Abstract
We address the critically important ionization properties of histidine side chains of membrane proteins, when exposed directly to lipid acyl chains within lipid bilayer membranes. The problem is important for addressing general principles that may underlie membrane protein function. To this end, we have employed a favorable host peptide framework provided by GWALP23 (acetyl-GGALW5LALALALALALALW19LAGA-amide). We inserted His residues into position 12 or 14 of GWALP23 (replacing either Leu12 or Leu14) and incorporated specific [2H]Ala labels within the helical core sequence. Solid-state 2H NMR spectra report the folding and orientation of the core sequence, revealing marked differences in the histidine-containing transmembrane helix behavior between acidic and neutral pH conditions. At neutral pH, the GWALP23-H12 and GWALP23-H14 helices exhibit well defined tilted transmembrane orientations in dioleoylphosphatidylcholine (DOPC)and dilauroylphosphatidylcholine (DLPC) bilayer membranes. Under acidic conditions, when His12 is protonated and charged, the GWALP23-H12 helix exhibits a major population that moves to the DOPC bilayer surface and a minor population that occupies multiple transmembrane states. The response to protonation of His14 is an increase in helix tilt, but GWALP23-H14 remains in a transmembrane orientation. The results suggest pKa values of less than 3 for His12 and about 3–5 for His14 in DOPC membranes. In the thinner DLPC bilayers, with increased water access, the helices are less responsive to changes in pH. The combined results enable us to compare the ionization properties of lipid-exposed His, Lys, and Arg side chains in lipid bilayer membranes.
Keywords: histidine, lipid bilayer, membrane protein, solid state NMR, transmembrane domain, GWALP23, deuterium quadrupole splitting, imidazole ionization constant
Introduction
Since the identification of histidine in the active site of chymotrypsin (1, 2) and the formulation of the “charge-relay” (or catalytic triad) hypothesis (3), the importance of the imidazole side chain for acid-base catalysis has been widely recognized. Imidazole is the only group on a protein side chain with a pKa around neutrality in aqueous physiological buffer (2). It is therefore vitally important to also know the ionization properties of the His side chain at various locations within lipid bilayer membranes. Indeed, as with soluble proteins, many membrane proteins contain functionally important His residues within their transmembrane domains: for example, those of the photosynthetic reaction center (4), cytochrome c oxidase (5, 6), influenza A M2 channel (7–10), and other transporters and channels (11–14). Given that proton conduction may require clusters of multiple residues (5, 6), it is important to know the limits for the titration properties of candidate residues such as histidines and carboxyl groups, among others, in bilayer membranes.
Polar residues within the membrane core of α-helical proteins, although sparse, are highly conserved. In many cases, this conservation is due to the direct involvement of these residues to the function of the protein (15). Numerous membrane proteins display pH-dependent behavior, and often a histidine residue within the membrane core acts as a pH “sensor” for the protein. For example, the protonation of a single His residue in the diphtheria toxin T domain results in a major conformational change leading eventually to the translocation of the catalytic C domain across the endosomal membrane. Interestingly, replacement of this His residue with Arg results in significant protein unfolding at neutral pH (16). Histidine is also a key residue in the pH-dependent M2 channel of the influenza A virus, where a single residue, His37, is responsible for channel activation (10) and the proton-selective conductance of the channel (17).
Some of the pKa values for crucial residues in membrane proteins are likely to differ substantially from the canonical values that are observed in aqueous solution (5, 6). A key advance toward understanding ionization behavior in lipid membranes was made by MacCallum et al. (18), who employed molecular dynamics simulations to calculate the partitioning of the neutral and charged forms of the side chains of Glu, Asp, Lys, and Arg between bulk water and a lipid bilayer of dioleoylphosphatidylcholine (DOPC).2 Interestingly, histidine was not modeled because “His is difficult to treat accurately due to its multiple possible protonation states” (18), namely the possibility of deprotonation from either of the two NH groups of charged imidazolium to give two different neutral isomers. With the dearth of modeling results, it becomes ever more important to have experimental results about the pKa of the His imidazole side chain in bilayer membranes.
An avenue for measuring side chain pKa values in the lipid membrane environment is provided by the framework of GWALP23, acetyl-GGALW(LA)6LWLAGA-[ethanol]amide (19, 20), which possesses two interfacial Trp residues and maintains a preferred and well defined tilted transmembrane orientation with low dynamic averaging (20–23). Indeed, GWALP23 (or its cousin having Tyr5 instead of Trp5) has been employed to determine a pKa of 6.5 at 37 °C for a lipid-exposed lysine side chain located about 8 Å from the center of DOPC bilayer membranes (24). Within the same framework, arginine is observed not to titrate below pH 9 (24). In agreement with molecular simulations (18), the Arg guanidinium group would prefer to exit the lipid bilayer rather than to deprotonate (25). It is furthermore striking that the pKa of Lys is altered more by the bilayer environment than is the pKa of Arg (24), when compared with a bulk water environment.
With the results for Lys and Arg in hand, in this study we employ the GWALP23 framework to examine the pKa values for His side chains at two locations in bilayer membranes of differing lipid thickness, namely DOPC and DLPC. In parallel with observations for lysine, the lipid bilayer environment tends to favor the neutral unprotonated state of histidine. Specifically, we find that the pKa of a lipid-exposed His imidazole side chain depends upon its bilayer location and its accessibility to the bulk aqueous solution. The results then assume a more general significance because they enable comparisons of the relative influence of the bilayer membrane environment on the pKa values for the side chains of arginine, lysine, and histidine.
Results
To probe the influence of the hydrophobic environment of the lipid bilayer on the ionization behavior of the histidine imidazole side chain, single His residues were introduced into the GWALP23 sequence (Table 1) at position 12, located directly between the two anchoring Trp residues, on the same helix face, separated by seven residues on both sides, or at position 14, located on the opposite face of the helix (Fig. 1). Using peptides with specifically labeled [2H]alanine residues, we employed solid-state NMR spectroscopy as a means to characterize peptide behavior in aligned lipid bilayer membranes. The repeating Leu-Ala core sequence of GWALP23 favors peptide folding into α-helical secondary structure within the hydrophobic interior of the lipid bilayer. To verify that the peptides retain their α-helical character with the addition of His residues, circular dichroism (CD) spectra were recorded. Indeed, the CD spectra for both GWALP23-H12 and GWALP23-H14 show a minimum near 208 nm and a broad shoulder at 222 nm, indicative of α-helical secondary structure. The bilayers also were well aligned, as indicated by the 31P NMR spectra of the head groups.
TABLE 1.
Sequences of GWALP23 peptides with single His, Arg or Lys substitutions
| Name | Sequence | Reference |
|---|---|---|
| GWALP23 | Acetyl-GGALWLALALALALALALWLAGA-amide | 19, 20 |
| GWALP23-H12 | Acetyl-GGALWLALALAH12ALALALWLAGA-amide | This work |
| GWALP23-H14 | Acetyl-GGALWLALALALAH14ALALWLAGA-amide | This work |
| GWALP23-R12 | Acetyl-GGALWLALALAR12ALALALWLAGA-amide | 25 |
| GWALP23-R14 | Acetyl-GGALWLALALALAR14ALALWLAGA-amide | 25 |
| Y5GWALP23-K12 | Acetyl-GGALYLALALAK12ALALALWLAGA-amide | 24 |
| Y5GWALP23-K14 | Acetyl-GGALYLALALALAK14ALALWLAGA-amide | 24 |
FIGURE 1.
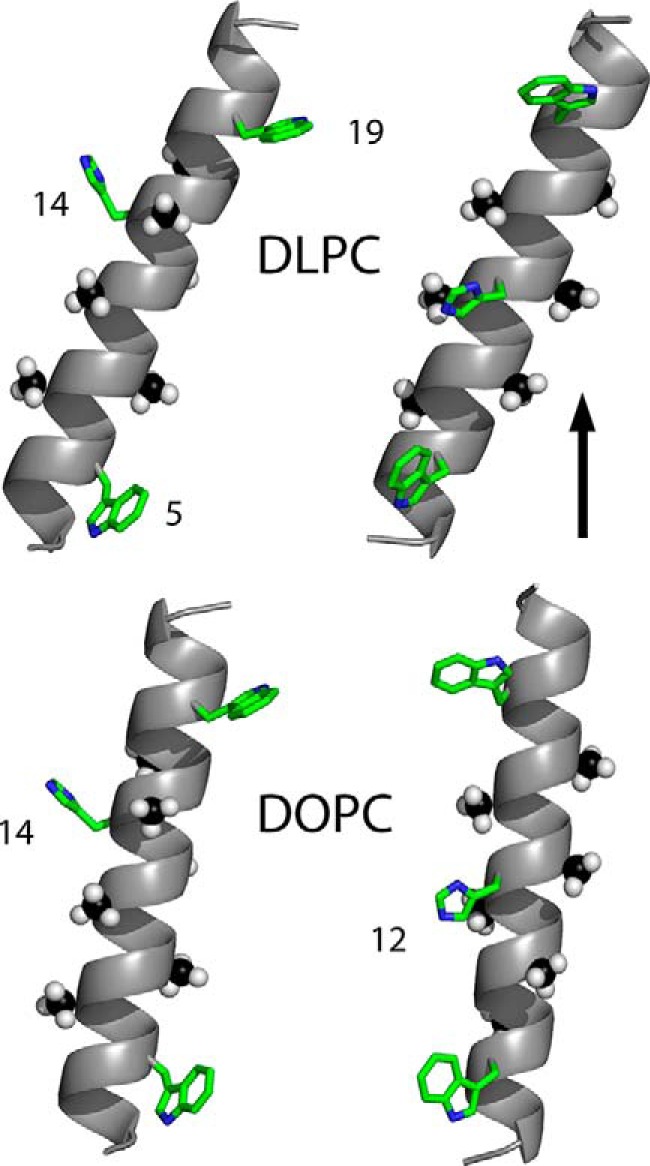
Models to illustrate the experimental tilted orientations of charged GWALP23-H14 (left) or neutral GWALP23-H12 (right), with respect to a vertical bilayer normal (see arrow) of DLPC or DOPC lipid bilayer membranes. The peptide with the charged His14 side chain is more tilted than is GWALP23-H12 in both membranes. In addition to rotational differences, as illustrated, each peptide helix is more tilted in the thinner DLPC bilayer (upper models) than in the thicker DOPC bilayer. The numbers indicate tryptophans 5 and 19, and histidines 12 and 14, in the peptide sequences. The six deuterated alanine methyl groups that underlie the tilt analysis are shown as space filling. Carbons are either green or black, nitrogens are blue, and hydrogens are white. The experimental helix tilt magnitudes and the pKa values of the histidine side chains are explained under “Results.”
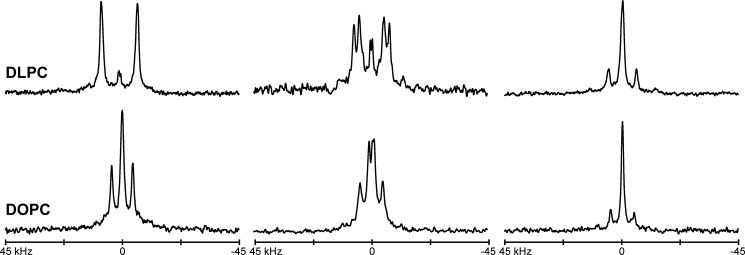
To assess the helix orientations, the 2H NMR spectra of Ala-d4-labeled GWALP23-H12 and GWALP23-H14 peptides were recorded in DLPC and DOPC bilayer-incorporated samples, hydrated with 10 mm buffer at a variety of pH conditions. The 2H NMR spectra for the aligned samples reveal significant differences in peptide behavior between the GWALP23-H12 and GWALP23-H14 isomers. Under neutral pH conditions, the spectra of the -H12 peptide in bilayers of DOPC or DLPC exhibit distinct signals for each CD3 methyl side chain of the six core alanine residues, consistent with dynamic averaging about a predominant and well defined tilted transmembrane orientation in each of the lipid bilayer membranes (Fig. 2). Spectra of GWALP23-H14 in DLPC bilayers, hydrated with pH 6 buffer, also display sharp, well defined quadrupolar splittings. In DOPC bilayers, however, spectra of the -H14 peptide appear to have somewhat broader signals with lower signal to noise ratio. When the pH is lowered, the spectral quality improves for GWALP23-H14 (see below), whereas GWALP23-H12 displays multi-state behavior at low pH.
FIGURE 2.
Deuterium NMR spectra for six labeled alanine residues in GWALP23-H12 in DLPC and DOPC bilayers, hydrated with 10 mm buffer at pH 4, showing β = 90° sample orientation. The peptide/lipid ratio is 1/60 at a temperature of 50 °C.
As indicated (Fig. 2), above pH 4, the 2H NMR spectra of the core alanine methyl groups of GWALP23-H12 show well defined signals in DLPC and DOPC bilayers, with quadrupolar splitting magnitudes nearly identical to those of the host peptide, GWALP23 (Table 2). Importantly, the peptide orientation does not change when Leu is replaced with a polar (yet neutral) His residue at position 12, located between Trp5 and Trp19. When the pH is lowered to 2, the His residue becomes charged, leading to spectra that exhibit multiple weak resonances, similar to those of Y5GWALP23-K+12 and GWALP23-R+12. Furthermore, we also observe the emergence of an additional set of strong resonances, as evident from Fig. 3. These signals likely correspond to a new predominant orientation of the -H+12 peptide helix, occupied by ∼70% of the population, in addition to multiple minor states. It is possible, therefore, to monitor the disappearance of the sharp signals observed at pH 4 and the appearance of a new signal at low pH to estimate the filling of this new state as well as the disappearance of the original state as a function of pH. The major tilted transmembrane orientation of GWALP23-H12 is 50% populated at pH 2.6, and the new orientation is 50% populated at pH 2.3 (Fig. 4). These observations are consistent with the fact that the ionization of the His residue of the -H012 peptide does not follow a simple two-state equilibrium. Because the -H012 population interconverts with the new major orientation and the multiple minor states at pH 2.6, the single-state, tilted transmembrane population of the -H012 peptide will be greater than that of the new major orientation or any of the minor states. Also apparent from Fig. 3 is the emergence at low pH of new sets of 2H resonances with large Δνq magnitudes, which likely arise from backbone Cα deuterons. We note that our d4-Ala residues always contain a Cα deuteron, and resonances from backbone deuterons have often been observed in arginine-containing peptides (25, 26). We do not yet understand the molecular properties or environmental conditions that govern the emergence of these signals within the context of the quadrupolar echo pulse sequence. In DLPC bilayers, the spectra of GWALP23-H12 retain strong, well defined signals under low pH conditions.
TABLE 2.
2H NMR quadrupolar splitting magnitudes (|Δνq|, in kHz) for labeled core alanine CD3 groups in GWALP23 peptides with single residue replacements
Sample orientation is β = 0°. Each value (in kHz) is the average of the magnitude observed at β = 0° and twice the magnitude observed for a β = 90° sample orientation. The position of each labeled alanine is identified. The positions of the variable amino acid residues are indicated as His12, His14, Arg12, Arg14, Lys12, or Lys14; with the side-chain charge indicated, if known. The label L12,14 refers to host peptide, GWALP23. Dashes indicate that the pH was not buffered, or that particular data points were not measured.
| Lipid and peptide | pH | Alanine CD3 position |
Reference | |||||
|---|---|---|---|---|---|---|---|---|
| 7 | 9 | 11 | 13 | 15 | 17 | |||
| DLPC | ||||||||
| H+/014 | 2.0–8.2a | 31.6 ± 0.6b | 16.6 | 21.8 | 11.7 | 1.8 | 30.6 | This work |
| R+14 | — | 33.0 | 21.1 | 25.7 | 9.3 | 6.8 | 30.8 | 26 |
| H+/012 | 2.0–8.2a | 28.3 | 28.3 | 27.0 | 19.2 | 22.4 | 1.6 | This work |
| L12,14 | — | 26.4 | 25.5 | 26.9 | 14.6 | 20.7 | 3.4 | 20, 23 |
| DOPC | ||||||||
| H+14 | 2.0 | 24.8 | 4.0 | 14.8 | 11.8 | 1.1 | 26.6 | This work |
| R+14 | — | 26.6 | 5.5 | 16.0 | 13.1 | 1.3 | 28.0 | 25 |
| K+14 | 5.2 | 20.2 | 3.2 | 8.4 | 19.7 | 10.5 | 29.8 | 24 |
| H014 | 6.0 | 19.4 | 1.6 | 15.0 | 10.0 | 1.2 | 22.0 | This work |
| K014 | 8.2 | 19.2 | 1.0 | 11.4 | 11.4 | 1.0 | 18.2 | 24 |
| H012 | 4.0 | 16.2 | 0.4 | 17.4 | 2.8 | 18.4 | 0.6 | This work |
| H+12 | 2.0 | 25 | 56 | 18.5 | 1.2 | — | — | This workc |
| L12,14 | — | 16.6 | 1.7 | 16.7 | 1.5 | 15.4 | 2.6 | 20, 23 |
a Results in DLPC are independent of pH between 2.0 and 8.2.
b Based on repeated measurements from multiple duplicated samples, the uncertainty in measuring |Δνq| is within a range of ± 0.6 kHz.
c Also observed quadrupolar splittings corresponding to the Cα-deuterons of Ala7 and Ala9 with magnitudes of 92 and 102 kHz, respectively, at β = 0° sample orientation.
FIGURE 3.
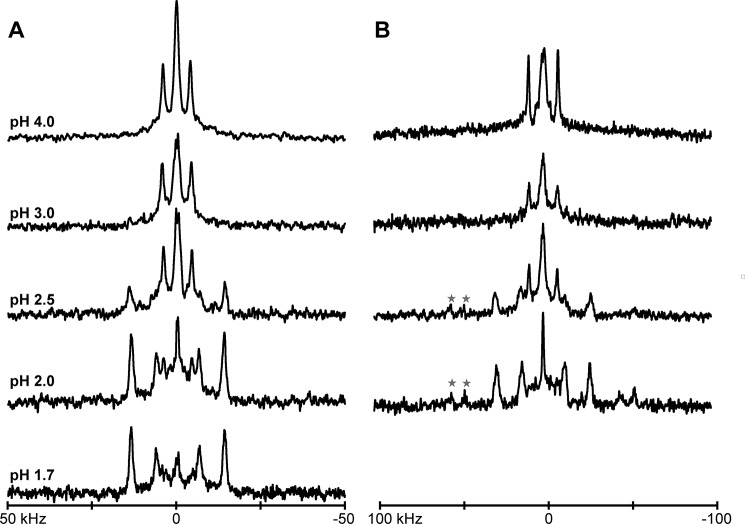
Selected deuterium NMR spectra for two labeled alanines (7 and 9) in GWALP23-H12 in DOPC bilayers, hydrated with 10 mm buffer at the indicated pH, showing (A) β = 90° and (B) β = 0° sample orientations. The difference in spectra from sharp, well resolved signals above pH 4, to spectra with multiple signals below pH 2.5, indicates that the His12 residue is charged only under strongly acidic conditions. Red stars in B indicate signals with large |Δνq|, which likely correspond to backbone Cα-D nuclei of labeled alanine residues. The peptide/lipid ratio is 1/60 at a temperature of 50 °C.
FIGURE 4.
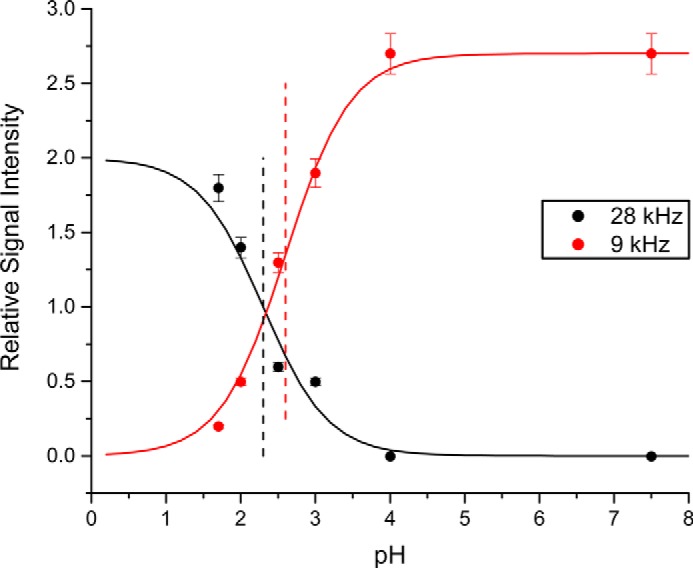
pH dependence of the signal intensities, ±5% estimated error, for the CD3 resonances of Ala7 (red circles, |Δνq| = 8 kHz) and Ala9 (black circles, |Δνq| = 28 kHz) of GWALP23-H12 in DOPC bilayers, at β = 90° sample orientation. The midpoints, shown by the dashed lines, correspond to 50% maximal peak intensity, observed at pH 2.3 for Ala9 or pH 2.6 for Ala7. Error bars indicate means ± 5% estimated error.
For the case of GWALP23-H14, when the pH is lowered from 6 to 2 in DOPC bilayers, the 2H NMR spectral quality improves. Furthermore, changes in |Δνq| for methyl groups of the core alanines, including Ala7 and Ala17, are also observed (Fig. 5). To confirm the spectral assignments of alanine residues when Ala15 and Ala17 are labeled, measurements were also recorded using a peptide with a single 2H-labeled Ala17. The collective changes in the |Δνq| magnitudes reveal a single well defined transmembrane orientation of GWALP23-H+14, with a different helix tilt than that of the neutral -H014 peptide. These results are analogous to those observed for GWALP23-K14 (24). By observing the 2H quadrupolar splittings of selected alanine CD3 groups as a function of pH, we obtained sets of titration curves for the histidine imidazole ring of GWALP23-H14 in DOPC bilayers (Fig. 6). It is important to note that the peptide contains no other ionizable groups. The titration curves for the pH dependence of the quadrupolar splittings of deuterated Ala7 and Ala17 both reveal a pKa of 4.1 for the buried His residue in GWALP23-H14 at the NMR experimental temperature of 50 °C (Fig. 6). Based on the reported temperature dependence of the ionization of the histidine imidazole side chain (27), we deduce pKa values of ∼4.4 at 25 °C and ∼4.3 at 37 °C for His14. Importantly, no titration is observed for the parent peptide with Leu14, as no ionizable groups are present in the peptide. In DOPC bilayer membranes at 37 °C, a pKa of 4.3 for His14 should be compared with values of 6.5 for Lys14 and >9 for Arg14 (24). It therefore appears that the pKa of bilayer-incorporated lysine is lowered (relative to the value in aqueous solution) to a greater extent than that of bilayer-incorporated arginine or histidine residue (see “Discussion”).
FIGURE 5.
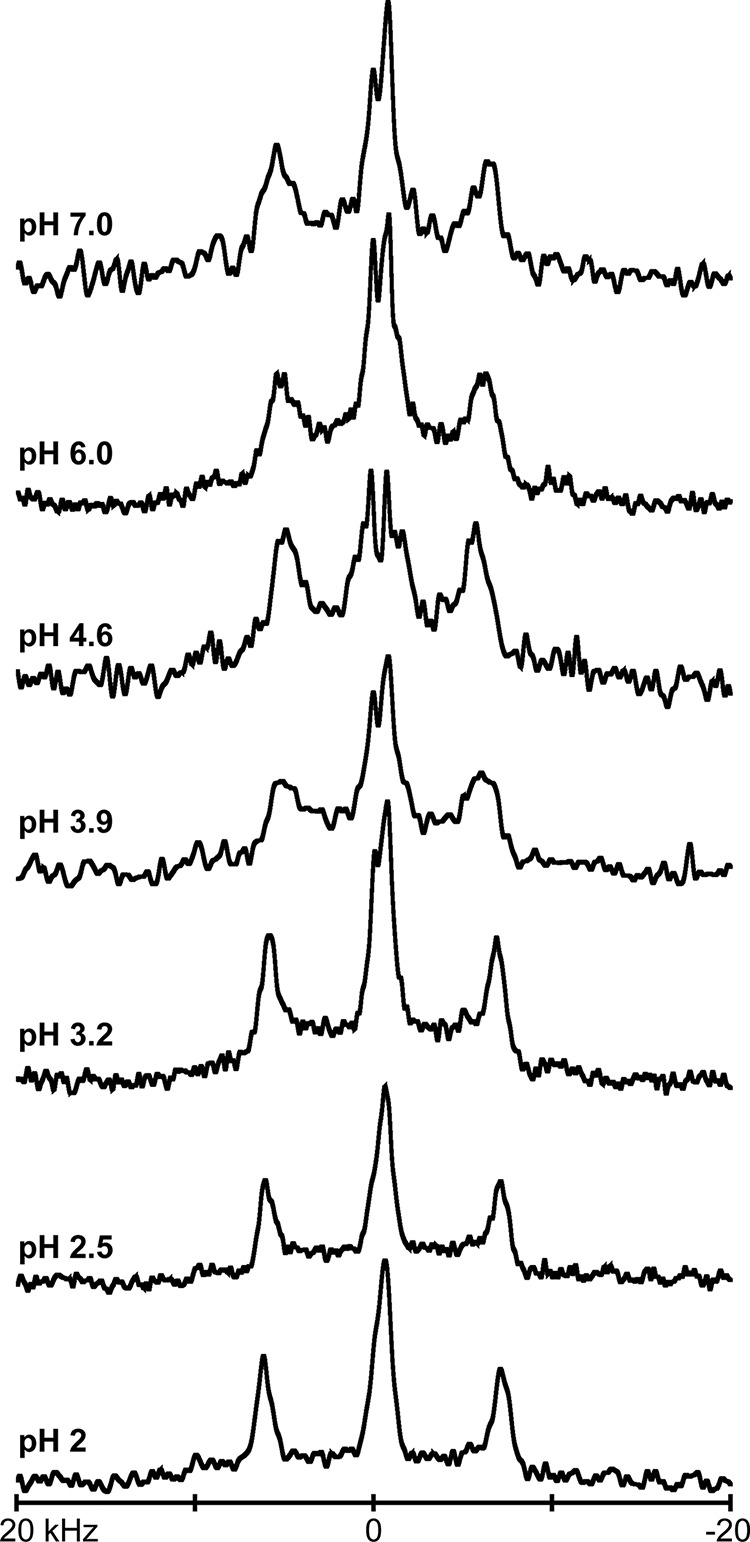
Selected deuterium NMR spectra for labeled alanines, 15 and 17 of GWALP23-H14 in DOPC bilayers, hydrated with 10 mm buffer at the indicated pH, showing β = 90° sample orientation, at a temperature of 50 °C.
FIGURE 6.
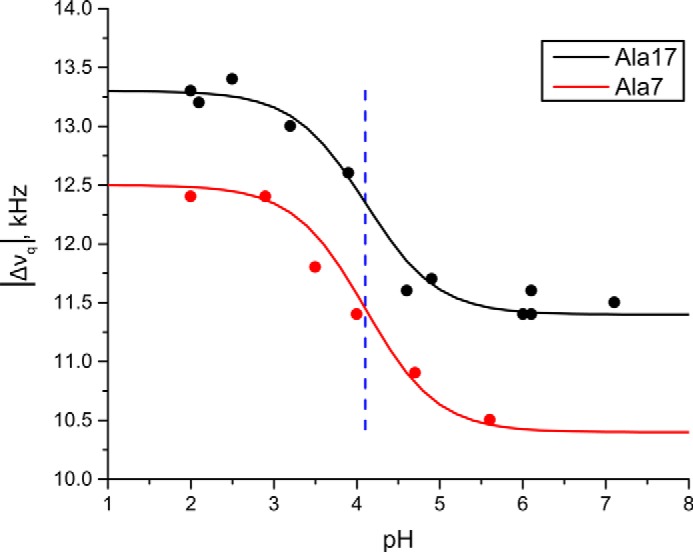
Titration curves for GWALP23-H14 in DOPC bilayers, indicated by the pH dependence of the Δνq values for the CD3 groups of Ala7 and Ala17 in the core helix. Both curves indicate a pKa value of 4.1 (blue dashed line).
Interestingly, similar titration experiments in DLPC bilayers show no changes in the quadrupolar splittings of the His14 or His12 peptides over a pH range of 2.5–12. Apparently, the histidines in DLPC are sufficiently exposed to the aqueous solvent that their titration does not influence the helix tilt of GWALP23-H14 or GWALP23-H12 in the thinner bilayers of DLPC. Nevertheless, the peptide behavior in DLPC is sensitive to the presence of the polar His14. Furthermore, some backbone Cα deuteron resonances are observed for GWALP23-H14 in DLPC (results not shown).
On the basis of the 2H quadrupolar splittings of the six core alanine methyl side chains for each of the transmembrane peptides, the helix tilt and rotation were analyzed using the “geometric analysis of labeled alanines” (GALA) (28, 29) approach (Table 3). The GALA method uses an α-helical geometry and a principal “semi-static” order parameter Szz to give an approximate treatment for describing the relative extent of overall motion of the helix. Based upon the 2H NMR quadrupolar splittings, this method uses the helix tilt (τ), the azimuthal rotation (ρ), and Szz as variables to find the lowest RMSD value. Alternatively, we also treated the helix dynamics using a modified Gaussian analysis to treat the widths of distributions of tilt (στ) and azimuthal rotation (σρ) (22, 23). Importantly, the different methods of analysis lead to the same conclusions about the transmembrane helix tilt of GWALP23-H12 and GWALP23-H14.
TABLE 3.
Semi-static GALA and modified Gaussian analysis of transmembrane orientations of related GWALP23 peptides
The parent GWALP23 sequence is acetyl-GGALWLALALAL12AL14ALALWLAGA-amide. In the noted examples, either residue Leu12 or Leu14 (but not both) was changed to His or Lys or Arg, as indicated, and the other residue remained a leucine.
| Lipid and peptide | pH | GALA fit results |
Modified Gaussian resultsa |
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| τ0 | ρ0 | Szz | RMSD (kHz) | τ0 | ρ0 | σρ | RMSD (kHz) | |||
| DLPC | ||||||||||
| H+/014 | 2.0–8.2b | 26.7° ± 5c | 253° ± 2 | 0.80 | 0.93 | 29° | 253° | 24° | 0.62 | This work |
| R+14 | —d | 26.7° | 260° | 0.83 | 1.58 | 26° | 260° | 0° | 1.65 | 26 |
| H+/012 | 2.0–8.2b | 23.3° ± 3 | 308° ± 2 | 0.70 | 0.66 | 18° | 305° | 15° | 1.34 | This work |
| L12,14 | — | 21.0° | 305° | 0.71 | 0.7 | 23° | 304° | 33° | 0.7 | 23 |
| DOPC | ||||||||||
| H+14 | 2.0 | 14.0° ± 3 | 246° ± 1 | 0.90 | 1.03 | 19° | 247° | 24° | 1.28 | This work |
| R+14 | —d | 15.0° | 247° | 0.93 | 0.89 | 19° | 246° | 17° | 1.33 | 25 |
| K+14 | 5.2 | 15.3° | 228° | 0.88 | 1.20 | 17° | 227° | 21° | 1.28 | 24 |
| H014 | 6.0 | 10.3° ± 1 | 248° ± 2 | 0.89 | 1.36 | 11° | 249° | 18° | 0.67 | This work |
| K014 | 8.2 | 9.0° | 244° | 0.86 | 0.31 | 10° | 243° | 18° | 0.36 | 24 |
| H012 | 4.0 | 6.0° ± 1 | 338° ± 3 | 0.93 | 0.74 | 10° | 332° | 48° | 0.89 | This work |
| H+12e | 2.0 | 81.0° ± 1 | 296° ± 1 | 0.85 | 0.70 | 81° | 297° | 5° | 0.88 | This worke |
| L12,14 | — | 6.0° | 323° | 0.87 | 0.6 | 9° | 321° | 48° | 0.7 | 23 |
a The modified Gaussian analysis followed Sparks et al. (23), with στ assigned a fixed finite value of 10°. στ and σρ are related to the widths of distributions of helix orientations, effectively indicating the uncertainties in τ0 and ρ0.
b In DLPC, the results with His12 and His14 do not depend on pH.
c Because τ0 and ρ0 are derived values from the GALA fits, we report ± numbers that correspond to those ranges where RMSD < 1.5 kHz.
d The results with -R14+ do not depend on pH (24, 25). For the L12,14 peptide, the pH was not buffered.
e Analysis of -H+12 orientation is based on quadrupolar splittings of labeled Ala7 and Ala9 Cα−deuterons along with labeled CD3 groups of Ala7, Ala9, Ala11, and Ala13. About 70% of the -H+12 population exists in this surface-bound state and the other portion remains transmembrane with multi-state behavior.
From the semi-static method, we observe distinctly different orientations in DLPC bilayers for the GWALP23-H12 and GWALP23-H14 peptides (Fig. 7), reflecting different ionization states of the His side chain. The tilt and rotation values of GWALP23-H12 are nearly identical to those of the host peptide, GWALP23 (Table 3; see also Ref. 20). Remarkably, there is no difference in peptide orientation when neutral (yet polar) histidine is substituted for neutral leucine at position 12, directly between the two interfacial Trp residues, on the same face of the helix. Interestingly, this is not the case for His at position 14, located on the opposite face of the helix. GALA analysis of GWALP23-H14 in DLPC bilayers indicates that the orientation of the His14 peptide differs from the host peptide, GWALP23, with Leu14, with a change of tilt Δτ of about 5° and a change of rotation Δρ of about 50° (Table 3). As can be seen in Fig. 7, the orientation for the helix with -H+14 is nearly identical to that of the -R+14 or -K+14 peptide (24).
FIGURE 7.
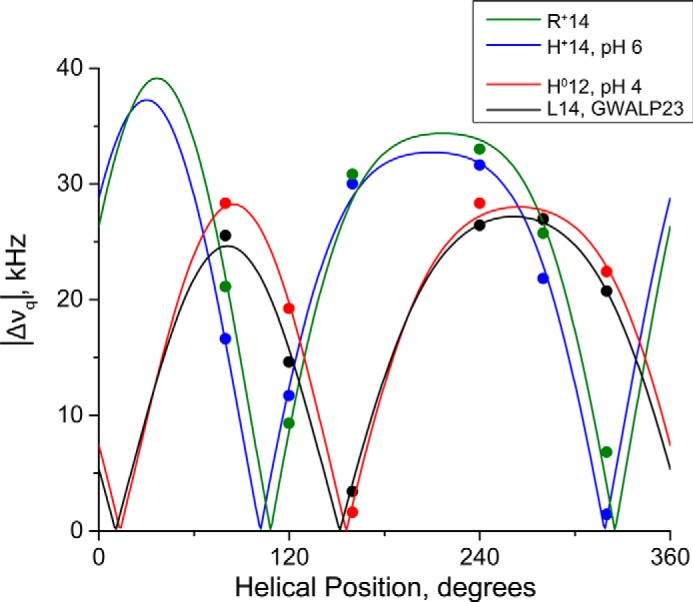
GALA quadrupolar wave plots of tilted transmembrane peptides in DLPC bilayers. GWALP23-H12 (red; tilt τ = 23°, rotation ρ = 308°, pH 4) is similar to GWALP23 (black; tilt τ = 21°, rotation ρ = 305°, irrespective of pH), indicating that the buried His12 residue is uncharged. GWALP23-H14 (blue; tilt τ = 27°, rotation ρ = 254°, pH 5.9) is similar to charged GWALP23-R14 (green; tilt τ = 27°, rotation ρ = 260°) (25), suggesting that the His14 residue is charged at pH 5.9.
Similar to the case of DLPC, in DOPC, the orientation of GWALP23-H012 once again is nearly identical to that of GWALP23 itself, as seen in Fig. 8. Interestingly, in addition to the multiple weak resonances, an additional set of signals is observed at low pH. The new peaks at low pH are due to changes in the population of helix orientations, not to changes in the lipid bilayer. As confirmation, no changes are observed in the NMR spectra for the peptide or the lipid for GWALP23 in DOPC at pH 2.0 (data not shown). GALA analysis of GWALP23-H12 at pH 2 reveals a new orientation, in addition to the multi-state behavior, for the charged GWALP23-H+12. This new orientation likely corresponds to a surface-bound state in which the charged -H+12 peptide comes out of the membrane, as the new tilt of the core helix is 81°. Nevertheless, in DOPC, -H014 serves to confer a different tilt from that of either the -R+14 peptide or the GWALP23 parent, as has been observed similarly when -K014 is present (24). GALA analysis of the -H+14 peptide in DOPC bilayers at pH 2 shows an increased tilt for the charged -H+14 peptide, when compared with the neutral -H014 peptide. This new orientation is similar to that observed for both the -R+14 and -K+14 peptides. By means of quadrupolar wave plots, Fig. 8 summarizes three different preferred tilted states for transmembrane helices that differ only with respect to whether the position 14 side chain is charged (cationic), neutral polar, or neutral hydrophobic.
FIGURE 8.
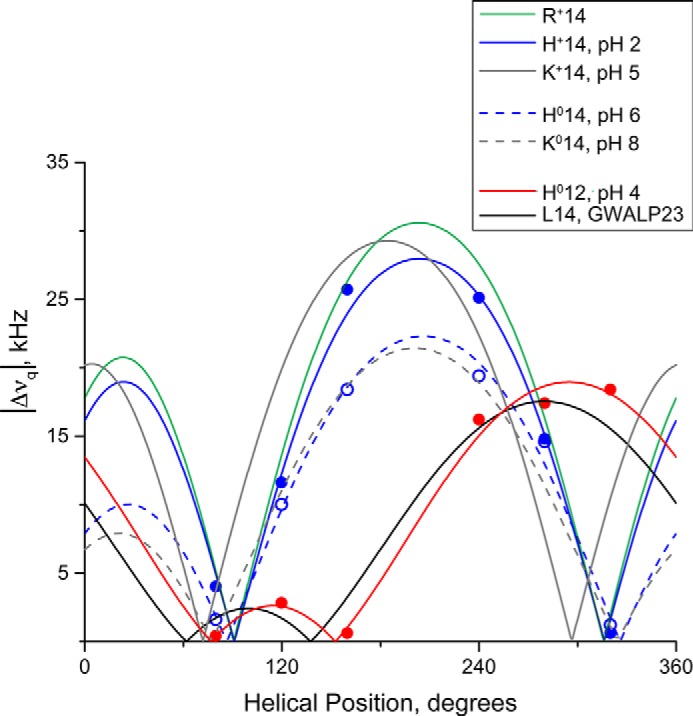
Quadrupolar wave analysis of tilted transmembrane peptides in DOPC bilayers. GWALP23-H14 (blue dashed line; tilt τ = 10°, rotation ρ = 250°, pH 5.9) is similar to neutral Y5GWALP23-K14 (gray dashed line; tilt τ = 9°, rotation ρ = 244°, pH 8.2) (24) indicating that the His14 residue is uncharged. At pH 2.0, the His residue of GWALP23-H14 (blue; tilt τ = 15°, rotation ρ = 247°) has a similar orientation to charged GWALP23-R14 (green; tilt τ = 15°, rotation ρ = 247°) (25) and charged Y5GWALP23-K14 (gray; tilt τ = 15°, rotation ρ = 228°, pH 5.2) (24), indicating that the His residue is charged. Note the similar rotation for all peptides with either Arg, His, or Lys at position 14, regardless of charge, indicating a change in rotation of ∼70° when compared with GWALP23 (black; tilt τ = 6°, rotation ρ = 323°) in response to the mutation of Leu14 to a polar residue. Changing the charge state of residue 14 is reflected by a change in peptide tilt in DOPC.
The accumulated results (Table 3) indicate that the transmembrane orientation of the core helix of GWALP23 depends upon not only the bilayer thickness, but also the identities and charge states of the side chains of residues 12 and 14. Importantly, the helix orientations do not depend upon the method for estimating the helix dynamics, whether semi-static or modified Gaussian. Indeed, the deduced values for the helix tilt (τ0) and azimuthal rotation (ρ0) agree for the two methods of analysis (Table 3). Other than the case of GWALP23-H012 in DOPC, for which the polar imidazole ring is located between two flanking Trp indole rings, the values of σρ from the modified Gaussian analysis (Table 3) are uniformly low, indicating only low to moderate dynamic averaging for each of the target transmembrane helices considered here. The somewhat higher value of σρ for GWALP23-H012 in thicker DOPC bilayers is not surprising, given that the protonation to yield GWALP23-H+12 leads to a major population exiting to the membrane surface along with multi-state behavior for the minor population of helices that remain in DOPC at low pH (see also Fig. 3).
The accumulated results furthermore illustrate several core principles. A neutral residue at position 12, between the flanking tryptophan indole rings 5 and 19 of GWALP23, whether leucine or neutral histidine or lysine, confers the same global orientation for the tilted transmembrane core helix. By contrast, a charged residue at position 12, whether arginine or charged histidine or lysine, confers multi-state behavior to the helix, in which a significant fraction of the population exits the membrane (25). Notably, residue 14 is located on the opposite face of the core helix from the flanking tryptophans 5 and 19. When the side chain of position 14 is altered, single-state behavior is always observed, but the favored state for the core helix takes on one of three choices (Fig. 8), depending upon whether the side chain of residue 14 is charged or neutral/polar or neutral/nonpolar. For the position 14 side chains as well as the position 12 side chains, the results are consistent among lysine, histidine, and arginine.
Discussion
We have examined the ionization properties of a lipid-exposed histidine residue incorporated into a transmembrane helix at two different positions, as well as the influence of histidine residue titration on the peptide orientations in the hydrophobic environment of the lipid bilayer. The results establish important limits for understanding membrane protein functions that involve proton-transfer reactions or other proton-mediated events. We will discuss the dependence of transmembrane helix orientations on His residue location and protonation state, the titration behavior of His12 and His14 in bilayer-incorporated GWALP23, the transmembrane to surface transition when His12 is protonated, and comparisons with the titration behavior of other histidines in soluble proteins or membrane environments. Finally, we will address the current status for understanding and comparing experimental or predicted differences in titration behavior when His or Lys as opposed to Arg is incorporated into a lipid bilayer membrane.
GWALP23-H012, with a neutral imidazole ring, is shown to have a transmembrane orientation identical to that of unmodified GWALP23 in both DLPC and DOPC bilayers. Notably, the helix orientation of GWALP23 is determined by the tryptophan residues, along with unwinding of the helix terminals (30), and the presence of neutral, polar His instead of Leu at position 12 does not alter this orientation. By contrast, GWALP23-H014 adopts a tilted transmembrane orientation distinct from that of the host peptide, similar to that observed (24) for Y5GWALP23-K014. Interestingly, the presence of neutral, polar His or Lys at position 14 results in a 40°-50° change in peptide rotation about the helix axis, suggesting that a polar residue instead of Leu at position 14 modulates the orientation of the peptide, along with the interfacial Trp residues and possibly the terminal fraying. Although there is little change in the overall helicity when Leu14 is substituted by -H014 or -K014, as confirmed by circular dichroism spectra (see also Ref. 23) and by the GALA curves (Figs. 7 and 8), admittedly there could be minor changes in the local helicity or terminal fraying that accompany the non-conservative replacement of Leu with His or Lys.
In DOPC bilayers, we observe major changes in peptide behavior as a function of pH. In the case of His12, under strongly acidic pH conditions, the His residue becomes charged and a significant population of GWALP23-H+12 abandons the transmembrane orientation and exits to the membrane surface. Conversely, protonation of His at position 14 results in a small increase in the peptide tilt and additional azimuthal rotation about the helix axis, with the peptide remaining in a transmembrane orientation. This new orientation most likely provides greater access to the interfacial region of the membrane for the charged His imidazolium ring, which allows the peptide helix to remain in a stable transmembrane orientation. The situation is similar for GWALP23-R+14 and GWALP23-K+14 (24, 25). Interestingly, the deuterium NMR spectra for GWALP23-H014 are slightly broadened when compared with those of GWALP23-H+14. The spectral differences suggest differences in peptide dynamics between the neutral and charged forms of the -H14 peptide. That is, the signal broadening observed for the GWALP23-H014 suggests that the neutral form may slow the motion of the peptide helix.
Our results are consistent with a low pKa for His12. Because of the multiple states associated with the -H+12 peptide, in equilibrium with a single transmembrane state for GWALP23-H012, it is not possible to directly assign a pKa value for bilayer-incorporated His12. Despite these limitations, one observes that the major tilted transmembrane orientation of GWALP23-H012 is 50% populated at pH 2.6, and the perpendicular interfacial orientation of GWALP23-H+12 is 50% populated at pH 2.3 (Fig. 4). Although not a two-state equilibrium, we suggest that these midpoints constitute upper and lower limits of the pKa for His12 in DOPC bilayers.
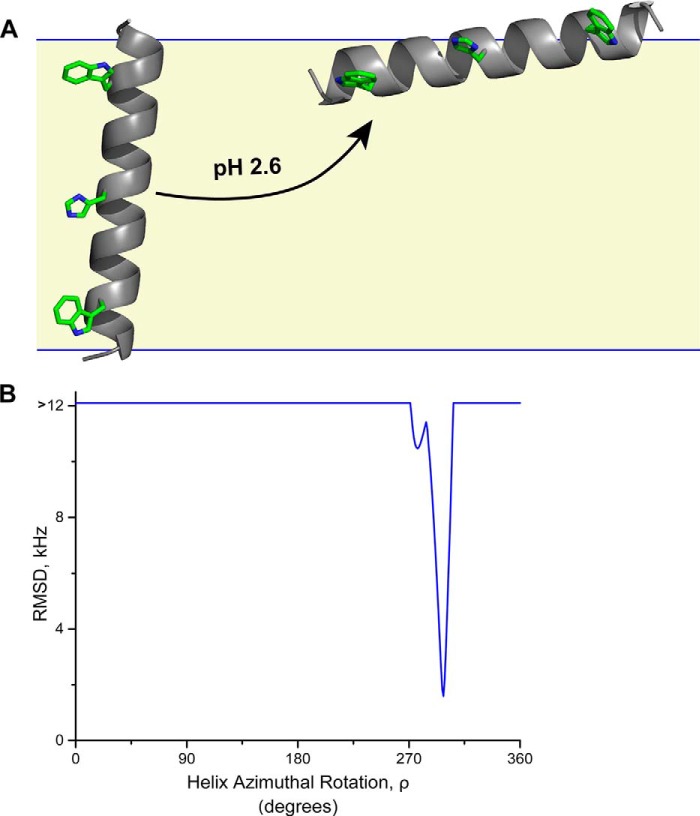
What is the surface orientation of GWALP23-H+12 at low pH? We have addressed this question using a semi-static GALA analysis with principal Szz order parameter, as well as a modified Gaussian analysis, employing Cα-deuteron quadrupolar splittings from Ala7 and Ala9 in addition to the available alanine methyl 2H quadrupolar splittings. Both methods yield the surface orientation depicted in Fig. 9, with tilt τ = 810 and azimuthal rotation ρ = 2960. (The extent of dynamic averaging is low, as σρ is 5° when στ is fixed at 5° in the modified Gaussian; or Szz is 0.85 in the semi-static treatment.) The azimuthal rotation, which fits the surface orientation of GWALP23-H+12 (Fig. 9), places the His and Trp side chains, on the same face of the helix, approximately parallel to the membrane surface, such that these side chains point neither into nor out of the membrane. The rotational preference is perhaps a compromise among the aromatic Trp and charged His side chains. Indeed, the rotational preference contrasts with that observed for several amphipathic surface-active antimicrobial peptides. For example, the amphipathic PGLa peptide binds to a bilayer surface so as to orient four lysine residues away from the bilayer, toward the aqueous solution (31). To explore the issue further, we searched for other possible minima in the rotation space for the surface-bound GWALP23-H+12. In this process, we found no solutions other than the helix orientation shown in Fig. 9A. For comparison, the dependence of RMSD of the fit to the 2H NMR data upon azimuthal rotation of the GWALP23-H+12 helix on the surface of a DOPC bilayer is illustrated in Fig. 9B.
FIGURE 9.
GWALP23-H+ 12 exits the bilayer. A, model to illustrate the transition of GWALP23-H+12 from its tilted transmembrane orientation (above pH 3) to a primary orientation at the surface of DOPC bilayer membranes (as the pH is lowered below 3). Carbons are green and nitrogens are blue. The midpoint for this transition is between pH 2.3 and 2.6 (see “Results”). The observed azimuthal rotation from 2H NMR experiments is illustrated for each of the helix orientations. B, dependence of RMSD of the fit to the 2H NMR data upon azimuthal rotation of the GWALP23-H+12 helix on the surface of a DOPC bilayer at low pH. The best fit occurs when the azimuthal rotation ρ is 296°, and the corresponding surface-oriented helix in A has a tilt τ of 81°.
In thinner DLPC bilayers, we observe different behavior in response to ionization of His12 in GWALP23. Interestingly, even at pH 2.1, the majority of the His12 population remains in the same tilted transmembrane orientation as observed at pH 8. We do, however, observe some signal broadening and the presence of weak multiple signals at pH 2.1. This could indicate the presence of some multi-state behavior for the -H+12 peptide; however, no signals corresponding to a perpendicular interfacial orientation are observed. It is possible that the pKa of His12 in DLPC bilayers is much lower than 2.1; however, another likely scenario is that water is able more easily to penetrate the thinner DLPC membrane and thereby satisfy the charge of His12, so that no major change in peptide helix orientation is observed when the imidazole ring titrates.
In soluble proteins, the depth of burial of a His imidazole ring and the polarity of the microenvironment influence the His side chain pKa (32). That is, buried histidine residues in soluble proteins are found in mixed polar/apolar environments, and therefore, the pKa values can vary tremendously depending on a range of factors. For example, His149 is completely buried within the hydrophobic core of xylanase and has a pKa < 2.3 (33), whereas His72 of bovine protein tyrosine phosphatase has a pKa of 9.2, due to electrostatic interactions between the buried imidazole ring and nearby negatively charged side chains (34). In staphylococcal nuclease, the pKa of His121 has been extensively studied. The observed pKa for this partially buried residue is 5.3; however, when nearby glutamic acid residue Glu75 is mutated to Ala, resulting in a more hydrophobic microenvironment for the His imidazole ring, the pKa of His121 drops to 4.0 (35).
A large range of pKa values is also observed for histidine residues of transmembrane domains. His37 is responsible for pH activation and selectivity of the tetrameric M2 ion channel of influenza A protein. A titration curve generated from 15N spectra of His37 yielded doubly degenerate pKa values of 7.6 and 4.5 (36). Single His residues engineered along the helical lining of the nicotinic acetylcholine receptor transmembrane pore have pKa values higher than 6.4, illustrating their accessibility to the aqueous phase (37). In a similar fashion, His residues in model helical peptides LAH2 and LAH4 incorporated in dodecylphosphocholine micelles have pKa values in a range from 4.9 to 6.6 (38), also suggesting aqueous access for the His side chains in the small micelles. These results are seemingly analogous to our observations for His12 of GWALP23 in the thin bilayers of DLPC. The extent of aqueous access, therefore, is likely to dictate the histidine pKa value in the membrane environment, which, in turn, will influence the pH dependence of membrane protein folding and function.
As an example of the folding/function paradigm, the human chloride intracellular channel 1 converts between cytosolic and membrane forms with changes in pH, with His74 and His185 providing major contributions to the pH-dependent conformational stability (39). Our observation of the helix repositioning as a function of the histidine protonation state (Fig. 9) is therefore emblematic of the potential for large-scale conformational rearrangements of membrane proteins in response to pH or other signal effectors. A further example is provided by residues 1–19 of the islet amyloid polypeptide (IAPP1–19), for which the rat and human sequences are identical, except that residue Arg18 in rat is replaced by His18 in human (40). At pH 7.3, the more toxic human IAPP1–19 with His18 (presumably neutral) inserts more deeply into lipid micelles, whereas the less toxic rat IAPP1–19 with Arg18 is surface-bound (40). At lower pH, the His18 and Arg18 peptides behave similarly (40). Similar to our observations with residue 12 in GWALP23 (Fig. 9), the charge status of residue 18 in IAPP1–19 seems to dictate the nature of the peptide/lipid binding and interaction. These observations illustrate the vital importance of having detailed direct comparisons among His, Lys, and Arg to define their influence on membrane protein structure and function.
The pH-dependent behavior that we observe for His12 and His14 in DOPC bilayers can be compared with results for buried Lys and Arg in analogous positions (25). For arginine, we are unable to observe loss of the positive charge due to deprotonation, up to at least pH 9 (24), and perhaps higher (41). Instead, the charged guanidinium group would rather exit the DOPC bilayer (25) than release a proton. Numerous molecular dynamics simulations are in agreement with this observation for arginine (18, 42–45). Arginine also remains cationic when buried in the hydrophobic interior of a folded protein (46). We note furthermore that the experimental value for the aqueous pKa of the Arg guanidinium side chain recently has been revised upward to 13.8 (47).
As noted, comparisons among histidine, lysine, and arginine in DOPC bilayer membranes are of interest. The lysine residue Lys14 has a pKa of 6.5 in DOPC (24), about 4 pH units below that of Lys in aqueous buffer. Residue Lys12 in the GWALP23 framework also experiences a pKa shift of at least 4 units, a lower limit for the extent of the shift (24). Although buried slightly more deeply than Lys14, and at least partially occluded from solvent access by Trp indole rings (26), one is unable to distinguish whether the pKa of Lys12 is actually lower than that of Lys14 (24). Nevertheless, the present results resolutely show that the pKa values for His12 and His14 on the GWALP23 framework are different from one another in DOPC bilayers. We observe at 50 °C a pKa value of 4.1 for His14 (Fig. 6; corresponding to 4.4 at 25 °C) and an even lower pKa between 2.3 and 2.6 for His12 (Fig. 4). The pKa shift is therefore position-dependent for the histidine side chain on the GWALP23 helix in DOPC. Notably, moreover, the extent of the pKa shift, relative to the aqueous value, follows a trend from arginine to histidine to lysine. For transmembrane GWALP23 in DOPC, a pKa shift is undetected for Arg14, and is about 2 pH units for His14 and 4 pH units for Lys14.
We likewise compare our results with recent simulations utilizing a constant pH molecular dynamics technique (48). The agreement between experiments and the computational predictions is quite good, as Panahi and Brooks (48) have calculated pKa values of 4.5 ± 0.3 for His14 and 4.0 ± 0.1 for His12 on the GWALP23 helix in DOPC bilayers. As a caveat, one notes that GWALP23-H+12 moves to primarily a surface orientation at low pH (Fig. 9), although the time scale for the exit of the helix from the bilayer should be slow relative to the time scale of the calculations (48).
Concluding Remarks
Histidine residues are significant for not only soluble protein function but also membrane protein function. Our results fill gaps in the knowledge of His imidazole ionization properties in lipid bilayer membranes. When attached to the transmembrane GWALP23 helix and buried within a bilayer membrane of DOPC, the His imidazole side chain displays position-dependent titration behavior. When located in the central position of the peptide sequence, His12 displays a pKa of about 2.3–2.6, perhaps influenced also by a tryptophan “cage” (26). The somewhat off-center His14 displays a pKa of about 4.1, in excellent agreement with computational predictions (48). The transmembrane helix responds to the histidine titration by changing its tilt when His14 is protonated or by exiting the bilayer when His12 is protonated. By contrast, in the thinner DLPC bilayer membranes, the more highly tilted transmembrane helices show little response to changes in pH.
The present results allow us to compare the ionization behavior of Arg, Lys, and His side chains in DOPC bilayer membranes. The summary findings are that the Arg guanidinium seeks hydration or exits the bilayer but does not release its proton, the Lys ammonium pKa is at least 4 pH units lower than its aqueous value, and the His imidazolium pKa is about 2–4 pH units lower, depending on its location in the bilayer. These results are of interest and importance for experiments as well as molecular dynamics simulations that address the properties of membrane proteins.
Experimental Procedures
Peptides were synthesized using solid-phase methods on a 0.1-mmol scale using an Applied Biosystems 433A synthesizer from Life Technologies. Protected amino acids were purchased from Novabiochem (San Diego, CA). Histidine and tryptophan side chains were protected with trityl and t-butoxycarbonyl protecting groups, respectively. Peptide cleavage from Rink amide resin was accomplished by treatment at 22 °C with a solution of trifluoroacetic acid/triisopropyl silane/water/phenol (85/5/5/5, v/v/v/w) over a 2-h period. The cleavage mixture was then filtered to separate the free peptide from the resin support. The crude peptide was precipitated using a 50/50 mixture of methyl-t-butyl ether and hexane and lyophilized from a 50/50 mixture of acetonitrile and water. Peptides were purified by reversed-phase HPLC on an octyl-silica column (Zorbax Rx-C8, 9.4 × 250 mm, 5-μm particle size; Agilent Technologies, Santa Clara, CA) using a gradient of 92–96% methanol, with 0.1% trifluoroacetic acid (v/v), over 24 min. Analytical HPLC and MALDI-TOF analyses were used to verify the peptide purity and identity.
Mechanically aligned samples (1:60, peptide:lipid) for solid-state 2H NMR experiments were prepared using DOPC or DLPC (Avanti Polar Lipids, Alabaster, AL) and hydrated (45%, w/w) with 10 mm glycine, acetate, or citrate buffer in deuterium-depleted water at specified pH values between pH 2 and pH 8. Bilayer alignment of each sample was confirmed by 31P NMR spectroscopy using a Bruker Avance 300 spectrometer (Billerica, MA). Deuterium NMR spectra were recorded using a Bruker Avance 300 spectrometer at 50 °C, at β = 90° or β = 0° macroscopic sample orientations, using a quadrupolar echo pulse sequence (49) with full phase cycling, a 90-ms recycle delay, 3.2-μs pulse length, and 115-μs echo delay. Between 0.7 and 1 million free induction decays were collected for each 2H experiment. Fourier transformation was accomplished using an exponential weighting function with 100-Hz line broadening.
The sealed, hydrated samples with ester lipids at pH 2.0 were found to be stable for several days. Samples above pH 4.0 are stable for several months. The intrinsic pKa of the DOPC lipid phosphate group has been measured to be about 0.8–1.0 (50). Furthermore, our control experiments with GWALP23 in ester lipids showed no changes in the 2H or 31P NMR spectra at pH 2.0 when compared with pH 6.0, indicating that the changes we observe at pH 2 for the -H12 and -H14 peptides are solely a result of the changes in ionization state of the His residue.
Helix orientations were analyzed by means of the semi-static GALA method, using the average tilt τ of the helix axis, the azimuthal rotation ρ, and the principal order parameter Szz as variables (28, 29). Additionally, we employed a modified Gaussian approach based on τ, ρ, a distribution width σρ, and a fixed στ, as described previously (23).
Author Contributions
R. E. K. conceived and coordinated the study. A. N. M. and D. V. G. designed and performed the experiments. The manuscript was written through contributions of all authors. All authors have reviewed results and given approval to the final version of the manuscript.
Acknowledgments
The peptide, NMR, and mass spectrometry facilities were supported in part by National Institutes of Health Grants GM103429 and GM103450. We thank Vitaly Vostrikov for software for semi-static GALA and modified Gaussian methods for analysis of helix orientations and dynamics.
This work was supported in part by National Science Foundation Division of Molecular and Cellular Biosciences (NSF MCB) Grant 1327611, and by the Arkansas Biosciences Institute. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DOPC
- dioleoylphosphatidylcholine
- DLPC
- dilauroylphosphatidylcholine
- GWALP23
- acetyl-GGALW(LA)6LWLAGA-amide
- GALA
- geometric analysis of labeled alanines
- RMSD
- root mean square deviation.
References
- 1. Schoellmann G., and Shaw E. (1963) Direct evidence for presence of histidine in active center of chymotrypsin. Biochemistry 2, 252–255 [DOI] [PubMed] [Google Scholar]
- 2. Bender M. L., and Kezdy F. J. (1964) Current status of the α-chymotrypsin mechanism. J. Am. Chem. Soc. 86, 3704–3714 [Google Scholar]
- 3. Blow D. M., Birktoft J. J., and Hartley B. S. (1969) Role of a buried acid group in mechanism of action of chymotrypsin. Nature 221, 337–340 [DOI] [PubMed] [Google Scholar]
- 4. Paddock M. L., Sagle L., Tehrani A., Beatty J. T., Feher G., and Okamura M. Y. (2003) Mechanism of proton transfer inhibition by Cd2+ binding to bacterial reaction centers: determination of the pKa of functionally important histidine residues. Biochemistry 42, 9626–9632 [DOI] [PubMed] [Google Scholar]
- 5. Kaila V. R. I., Sharma V., and Wikström M. (2011) The identity of the transient proton loading site of the proton-pumping mechanism of cytochrome c oxidase. Biochim. Biophys. Acta 1807, 80–84 [DOI] [PubMed] [Google Scholar]
- 6. Lu J., and Gunner M. R. (2014) Characterizing the proton loading site in cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 111, 12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang C., Lamb R. A., and Pinto L. H. (1995) Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys. J. 69, 1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okada A., Miura T., and Takeuchi H. (2001) Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from influenza A virus. Biochemistry 40, 6053–6060 [DOI] [PubMed] [Google Scholar]
- 9. Hu J., Fu R., Nishimura K., Zhang L., Zhou H. X., Busath D. D., Vijayvergiya V., and Cross T. A. (2006) Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. U.S.A. 103, 6865–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu F., Schmidt-Rohr K., and Hong M. (2012) NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J. Am. Chem. Soc. 134, 3703–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rehwald M., Neuschäfer-Rube F., de Vries C., and Püschel G. P. (1999) Possible role for ligand binding of histidine 81 in the second transmembrane domain of the rat prostaglandin F2α receptor. FEBS Lett. 443, 357–362 [DOI] [PubMed] [Google Scholar]
- 12. Larson C. A., Adams P. L., Blair B. G., Safaei R., and Howell S. B. (2010) The role of the methionines and histidines in the transmembrane domain of mammalian copper transporter 1 in the cellular accumulation of cisplatin. Mol. Pharmacol. 78, 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paukert M., Chen X., Polleichtner G., Schindelin H., and Gründer S. (2008) Candidate amino acids involved in H+ gating of acid-sensing ion channel 1a. J. Biol. Chem. 283, 572–581 [DOI] [PubMed] [Google Scholar]
- 14. Chaloupka R., Courville P., Veyrier F., Knudsen B., Tompkins T. A., and Cellier M. F. M. (2005) Identification of functional amino acids in the Nramp family by a combination of evolutionary analysis and biophysical studies of metal and proton cotransport in vivo. Biochemistry 44, 726–733 [DOI] [PubMed] [Google Scholar]
- 15. Illergård K., Kauko A., and Elofsson A. (2011) Why are polar residues within the membrane core evolutionarily conserved? Proteins 79, 79–91 [DOI] [PubMed] [Google Scholar]
- 16. Rodnin M. V., Kyrychenko A., Kienker P., Sharma O., Posokhov Y. O., Collier R. J., Finkelstein A., and Ladokhin A. S. (2010) Conformational switching of the diphtheria toxin T domain. J. Mol. Biol. 402, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Venkataraman P., Lamb R. A., and Pinto L. H. (2005) Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J. Biol. Chem. 280, 21463–21472 [DOI] [PubMed] [Google Scholar]
- 18. MacCallum J. L., Bennett W. F. D., and Tieleman D. P. (2008) Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 94, 3393–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vostrikov V. V., Grant C. V., Daily A. E., Opella S. J., and Koeppe R. E. 2nd (2008) Comparison of “Polarization inversion with spin exchange at magic angle” and “geometric analysis of labeled alanines” methods for transmembrane helix alignment. J. Am. Chem. Soc. 130, 12584–12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vostrikov V. V., Daily A. E., Greathouse D. V., and Koeppe R. E. 2nd (2010) Charged or aromatic anchor residue dependence of transmembrane peptide tilt. J. Biol. Chem. 285, 31723–31730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vostrikov V. V., Grant C. V., Opella S. J., and Koeppe R. E. 2nd (2011) On the combined analysis of 2H and 15N/1H solid-state NMR data for determination of transmembrane peptide orientation and dynamics. Biophys. J. 101, 2939–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strandberg E., Esteban-Martín S., Ulrich A. S., and Salgado J. (2012) Hydrophobic mismatch of mobile transmembrane helices: merging theory and experiments. Biochim. Biophys. Acta 1818, 1242–1249 [DOI] [PubMed] [Google Scholar]
- 23. Sparks K. A., Gleason N. J., Gist R., Langston R., Greathouse D. V., and Koeppe R. E. 2nd (2014) Comparisons of interfacial Phe, Tyr, and Trp residues as determinants of orientation and dynamics for GWALP transmembrane peptides. Biochemistry 53, 3637–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gleason N. J., Vostrikov V. V., Greathouse D. V., and Koeppe R. E. 2nd (2013) Buried lysine, but not arginine, titrates and alters transmembrane helix tilt. Proc. Natl. Acad. Sci. U.S.A. 110, 1692–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vostrikov V. V., Hall B. A., Greathouse D. V., Koeppe R. E. 2nd, and Sansom M. S. P. (2010) Changes in transmembrane helix alignment by arginine residues revealed by solid-state NMR experiments and coarse-grained MD simulations. J. Am. Chem. Soc. 132, 5803–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vostrikov V. V., Hall B. A., Sansom M. S. P., and Koeppe R. E. 2nd (2012) Accommodation of a central arginine in a transmembrane peptide by changing the placement of anchor residues. J. Phys. Chem. B 116, 12980–12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nagai H., Kuwabara K., and Carta G. (2008) Temperature dependence of the dissociation constants of several amino acids. J. Chem. Eng. Data 53, 619–627 [Google Scholar]
- 28. van der Wel P. C. A., Strandberg E., Killian J. A., and Koeppe R. E. 2nd (2002) Geometry and intrinsic tilt of a tryptophan-anchored transmembrane α-helix determined by 2H NMR. Biophys. J. 83, 1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strandberg E., Ozdirekcan S., Rijkers D. T. S., van der Wel P. C. A., Koeppe R. E. 2nd, Liskamp R. M. J., and Killian J. A. (2004) Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys. J. 86, 3709–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mortazavi A., Rajagopalan V., Sparks K. A., Greathouse D. V., and Koeppe R. E. 2nd (2016) Juxta-terminal helix unwinding as a stabilizing factor to modulate the dynamics of transmembrane helices. ChemBioChem 17, 462–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reißer S., Strandberg E., Steinbrecher T., and Ulrich A. S. (2014) 3D hydrophobic moment vectors as a tool to characterize the surface polarity of amphiphilic peptides. Biophys. J. 106, 2385–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgcomb S. P., and Murphy K. P. (2002) Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins 49, 1–6 [DOI] [PubMed] [Google Scholar]
- 33. Plesniak L. A., Connelly G. P., Wakarchuk W. W., and McIntosh L. P. (1996) Characterization of a buried neutral histidine residue in Bacillus circulans xylanase: NMR assignments, pH titration, and hydrogen exchange. Protein Sci. 5, 2319–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tishmack P. A., Bashford D., Harms E., and Van Etten R. L. (1997) Use of 1H NMR spectroscopy and computer simulations to analyze histidine pKa changes in a protein tyrosine phosphatase: experimental and theoretical determination of electrostatic properties in a small protein. Biochemistry 36, 11984–11994 [DOI] [PubMed] [Google Scholar]
- 35. Baran K. L., Chimenti M. S., Schlessman J. L., Fitch C. A., Herbst K. J., and Garcia-Moreno B. E. (2008) Electrostatic effects in a network of polar and ionizable groups in staphylococcal nuclease. J. Mol. Biol. 379, 1045–1062 [DOI] [PubMed] [Google Scholar]
- 36. Colvin M. T., Andreas L. B., Chou J. J., and Griffin R. G. (2014) Proton association constants of His 37 in the influenza-A M218–60 dimer-of-dimers. Biochemistry 53, 5987–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cymes G. D., Ni Y., and Grosman C. (2005) Probing ion-channel pores one proton at a time. Nature 438, 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bechinger B. (1996) Towards membrane protein design: pH-sensitive topology of histidine-containing polypeptides. J. Mol. Biol. 263, 768–775 [DOI] [PubMed] [Google Scholar]
- 39. Achilonu I., Fanucchi S., Cross M., Fernandes M., and Dirr H. W. (2012) Role of individual histidines in the pH-dependent global stability of human chloride intracellular channel 1. Biochemistry 51, 995–1004 [DOI] [PubMed] [Google Scholar]
- 40. Nanga R. P. R., Brender J. R., Xu J., Veglia G., and Ramamoorthy A. (2008) Structures of rat and human islet amyloid polypeptide IAPP1–19 in micelles by NMR spectroscopy. Biochemistry 47, 12689–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thibado J. K., Martfeld A. N., Greathouse D. V., and Koeppe R. E. (2015) Influence of cholesterol on single arginine-containing transmembrane helical peptides. Biophys. J. 108, Supp. 1, p553a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roux B. (2007) Lonely arginine seeks friendly environment. J. Gen. Physiol. 130, 233–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dorairaj S., and Allen T. W. (2007) On the thermodynamic stability of a charged arginine side chain in a transmembrane helix. Proc. Natl. Acad. Sci. U.S.A. 104, 4943–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li L., Vorobyov I., MacKerell A. D. Jr., and Allen T. W. (2008) Is arginine charged in a membrane? Biophys. J. 94, L11–L13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoo J., and Cui Q. (2008) Does arginine remain protonated in the lipid membrane? Insights from microscopic pKa calculations. Biophys. J. 94, L61–L63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harms M. J., Schlessman J. L., Sue G. R., and García-Moreno B. (2011) Arginine residues at internal positions in a protein are always charged. Proc. Natl. Acad. Sci. U.S.A. 108, 18954–18959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fitch C. A., Platzer G., Okon M., Garcia-Moreno B. E., and McIntosh L. P. (2015) Arginine: its pKa value revisited. Protein Sci. 24, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Panahi A., and Brooks C. L. 3rd (2015) Membrane environment modulates the pKa values of transmembrane helices. J. Phys. Chem. B 119, 4601–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Davis J. H., Jeffrey K. R., Bloom M., Valic M. I., and Higgs T. P. (1976) Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem. Phys. Lett 42, 390–394 [Google Scholar]
- 50. Moncelli M. R., Becucci L., and Guidelli R. (1994) The intrinsic pKa values for phosphatidylcholine, phosphatidylethanolamine and phosphatidylserine in monolayers deposited on mercury electrodes. Biophys. J. 66, 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]