Abstract
Murine natural killer (NK) cells are regulated by the interaction of Ly49 receptors with major histocompatibility complex class I molecules (MHC-I). Although the ligands for inhibitory Ly49 were considered to be restricted to classical MHC (MHC-Ia), we have shown that the non-classical MHC molecule (MHC-Ib) H2-M3 was a ligand for the inhibitory Ly49A. Here we establish that another MHC-Ib, H2-Q10, is a bona fide ligand for the inhibitory Ly49C receptor. H2-Q10 bound to Ly49C with a marginally lower affinity (∼5 μm) than that observed between Ly49C and MHC-Ia (H-2Kb/H-2Dd, both ∼1 μm), and this recognition could be prevented by cis interactions with H-2K in situ. To understand the molecular details underpinning Ly49·MHC-Ib recognition, we determined the crystal structures of H2-Q10 and Ly49C bound H2-Q10. Unliganded H2-Q10 adopted a classical MHC-I fold and possessed a peptide-binding groove that exhibited features similar to those found in MHC-Ia, explaining the diverse peptide binding repertoire of H2-Q10. Ly49C bound to H2-Q10 underneath the peptide binding platform to a region that encompassed residues from the α1, α2, and α3 domains, as well as the associated β2-microglobulin subunit. This docking mode was conserved with that previously observed for Ly49C·H-2Kb. Indeed, structure-guided mutation of Ly49C indicated that Ly49C·H2-Q10 and Ly49C·H-2Kb possess similar energetic footprints focused around residues located within the Ly49C β4-stand and L5 loop, which contact the underside of the peptide-binding platform floor. Our data provide a structural basis for Ly49·MHC-Ib recognition and demonstrate that MHC-Ib represent an extended family of ligands for Ly49 molecules.
Keywords: crystal structure, innate immunity, major histocompatibility complex (MHC), natural killer cells (NK cells), receptor structure-function
Introduction
The primary function of major histocompatibility complex class I molecules (MHC-I)5 is to capture peptides derived from self- and pathogen-derived proteins to display them on the cell surface for immune surveillance by CD8+ T cells. There are two broad types of MHC-I. “Classical” or “class Ia” MHC (MHC-Ia) are ubiquitously expressed on all nucleated cells and are highly polymorphic in nature, thereby allowing them to present a diverse array of peptides to CD8+ T cells and ensure immunity to a wide range of infections (1). In addition to the polymorphic MHC class Ia molecules, the MHC gene cluster also encodes a number of “non-classical” or “class Ib” MHC molecules (MHC-Ib). MHC-Ib share structural similarity with their MHC-Ia counterparts, but have limited polymorphism and often display a tissue-specific pattern of expression (2). In contrast to humans, where MHC-Ib are limited to three genes encoding HLA-E, HLA-F, and HLA-G, MHC-Ib genes in mice are highly divergent, with most mouse strains encoding dozens of genes in the H2-Q, H2-T, and H2-M regions (3). However, the function of many of the murine MHC-Ib currently remains unknown.
In addition to their role in activating CD8+ T cells, MHC-Ia also serve as ligands for receptor families that regulate the function of natural killer (NK) cells. For example, groups of HLA-B and HLA-C alleles in humans are recognized by inhibitory KIR3DL1 and KIR2DL1/2/3, respectively (4). Down-regulation of surface MHC-Ia, as may occur during viral infection or following malignant transformation, results in a loss of inhibitory signals and can trigger NK cell activation, a process termed “missing-self” (5). Furthermore, recognition of self-MHC-Ia by inhibitory KIR also plays a key role in the generation of NK cells capable of responding to cells that lack MHC-Ia. Indeed, NK cells expressing inhibitory receptors that have specificity for self-MHC-I proteins have elevated responses to MHC-I-deficient target cells when compared with those that lack such receptors, a process called licensing (6).
The functional homologue of KIR in mice is the Ly49 family of NK cell receptors. Like KIR, the highly polymorphic Ly49 family of ligands encodes for both activating and inhibitory isoforms that recognize MHC-I or MHC-I-like ligands (7). Also similar to KIR, the established ligands for the Ly49 family are composed of MHC-Ia. These interactions include recognition of H-2Dd by Ly49A, H-2Dk by Ly49G, and H-2Kb by Ly49C (8, 9). The ternary structures of H-2Dd·Ly49A and H-2Kb·Ly49C have been solved, providing molecular details of the interaction between Ly49 and MHC-Ia. Although the human KIR receptors bind over the α1 and α2 helices of the MHC-Ia, positioned over the C-terminal end of the peptide (10, 11), the Ly49·MHC-Ia crystal structures revealed a completely different mode of recognition. In both the Ly49A·H-2Dd and the Ly49C·H-2Kb structures, the Ly49 was bound to a concave groove underneath the peptide-binding platform (12), a location that precludes any direct contact with the peptide. Although the overall docking mode was conserved, the Ly49A·H-2Dd and Ly49C·H-2Kb structures did differ in the configuration of the Ly49 homo-dimer and the stoichiometry of the Ly49·MHC-Ia complex. These differences likely relate to the capacity of Ly49 to recognize MHC-I within the plane of the same membrane (in cis) and also on opposing cell membranes (in trans). Although cis interactions do not lead to productive signaling, they are proposed to restrict NK inhibition, thus facilitating NK effector functions (13).
MHC-Ib in humans and mice can perform specialized functions, including in the regulation of NK cells. Mouse Qa-1b and its human homologue, HLA-E, play a dual role in immunity by presenting peptides to receptors expressed on NK cells as well as to the T cell receptor of CD8+ cells (14). In humans, the MHC-Ib HLA-G exhibits a highly restricted pattern of tissue expression, largely limited to developing trophoblast cells in the placenta (15, 16). Hence HLA-G has been proposed to play a specialized role in the maintenance of pregnancy by modulating leukocyte function, acting as a ligand for receptors of the leukocyte immunoglobulin-like receptor (LILR) and/or KIR receptor families (17). Furthermore, some HLA-G isoforms include secreted forms of the molecule (sHLA-G). In addition to the role that sHLA-G might play in the regulation of NK cell function (18), it has also been shown to modulate chemokine receptor expression on certain CD4 and CD8 T cell populations (19).
Although a direct homologue of HLA-G has yet to be confirmed in mice, the murine MHC-Ib H2-Q10 is the only murine MHC-I found in the serum in appreciable concentrations (20). H2-Q10 has a truncation in the C terminus and the presence of several substitutions in the transmembrane region that are responsible for the soluble nature of this protein. Like HLA-G, H2-Q10 also shows a restricted pattern of expression, being overexpressed in the liver when compared with other tissues (21). In addition, both H2-Q10 (22) and HLA-G (23) have a more diverse peptide binding repertoire than many other MHC-Ib such as Qa-1b, HLA-E, and H2-M3, where the peptide binding repertoires are highly restricted (22). These factors suggest that H2-Q10 may play a highly specialized role in immunity, yet its precise function currently remains unknown.
It was recently demonstrated that the MHC-Ib, H2-M3, is a functional, high affinity ligand for Ly49A (24). Moreover, H2-M3 played a role in licensing Ly49A-expressing NK cells. This provided the first evidence that MHC-Ib could act as ligands for Ly49 family members. However, whether additional Ly49 ligands exist within the MHC-Ib family, as well as the molecular basis underpinning these interactions, remains unclear. Here, we demonstrate that H2-Q10 is a bona fide ligand for Ly49C. Furthermore, we report the first crystal structures of H2-Q10 and the Ly49C·H2-Q10 complex. H2-Q10 has a structural and energetic footprint on Ly49C that overlaps significantly with that of H-2Kb. These results extend the known ligands for Ly49 receptors and suggest that the family of MHC-Ib play an important role in the shaping of NK cell responses.
Results
H2-Q10 Binds Specifically to Ly49C
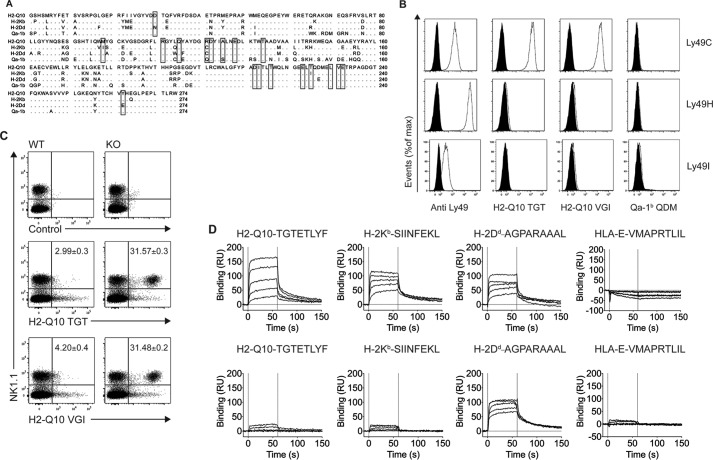
Alignment of the sequence of the H2-Q10 heavy chain with classical MHC-encoded heavy chains identified regions of sequence homology between H2-Q10 (National Center for Biotechnology Information (NCBI) accession code AAH11215.1), H-2Kb (NCBI accession code 1VAC_A) and H-2Dd (NCBI accession code 1BII_A), both of which are known ligands for Ly49C (Fig. 1A). This prompted us to determine whether H2-Q10 could also act as a ligand for Ly49C. Initially, the binding of H2-Q10 tetramers loaded with peptides derived from either T cell receptor (TCR) Vβ chain (TGTETLYF) or ribophorin (VGITNVDL) to a panel of CHO cells expressing Ly49CB6, Ly49HB6, and Ly49IB6 was assessed. Among these closely related Ly49 molecules, H2-Q10 bound to Ly49C, but not Ly49H or Ly49I, in a manner that was independent of the sequence of the bound peptide (Fig. 1B). In contrast, Qa-1b did not bind appreciably to any of the Ly49-expressing cells. These data provide the first evidence that Ly49C is able to specifically recognize H2-Q10.
FIGURE 1.
H2-Q10 is a bone-fide ligand for C57BL/6 Ly49C. A, sequence alignment of H2-Q10, H-2Kb, H-2Dd, and Qa-1b. Residues conserved in at least three of the sequences are shown as dots. Binding sites for Ly49C on H-2Kb are shown within boxes. B, tetramer staining of CHO cells expressing Ly49C, Ly49H, and Ly49I. Open curves show staining with Ly49-specific antibody (Anti Ly49), H2-Q10 loaded with TGTETLYF (H2-Q10 TGT), H2-Q10 loaded with VGITNVDL (H2-Q10 VGI), or Qa-1b loaded with AMAPRTLLL (Qa-1b QDM); black filled curves show isotype control antibody (for anti-Ly49) or irrelevant human HLA-E tetramer (for H2-Q10). Data are representative of at least 5 independent experiments. C, H2-Q10 binding to Ly49C ex vivo is impaired by cis interactions with MHC-I. Splenocytes were prepared from WT and H-2Kb/Db double-deficient mice. Cells were stained with NK1.1, CD3, an irrelevant human HLA-E tetramer (control), H2-Q10 tetramer loaded with TGTETLYF (H2-Q10 TGT), or H2-Q10 loaded with VGITNVDL (H2-Q10 VGI). The dot plots show events that have been electronically gated as singlets, live and CD3− (not shown), and presented as NK1.1 (vertical) versus tetramer (horizontal). Numbers in the top right quadrant of the H2-Q10-stained panels represent the percentage of H2-Q10+ NK cells and are presented as mean ± S.E. of four mice. Data are representative of 2 independent experiments using four mice per experiment (n = 8). D, Ly49C binds with similar affinity to H2-Q10, H-2Kb, and H-2Dd. Binding decreasing concentrations (30, 12, 4.8, 1.92, 0.77 μm; solid lines top to bottom) of Ly49C (upper) or Ly49A (lower) to streptavidin-immobilized H2-Q10-TGTETLYF, H-2Kb-SIINFEKL, H-2Dd-AGPARAAAL, or HLA-E-VMAPRTLIL is shown. Results are presented in response units (RU) after subtraction of the baseline values. Solid vertical lines at 0 and 60 s indicate injection start and stop, respectively. Binding affinity (determined by equilibrium analysis): H2-Q10-TGTETLYF-Ly49C KD = 5.0 ± 0.5 μm; H-2Kb-SIINFEKL-Ly49C, KD = 1.0 ± 0.2 μm; H2-Dd-AGAPARAAAL-Ly49C, KD = 1.3 ± 0.5 μm; H2-Dd-AGAPARAAAL-Ly49A KD = 0.5 ± 0.1 μm. Responses for HLA-E with Ly49A and Ly49C, H2-Q10 with Ly49A, and H-2Kb with Ly49A are below the limit of detection.
cis Interactions with H-2K Prevent H2-Q10 Binding to Ly49C
In addition to recognizing H-2Kb in trans (on an opposing cell membrane), Ly49C is known to interact with H-2Kb through cis interactions (9). This cis binding can alter the threshold for NK cell activation by making inhibitory receptors unavailable for binding and may also be important in the education of murine NK cells (25). We next sought to determine whether H2-Q10 binding to Ly49C could be regulated by H-2K molecules expressed on the surface of NK cells from wild-type mice where cis interactions are present or H-2KbDb-deficient mice in which cis interactions with MHC-I are largely absent. Although we were unable to detect binding of H2-Q10 tetramers to NK cells from wild-type mice, a significant proportion of NK cells isolated from H-2Kb/Db-deficient mice was stained by H2-Q10 tetramers (Fig. 1C). Similar to the results obtained for CHO cells (Fig. 1B), both tetramers yielded similar results irrespective of differences in the peptide sequence. These results demonstrate that cis interaction of Ly49C with H-2K molecules prevents recognition of H2-Q10 tetramers in trans.
H2-Q10 Binds Ly49C but Not Ly49A
Given the inability of tetrameric forms H2-Q10 to effectively compete with Ly49C ligands bound in cis, we sought to determine the affinity of the Ly49C·H2-Q10 interaction using SPR. As positive controls, we included known Ly49C ligands H-2Kb-SIINFEKL and H-2Dd-AGPARAAAL, both of which bound to Ly49C with high affinity (1.0 ± 0.2 and 1.4 ± 0.5 μm, respectively, Fig. 1D). Using the same conditions, H2-Q10 also bound Ly49C, albeit with slightly weaker affinity (5.0 ± 0.5 μm). As a further control, we also assessed the binding of Ly49A to the various MHC-I. Although Ly49A bound to H-2Dd with high affinity (0.5 ± 0.1 μm, Fig. 1D) in line with previously reported data (43), it did not bind to either H-2Kb or H2-Q10. Moreover, neither Ly49A nor Ly49C bound appreciably to the human MHC class Ib molecule HLA-E (Fig. 1). Overall, these data indicate that the Ly49C·H2-Q10 interaction is well within the range typically observed for Ly49 receptors interacting with class Ia MHC.
Structure of Unliganded H2-Q10
To understand the structural basis for Ly49-mediated recognition of H2-Q10, we firstly determined the structure of H2-Q10 presenting a ribophorin-derived peptide (VGITNVDL) to 2.3 Å resolution (Table 1). Despite reports that it is detectable in the serum as a multivalent complex of 200–300 kDa (20, 26), H2-Q10-VGITNVDL eluted as a monomer during size-exclusion chromatography (not shown) and the crystal packing was not suggestive of higher order assemblies. The overall structure of H2-Q10 is similar to other MHC-Ia and MHC-Ib, comprising a heavy chain of α1, α2, and α3 domains that is non-covalently associated with a β2M subunit (Fig. 2A). The root mean square deviation (r.m.s.d) between H2-Q10 and H-2Kb, HLA-A2, H2-M3, and HLA-G was 1.87, 0.94, 0.77, and 1.02 Å over all Cα atoms, respectively, further highlighting the high degree of structural conservation between these related molecules.
TABLE 1.
Data collection and refinement statistics
| Ly49C-Q10 | Q10 | |
|---|---|---|
| Data collection statistics | ||
| Temperature (K) | 100 | 100 |
| X-ray source | MX2 Australian Synchrotron | MX1 Australian Synchrotron |
| Space group | I2 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 101.0, 57.8, 210.7 | 55.4, 81.7, 97.9 |
| α, β, γ (°) | 90.0, 96.15, 90.0 | 90, 90, 90 |
| Resolution (Å) | 60.45–3.3 (3.56–3.3) | 42.0–2.3 (2.42–2.3) |
| Total no. of observations | 64879 (13432) | 136962 (19918) |
| No. of unique observations | 18565 (3785) | 20349 (2894) |
| Multiplicity | 3.5 (3.5) | 6.7 (6.9) |
| Data completeness (%) | 99.9 (100) | 99.8 (99.6) |
| I/σI | 5.8 (2.1) | 6.6 (2.2) |
| Rmergea (%) | 27.3 (69.1) | 24.1 (95.3) |
| Rpim (%) | 21.0 (52.1) | 9.2 (36.0) |
| Refinement statistics | ||
| Non-hydrogen atoms | ||
| Protein | 7879 | 3126 |
| Water | 0 | 136 |
| Sugar | 0 | 0 |
| Rfactorb (%) | 19.6 | 17.7 |
| Rfree (%) | 25.7 | 24.7 |
| r.m.s.d from ideality | ||
| Bond lengths (Å) | 0.009 | 0.01 |
| Bond angles (°) | 1.05 | 1.09 |
| Ramachandran plot | ||
| Favored regions (%) | 92.8 | 96.8 |
| Allowed regions (%) | 6.2 | 2.9 |
| Disallowed regions (%) | 1.0 | 0.3 |
a Rmerge = Σhkl Σj |Ihkl,j − 〈 Ihkl 〉 |/Σhkl Σj Ihkl,j.
b Rfactor = Σhkl‖Fo| − |Fc‖/Σhkl|Fo| for all data excluding the 5% that comprised the Rfree used for cross-validation.
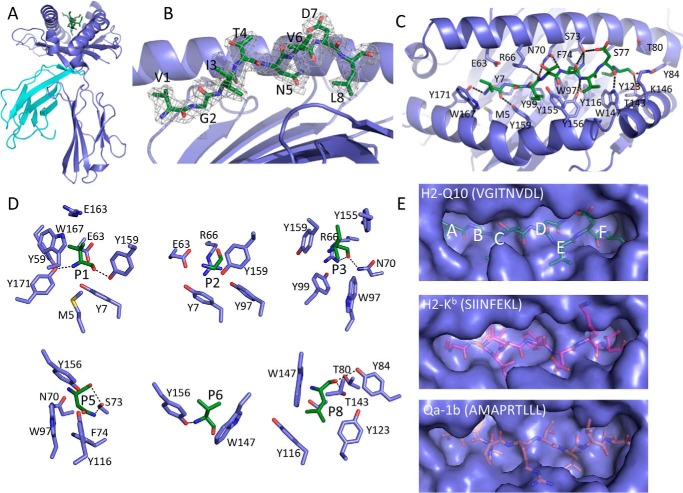
FIGURE 2.
Structure of unliganded H2-Q10. A, overview of the unliganded H2-Q10 (VGITNVDL) complex. The heavy chain is shown in violet, β2M is in cyan, and the VGITNVDL peptide is in green. B, the conformation of the bound peptide (stick representation). The corresponding 2Fo − Fc electron density is shown in mesh format contoured at 1σ. C, top view showing interactions between the H2-Q10 heavy chain (violet) and the bound peptide (green sticks). D, interactions of each peptide anchor position with H2-Q10. Hydrogen bonds are represented by dashed lines. E, comparison of the peptide-binding groove of H2-Q10 with other MHC-Ia (H2-Kb) and MHC-Ib (Qa-1b). Pocket positions of H2-Q10 are labeled.
The H2-Q10 peptide-binding groove is formed by an antiparallel β-sheet floor flanked by α1 and α2 helices, and is lined with a myriad of aromatic side chains that support the VGITNVDL peptide in an extended conformation (Fig. 2, B and C). Previous peptide elution and substitution experiments have identified that H2-Q10 binds a heterogeneous repertoire of peptides with a preference for Gly at P2, Leu/Val at P6, and Phe/Leu at Pω (22). Accordingly, the H2-Q10 peptide-binding groove is considered to resemble that of MHC-Ia rather than those of other more specialized MHC-Ib such as Qa-1b (27), H2-M3 (28), or HLA-E (29, 30). Overall, the unliganded H2-Q10 structure supports this proposition. For example, the main chain amine and carboxyl groups of P1-Val are hydrogen-bonded to the side chains of Tyr-171 and Tyr-159 (Fig. 2, C and D). These interactions are conserved in MHC-Ia structures (31). Moreover, the C terminus of P8-Leu is similarly tethered by hydrogen bonds to Tyr-84 and Thr-143, and is tightly enshrined by Thr-143 and Trp-147, a feature of MHC-Ia that directly contrasts to the looser fit of PΩ observed in other MHC-Ib including Qa-1 and HLA-E (Fig. 2, C–E) (27, 29). In addition to the hydrogen bonding of the peptide termini, H2-Q10 has five hydrophobic pockets that serve to anchor the peptide at P2-Gly, P3-Ile, P5-Asn, P6-Val, and P8-Leu (Fig. 2, D and E). Although the F pocket is deep to accommodate the Leu or Phe residues that are favored by H2-Q10 at PΩ, the remainder of the peptide-binding pockets are relatively shallow (Fig. 2E). In particular, the B pocket is particularly small, consisting of a high “floor” provided by Tyr-7 and narrow “walls” formed by Arg-66 and Tyr-159. Together, these residues dictate the absolute requirement for a Gly residue at P2 (22). The C and E pockets are similarly shallow, but relatively wide, thereby allowing P3-Ile and P6-Val to point laterally to the side of the groove. With the exception of P8-Leu, the only other peptide side chain residue that points directly into the H2-Q10 groove is P5-Asn, which makes extensive van der Waals interactions with Trp-97 and Phe-74, as well as hydrogen bonds to Ser-73 and Tyr-116 (Fig. 2, D and E). The extent of these interactions is somewhat surprising, because peptide elution studies suggest that H2-Q10 can accommodate a variety of amino acids at this position (22). In addition to these anchor residues, positions P4-Thr, P6-Val and P7-Asp are solvent-exposed and thus may represent candidate T cell receptor contact sites.
Overview of the Ly49C·H2-Q10 Structure
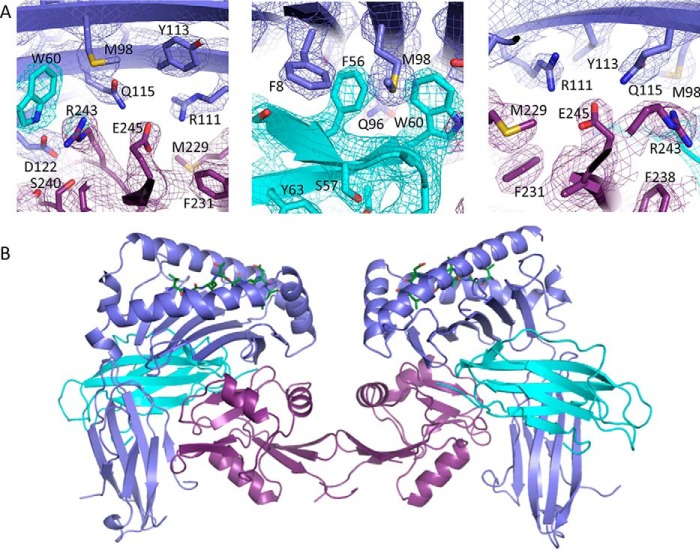
Next we determined the structure of H2-Q10-VGITNVDL bound to the lectin domain of Ly49CB6 to 3.3 Å resolution. The structure refined well, to an Rfactor and Rfree of 22.9 and 26.6%, respectively, and the electron density at the Ly49·Q10 interface was, for the most part, unambiguous (Fig. 3A), permitting a detailed understanding of the molecular interactions that underpin Ly49C·H2-Q10 recognition. During gel filtration chromatography, Ly49C eluted at a position consistent with a dimer (not shown), and in the crystal structure, two Ly49C lectin domains were related by non-crystallographic symmetry to form a homo-dimer. Our observation that Ly49C forms a homo-dimer is in line with previous studies regarding the oligomeric status of this receptor both in solution (32) and on the cell surface (33). In the structure, each of the Ly49 lectin domains engaged H2-Q10 via a concave surface underneath the peptide-binding platform that encompassed the α1, α2, α3, and β2M subunits of the MHC-Ib (Fig. 3B). Accordingly, the entire complex comprised one Ly49 dimer bound to two H2-Q10 molecules in a butterfly arrangement that is reminiscent of the Ly49C·H-2Kb structure (32) (Fig. 3B). As observed in previous Ly49·MHC-Ia structures, the receptor does not make any direct contacts with the bound peptide (12, 32).
FIGURE 3.
Overview of the Ly49C H2-Q10 complex. A, the 2Fo − Fc electron density map is shown contoured at 1σ and colored according to chain (H2-Q10 (violet); β2M (cyan); Ly49C (magenta)). B, the Ly49C.H2-Q10 complex visible within the crystallographic asymmetric unit is shown in ribbon representation. Within the Ly49C homo-dimer (magenta), each Ly49C monomer interacts with one H2-Q10 molecule (H2-Q10 violet; β2m (cyan)).
Variable Ly49 Configuration
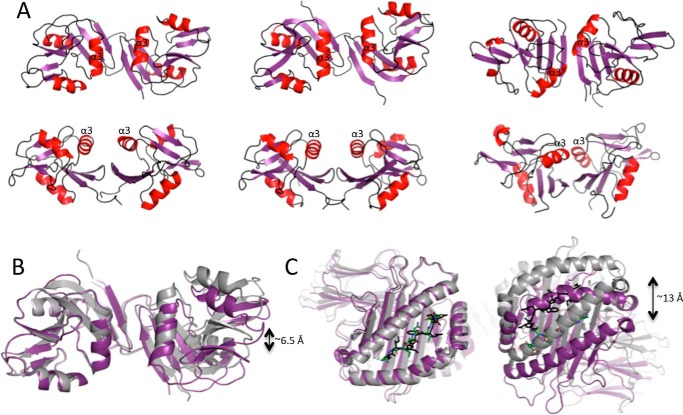
Ly49 receptors can adopt markedly different configurations to recognize MHC-I within the plane of the same membrane (in cis) or on opposing cells (in trans) (33). To recognize MHC-I in trans, Ly49 molecules have adopted an “open” conformation whereby the α2 helices are splayed apart in a configuration that allows the Ly49 stalks to backfold and interject the Ly49 homo-dimer interface, thereby orienting the lectin domains in a manner that enables simultaneous recognition of two MHC-I (as observed for Ly49C-mediated recognition of H-2Kb). However, in the “closed” cis binding conformation (as observed in the Ly49A·H-2Dd structure), the Ly49 monomers are tightly juxtaposed in a manner that precludes simultaneous engagement of two MHC-I ligands due to steric clashes. Within the Ly49C·H2-Q10 structure, the Ly49 homo-dimer most closely resembles the open conformation, in that the α2 helices are separated by a gap of ∼11.5 Å (Fig. 4A).
FIGURE 4.
Variations in Ly49 configuration. A, the configuration of the Ly49 homo-dimer from the following structures is shown in ribbon representation with the α-helices in red and in two different orientations: Ly49C·H2-Q10 (left), Ly49C·H-2Kb (middle), and Ly49A·H2-Dd (right). B, overlay of the Ly49C homo-dimer in the H2-Q10 bound (magenta) and H2-Kb bound (gray) states showing the relative difference in domain juxtapositioning. The molecules were aligned based on the left-hand side monomer. C, the same structural alignment as described in B, but in this case, only the MHC-I are shown. The peptides are shown as sticks in green (Ly49C·H2-Q10) or black (Ly49C·H-2Kb).
Although the Ly49C configuration is clearly open, the precise domain juxtapositioning differs by a rotation of ∼9° from that observed in the unliganded and H-2Kb bound forms of Ly49C, such that the atoms on the outer surface of the homo-dimer are translated by up to ∼6.5 Å (Fig. 4B). This effect is magnified at the level of the H2-Q10 molecule, which is translated by up to 13 Å at the N terminus of the α1 helix when compared with the Ly49C·H-2Kb structure (Fig. 4C). Thus, it may be that H2-Q10 binding, but not the H-2Kb interaction, triggers a change in configuration of the Ly49 homo-dimer. However, we cannot exclude the possibility that the observed differences are due to inherent plasticity at the Ly49 homo-dimer interface.
The Ly49C·H2-Q10 Interface
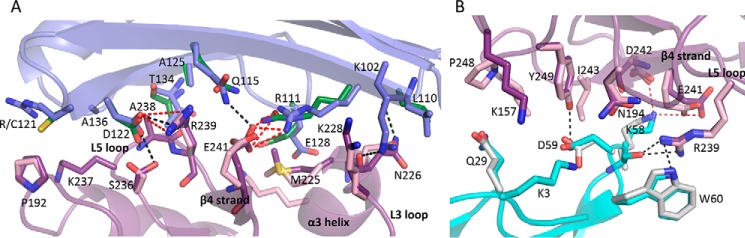
Each Ly49C monomer interacts with a broad region of the H2-Q10 molecule, burying a total solvent-accessible surface area of 2,200 Å2. Of this, 50% is accounted for by Ly49C, 39% is derived from the H2-Q10 heavy chain, and the remaining 11% is attributed to the β2M subunit. The total buried surface area is less than that observed for Ly49C·H-2Kb (2,400 Å2) and Ly49A·H-2Dd (2,800 Å2). Overall, the Ly49C·H2-Q10 interface is polar in nature, comprising 13 H-bonds and 9 salt bridges (Table 2), and is characterized by good charge complementarity (Fig. 5). Although the H2-Q10 surface is primarily electronegative and the Ly49C surface is overwhelmingly electropositive, most of the ionic interactions at the interface are attributed to a narrow negatively charged strip that runs down the Ly49C surface and comprises Glu-241, Asp-242, and Asp-244. Together, these residues form a network of salt bridges with an electropositive region of the MHC-Ib that includes residues from both the H2-Q10 heavy chain (Arg-111 and Lys-243) and the β2M subunit (Lys-58) (Fig. 5).
TABLE 2.
Comparison between Ly49C·H2-Q10 and Ly49C·H-2Kb contacts
Atomic contacts for the Ly49C·H2-Q10 structure were determined with the CCP4i implementation of CONTACT and are shown alongside those previously reported for the Ly49C·H-2Kb structure (34) (PDB ID: 3C8K). van der Waals interactions were defined as non-hydrogen bond contact distances of 4 Å or less. Hydrogen-bond interactions were defined as contact distances of 3.4 Å or less. Only amino acid residues that are well defined in electron density are included.
| Ly49C·H2-Q10 |
Ly49C·H-2Kb |
Type | ||||
|---|---|---|---|---|---|---|
| Ly49C | H2-Q10 | β2M | Ly49C | H2-Kb | β2m | |
| Lys-157 | Lys-3 | VDW | ||||
| Ser-161 | Gln-226 | Ser-161 | Ile-225 | VDW | ||
| Pro-192 | Arg-121 | Pro-192 | Cys-121 | VDW | ||
| Asn-194 | Asp-59 | Asn-194 | Asp-59 | VDW | ||
| Asn-194 | Lys-3 | VDW | ||||
| Lys-221 | Glu-128 | VDW | ||||
| Lys-221 | Glu-128 | Lys-221 | Glu-128 | H-bond | ||
| Lys-224 | Arg-111 | VDW | ||||
| Lys-224 | Glu-128 | VDW | ||||
| Met-225 | Arg-111 | Met-225 | Arg-111 | VDW | ||
| Met-225 | Glu-128 | Met-225 | Glu-128 | VDW | ||
| Asn-226 | Lys-102 | VDW | ||||
| Asn-226 | Lys-102 | H-bond | ||||
| Asn-226 | Leu-110 | Asn-226 | Leu-110 | VDW | ||
| Asn-226 | Leu-110 | H-bond | ||||
| Phe-227 | Lys-102 | VDW | ||||
| Lys-228 | Lys-102 | Lys-228 | Asp-30, Asp-212 | VDW | ||
| Arg-230 | Asp-212, Ile-213 | VDW | ||||
| Arg-230 | Asp-212, Ile-213 | H-bond | ||||
| Arg-230 | Asp-212 | Salt bridge | ||||
| Ser-236 | Asp-122 | Ser-236 | Asp-122 | VDW | ||
| Ser-236 | Asp-122 | Ser-236 | Asp-122 | H-bond | ||
| Lys-237 | Asp-122 | Lys-237 | Asp-122 | VDW | ||
| Lys-237 | Asp-122 | H-bond | ||||
| Lys-237 | Ala-136 | VDW | ||||
| Ala-238 | Asp-122 | Ala-238 | Asp-122 | VDW | ||
| Ala-238 | Asp-122 | Ala-238 | Asp-122 | H-bond | ||
| Ala-238 | Thr-134 | Ala-238 | Thr-134, Val-135 | VDW | ||
| Arg-239 | Asp-122 | Arg-239 | Asp-122 | H-bond | ||
| Arg-239 | Ile-98 | VDW | ||||
| Arg-239 | Asp-122 | Arg-239 | Asp-122 | VDW | ||
| Arg-239 | Asp-122 | Salt bridge | ||||
| Arg-239 | Gln-115 | VDW | ||||
| Arg-239 | Gln-115 | Arg-239 | Gln-115 | H-bond | ||
| Arg-239 | Ala-125 | VDW | ||||
| Arg-239 | Lys-58 | Arg-239 | Lys-58, Asp-59 | VDW | ||
| Arg-239 | Lys-58 | Arg-239 | Lys-58 | H-bond | ||
| Arg-239 | Trp-60 | Arg-239 | Trp-60 | VDW | ||
| Arg-239 | Trp-60 | Arg-239 | Trp-60 | H-bond | ||
| Glu-241 | Arg-111 | Glu-241 | Arg-111 | VDW | ||
| Glu-241 | Arg-111 | Glu-241 | Arg-111 | Salt bridge | ||
| Glu-241 | Lys-58 | VDW | ||||
| Glu-241 | Lys-58 | Glu-241 | Lys-58 | Salt bridge | ||
| Glu-241 | Gln-115 | Glu-241 | Gln-115 | VDW | ||
| Glu-241 | Gln-115 | H-bond | ||||
| Asp-242 | Lys-58 | VDW | ||||
| Asp-242 | Lys-58 | Asp-242 | Lys-58 | Salt bridge | ||
| Ile-243 | Asp-59 | Ile-243 | Lys-58 | VDW | ||
| Asp-244 | Lys-243 | Salt bridge | ||||
| Asn-246 | Leu-230 | Asn-246 | Leu-230, Val-231 | VDW | ||
| Asn-246 | Leu-230 | Asn-246 | Leu-230 | H-bond | ||
| Asn-246 | Glu-232 | Asn-246 | Glu-232 | VDW | ||
| Ile-247 | Glu-232 | Ile-247 | Gln-29 | VDW | ||
| Ile-247 | Gln-29 | VDW | ||||
| Pro-248 | Gln-29 | Pro-248 | Gln-29 | VDW | ||
| Tyr-249 | Asp-59 | Tyr-249 | Asp-59 | VDW | ||
| Tyr-249 | Asp-59 | Tyr-249 | Asp-59 | H-bond | ||
FIGURE 5.
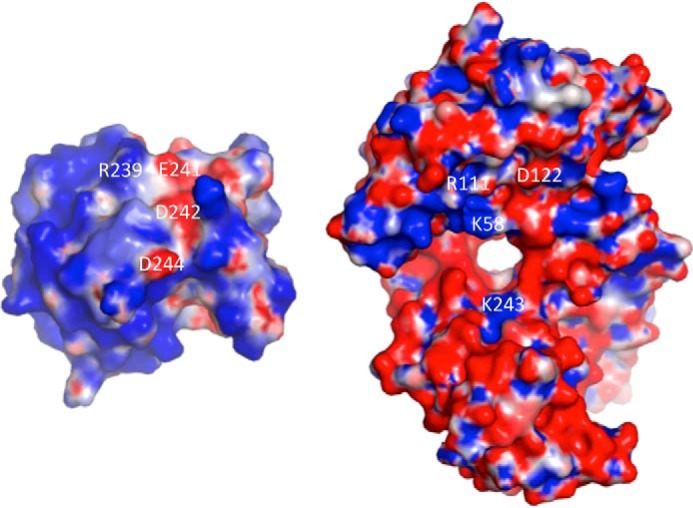
The molecular surface at the Ly49C·H2-Q10 interface. Electrostatic complementarity at the Ly49C·H2-Q10 interface is shown. Positive surfaces of Ly49C (left) and H2-Q10 (right) are shown in red (−5 kiloteslas), neutral surfaces are in white (0 kiloteslas), and negative surfaces in blue (+5 kiloteslas). Charge potentials were calculated with APBS (42) and are visualized in PYMOL. Amino acid residues that form salt bridges between Ly49C and H2-Q10 are labeled.
The majority of the contacts at the Ly49C·H2-Q10 interface are derived from the Ly49C L3 and L5 loops as well as the β4-strand. In particular, Glu-241 from the β4-strand lies at the heart of the interface and provides a central focal point for a triad of residues from the H2-Q10 heavy chain (Arg-111 and Gln-115) and β2M subunit (Lys-58) (Fig. 6). Adjacent to Glu-241 lies another prominent residue, Arg-239 from the L5 loop, which inserts into the H2-Q10/β2M juncture, forming an extensive network of both polar and non-polar interactions with Asp-122 and Gln-115 from H2-Q10 and the main chain of Lys-58 from the β2M subunit. These central, predominantly polar interactions are supplemented by a number of additional contacts derived from the L5 loop (Ser-236, Lys-237, and Ala-238) that are primarily non-polar in nature with the exception of a hydrogen bond from the hydroxyl group of Ser-236 to Asp-122 of H2-Q10. In contrast to the interactions with the H2-Q10 α1/α2 domain and the β2M subunit, the interface between Ly49C and the H2-Q10 α3 domain is markedly less well defined, both in the current structure and in the 2.9 Å resolution structure of Ly49C·H-2Kb (Protein Data Bank (PDB) ID: 3C8K) (34). In particular, the electron density for some of the larger Ly49C side chain residues (e.g. Lys-203 and Arg-230) is poorly defined, suggesting that these residues may be flexible and not contribute significantly to the interaction.
FIGURE 6.
Molecular interaction at the Ly49C·H2-Q10 interface. The interactions of Ly49C (magenta) with the H2-Q10 α1/α2 domain (violet) (A) and the β2M subunit (cyan) (B) are shown. For comparison, the equivalent side chain residues in the Ly49C·H-2Kb structure (PDB ID: 3C8K) are overlaid in pink (for Ly49C), green (for the H2-Kb heavy chain), and gray (for the β2M subunit). Hydrogen bonds and salt-bridge interactions are represented by black and red dashed lines, respectively, and are shown only for the Ly49C·H2-Q10 structure.
Comparison with Ly49C·H-2Kb Structure
Overall, the docking mode of H2-Q10 on Ly49C is highly similar to that of H-2Kb (r.m.s.d 0.89 Å over 470 Cα atoms, based on alignment of one Ly49C monomer with one H2-Q10/H-2Kb molecule). Moreover, the interactions at the Ly49C·H2-Q10 interface mirror those found within the Ly49C·H-2Kb complex, further emphasizing the conserved nature of these related MHC molecules (Fig. 7A).
FIGURE 7.
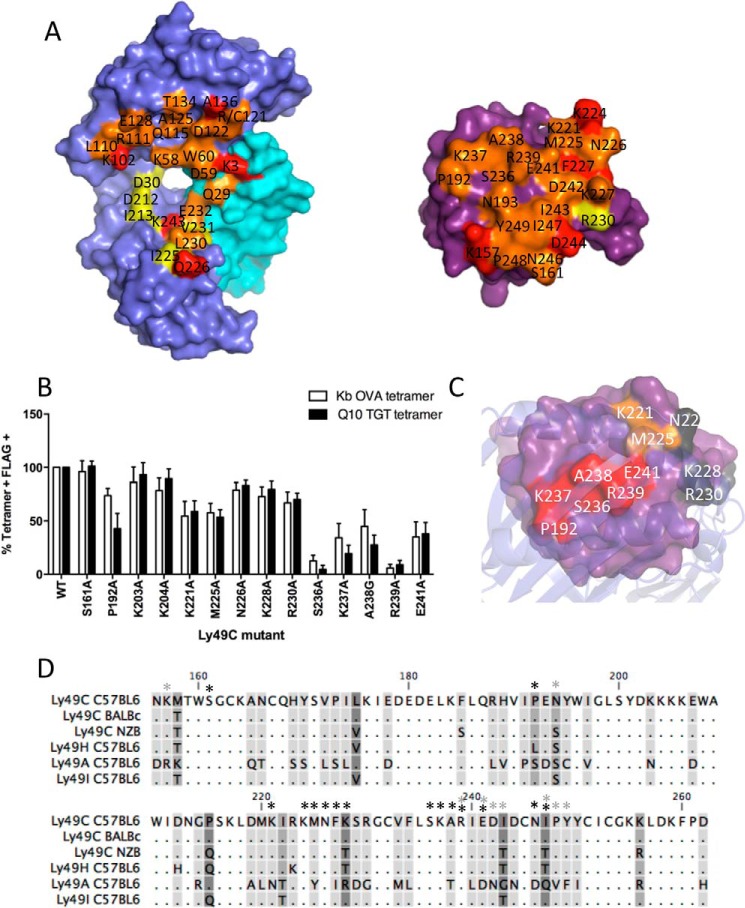
Comparison of the structure of H2-Q10 and H-2Kb on Ly49C. A, comparison of the binding footprint on H2-Q10/H2-Kb (left) and Ly49C (right). Contacting surface residues unique to Ly49C·H2-Q10 and Ly49C·H-2Kb are shown in red and yellow, respectively. Surface contacts that are conserved in both Ly49C interfaces with H2-Q10 and H2-Kb are colored orange. B, comparison of the energetic footprint of Ly49C on H-2Kb and H2-Q10. Full-length C57/BL6 Ly49C-FLAG was mutated to Ala (or Gly were an Ala was present) at residues known to bind H-2Kb, and wild-type or mutant Ly49C was transiently transfected into HEK-293T cells. Tetramers of H2-Q10-TGTETLYF (Q10 TGT), H-2Kb-SIINFEKL (Kb OVA), and HLA-E-VMAPRTLIL were used to stain transfected 293T together with an anti-FLAG mAb and analyzed by flow cytometry. Results show tetramer binding (percentage of tetramer-positive, FLAG-positive (% Tetramer + FLAG +)of 3 independent transfections normalized to binding of wild-type Ly49C (showing means ± S.E.) C, energetic hotspot for Ly49C binding to H2-Q10. The surface of Ly49C is shown in purple with residues that severely (<50% of wild type), moderately (between 50 and 70% of wild type), or do not (between 70 and 100% of wild type) impact H2-Q10 binding shown in red, orange, and gray, respectively. The view shown is looking down from the top of the H2-Q10 peptide-binding platform. D, sequence alignment of Ly49 receptors from different mouse strains. Dots indicate conserved residues. Conservation of residues between Ly49s is shown on a scale, whereby residues that are shared between all Ly49s are unmarked and the most variable residues are shown in dark gray. Asterisks indicate residues of C57BL/6 Ly49C that interact with the H2-Q10 heavy chain (black) or β2M (gray) as determined by crystallographic analysis.
Although similar, a comparison of the Ly49C·H2-Q10 and Ly49C·H-2Kb binding footprint suggests that the Ly49C contacts with the α1/α2 chains and the β2M subunit are particularly well conserved (Fig. 7A). For example, of the nine contact residues within the H2-Q10 α1/α2 region, seven are absolutely conserved and make similar interactions in the Ly49C·H-2Kb structure, one (Arg-121) differs in sequence (Cys-121 in H-2Kb) but maintains a contact with Ly49C, and only one (Asp-30) does not contact Ly49C. Notably, these conserved interactions include residues from the Ly49C β4-strand and L5 loop that, as discussed above, are centrally located at the heart of the Ly49C·H2-Q10 interface. By comparison, the physio-chemical properties of Qa-1b in this region differ substantially, with changes in amino acid charge (e.g. K102D and R121Q) and hydrophobicity (e.g. A125S and A136N) (Fig. 1A) providing a basis for the inability of this related MHC-Ib to bind to Ly49C. The Ly49C footprint on H2-Q10 is however substantially different from that of H-2Kb within the α3 region, where there are sequence differences in a number of contact residues including 212, 213, and 225 (Fig. 7A).
Comparison of the Energetic Footprint of Ly49C on H-2Kb and H2-Q10
Structural analysis indicated that the footprints of H-2Kb and H2-Q10 on Ly49C are largely overlapping, and our biophysical data indicated that H2-Q10 bound with only a marginally lower affinity. Hence we sought to assess whether the energetic footprints of both ligands on Ly49C were also similar. We generated 14 Ly49C transfectants that contained single site mutations at residues directly involved in interactions with H-2Kb and analyzed binding of either H-2Kb or H2-Q10 tetramers by flow cytometry. The data showed that that the energetic footprints of H-2Kb and H2-Q10 on Ly49C are largely overlapping, correlating well with the structural analysis. Notably, mutation of Ly49C residues located at the binding interface (Glu-241, Ser-236, Lys-237, Ala-238, and Arg-239) severely impacted H2-Q10 and H-2Kb binding to <50% of wild type (Fig. 7B), suggesting that this region represents the major energetic driving force for both interactions (Fig. 7C). Furthermore, mutation of Ly49C residues involved in differential contacts in the α3 region between H-2Kb and H2-Q10 (Lys-203 and Arg-230) had little effect on binding of either tetramer (Fig. 7A). Accordingly, although some differences exist at the Ly49C-α3 interface, these can be well tolerated as this region contributes little to the binding energy of the interaction. Therefore, in conclusion, our data demonstrate that Ly49C has an equivalent structural and energetic footprint on H-2Kb and H2-Q10. These data add great credence to the notion that H2-Q10 represents a genuine ligand for Ly49C.
Discussion
The control of murine NK cells through ligation of the inhibitory Ly49 receptors has typically been composed of interactions with classical MHC-Ia ligands. Previously, we had shown that the highly conserved MHC-Ib, H2-M3, interacts with the NK cell receptor Ly49A (24). Here we find that H2-Q10 binds to Ly49C, likely expanding the role of MHC-Ib in regulating murine NK cell responses. Our data also provide the first structure of unliganded H2-Q10, as well as the first structure of a murine NK cell receptor with an MHC-Ib ligand.
To date, the unliganded structures of several MHC-Ib have been described, including human HLA-E (29, 30) and HLA-G (35), and murine Qa-1b (27), Qa-2 (Q9 (36)), and H2-M3 (28). At a population level, the limited polymorphism in MHC-Ib means that they present a more limited peptide repertoire than the class Ia counterparts. However, the nature of the peptide-binding groove itself further limits the repertoire in many instances. For example, although there are structural differences within the peptide-binding grooves of HLA-E and Qa-1b, both molecules harbor five anchor positions, serving to effectively restrict the number of peptides that can be accommodated. In contrast, H2-M3 and Q9 show only one and two anchor sites, respectively, but at rather unusual positions, namely at P1 in H2-M3 and at P7 and P9 in Q9. Although HLA-G possesses five anchor sites, it has a peptide binding repertoire that is less restricted than HLA-E. Curiously, we have shown a similar situation in the H2-Q10 peptide-binding grove, which also showed five anchor positions at P2-Gly, P3-Ile, P5-Asn, P6-Val, and P8-Leu. Interestingly, HLA-G may serve a highly specialized role at the fetal/maternal interface (17). It is possible that H2-Q10 shares an analogous role in controlling liver NK cell function.
Our data indicate that the mode of binding of Ly49C to classical (H-2Kb) and non-classical (H2-Q10) MHC is largely conserved. The key features that allow binding to both molecules, namely extensive contacts in the α1/α2 region and the lack of peptide contact, could potentially permit the interaction of Ly49C with a wide range of MHC-I. The residues of Ly49C involved in binding the H2-Q10 heavy chain are highly conserved across mouse strains (Fig. 7D), with 14/16 amino acids conserved between C57BL/6, BALB/c, and NZB strains. Furthermore, position 228, which is a Lys in C57BL/6 and a Thr in NZB Ly49C, did not contribute energetically to the interaction with H2-Q10. The other different amino acid (position 247, which is an Ile in C57BL/6 and a Thr in NZB Ly49C) made only non-polar interactions with Glu-232 of H2-Q10 and is not conserved in the H-2Kb·Ly49C interface. Hence Ly49C position 247 is unlikely to be critical to the interaction with the MHC-I heavy chain. Interestingly, the same amino acids at positions 228 and 247 (a Thr in both cases) are shared between NZB Ly49C and C57BL/6 Ly49H, yet our tetramer binding data indicate that C57BL/6 Ly49H does not interact with H2-Q10 (Fig. 1). However, C57BL/6 Ly49H also differs from Ly49C at position 192, where this is a Pro in Ly49C and a Leu in Ly49H. Intriguingly, Pro-192 made a significant contribution to the binding energy between Ly49C and H2-Q10, with mutation of this residue resulting in an ∼50% loss of tetramer binding. Therefore, Pro-192 in Ly49C may be important in allowing H2-Q10 to bind to Ly49C, whereas Leu in this position prevents H2-Q10 from binding to Ly49H. These observations emphasize the impact that even fine molecular alterations can have on Ly49 receptor-ligand specificity. Furthermore, it is not immediately apparent why H2-Q10 does not bind to Ly49I, as all of the residues that contact the H2-Q10 heavy chain are conserved between Ly49C and Ly49I, although there are some minor differences in residues that contact the β2M subunit (e.g. the substitution of Arg-194 in Ly49C to Ser and of Ile-243 in Ly49C to Thr). Curiously, previous studies investigating Ly49I report only very weak interactions with MHC (9). It is possible that these minor differences in the residues that contact β2M prevent the interaction of Ly49I with MHC or alternatively that Ly49I has a completely different mode of binding, such as that seen between Ly49H and m157 (37).
In contrast, only 8/16 amino acids of Ly49C that bind H2-Q10 are conserved with C57BL/6 Ly49A. Furthermore, five of these amino acids made significant energetic contributions to the interaction between Ly49C and H2-Q10 (Pro-192, Lys-221, Met-225, Ala-238, and Glu-241), most likely accounting for the inability of H2-Q10 to interact with Ly49A. Taken together, our results indicate that H2-Q10 can specifically interact with the Ly49C receptor, and this interaction is most likely conserved between different alleles of Ly49C.
Previously, studies on the role of MHC-Ib in the control of murine NK cells have been almost exclusively limited to the interaction of CD94-NKG2 receptors with Qa-1b. Interestingly, we observed that Qa-1b tetramers do not bind to Ly49C-expressing CHO cells. Given our structural analysis of the footprint of Ly49C on H-2Kb and H2-Q10, it is likely that differences in the α2 region between Qa-1b and H2-Q10/H-2Kb may account for this finding. For example, Ala-125 and Ala-136 in H2-Q10/H-2Kb both interact with energetically critical residues of Ly49C (Arg-239 and Lys-237, respectively) and are not conserved in the Qa-1b sequence (residues Ser-125 and Asn-136). It is tempting to speculate that functionally analogous systems exist in mice and humans, whereby the CD94-NKG2 receptors exclusively bind HLA-E/Qa-1b, whereas Ly49/KIR bind a more diverse but structurally related group of molecules extending into non-classical MHC ligands. It is interesting to note recent evidence suggests that the MHC-Ib HLA-F is a ligand for KIR3DL2 and KIR2DS4 (38). It is also possible that unlike MHC-Ia, where the need to present diverse peptide repertoires for T cell recognition has driven the acquisition of polymorphism, the capacity of a number of MHC-Ib to interact with immunoreceptors such as those of the Ly49 family may have served to limit their genetic diversification.
Although we have identified a role for H2-Q10 in NK cell interactions, our data do not necessarily preclude a function for this molecule in mediating other immune responses. Previous peptide elution and substitution experiments have identified that H2-Q10 binds a heterogeneous repertoire of peptides with a preference for Gly at P2, Leu/Val at P6, and Phe/Leu at Pω (22). Accordingly, the H2-Q10 peptide-binding groove is considered to resemble that of MHC-Ia rather than of other more specialized MHC-Ib such as Qa-1b, H2-M3, or HLA-E. Overall, the unliganded H2-Q10 structure supports this proposition, and in addition, peptide residues P4-Thr, P6-Val, and P7-Asp are solvent-exposed and thus could represent candidate T cell receptor contact sites. Secretion of H2-Q10 was down-regulated following liver injury (20), perhaps suggesting that this molecule plays a role in mediating NK or T cell responses following cellular stress. Indeed, the biology, regulation, and precise function of this unusual MHC molecule require further investigation.
Experimental Procedures
Mice
C57BL/6 and H-2KbDb knock-out (39) mice were from the Department of Microbiology, University of Melbourne. All mice used were 6–10 weeks of age. All experiments were performed in accordance with the animal ethics guidelines of the National Health and Medical Research Council of Australia. All experiments were approved by the University of Melbourne Animal Ethics Committee.
Flow Cytometry
For NK cells, spleens were collected from wild-type and H-2Kb/Db-deficient mice and red blood cells removed using ACK (ammonium-chloride-potassium) lysing buffer. Nonspecific receptors were then blocked with 2.4G2 antibody. Cells (5 × 106) were stained with NK1.1 (PK136; eBioscience), CD3 (17A2; BioLegend), H2-Q10 and HLA-E tetramers prior to fixation with 2% paraformaldehyde. Following washing, 5 × 103 NK cell (NK1.1+/CD3−) events were electronically collected using an LSR-II (BD Biosciences).
For CHO cells expressing Ly49, cells were cultured for 1 day at a density of 2 × 105 cells/ml and removed from the tissue culture plastic with 50 mm EDTA. Following blockade of nonspecific binding by 2.4G2 antibody, cells were stained with antibody to Ly49 or H2-Q10 tetramers. Following fixation with 2% paraformaldehyde and subsequent washing, 5 × 104 events were electronically collected using an LSR-II or FACSCanto (BD Biosciences).
Cloning and Expression of Recombinant Proteins
For mammalian expression, the entire extracellular domains of Ly49A and Ly49C (Ly49-EC) from C57BL/6 mice were cloned into the pHLsec vector downstream of a signal peptide, His tag, and BirA site, as described previously (24, 37). For expression in mammalian cells, secreted Ly49-EC produced from transiently transfected HEK-293T cells was harvested from the culture medium 3 days after transfection, dialyzed against 10 mm Tris, pH 8.0, 500 mm NaCl, and purified using nickel affinity resin. The Ly49-EC was eluted from the nickel resin with 10 mm Tris, pH 8.0, 500 mm NaCl, 500 mm imidazole, and the protein was purified by gel filtration chromatography using an S200 16/60 column (GE Healthcare).
For bacterial expression, the entire Ly49C EC or lectin-like domain (starting from Ser-132) was cloned into the pET30 vector and expressed in BL21 Escherichia coli cells as described previously (37). Inclusion bodies harboring the Ly49 protein were resuspended in 6 m guanidine, 0.5 mm Na-EDTA, and 1 mm DTT and refolded by flash dilution in a solution containing 100 mm Tris-HCl (pH 8.0), 400 mm l-arginine-HCl, 5 mm reduced glutathione, and 0.5 mm oxidized glutathione. The refolding solution was then dialyzed against 25 mm HEPES (pH 7.5), followed by purification by cation exchange (carboxymethyl and sulphopropyl resin) and size-exclusion chromatography.
cDNA encoding residues 1–274 of H2-Q10 was generated by GenScript (Piscataway, NJ) and cloned into a pUC57 vector. H2-Q10 was then subcloned into a pET-30-based vector that allowed for an in-frame fusion of a substrate peptide for the enzyme BirA. The H2-Q10, H-2Kb, H-2Dd, Qa-1b, or HLA-E heavy chains and murine β2-microglobulin were expressed separately in E. coli, purified from inclusion bodies, and refolded in the presence of either TCR Vβ chain (TGTETLYF) or ribophorin (VGITNVDL) peptides for H2-Q10, OVA (SIINFEKL) for H-2Kb, motif peptide (AGPARAAAL) for H-2Dd, QDM (AMAPRTLLL) for Qa-1b, or VMAPRTLIL for HLA-E and purified essentially as described previously (40).
Surface Plasmon Resonance
A ProteOn XPR36 protein interaction array system (Bio-Rad) was used for surface plasmon resonance (SPR) as described (24). An NLC (NeutrAvidin coated sensor chip, Bio-Rad) was used to capture ∼300 response units of biotinylated H2-Q10-TGTETLYF, H-2Kb SIINFEKL, H-2Dd-AGPARAAAL, and HLA-E-VMAPRTLIL. An empty flow cell containing NeutrAvidin alone served as a control surface. Serially diluted (30–0.8 μm) Ly49A or Ly49C was injected over test and control surfaces at a rate of 100 μl/min for 60 s. After subtraction of the response from the control flow cell, KD was measured by non-linear regression at equilibrium using Prism (GraphPad) and ProteOn Manager software (version 3.0.1). All interactions were tested minimally in duplicate.
Crystallization, Data Collection, Structure Determination, and Refinement
All crystals were obtained at 20 °C using the hanging drop vapor diffusion method. Unliganded H2-Q10 in complex with a ribophorin peptide (VGITNVDL, VGI) was crystallized at 6.8 mg/ml from a solution containing 8% PEG 3350, 0.1 m sodium malonate, pH 5.0, and 0.01 m β-nicotinamide adenine dinucleotide hydrate. For Ly49C·H2-Q10-VGI, individual proteins were mixed in a 1 Ly49 dimer/2 H2-Q10VGI molar ratio at a total protein concentration of 6.8 mg/ml and crystallized from a solution containing 12% PEG 8000, 10% PEG 400, 0.1 m HEPES, pH 7.2, and 0.2 m NaCl. Prior to data collection, crystals were equilibrated in a crystallization solution supplemented with 30% PEG 3350 (H2-Q10VGI) or 20% PEG 8000 (Ly49C·H2-Q10VGI) for cryoprotection. Crystals were flash-cooled using liquid nitrogen, and x-ray diffraction data were recorded on a Quantum-315 CCD detector at the MX1 (H2-Q10-VGI) or MX2 (Ly49C·H2-Q10VGI) beamlines of the Australian Synchrotron. Data were integrated using MOSFLM and scaled using SCALA from the CCP4 program suite.
The first unliganded crystal structure of H2-Q10VGI was resolved to 2.3 Å by molecular replacement using PHASER with a search model comprising the heavy chain and β2M subunit from H-2Db (PDB code 1YN6). Following subsequent rounds of model building in COOT and refinement using Buster, the completed structure comprised residues 1–276 of H2-Q10, 1–99 of β2M, and 1–8 of the peptide and refined to an Rfactor/Rfree of 17.7/24.7. The 3.3 Å structure of the Ly49C·H2-Q10VGI complex was also determined by PHASER using the unliganded H2-Q10VGI structure (described above) and the lectin domain of Ly49C (PDB code 3C8J) as search models. The asymmetric unit comprised two molecules of H2-Q10VGI and two Ly49C monomers. The contents of the asymmetric unit were reorganized using the ACHESYM algorithm (41), revealing that the Ly49C monomers formed a dimer. Thus, the entire complex was composed of one Ly49C dimer and two molecules of H2-Q10VGI. Following refinement using Buster, the completed structure comprised residues 1–277 of H2-Q10, 1–99 of β2M, 1–8 of the peptide, and 141–262 of Ly49C. Details of the data processing and refinement statistics are given in Table 1.
Mutagenesis
cDNA encoding entire C57/BL6 Ly49C fused to a C-terminal FLAG motif was generated by GenScript and cloned into a pMIG plasmid for transient transfection into HEK-293T cells. Site-directed mutants of Ly49C were generated by the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), and the sequence of the mutated cDNA was verified by DNA sequencing. The residues chosen for mutagenesis were chosen by structural analysis of the Ly49·H-2Kb complex (34) and were mutated to Ala (or Gly where an Ala was present). Wild-type or mutant Ly49C pMIG constructs were transiently transfected into HEK-293T (FuGENE), and expression of Ly49C was confirmed by an antibody to FLAG and/or Ly49C. Biotinylated monomers of H2-Q10-TGTETLYF, H-2Kb-SIINFEKL, and HLA-E-VMAPRTLIL were conjugated with streptavidin-phycoerythrin and used to stain transfected 293T together with the anti-FLAG mAb and analyzed by flow cytometry. Results were analyzed by FlowJo (TreeStar).
Author Contributions
L. C. S. made tetramers, recombinant proteins, and mutant Ly49C transfectants and conducted the SPR and mutagenesis experiments, analyzed the results, and wrote the manuscript. R. B. made recombinant proteins and conducted all of the crystallographic analysis, data collection, and structural refinement and wrote the paper. N. S. and J. M. L. W. assisted in cloning, mutagenesis, and protein production. N .L. L. and M. M. provided H-2KbDb-deficient mice. F. A. D. and G. R. B. assisted with cloning, produced recombinant protein, and performed crystallization experiments. J. A. T., J. R., and A. G. B assisted in drafting the manuscript and provided important intellectual input. D. M. A. conceived the idea for the project, performed flow cytometry experiments and wrote the paper with L. C. S. and R. B. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank the staff at the Monash Macromolecular Crystallisation Facility and the Australian Synchrotron MX1 and MX2 beamlines for their expert assistance.
This work was supported by National Health and Medical Research Council (NHMRC) Career Development Awards (to L. C. S., R. B, and D. M. A.), the Sylvia and Charles Viertel Charitable Foundation (to N. L. L.), and by an NHMRC Australia Fellowship (to J. R.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5J6H and 5J6G) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MHC-I
- major histocompatibility complex class I molecule(s)
- MHC-Ia
- class Ia MHC molecule(s)
- MHC-Ib
- class Ib MHC molecule(s)
- NK
- natural killer
- KIR
- killer cell immunoglobulin-like receptor
- r.m.s.d.
- root mean square deviation
- VDW
- van der Waals
- EC
- extracellular domain
- NZB
- New Zealand Black
- β2M
- β2 microglobulin.
References
- 1. Rossjohn J., Gras S., Miles J. J., Turner S. J., Godfrey D. I., and McCluskey J. (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 [DOI] [PubMed] [Google Scholar]
- 2. Shawar S. M., Vyas J. M., Rodgers J. R., and Rich R. R. (1994) Antigen presentation by major histocompatibility complex class I-B molecules. Annu. Rev. Immunol. 12, 839–880 [DOI] [PubMed] [Google Scholar]
- 3. Takada T., Kumánovics A., Amadou C., Yoshino M., Jones E. P., Athanasiou M., Evans G. A., and Fischer Lindahl K. (2003) Species-specific class I gene expansions formed the telomeric 1 Mb of the mouse major histocompatibility complex. Genome Res. 13, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saunders P. M., Vivian J. P., O'Connor G. M., Sullivan L. C., Pymm P., Rossjohn J., and Brooks A. G. (2015) A bird's eye view of NK cell receptor interactions with their MHC class I ligands. Immunol. Rev. 267, 148–166 [DOI] [PubMed] [Google Scholar]
- 5. Kärre K., Ljunggren H. G., Piontek G., and Kiessling R. (1986) Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319, 675–678 [DOI] [PubMed] [Google Scholar]
- 6. Kim S., Sunwoo J. B., Yang L., Choi T., Song Y. J., French A. R., Vlahiotis A., Piccirillo J. F., Cella M., Colonna M., Mohanakumar T., Hsu K. C., Dupont B., and Yokoyama W. M. (2008) HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. U.S.A. 105, 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dimasi N., and Biassoni R. (2005) Structural and functional aspects of the Ly49 natural killer cell receptors. Immunol. Cell Biol. 83, 1–8 [DOI] [PubMed] [Google Scholar]
- 8. Hanke T., Takizawa H., McMahon C. W., Busch D. H., Pamer E. G., Miller J. D., Altman J. D., Liu Y., Cado D., Lemonnier F. A., Bjorkman P. J., and Raulet D. H. (1999) Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity 11, 67–77 [DOI] [PubMed] [Google Scholar]
- 9. Scarpellino L., Oeschger F., Guillaume P., Coudert J. D., Lévy F., Leclercq G., and Held W. (2007) Interactions of Ly49 family receptors with MHC class I ligands in trans and cis. J. Immunol. 178, 1277–1284 [DOI] [PubMed] [Google Scholar]
- 10. Boyington J. C., Motyka S. A., Schuck P., Brooks A. G., and Sun P. D. (2000) Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 405, 537–543 [DOI] [PubMed] [Google Scholar]
- 11. Vivian J. P., Duncan R. C., Berry R., O'Connor G. M., Reid H. H., Beddoe T., Gras S., Saunders P. M., Olshina M. A., Widjaja J. M., Harpur C. M., Lin J., Maloveste S. M., Price D. A., Lafont B. A., et al. (2011) Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tormo J., Natarajan K., Margulies D. H., and Mariuzza R. A. (1999) Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature 402, 623–631 [DOI] [PubMed] [Google Scholar]
- 13. Doucey M. A., Scarpellino L., Zimmer J., Guillaume P., Luescher I. F., Bron C., and Held W. (2004) Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat. Immunol. 5, 328–336 [DOI] [PubMed] [Google Scholar]
- 14. Sullivan L. C., Clements C. S., Rossjohn J., and Brooks A. G. (2008) The major histocompatibility complex class Ib molecule HLA-E at the interface between innate and adaptive immunity. Tissue Antigens 72, 415–424 [DOI] [PubMed] [Google Scholar]
- 15. Ishitani A., Sageshima N., Lee N., Dorofeeva N., Hatake K., Marquardt H., and Geraghty D. E. (2003) Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J. Immunol. 171, 1376–1384 [DOI] [PubMed] [Google Scholar]
- 16. LeMaoult J., Le Discorde M., Rouas-Freiss N., Moreau P., Menier C., McCluskey J., and Carosella E. D. (2003) Biology and functions of human leukocyte antigen-G in health and sickness. Tissue Antigens 62, 273–284 [DOI] [PubMed] [Google Scholar]
- 17. Rajagopalan S. (2014) HLA-G-mediated NK cell senescence promotes vascular remodeling: implications for reproduction. Cell. Mol. Immunol. 11, 460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morandi F., Ferretti E., Castriconi R., Dondero A., Petretto A., Bottino C., and Pistoia V. (2011) Soluble HLA-G dampens CD94/NKG2A expression and function and differentially modulates chemotaxis and cytokine and chemokine secretion in CD56bright and CD56dim NK cells. Blood 118, 5840–5850 [DOI] [PubMed] [Google Scholar]
- 19. Morandi F., Ferretti E., Bocca P., Prigione I., Raffaghello L., and Pistoia V. (2010) A novel mechanism of soluble HLA-G mediated immune modulation: downregulation of T cell chemokine receptor expression and impairment of chemotaxis. PLoS ONE 5, e11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lew A. M., Maloy W. L., and Coligan J. E. (1986) Characteristics of the expression of the murine soluble class I molecule (Q10). J. Immunol. 136, 254–258 [PubMed] [Google Scholar]
- 21. Cosman D., Kress M., Khoury G., and Jay G. (1982) Tissue-specific expression of an unusual H-2 (class I)-related gene. Proc. Natl. Acad. Sci. U.S.A. 79, 4947–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zappacosta F., Tabaczewski P., Parker K. C., Coligan J. E., and Stroynowski I. (2000) The murine liver-specific nonclassical MHC class I molecule Q10 binds a classical peptide repertoire. J. Immunol. 164, 1906–1915 [DOI] [PubMed] [Google Scholar]
- 23. Lee N., Malacko A. R., Ishitani A., Chen M. C., Bajorath J., Marquardt H., and Geraghty D. E. (1995) The membrane-bound and soluble forms of HLA-G bind identical sets of endogenous peptides but differ with respect to TAP association. Immunity 3, 591–600 [DOI] [PubMed] [Google Scholar]
- 24. Andrews D. M., Sullivan L. C., Baschuk N., Chan C. J., Berry R., Cotterell C. L., Lin J., Halse H., Watt S. V., Poursine-Laurent J., Wang C. R., Scalzo A. A., Yokoyama W. M., Rossjohn J., Brooks A. G., and Smyth M. J. (2012) Recognition of the nonclassical MHC class I molecule H2-M3 by the receptor Ly49A regulates the licensing and activation of NK cells. Nat. Immunol. 13, 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bessoles S., Angelov G. S., Back J., Leclercq G., Vivier E., and Held W. (2013) Education of murine NK cells requires both cis and trans recognition of MHC class I molecules. J. Immunol. 191, 5044–5051 [DOI] [PubMed] [Google Scholar]
- 26. Kress M., Cosman D., Khoury G., and Jay G. (1983) Secretion of a transplantation-related antigen. Cell 34, 189–196 [DOI] [PubMed] [Google Scholar]
- 27. Zeng L., Sullivan L. C., Vivian J. P., Walpole N. G., Harpur C. M., Rossjohn J., Clements C. S., and Brooks A. G. (2012) A structural basis for antigen presentation by the MHC class Ib molecule, Qa-1b. J. Immunol. 188, 302–310 [DOI] [PubMed] [Google Scholar]
- 28. Wang C. R., Castaño A. R., Peterson P. A., Slaughter C., Lindahl K. F., and Deisenhofer J. (1995) Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell 82, 655–664 [DOI] [PubMed] [Google Scholar]
- 29. Hoare H. L., Sullivan L. C., Clements C. S., Ely L. K., Beddoe T., Henderson K. N., Lin J., Reid H. H., Brooks A. G., and Rossjohn J. (2008) Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors. J. Mol. Biol. 377, 1297–1303 [DOI] [PubMed] [Google Scholar]
- 30. O'Callaghan C. A., Tormo J., Willcox B. E., Braud V. M., Jakobsen B. K., Stuart D. I., McMichael A. J., Bell J. I., and Jones E. Y. (1998) Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol. Cell 1, 531–541 [DOI] [PubMed] [Google Scholar]
- 31. Madden D. R., Garboczi D. N., and Wiley D. C. (1993) The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell 75, 693–708 [DOI] [PubMed] [Google Scholar]
- 32. Dam J., Guan R., Natarajan K., Dimasi N., Chlewicki L. K., Kranz D. M., Schuck P., Margulies D. H., and Mariuzza R. A. (2003) Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2Kb. Nat. Immunol. 4, 1213–1222 [DOI] [PubMed] [Google Scholar]
- 33. Back J., Malchiodi E. L., Cho S., Scarpellino L., Schneider P., Kerzic M. C., Mariuzza R. A., and Held W. (2009) Distinct conformations of Ly49 natural killer cell receptors mediate MHC class I recognition in trans and cis. Immunity 31, 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deng L., Cho S., Malchiodi E. L., Kerzic M. C., Dam J., and Mariuzza R. A. (2008) Molecular architecture of the major histocompatibility complex class I-binding site of Ly49 natural killer cell receptors. J. Biol. Chem. 283, 16840–16849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clements C. S., Kjer-Nielsen L., Kostenko L., Hoare H. L., Dunstone M. A., Moses E., Freed K., Brooks A. G., Rossjohn J., and McCluskey J. (2005) Crystal structure of HLA-G: a nonclassical MHC class I molecule expressed at the fetal-maternal interface. Proc. Natl. Acad. Sci. U.S.A. 102, 3360–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He X., Tabaczewski P., Ho J., Stroynowski I., and Garcia K. C. (2001) Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation. Structure 9, 1213–1224 [DOI] [PubMed] [Google Scholar]
- 37. Berry R., Ng N., Saunders P. M., Vivian J. P., Lin J., Deuss F. A., Corbett A. J., Forbes C. A., Widjaja J. M., Sullivan L. C., McAlister A. D., Perugini M. A., Call M. J., Scalzo A. A., Degli-Esposti M. A., et al. (2013) Targeting of a natural killer cell receptor family by a viral immunoevasin. Nat. Immunol. 14, 699–705 [DOI] [PubMed] [Google Scholar]
- 38. Goodridge J. P., Burian A., Lee N., and Geraghty D. E. (2013) HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J. Immunol. 191, 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pérarnau B., Saron M. F., Reina San Martin B., Bervas N., Ong H., Soloski M. J., Smith A. G., Ure J. M., Gairin J. E., and Lemonnier F. A. (1999) Single H2Kb, H2Db and double H2KbDb knockout mice: peripheral CD8+ T cell repertoire and anti-lymphocytic choriomeningitis virus cytolytic responses. Eur. J. Immunol. 29, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 40. Hoare H. L., Sullivan L. C., Pietra G., Clements C. S., Lee E. J., Ely L. K., Beddoe T., Falco M., Kjer-Nielsen L., Reid H. H., McCluskey J., Moretta L., Rossjohn J., and Brooks A. G. (2006) Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol. 7, 256–264 [DOI] [PubMed] [Google Scholar]
- 41. Kowiel M., Jaskolski M., and Dauter Z. (2014) ACHESYM: an algorithm and server for standardized placement of macromolecular models in the unit cell. Acta Crystallogr. D Biol. Crystallogr. 70, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andrews D. M., Sullivan L. C., Baschuk N., Chan C. J., Berry R., Cotterell C. L., Lin J., Halse H., Watt S. V., Poursine-Laurent J., Wang C. R., Scalzo A. A., Yokoyama W. M., Rossjohn J., Brooks A. G., and Smyth M. J. (2012) Recognition of the non-classical MHC class I molecule H2-M3 by the receptor Ly49A regulates licensing and activation of NK cells. Nat. Immunol. 13, 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]