell 21, 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stöckel J., Bennewitz S., Hein P., and Oelmüller R. (2006) The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilde A., Lünser K., Ossenbühl F., Nickelsen J., and Börner T. (2001) Characterization of the cyanobacterial ycf37: mutation decreases the photosystem I content. Biochem. J. 357, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Albus C. A., Ruf S., Schöttler M. A., Lein W., Kehr J., and Bock R. (2010) Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22, 2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J., Yang H., Lu Q., Wen X., Chen F., Peng L., Zhang L., and Lu C. (2012) PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24, 4992–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fristedt R., Williams-Carrier R., Merchant S. S., and Barkan A. (2014) A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants. J. Biol. Chem. 289, 30657–30667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen G., Antonkine M. L., van der Est A., Vassiliev I. R., Brettel K., Bittl R., Zech S. G., Zhao J., Stehlik D., Bryant D. A., and Golbeck J. H. (2002) Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of FX in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 277, 20355–20366 [DOI] [PubMed] [Google Scholar]
- 20. Shen G., Zhao J., Reimer S. K., Antonkine M. L., Cai Q., Weiland S. M., Golbeck J. H., and Bryant D. A. (2002) Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 277, 20343–20354 [DOI] [PubMed] [Google Scholar]
- 21. Schwenkert S., Netz D. J., Frazzon J., Pierik A. J., Bill E., Gross J., Lill R., and Meurer J. (2010) Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stöckel J., and Oelmüller R. (2004) A novel protein for photosystem I biogenesis. J. Biol. Chem. 279, 10243–10251 [DOI] [PubMed] [Google Scholar]
- 23. Touraine B., Boutin J.-P., Marion-Poll A., Briat J.-F., Peltier G., and Lobréaux S. (2004) Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 40, 101–111 [DOI] [PubMed] [Google Scholar]
- 24. Yabe T., Morimoto K., Kikuchi S., Nishio K., Terashima I., and Nakai M. (2004) The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16, 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartsevich V. V., and Pakrasi H. B. (1997) Molecular identification of a novel protein that regulates biogenesis of photosystem I, a membrane protein complex. J. Biol. Chem. 272, 6382–6387 [DOI] [PubMed] [Google Scholar]
- 26. Zak E., and Pakrasi H. B. (2000) The BtpA protein stabilizes the reaction center proteins of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803 at low temperature. Plant Physiol. 123, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rochaix J.-D. (2011) Assembly of the photosynthetic apparatus. Plant Physiol. 155, 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chi W., Ma J., and Zhang L. (2012) Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 3420–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Srivastava R., Pisareva T., and Norling B. (2005) Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5, 4905–4916 [DOI] [PubMed] [Google Scholar]
- 30. Liberton M., Saha R., Jacobs J. M., Nguyen A. Y., Gritsenko M. A., Smith R. D., Koppenaal D. W., and Pakrasi H. B. (2016) Global proteomic analysis reveals an exclusive role of thylakoid membranes in bioenergetics of a model cyanobacterium. Mol. Cell. Proteomics 15, 2021–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng A. H., Berla B. M., and Pakrasi H. B. (2015) Fine-tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 81, 6857–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen G., Boussiba S., and Vermaas W. F. (1993) Synechocystis sp. PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5, 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilde A., Härtel H., Hübschmann T., Hoffmann P., Shestakov S. V., and Börner T. (1995) Inactivation of a Synechocystis sp. strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Norling B., Zak E., Andersson B., and Pakrasi H. (1998) 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 436, 189–192 [DOI] [PubMed] [Google Scholar]
- 35. Chitnis P. R., Reilly P. A., Miedel M. C., and Nelson N. (1989) Structure and targeted mutagenesis of the gene encoding 8-kDa subunit of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 264, 18374–18380 [PubMed] [Google Scholar]
- 36. Lyska D., Meierhoff K., and Westhoff P. (2013) How to build functional thylakoid membranes: from plastid transcription to protein complex assembly. Planta 237, 413–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vermaas W. F., Timlin J. A., Jones H. D., Sinclair M. B., Nieman L. T., Hamad S. W., Melgaard D. K., and Haaland D. M. (2008) In vivo hyperspectral confocal fluorescence imaging to determine pigment localization and distribution in cyanobacterial cells. Proc. Natl. Acad. Sci. U.S.A. 105, 4050–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang S., Shen G., Li Z., Golbeck J. H., and Bryant D. A. (2014) Vipp1 is essential for the biogenesis of photosystem I but not thylakoid membranes in Synechococcus sp. PCC 7002. J. Biol. Chem. 289, 15904–15914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu H., Zhang H., Weisz D. A., Vidavsky I., Gross M. L., and Pakrasi H. B. (2014) MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc. Natl. Acad. Sci. U.S.A. 111, 4638–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Allen M. M. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4, 1–4 [DOI] [PubMed] [Google Scholar]
- 41. Quan J., and Tian J. (2011) Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 6, 242–251 [DOI] [PubMed] [Google Scholar]
- 42. Arnon D. I., McSwain B. D., Tsujimoto H. Y., and Wada K. (1974) Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim. Biophys. Acta 357, 231–245 [DOI] [PubMed] [Google Scholar]
- 43. Porra R. J., Thompson W. A., and Kriedemann P. E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 44. Kubota H., Sakurai I., Katayama K., Mizusawa N., Ohashi S., Kobayashi M., Zhang P., Aro E.-M., and Wada H. (2010) Purification and characterization of photosystem I complex from Synechocystis sp. PCC 6803 by expressing histidine-tagged subunits. Biochim. Biophys. Acta 1797, 98–105 [DOI] [PubMed] [Google Scholar]
- 45. Cameron J. C., and Pakrasi H. B. (2011) Glutathione in Synechocystis 6803: a closer look into the physiology of a gshB mutant. Plant Signal. Behav. 6, 89–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., and Pakrasi H. B. (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 [DOI] [PubMed] [Google Scholar]
- 47. Dühring U., Ossenbühl F., and Wilde A. (2007) Late assembly steps and dynamics of the cyanobacterial photosystem I. J. Biol. Chem. 282, 10915–10921 [DOI] [PubMed] [Google Scholar]
- 48. Schottkowski M., Ratke J., Oster U., Nowaczyk M., and Nickelsen J. (2009) Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Mol. Plant 2, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 49. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saitou N., and Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 51. Liberton M., Howard Berg R., Heuser J., Roth R., and Pakrasi H. B. (2006) Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227, 129–138 [DOI] [PubMed] [Google Scholar]
- 52. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]