Abstract
Maintaining protein homeostasis is critical for survival at the cellular and organismal level (Morimoto, R. I. (2011) Cold Spring Harb. Symp. Quant. Biol. 76, 91–99). Cells express a family of molecular chaperones, the heat shock proteins, during times of oxidative stress to protect against proteotoxicity. We have identified a second stress responsive transcription factor, dFOXO, that works alongside the heat shock transcription factor to activate transcription of both the small heat shock protein and the large heat shock protein genes. This expression likely protects cells from protein misfolding associated with oxidative stress. Here we identify the regions of the Hsp70 promoter essential for FOXO-dependent transcription using in vitro methods and find a physiological role for FOXO-dependent expression of heat shock proteins in vivo.
Keywords: 70-kilodalton heat shock protein (Hsp70), Drosophila, FOXO, oxidative stress, small heat shock protein (sHsp), transcription
Introduction
The forkhead box (fox)3 superfamily of transcription factors is defined by a DNA binding domain structurally related to the forkhead protein (2). The subfamily O (FOXO) group is distinct because of the presence of a five amino acid insert between helix 3 and helix 4 compared with other Fox family proteins. This family is conserved from Caenorhabditis elegans to mammals.
Invertebrates have a single FOXO gene (daf-16 in worms, dFOXO in flies). In mammals, the family has expanded to include four different FOXO genes (FOXO1, FOXO3, FOXO4, and FOXO6). In all organisms studied, FOXO family transcription factors play an important physiological role in protecting organisms against stress (3).
Although the best studied pathway controlling the FOXO family is insulin signaling, it is now clear that multiple stressors can activate FOXO (3). In Drosophila, dFOXO regulates the transcription of genes that promote survival under conditions of oxidative stress, changes in cellular metabolism, and unfolded proteins (4–6). Increased dFOXO activity extends lifespan in Drosophila by changing the transcriptional landscape (7–9). Both cell autonomous and non-autonomous roles for dFOXO have been identified (10). The cell autonomous role for dFOXO is likely derived from the activation of genes with protective functions. Thus there is great interest in understanding the genes under the dFOXO regulon.
The same cellular stress conditions that activate the FOXO family often result in proteotoxicity, the accumulation of toxic protein species. Proteotoxicity results from misfolded or aggregated proteins, which can arise from acute oxidative stress, heat shock, and age (1). Unchecked, this can result in cell death, aging, and disease at the organismal level (11).
The cell uses many mechanisms to protect the proteome and maintain proteostasis. One important mechanism is the induction of expression of the heat shock proteins (Hsps) under conditions with the potential to promote proteotoxicity. The Hsps help to maintain proteostasis by acting as molecular chaperones during times of acute cellular stress or over the course of organismal aging (1, 11).
There are two families of inducible Hsps with distinct protective mechanisms and functions, the small and large Hsps. The small Hsps (sHsps) are members of the Hsp20/α-crystallin family of chaperones whose main function is to prevent the formation of denatured protein aggregates in the cell. Drosophila melanogaster species has four major small heat shock proteins Hsp22, Hsp23, Hsp26 and Hsp27 (12). The large heat shock protein family, which includes Hsp70, has ATP-dependent chaperone activity and acts to properly fold nascent polypeptide chains and improperly folded, yet soluble, proteins (13, 14). Thus, both families of Hsps contribute to proteostasis using distinct mechanisms targeting different types of protein damage, so it would be advantageous to activate both pathways in response to stress.
Stress-inducible Hsps are distinct from their constitutive family members, such as the Hsp90 chaperones, in that they are specifically expressed during times of cellular stress, whereas the constitutive Hsps maintain proteostasis under normal conditions. D. melanogaster species has six stress-inducible Hsp70 genes that have high sequence identity both in their promoters and within their open reading frames. The expression of the Hsps in Drosophila is necessary for proper stress resistance, and the overexpression of these genes can increase lifespan (15–17).
Heat shock transcription factor (HSF) regulates the expression of the Hsps during times of stress and during the heat shock response. HSF binds to heat shock elements (HSEs) within the promoter regions of the sHsps and Hsp70 in response to heat stress (18). To date, HSF is the best-characterized factor that influences Hsp expression, although the FOXO family member DAF-16 is known to specifically activate expression of only the sHsps in C. elegans (19).
DAF-16 has a role in maintaining proteostasis in C. elegans by transcriptionally up-regulating a subset of sHsp genes (20). A number of these genes play a role in DAF-16-dependent lifespan extension and contains DAF-16 recognition sequences within their promoter (19).
Consistent with the results from C. elegans, here we show a direct role for Drosophila dFOXO in the expression of the inducible sHsps. The sHsps are activated when dFOXO activity is increased by oxidative stress. In contrast to the results in C. elegans, we also establish the large Hsp, Hsp70, as a direct dFOXO target and determine the promoter sequence elements required for dFOXO activation. We show a physiological role for dFOXO-induced transcription of Hsp70 in the oxidative stress response. In Drosophila, dFOXO activates transcription of both classes of protein chaperones, providing a broader network of transcriptional targets to better protect cells against stress and proteotoxicity.
Results
Heat Shock Proteins Are Targets of dFOXO
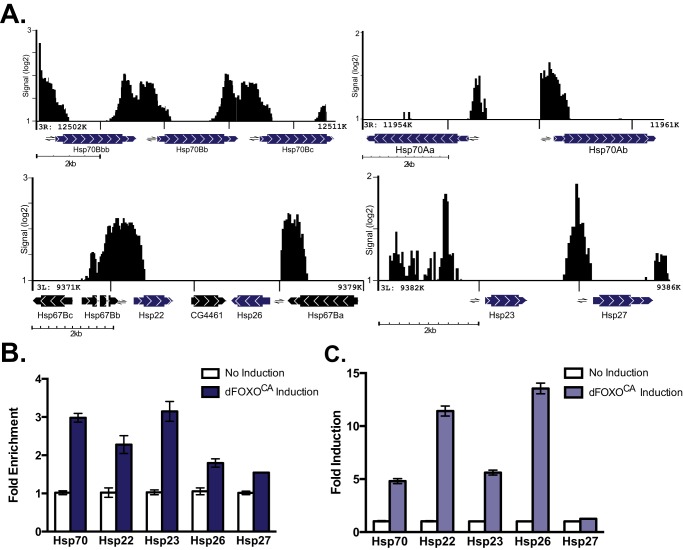
We recently identified genomic targets of constitutively active Drosophila FOXO (dFOXOCA) by chromatin immunoprecipitation (ChIP) followed by microarray analysis (21). Gene ontology analysis showed the genes involved in the heat shock response were significantly enriched (p = 2.8e−2). Specifically, HSF and the stress-inducible Hsps have dFOXO bound at their promoters (Figs. 1A and 2A) (21). Interestingly, and contrasting work done in C. elegans, we identified the stress-inducible Hsp70 family of protein chaperones as a dFOXO target. Consistent with results in C. elegans and early work in Drosophila, we also saw enrichment of dFOXO binding to the Drosophila sHsps (Hsp22, Hsp23, Hsp26, and Hsp27) promoter regions (Fig. 1A) (4, 19).
FIGURE 1.
Constitutively active dFOXO binds heat shock protein promoters and induces expression in Drosophila cells. A, ChIP-chip data from Drosophila S2 cells expressing constitutively active dFOXO (dFOXOCA). The data are plotted as the signal enrichment input on a log2 scale, and the x axis denotes the position in the genome. The genes are indicated by bars with arrowheads, where the thinner portion of the bar indicates the untranslated region and the thicker bar indicates the coding region. Lines indicate introns (if present). The arrowheads indicate the direction of the gene, and we define the promoter as within 2 kb of the transcription start site. Arrows denote ChIP primers used in B for each gene. B, ChIP data from cells overexpressing dFOXOCA. The data are shown as an enrichment of dFOXOCA binding normalized to the control 28S gene region enrichment (error bars represent S.E.). C, RT-qPCR from cells with or without induction of dFOXOCA. -Fold induction was calculated by normalizing to expression of rp49 (error bars represent S.E.).
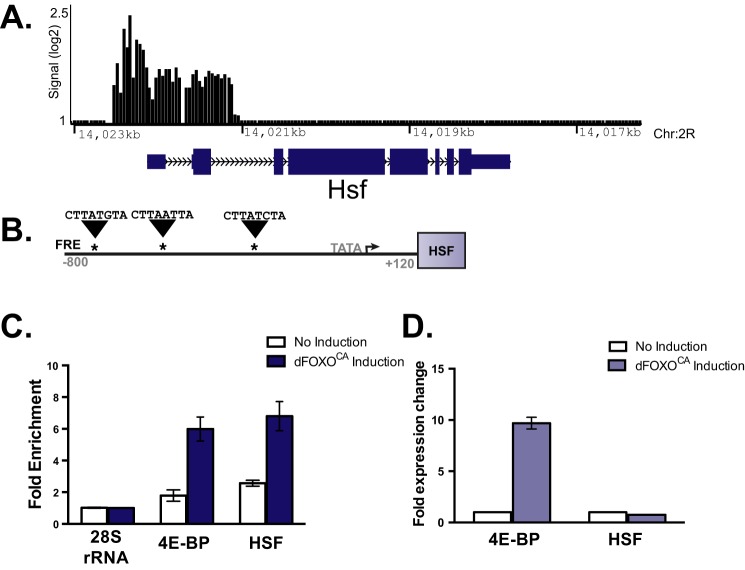
FIGURE 2.
dFOXO binds to the HSF promoter region but does not change HSF expression levels. A, ChIP-chip data from Drosophila S2 cells expressing dFOXOCA. The data are plotted as the signal on a log2 scale at the HSF promoter region. B, schematic of HSF promoter. The putative FREs are denoted by asterisks. The arrow denotes the transcription start site. C, ChIP-qPCR done in S2 cells expressing dFOXOCA. There is enrichment of dFOXOCA at the HSF gene and at a known FOXO target (4E-BP) compared with 28S rRNA gene (error bars represent S.E.). D, RT-qPCR was done in cells expressing dFOXOCA. The bona fide FOXO target, 4E-BP, has an increase in mRNA levels in response to dFOXO, but HSF does not have a FOXO-dependent change in expression levels (error bars represent S.E.).
To validate these results, we measured dFOXOCA binding to the Hsp promoters by performing ChIP-qPCR. As seen with the microarray experiments, there is enrichment of dFOXO binding at HSF and all of the stress-inducible Hsps that we examined (Figs. 1B and 2C). We utilized RT-qPCR to measure the effect of dFOXOCA overexpression on HSF and Hsp transcript levels. Both Hsp70 and the sHsps are induced by dFOXOCA expression (Fig. 1C). By contrast, under these conditions, dFOXO does not affect HSF transcript or protein levels despite binding to the promoter of HSF (Fig. 2D; see Fig. 5B). These findings demonstrate that the inducible sHsp genes are a conserved set of FOXO family targets between C. elegans and D. melanogaster. In addition, the stress-inducible Hsp70 genes are dFOXO targets in Drosophila.
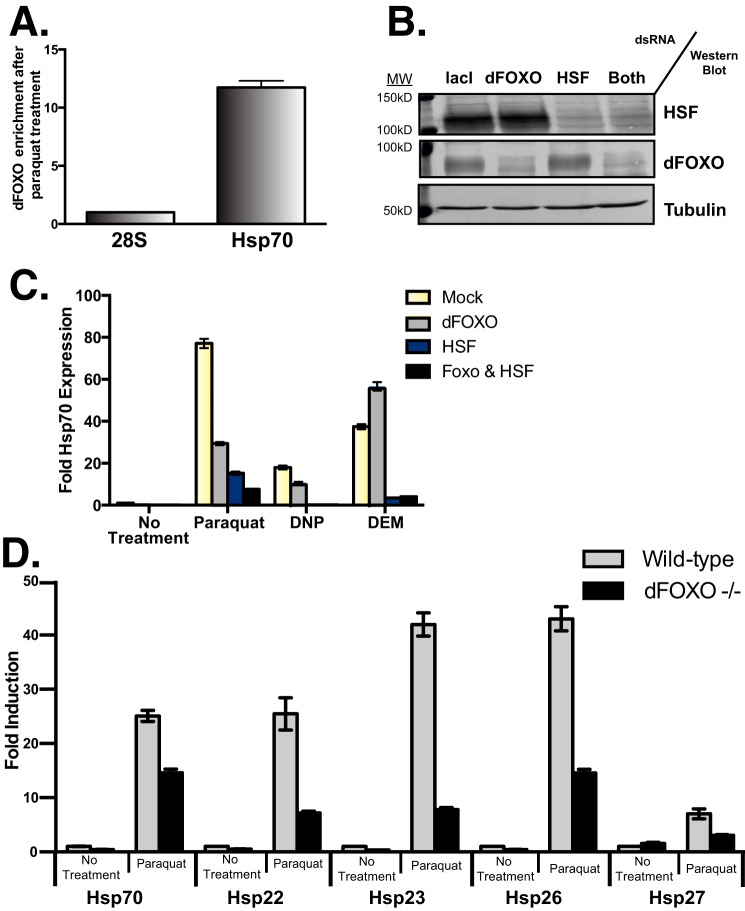
FIGURE 5.
Oxidative stress activates dFOXO in Drosophila cells and in vivo resulting in Hsp gene transcription. A, enrichment of endogenous dFOXO binding to the Hsp70 promoter region by ChIP in response to oxidative stress caused by paraquat treatment. Enrichment is calculation as a fraction of 28S rRNA gene (the error bar represents S.E.). B, Western blots showing the knockdown of dFOXO and HSF in Drosophila S2 cells. C, -fold induction of Hsp70 transcript levels in S2 cells in response to 50 mm paraquat or 1 mm DNP for 4 h or 0.1% diethyl maleate (DEM) for 24 h after knockdown. S2 cells treated with double-stranded RNA against lacI, dFOXO, or HSF for 72 h before paraquat treatment. RT-qPCR was done to detect transcript levels of Hsp70 compared with the control rp49 transcript. Data are plotted as Hsp70 transcript level over rp49 normalized to non-treated lacI (error bars represent S.E.). D, -fold induction of Hsp transcript levels in 7-day-old adult male flies in response to paraquat feeding. RT-qPCR was done to detect transcript levels of Hsps compared with the control rp49 transcript. Data are plotted as Hsps transcript level over rp49 normalized to wild-type mock treated flies (error bars represent S.E.).
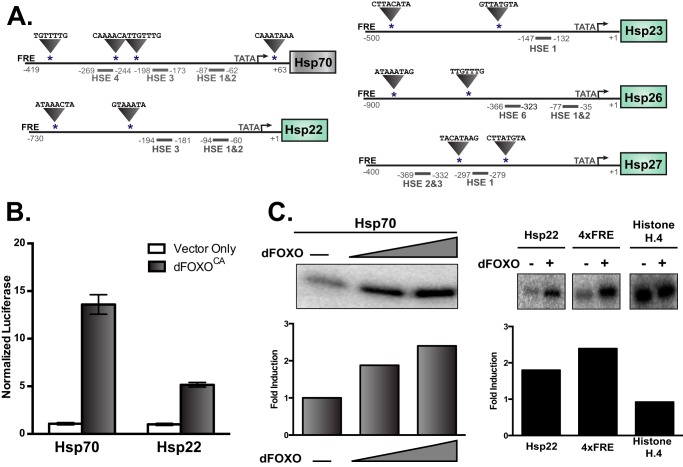
These results identified Hsps as a direct target of dFOXO, so we looked for potential FOXO-response elements (FREs) within the promoter regions. All of the inducible Hsp promoters contain multiple FREs (Fig. 3A). To help determine the promoter regions that respond to dFOXO, we put firefly luciferase under the control of the promoter regions of Hsp70Bb (−419 to +63) or Hsp22 (−730 to +98) (representative of the sHSPs) and co-transfected dFOXOCA in Drosophila cells. Although controlling for luciferase transfection efficiency using a constitutive Renilla reporter, we measured the effect of dFOXOCA expression on the activity of these promoters. We found Hsp70 and Hsp22 are both activated by dFOXOCA in the culture model, indicating these regions contain FOXO responsive sequences (Fig. 3B).
FIGURE 3.
Large and small heat shock proteins are direct transcriptional targets of dFOXO in Drosophila. A, schematic of Hsp promoters. The putative FREs are denoted by asterisks, and the HSEs are mapped. The arrow denotes the transcription start site. B, dual luciferase reporter assays where dFOXOCA was co-transfected into Drosophila S2 cells along with Hsp-firefly luciferase reporters. Data are plotted as firefly luciferase over renilla luciferase transfection control, normalized to vector-only control (error bars represent S.E.). C, primer extension analysis of in vitro transcription reactions containing recombinant dFOXO and Drosophila embryo nuclear extracts is shown. The promoter driving the transcription of the same reporter is indicated at the top of each reaction. The 4xFRE reporter serves as a positive control, and the histone H.4 promoter serves as a negative control. RNA produced by the reactions was measured by primer extension. Raw phosphorimaging units were used in the quantitation of -fold induction. All reactions were in the linear range of the phosphorimager.
ChIP data identify Hsp70 genes and sHsps as direct targets of dFOXO, and we observed that expression of dFOXOCA increases expression of both Hsp70 and sHsps (Fig. 1, B and C). Because in cell-based assays it is difficult to differentiate direct from indirect effects, we then performed in vitro transcription assays using the promoter regions we have identified as responding to dFOXO.
We tested dFOXO's ability to directly activate the transcription of the Hsp70 and Hsp22 promoters. The addition of recombinant dFOXO to Drosophila nuclear extract activates transcription of both the Hsp70 and Hsp22 constructs (Fig. 3C). The response is comparable to a synthetic reporter containing four copies of a consensus FRE (4×FRE) described previously (5). The addition of dFOXO to the extract does not effect the transcription from the histone H.4 promoter (Fig. 3C), indicating the response is specific to the Hsp promoters. These results show dFOXO is capable of directly activating the Hsp70 and Hsp22 promoters.
dFOXO Binds the Hsp70 Promoter Upstream of the HSEs
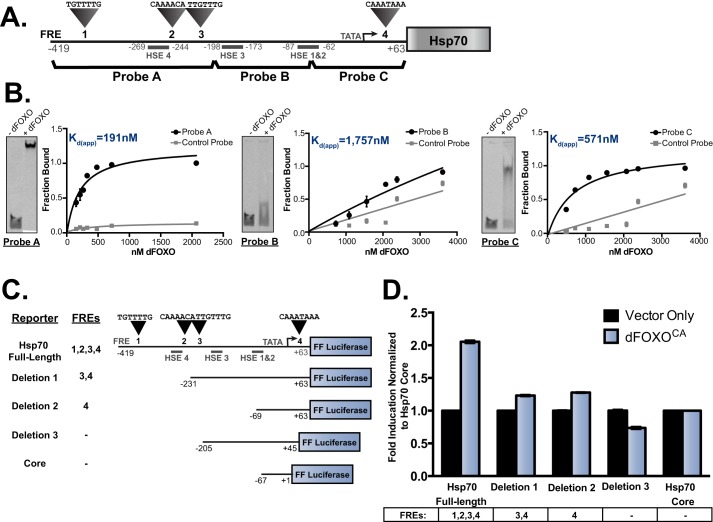
The heat shock elements to which HSF binds in the Hsp70 promoter are well characterized (22, 23); however, the required sequences for dFOXO activation are unknown. The Hsp70 promoter contains four HSEs as well as four putative FREs (TGTTTT or TGTTTAT). The FREs contained in this promoter are variants of the sequences enriched in dFOXO-bound regions we previously identified (21) (1–4 in Fig. 4A). These sequences are very similar to those found upstream of 4E-BP, which is one of the best-characterized dFOXO targets (5, 24). To determine which promoter regions dFOXO is capable of binding, we performed electrophoretic mobility shift assays where regions of the promoter were labeled and incubated in the presence recombinant dFOXO. We plotted the fraction bound against the concentration of dFOXO in the reaction (Fig. 4B) to determine an apparent disassociation constant (Kd). We fit the data to a nonlinear regression, assuming one binding site. Probe A has the lowest apparent Kd followed by Probe C, whereas Probe B is bound only slightly better than nonspecific DNA. These results indicate that the sequences contained in Probes A and C can be bound by dFOXO directly, potentially resulting in FOXO-dependent activation of Hsp70. We decided to test whether these regions also correlate with FOXO-dependent transcriptional activity.
FIGURE 4.
dFOXO binds to FREs upstream of the transcription start site in the Hsp70 promoter. A, schematic of the Hsp70 promoter including putative FREs and defined HSEs. The regions corresponding to probes for EMSA assays are indicated. B, representative images of FOXO-dependent shifts of probes are shown along with binding curves. Data from EMSA assays are quantitated by plotting the fraction of the probe bound by the concentration of recombinant dFOXO in the reaction (error bars represent S.E.). Apparent Kd values were calculated by assuming there is a single, specific binding site. C, promoter deletion constructs used for dual luciferase assays are shown. The putative FREs are numbered, and the FREs contained in each constructed are denoted. D, dual luciferase reporter assays comparing the full-length Hsp70 promoter activation to the deletion constructs. The data are plotted as -fold induction when dFOXOCA is expressed normalized to the vector-only or dFOXOCA-transfected Hsp70 core promoter construct (error bars represent S.E.).
To better characterize whether the FOXO-dependent activity requires the identified dFOXO binding sites, we carried out a promoter deletion analysis and compared these to the full-length Hsp70 reporter containing all HSEs and putative FREs. Deletion 1 removes FRE 1 and 2. Deletion 2 contains only FRE 4, and deletion 3 does not contain any FRE-containing sequences (Fig. 4C).
We performed a dual luciferase assay after transfection of Drosophila cells with these constructs and dFOXOCA or a vector-only control. We normalized the response to a construct that contains the minimal Hsp70 core promoter element lacking all HSEs and FREs (−67 to +1). We found that deletions 1 and 2 have less FOXO-dependent activity than the full-length Hsp70 promoter. The construct that contains no FREs, deletion 3, has no FOXO-dependent expression (Fig. 4D). These results indicate that the putative FRE-containing sequences that dFOXO bound in vitro are necessary for FOXO-dependent activation of the Hsp70 promoter.
Oxidative Stress Activates Endogenous dFOXO Resulting in Hsp Transcription
Our in vitro data and the data collected using the expression of constitutively active dFOXO identified Hsp70 as a dFOXO target. The question remains of what conditions result in endogenous dFOXO activating transcription of the Hsps. A likely candidate is oxidative stress. dFOXO is activated in response to cellular oxidative stress by JNK (4). Hsp expression increases in response to oxidative stress, but this transcriptional activation has previously been attributed exclusively to HSF activity (25).
To test the role of dFOXO in Hsp expression in response to oxidative stress, we first used the compound paraquat. Paraquat creates intracellular reactive oxygen species by undergoing reduction into the superoxide free radical in the mitochondria. It has been used previously to activate FOXO-mediated transcription (4, 26, 27). To confirm our ChIP data using constitutively active dFOXO, we also performed ChIP for endogenous dFOXO in response to paraquat treatment. dFOXO is enriched ∼12-fold at the Hsp70 promoter with paraquat treatment (Fig. 5A). Thus, oxidative stress results in dFOXO binding to the Hsp70 promoter.
To determine the relative contribution of HSF or dFOXO to Hsp70 activation, we knocked down dFOXO, HSF, or both in cultured cells using RNA interference (Fig. 5B). In addition, we treated cells with compounds that create intracellular oxidative stress through different mechanisms. We used paraquat, the mitochondrial uncoupler 2,4-dinitrophenol (DNP), the first compound shown to induce the heat shock response (28), and diethyl maleate, which depletes glutathione.
We incubated cells with nonspecific double-stranded RNA or double-stranded RNA directed against dFOXO, HSF, or both. After 72 h, the cells were incubated with paraquat, DNP, or diethyl maleate. RT-qPCR was performed to quantitate the relative expression of Hsp70 for each condition. Both dFOXO and HSF are required for the full activation of Hsp70 in response to oxidative stress created by paraquat or DNP treatment (Fig. 5C). However, HSF is solely responsible for the Hsp70 induction in response to diethyl maleate (DEM; Fig. 5C). These results show endogenous dFOXO and HSF are responsible for activating transcription of Hsp70 in response to intracellular reactive oxygen species.
Because both dFOXO and Hsp70 are required for adult flies to survive paraquat treatment (24, 29), we used paraquat to test if dFOXO activation of Hsp70 is relevant in vivo. We subjected a fly line containing a disruption of the dFOXO gene (−/− dFOXO Δ94) and the isogenic parental line (wDAH) to paraquat treatment and measured the response of the Hsp genes. We starved 7-day-old adult male flies for 5 h, then provided them Schneider's media with or without paraquat, and the flies were allowed to consume the solution for 24 h. Total RNA was extracted from the whole fly, and RT-qPCR was performed to measure the relative Hsp transcripts. In response to intracellular reactive oxygen species production, the dFOXO-null flies had less expression of Hsp22, Hsp23, Hsp26, Hsp27, and Hsp70 (Fig. 5D). This result indicates that dFOXO mediates expression of sHsps and the large heat shock protein Hsp70 in response to oxidative stress in vivo and that HSF alone is insufficient for the response.
Discussion
In the work described above we have identified an expanded set of Hsp transcriptional targets for Drosophila FOXO. The FOXO-dependent expression of the Hsps expands the role for dFOXO in maintaining proteostasis in response to stress and identifies a new transcriptional activator for the Hsp70 genes. Hsp70 properly folds nascent proteins and re-folds soluble proteins, whereas sHsps disassemble aggregates; both work to correct non-native protein interactions. Thus, dFOXO activation of both chaperones protects against proteotoxicity at two separate stages of protein stabilization.
Specifically, in response to oxidative stress, dFOXO is a necessary contributor to increased Hsp transcript levels. Furthermore, our data indicate that dFOXO is the major contributor to the response for the sHsps. Together with HSF, dFOXO is able to mount a transcriptional response that allows cells to survive acute stress. We propose that having two transcription factors that can activate stress response genes is advantageous to the cell because maintaining proteostasis is imperative for cell survival when the accumulation of oxidized, misfolded, or aggregated proteins results in proteotoxicity (Fig. 6).
FIGURE 6.
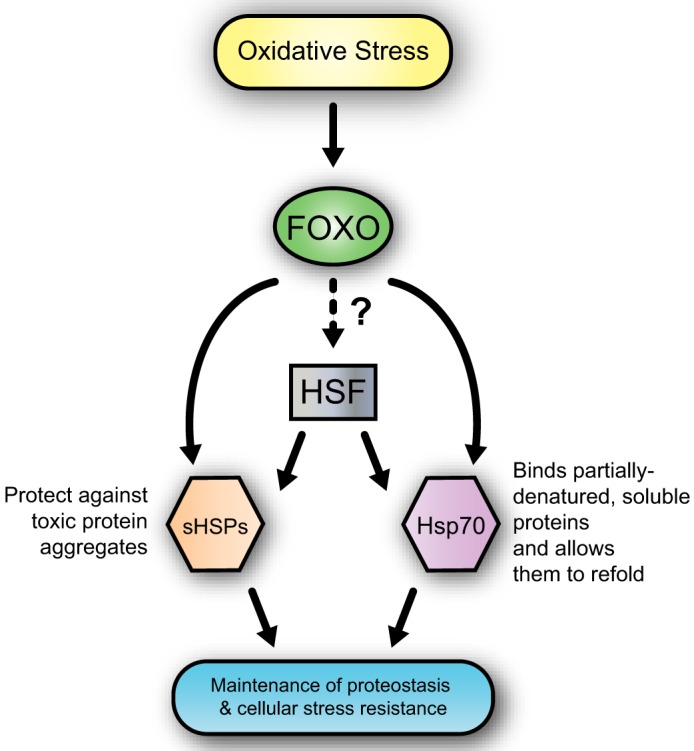
Model of FOXO's contribution to maintaining proteostasis by controlling expression of both families of Hsp chaperones. Acute oxidative stress activates FOXO, and dFOXO directly transcriptionally up-regulates the sHsps and Hsp70 genes. The two families of chaperones protect against proteotoxicity using separate mechanisms. The role for dFOXO in transcriptional regulation of HSF has not been fully elucidated.
In Drosophila, dFOXO binds the promoters of more than a thousand genes, and identifying the subset of genes that reduces cellular damage is ongoing within the field (21, 30–33). Drosophila FOXO was previously shown to influence the expression of a sHsp-like gene, and there have been previous attempts to show Drosophila FOXO-dependent Hsp70 expression; however, the FREs disrupted in that study do not match those identified here and oddly included the TATA core, abrogating promoter function and complicating the interpretation of the results (4, 9). This work defines a comprehensive set of Hsps that are direct targets of Drosophila FOXO that help to maintain protein homeostasis.
The differential requirements for HSF and dFOXO in response to specific oxidative stress inducing agents was an unexpected result. The Hsp70 transcriptional response to paraquat and DNP requires dFOXO, and contrast, diethyl maleate, which depletes glutathione from the cells, does not require dFOXO for full activation of Hsp70 (Fig. 5C). Because of the differential response, it is plausible that they respond to different signals; HSF directly senses misfolded proteins and drives expression of Hsps, whereas dFOXO may sense the stress indirectly (34, 35). A possible explanation implicates the mitochondria as a dFOXO-specific sensor. Paraquat undergoes reduction within and damages the mitochondria (36), and DNP is a mitochondrial uncoupler. Our work suggests that HSF responds to all oxidative stress, but FOXO responds to selective types of oxidative stress, perhaps through the mitochondria.
We also identified the HSF gene as bound by dFOXO. However, we could find no effect on HSF transcription or HSF protein levels under the experimental conditions used here despite the fact that dFOXO was found reproducibly bound to the HSF promoter. This might indicate there is another signaling event required for FOXO-mediated regulation of HSF that is missing from our experimental approach. This is intriguing because a close relationship between HSF and FOXO in stress responses and lifespan regulation has been proposed in C. elegans (19, 37). daf-16 is required for hsf-1 to extend lifespan (19). There is evidence to support that HSF and DAF-16 affect each other's activity, and DAF-16 and HSF have a set of overlapping targets but do not require always require each other for transcription of their target genes (19, 37–39). Future work should be aimed at identifying the pathways whose cross-talk is required for the connection between FOXO and HSF regulation.
Much of the work on the FOXO family has focused on its role in modulating aging and lifespan. We suggest another physiological role for FOXO-dependent transcription of Hsp70 may occur as the organism ages. Because acute oxidative stress has a transcriptional profile similar to aging (25), we propose that dFOXO may play a role in Hsp transcription over the course of aging. The accumulation of free radicals as well as aging results in an increase in Hsp22 and Hsp70 transcription (25). Increased expression of these genes is also predictive of improved survival rate in response to stress (40). Previously, the aging-dependent expression of Hsp70 was attributed solely to HSF. However, it seems likely that dFOXO's contribution was overlooked (41). Based on our current results, we propose a role for FOXO-dependent activation of the large heat shock protein family during aging. Both dFOXO and HSF can potentially activate the expression of both families of heat shock protein genes in D. melanogaster in response to both oxidative stress and aging, increasing survival. Further work will be required to determine if this role is conserved in higher animals.
Experimental Procedures
Fly Lines, Constructs, and Antibodies
The wDAH and dFOXO-null (Δ94/Δ94) fly lines have been previously described (21). pAc5V5-dFOXOCA and pGL4xFRE were previously described (21). Promoter regions from genomic Hsp70Bb, Hsp22, and histone H4 were cloned into pGLbasic vector, and these were used in dual-luciferase assays and as templates for in vitro transcription. pGLHsp70Bb was used for cloning deletion constructs and band shift probes. For Western blotting and ChIP, antibodies against tubulin (DSHB Hybridoma Product E7), dFOXO (21), and HSF (42) were used.
Cell Culture, RNAi, RNA Extraction, and RT-qPCR
For dFOXO overexpression experiments, a Drosophila S2 cell line (321) that contains a stable transfection of pMTdFOXOA3 were induced with 500 μm CuSO4 for 16 h (5). For all RNAi experiments, a clonal S2 cell line (S2C1) cultured in Schneider's media supplemented with 10% FBS was used. Cells were incubated in serum-free media with 15 μg/ml double-stranded RNA for 1 h before the addition of media with serum. After 72 h the cells were treated with 50 mm paraquat (methyl viologen dichloride hydrate, Sigma) for 4 h, 1 mm 2,4-dinitrophenol (Sigma), or 0.1% diethyl maleate (Sigma) for 24 h, or left untreated. RNA isolation and RT-qPCR were performed as previously described (26) using primers in Table 1.
TABLE 1.
Primers used for RT-qPCR and ChIP-qPCR
| Gene | Forward | Reverse |
|---|---|---|
| Quantitative RT-PCR primers | ||
| rp49 | CGCCACCAGTCGGATCGATATGC | GTTCTCTTGAGAACGCAGGCGACC |
| Hsp70 | CAAAGTTGTAAGCGACGGCGGAA | TGTCTCCGGCTGTGGAGCGCA |
| Hsp22 | ACGCCTCTCCTCGCCCTTTCACG | GGAAGTGCCTGGAGCTATAGCCAC |
| Hsp23 | GAAAGGATGGCTTCCAGGTC | GCCTCCTTGGGATTCTCCTT |
| Hsp26 | CAAGGTTCCCGATGGCTACAAGGC | GCCTGCGGCTTGGGAATACTGA |
| Hsp27 | GACTTCGGTTTTGGCGTCCA | ACTTGGCCTGTTCCTTGCTG |
| HSF | TGGCCAGCTTCATAAGGCAA | ACCCCGCATGACTTTCACAT |
| 4E-BP | GTTTGGTGCCTCCAGGAGTGG | CGTCCAGCGGAAAGTTTTCG |
| ChIP qPCR primers | ||
| 28S rRNA | GAGTAGGAAGGTACAATGGTATGC | GAACCGTATTCCCTTTCGTTCAA |
| Hsp70 | TCTCTGGCCGTTATTCGTTATTC | GCTGCGCTTGTTTATTTGCTTA |
| Hsp22 | CTGGAATGAATGGGCTCAAA | CCTTGGGGAAGGTCAGTTTC |
| Hsp23 | CCAGGCCTTTTCATTCCCAC | GGGGCACAAACATCGACA |
| Hsp26 | GTGCGCCTGTATGAGTGAGA | GTGGGAGATTGCTGGCGTTA |
| Hsp27 | GGGCGTATTCAAAGGGGCTT | GCAACCAGGGACGGAACTTT |
| HSF | GCAAGTTGGTGGCTCGAATG | CTGCACACCAACACCAACAC |
| 4E-BP | CGAGGTGTGCAGGCTGC | CACACTCGATATGGCTGCGAC |
ChIP
The 321 stable line was induced with 500 μm CuSO4 for 16 h. ChIP was performed as previously described (20). The immunoprecipitated DNA was assayed by qPCR using primers for the control rRNA gene and the promoter regions of HSF, Hsp22, Hsp23, Hsp26, Hsp27, and Hsp70 (Table 1).
In Vitro Transcription and His-tagged dFOXO Purification
Nuclear extracts were prepared from Drosophila embryos as previously described (43). Promoter templates (pGLHsp70Bb, pGLHsp22, pGL4xFRE, and pGLH4), recombinant His-dFOXO, and recombinant NTPs were added to the extracts. dFOXO purification and primers extension assays were performed as previously described (5).
Transient Transfection and Dual-luciferase Assay
S2C1 cells were plated at a 0.5 × 106 cells/ml in 24-well plates. The cells were transfected with reporter plasmid and expression plasmid at a ratio of 1:10 using the Effectene protocol (Qiagen). Twenty hours post-transfection the cells were harvested according to the passive lysis protocol for the Promega Dual-Luciferase reporter assay system. The expression of firefly and renilla luciferase was measured either using the dual-luciferase reporter assay system (Promega), or firefly luciferase was measured in 75 mm HEPES, pH 8.0, 20 mm DTT, 5 mm MgSO4, 530 μm ATP, 500 μm coenzyme A, 500 μm D-luciferin, and 100 μm EDTA, and Renilla luciferase was measured by adding an equal volume of 1.0 m NaCl, 0.5 m Na2SO4, 25 mm Na4PPi, 15 mm EDTA, 10 mm NaOAc, and 0.1 mm coelenterazine.
Band-shift Assay
Regions of the Drosophila Hsp70 promoter were cloned into pBC (Stratagene), and the probes were made by PCR with primers labeled with Dylight 680 fluorophore (Thermo). The band-shifts were done as previously described (5). Recombinant dFOXO was incubated with labeled probes and separated on a 5% acrylamide, 1×TGE (25 mm Tris, 190 mm glycine, 1 mm EDTA), 4 mm MgCl2, 2.5% glycerol gel. Gels were imaged using LI-COR Odyssey. The data were plotted as the fraction of probe bound to final concentration of recombinant dFOXO in the binding reaction. The data were fit to a nonlinear regression with the assumption of a single, specific binding site, and the apparent disassociation constant (Kd) was calculated.
Paraquat Feeding
Adult fly feeding protocol was described previously (26). RT-qPCR was performed to determine the relative levels of transcription of both the control rp49 and Hsps.
Gene Ontology Analysis
Genes previously described as enriched for dFOXO binding (21) were analyzed using the DAVID Bioinformatics Resources available online (david.ncifcrf.gov).
Author Contributions
M. T. M. and M. R. D. conceived the study and wrote the paper. M. T. M. and M. R. D. provided the original reagents. M. R. D. performed the experiments. M. T. M. and M. R. D. analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Linda Partridge's laboratory for the Δ94 heterozygous fly lines and John Lis's laboratory for the HSF antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM11703 (to M. T. M.). This work was also supported by a New Scholar in Aging Award from the Ellison Medical Foundation (to M. T. M.). The authors declare that they have no conflicts of interest with the contents of this paper. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- fox
- forkhead box
- FOXO
- fox subfamily O
- Hsp
- heat shock protein
- sHsp
- small Hsp
- HSF
- heat shock transcription factor
- HSE
- heat shock element
- qPCR
- quantitative PCR
- FRE
- FOXO-response element
- DNP
- 2,4-dinitrophenol.
References
- 1. Morimoto R. I. (2011) The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 76, 91–99 [DOI] [PubMed] [Google Scholar]
- 2. Hannenhalli S., and Kaestner K. H. (2009) The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eijkelenboom A., and Burgering B. M. (2013) FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14, 83–97 [DOI] [PubMed] [Google Scholar]
- 4. Wang M. C., Bohmann D., and Jasper H. (2005) JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121, 115–125 [DOI] [PubMed] [Google Scholar]
- 5. Puig O., Marr M. T., Ruhf M. L., and Tjian R. (2003) Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W., Hietakangas V., Wee S., Lim S. C., Gunaratne J., and Cohen S. M. (2013) ER stress potentiates insulin resistance through PERK-mediated FOXO phosphorylation. Genes Dev. 27, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giannakou M. E., Goss M., Jacobson J., Vinti G., Leevers S. J., and Partridge L. (2007) Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell 6, 429–438 [DOI] [PubMed] [Google Scholar]
- 8. Hwangbo D. S., Gershman B., Gersham B., Tu M. P., Palmer M., and Tatar M. (2004) Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429, 562–566 [DOI] [PubMed] [Google Scholar]
- 9. Demontis F., and Perrimon N. (2010) FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alic N., Tullet J. M., Niccoli T., Broughton S., Hoddinott M. P., Slack C., Gems D., and Partridge L. (2014) Cell-nonautonomous effects of dFOXO/DAF-16 in aging. Cell Rep. 6, 608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balch W. E., Morimoto R. I., Dillin A., and Kelly J. W. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 12. Ingolia T. D., and Craig E. A. (1982) Four small Drosophila heat shock proteins are related to each other and to mammalian α-crystallin. Proc. Natl. Acad. Sci. U.S.A. 79, 2360–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Langer T., Lu C., Echols H., Flanagan J., Hayer M. K., and Hartl F. U. (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356, 683–689 [DOI] [PubMed] [Google Scholar]
- 14. Hartl F. U. (1996) Molecular chaperones in cellular protein folding. Nature 381, 571–579 [DOI] [PubMed] [Google Scholar]
- 15. Tower J. (2011) Heat shock proteins and Drosophila aging. Exp. Gerontol. 46, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morrow G., Battistini S., Zhang P., and Tanguay R. M. (2004) Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J. Biol. Chem. 279, 43382–43385 [DOI] [PubMed] [Google Scholar]
- 17. Tatar M., Khazaeli A. A., and Curtsinger J. W. (1997) Chaperoning extended life. Nature 390, 30. [DOI] [PubMed] [Google Scholar]
- 18. Fernandes M., Xiao H., and Lis J. T. (1995) Binding of heat shock factor to and transcriptional activation of heat shock genes in Drosophila. Nucleic Acids Res. 23, 4799–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu A. L., Murphy C. T., and Kenyon C. (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145 [DOI] [PubMed] [Google Scholar]
- 20. Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., and Kenyon C. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 [DOI] [PubMed] [Google Scholar]
- 21. Spellberg M. J., and Marr M. T. 2nd. (2015) FOXO regulates RNA interference in Drosophila and protects from RNA virus infection. Proc. Natl. Acad. Sci. U.S.A. 112, 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pelham H. R. (1982) A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell 30, 517–528 [DOI] [PubMed] [Google Scholar]
- 23. Perisic O., Xiao H., and Lis J. T. (1989) Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell 59, 797–806 [DOI] [PubMed] [Google Scholar]
- 24. Jünger M. A., Rintelen F., Stocker H., Wasserman J. D., Végh M., Radimerski T., Greenberg M. E., and Hafen E. (2003) The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landis G. N., Abdueva D., Skvortsov D., Yang J., Rabin B. E., Carrick J., Tavaré S., and Tower J. (2004) Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 101, 7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olson C. M., Donovan M. R., Spellberg M. J., and Marr M. T. 2nd. (2013) The insulin receptor cellular IRES confers resistance to eIF4A inhibition. Elife 2, e00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arking R., Buck S., Berrios A., Dwyer S., and Baker G. T. 3rd. (1991) Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev. Genet. 12, 362–370 [DOI] [PubMed] [Google Scholar]
- 28. Ritossa F. (1962) New puffing pattern induced by temperature shock and Dnp in Drosophila. Experientia 18, 571–573 [Google Scholar]
- 29. Shukla A. K., Pragya P., Chaouhan H. S., Tiwari A. K., Patel D. K., Abdin M. Z., and Chowdhuri D. K. (2014) Heat shock protein-70 (Hsp-70) suppresses paraquat-induced neurodegeneration by inhibiting JNK and caspase-3 activation in Drosophila model of Parkinson's disease. PLoS ONE 9, e98886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teleman A. A., Hietakangas V., Sayadian A. C., and Cohen S. M. (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 7, 21–32 [DOI] [PubMed] [Google Scholar]
- 31. Alic N., Andrews T. D., Giannakou M. E., Papatheodorou I., Slack C., Hoddinott M. P., Cochemé H. M., Schuster E. F., Thornton J. M., and Partridge L. (2011) Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol. Syst. Biol. 7, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alic N., Giannakou M. E., Papatheodorou I., Hoddinott M. P., Andrews T. D., Bolukbasi E., and Partridge L. (2014) Interplay of dFOXO and two ETS-family transcription factors determines lifespan in Drosophila melanogaster. PLoS Genet. 10, e1004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gershman B., Puig O., Hang L., Peitzsch R. M., Tatar M., and Garofalo R. S. (2007) High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol Genomics 29, 24–34 [DOI] [PubMed] [Google Scholar]
- 34. Zou J., Guo Y., Guettouche T., Smith D. F., and Voellmy R. (1998) Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471–480 [DOI] [PubMed] [Google Scholar]
- 35. Becker J., Mezger V., Courgeon A. M., and Best-Belpomme M. (1990) Hydrogen peroxide activates immediate binding of a Drosophila factor to DNA heat-shock regulatory element in vivo and in vitro. Eur. J. Biochem. 189, 553–558 [DOI] [PubMed] [Google Scholar]
- 36. Cochemé H. M., and Murphy M. P. (2008) Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 283, 1786–1798 [DOI] [PubMed] [Google Scholar]
- 37. Douglas P. M., Baird N. A., Simic M. S., Uhlein S., McCormick M. A., Wolff S. C., Kennedy B. K., and Dillin A. (2015) Heterotypic signals from neural HSF-1 separate thermotolerance from longevity. Cell Rep. 12, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McColl G., Rogers A. N., Alavez S., Hubbard A. E., Melov S., Link C. D., Bush A. I., Kapahi P., and Lithgow G. J. (2010) Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 12, 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh V., and Aballay A. (2006) Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 13092–13097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang J., and Tower J. (2009) Expression of hsp22 and hsp70 transgenes is partially predictive of Drosophila survival under normal and stress conditions. J. Gerontol. A Biol. Sci. Med. Sci. 64, 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wheeler J. C., King V., and Tower J. (1999) Sequence requirements for upregulated expression of Drosophila hsp70 transgenes during aging. Neurobiol. Aging 20, 545–553 [DOI] [PubMed] [Google Scholar]
- 42. Guertin M. J., and Lis J. T. (2010) Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 6, e1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biggin M. D., and Tjian R. (1988) Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53, 699–711 [DOI] [PubMed] [Google Scholar]