Abstract
Background
Although individual urine biomarkers are associated with chronic kidney disease (CKD) incidence and all-cause mortality in the setting of HIV infection, their combined utility for prediction remains unknown.
Methods
We measured eight urine biomarkers shown previously to be associated with incident CKD and mortality risk among 902 HIV-infected women in the Women's Interagency HIV Study: N-acetyl-β-d-glucosaminidase (NAG), kidney injury molecule-1 (KIM-1), alpha-1 microglobulin (α1m), interleukin 18, neutrophil gelatinase-associated lipocalin, albumin-to-creatinine ratio, liver fatty acid-binding protein and α-1-acid-glycoprotein. A group-based cluster method classified participants into three distinct clusters using the three most distinguishing biomarkers (NAG, KIM-1 and α1m), independent of the study outcomes. We then evaluated associations of each cluster with incident CKD (estimated glomerular filtration rate <60 mL/min/1.73 m2 by cystatin C) and all-cause mortality, adjusting for traditional and HIV-related risk factors.
Results
Over 8 years of follow-up, 177 CKD events and 128 deaths occurred. The first set of clusters partitioned women into three groups, containing 301 (Cluster 1), 470 (Cluster 2) and 131 (Cluster 3) participants. The rate of CKD incidence was 13, 21 and 50% across the three clusters; mortality rates were 7.3, 13 and 34%. After multivariable adjustment, Cluster 3 remained associated with a nearly 3-fold increased risk of both CKD and mortality, relative to Cluster 1 (both P < 0.001). The addition of the multi-biomarker cluster to the multivariable model improved discrimination for CKD (c-statistic = 0.72–0.76, P = 0.0029), but only modestly for mortality (c = 0.79–0.80, P = 0.099). Clusters derived with all eight markers were no better for discrimination than the three-biomarker clusters.
Conclusions
For predicting incident CKD in HIV-infected women, clusters developed from three urine-based kidney disease biomarkers were as effective as an eight-marker panel in improving risk discrimination.
Keywords: biomarker, chronic kidney disease, cluster analysis, HIV, risk discrimination
INTRODUCTION
HIV infection is associated with early onset of kidney disease [1], despite the widespread use of highly active antiretroviral therapies (HAART). The current surveillance strategy based on serum creatinine and urine dipstick for proteinuria is too insensitive [2] to capture the early stages of kidney disease. Although creatinine-based estimated glomerular filtration rate (eGFR) and albuminuria can predict mortality and end-stage renal disease [3], these markers lack the specificity to discern the etiology of kidney disease, and only reflect glomerular disease.
Better diagnostic methods may be useful to detect early kidney disease, and to determine whether it is related to specific risk factor exposures. The urine albumin-to-creatinine ratio (ACR) is an established marker of glomerular injury, but it may not capture renal injury at other sites. We previously have shown that novel urine biomarkers of tubule damage and dysfunction are associated with incident chronic kidney disease (CKD) and mortality risk among HIV-infected women enrolled in the Women's Interagency HIV Study (WIHS) [4–6].
The challenge of utilizing several biomarkers, however, is that they may offer contradictory findings, and there is no established method to integrate the results [7]. CKD is a heterogeneous condition, especially in the setting of HIV infection, where the clinical presentation and severity can vary by cause and the presence of comorbidities. Classifying patients beyond their traditional risk factors using a biomarker-generated ‘phenotype’ could improve diagnosis and determination of prognosis. In this study, we utilized an unsupervised cluster approach (i.e. uninformed by the outcome variable) to group participants based on the results of multiple urine biomarkers of tubular injury and function, and we replicated these methods using three versus eight biomarkers. Our goal was to identify a parsimonious set of markers to obtain maximum utility from the multiple urine biomarkers. We hypothesized that in combination, a complementary, parsimonious set of biomarkers would improve prediction above readily available, traditional kidney risk factors and HIV-related risk factors.
MATERIALS AND METHODS
Study population
The WIHS is an ongoing multicenter, prospective cohort study that enrolled 3067 HIV-infected and 1070 HIV-uninfected women from six US locations: Bronx, Brooklyn, Chicago, Los Angeles, San Francisco and Washington, DC in 1994–95, 2001–02 and 2011–12. Details of study design, data collection methods and baseline characteristics are published elsewhere [8, 9]. Participants undergo semiannual visits that include an interviewer-administered questionnaire, a physical examination and collection of laboratory specimens.
The WIHS Kidney Aging Study was designed as a nested cohort study (from the original 1994–95 cohort) to investigate the onset of kidney disease in the setting of HIV infection, utilizing stored urine and serum specimens. Characteristics of the original cohort, which tended to have worse disease characteristics and less HAART use compared with the later cohorts, are described elsewhere [10]. The baseline visit for this ancillary study was conducted from October 1999 to March 2000. One thousand HIV-infected and 250 uninfected women were included. Of these women, 450 were sampled from the WIHS bone sub-study and 800 were selected at random including age-/race-matched uninfected controls. There were no exclusions based on race or ethnicity. This analysis included HIV-infected women exclusively. For this study, 908 HIV-infected women who had stored urine available and at least one follow-up visit were included. Six participants with missing urine biomarker measurements were excluded from the present analysis, leaving a final sample size of 902 HIV-infected women.
The institutional review boards of participating institutions approved the study protocol at all WIHS study sites, and informed consent was obtained from all study participants. This study of kidney injury was also approved by the University of California, San Francisco, San Francisco VA Medical Center and Yale committees on human research.
Urine biomarkers
Urine kidney injury biomarkers measured in this study included: ACR, interleukin 18 (IL-18), neutrophil gelatinase-associated lipocalin (NGAL), kidney injury molecule-1 (KIM-1), liver fatty acid-binding protein (L-FABP), α-1-acid-glycoprotein (AAG), α-1-microglobulin (α1m) and N-acetyl-β-d-glucosaminidase (NAG). All urinary biomarkers were measured at the Cincinnati Children's Hospital Medical Center Biomarker Laboratory. Urine α1m was measured by a commercially available assay (Siemens BNII Nephelometer; Siemens, Munich, Germany). Urine NAG activity was measured using a colorimetric assay (Roche Diagnostics) as described in detail elsewhere [11]. Urine KIM-1 was measured using a commercially available ELISA (R&D Systems, Inc., Minneapolis, MN, USA) [12]. Urine albumin and creatinine were measured by immunoturbidimetry and colorimetric enzyme assay, respectively, using a Siemens Dimension Xpand plus HM clinical analyzer (Siemens). Urine IL-18 was measured using a commercially available ELISA kit (Medical and Biological Laboratories Co., Nagoya, Japan). Urine NGAL was assayed using a human-specific commercially available ELISA kit (AntibodyShop, Grusbakken, Denmark) [13]. Urine L-FABP was measured using a commercially available ELISA kit (CMIC Co., Tokyo, Japan) as per manufacturer's instructions. Urine AAG was measured using an ELISA kit (Human Orosomucoid ELISA Quantitation Kit; Genway Biotech, Inc., San Diego, CA, USA).
All urine specimens were in continuous storage at −80°C without prior freeze–thaw. Laboratory personnel were blinded to clinical information about the participants, including HIV status, and specimens were evaluated in random order. Coefficients of variation (CVs) for the urine measures based on replicate samples were: α1m, 5.2%; KIM-1, 5.2%; albumin, 5.9%; creatinine, 4.1%; IL-18, 7.2%; NGAL, 5.4%; L-FABP, 8.9%; AAG, 8.5%.
Forty percent of participants had undetectable urine α1m (<0.6 mg/dL), and the distribution among those with detectable α1m was right-skewed. Because of the left-censored nature of this data, we analyzed α1m using different approaches: as a dichotomized variable (detectable or undetectable), as a log-transformed continuous variable (replacing below detectable values with the limit of detection) and as an ordinal variable with three categories of urine α1m.
Outcomes
Kidney function was estimated using the CKD-EPI equation for serum cystatin C (eGFRCys), as in our prior studies [14, 15]. We chose a priori to estimate GFR by cystatin C rather than creatinine, because it is less susceptible to bias by muscle mass and health status [16]. Cystatin C was measured centrally at the UCLA Clinical Immunology Research Laboratory using a particle-enhanced immunoturbidimetric assay (Gentian, Moss, Norway), which has been calibrated against the new World Standard Reference material ERM-DA471/IFCC [14]. The assay was run concurrently from all three clinic visits, minimizing concerns for drift in the assay over time. Intra-assay CVs, based on 10 replicates, were <2% at serum concentrations of 0.7 and 1.1 mg/L. Inter-assay CVs were 4.4 and 3.9% at serum concentrations of 0.8 and 2.2 mg/L, respectively.
The primary outcomes of this study were incident CKD (defined as eGFRCys <60 mL/min/1.73 m2 at either of two follow-up visits among HIV-infected women with baseline eGFRCys ≥60 mL/min/1.73 m2) and all-cause mortality. Median follow-up time for incident CKD was 7.9 years, during which 177 cases of CKD occurred. For incident CKD cases, median eGFRCys was 79 [interquartile range (IQR): 71–88] at baseline and 52 (IQR: 43–59) at follow-up. As a sensitivity analysis, we defined incident CKD using eGFR by creatinine, calculated using the CKD-EPI equation [17] as eGFR <60 mL/min/1.73 m2 at any two consecutive visits within 8 years after baseline. Vital status was ascertained over 8 years of follow-up, during which 128 deaths occurred. Vital status and date of death were determined using the National Death Index and data from medical records and providers. Detailed methods have been described in prior studies [18–20].
Statistical analysis
The goal of our clustering procedure was to simplify the data from eight distinct biomarkers to partition subjects into a small number of groups based on the totality of biomarker information. This grouping was based solely on the aggregate biomarker data and was separate from clinical characteristics or subsequent outcomes. We first examined unadjusted Spearman correlations between markers. We then performed unsupervised group-based clustering of biomarkers by adopting the group-based trajectory approach used by Jones and Nagin [21] for longitudinal outcomes. While Jones and Nagin grouped repeated measures over time into different trajectory patterns, here we grouped multiple biomarkers into clusters according to their joint response pattern. Groups of participants having similar biomarker patterns can be identified as clusters. This method has the advantage of allowing biomarkers to be clustered based on unspecified, distinct patterns (e.g. low level in one biomarker and high level in other) instead of grouping only similar biomarkers into the same cluster.
Because the urine biomarkers were right-skewed, we log-transformed each marker to normalize its distribution. Clusters were constructed using SAS Proc NLMIXED, using the Bayesian Information Criteria to guide the selection of the number of clusters. Clusters were derived using biomarker values, independent of the study outcomes or other clinical information. We first used all eight biomarkers to construct clusters. We then identified a parsimonious set of biomarkers to construct clusters, defining the three most distinguishing biomarkers as those with the highest ratio of between- to within-group variance in biomarker levels. The final stratification into three clusters was based on the three most distinguishing biomarkers.
Baseline clinical and demographic characteristics were compared across clusters using χ2 and Kruskal–Wallis tests for categorical and continuous variables, respectively. As in our previous studies, relative risk regression (using a modified Poisson approach) and Cox proportional hazards regression were used to examine associations with incident CKD and all-cause mortality, respectively [5]. Covariates from the baseline examination included age, ethnicity, smoking, hypertension, diabetes, CD4 count, HIVRNA and hepatitis C (defined as HCV antibody positive, excluding those who resolved their HCV infection by HCVRNA testing). Additionally, we included baseline eGFR by cystatin as a covariate in models of mortality. We calculated risk ratios for each outcome using Cluster 1 as the reference category. Women with eGFRCys <60 mL/min/1.73 m2 at baseline were excluded from the incident CKD analysis.
We assessed model performance using discrimination, calibration, Nagelkerke's R2 (for overall performance) and net reclassification index (NRI) [22]. For survival models, Harrell's c was used to assess discrimination [23]. Three-category NRI was calculated using tertiles of predicted risk to compare changes in risk categories. All analyses were conducted using SAS version 9.4 (SAS Institute, Inc., Cary, NC, USA).
RESULTS
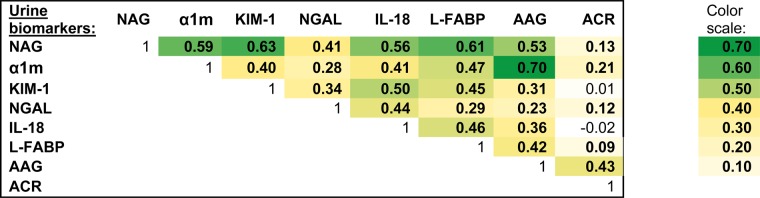
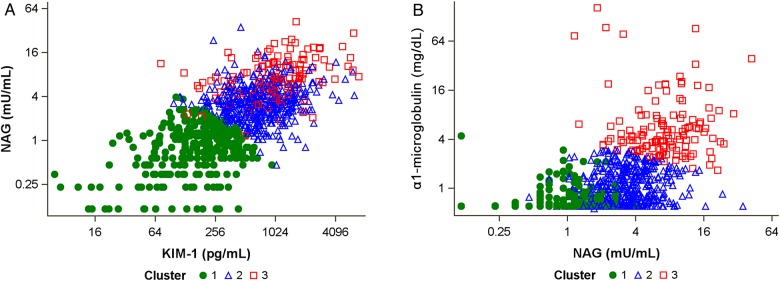
In unadjusted analysis, the urine biomarkers were moderately intercorrelated, although most measures had correlation coefficients of 0.6 or less (Figure 1). The strongest correlation was between α1m and AAG (r = 0.70), and the weakest correlation was between KIM-1 and ACR (r = 0.01). The final clusters were derived using NAG, KIM-1 and α1m, as they had the highest ratio of between- to within-group variance. A comparison of levels of these three markers found that NAG and KIM-1 were most able to distinguish Cluster 1 from Clusters 2 and 3, whereas α1m appeared to separate Cluster 3 from Clusters 1 and 2 (Figure 2).
FIGURE 1:
Correlations of urine biomarkers among HIV-infected women.
FIGURE 2:
Comparison of distinguishing urine biomarkers by cluster membership. (A) Scatterplot shows separation of Cluster 1 from Clusters 2 and 3 by NAG and KIM-1. (B) Scatterplot shows separation of Cluster 3 from Clusters 1 and 2 by NAG and α1m. Note that clusters were derived from NAG, KIM-1 and α1m, independent of study outcomes.
We then compared baseline demographic and clinical characteristics, stratified by cluster (Table 1). HIV-infected women in Cluster 3 were older and more often African-American, current smokers and hypertensive compared with those in Cluster 1. Markers of worse HIV infection status were seen in Cluster 3, including lower CD4 counts and lower body mass index (BMI), more detectable viremia and albuminuria, and less HAART use. Lower waist circumference, more frequent history of AIDS and coinfection with hepatitis C, and lower eGFRCys were also more prevalent in Cluster 3.
Table 1.
Baseline characteristics of HIV-infected women, stratified by biomarker-derived cluster
| Parameter | Biomarker-derived clustera |
P-value | ||
|---|---|---|---|---|
| 1 (N = 301) | 2 (N = 470) | 3 (N = 131) | ||
| Baseline age (years) | 40 (36–45) | 41 (36–45) | 43 (38–48) | 0.0003 |
| African-American | 152 (50%) | 272 (58%) | 94 (72%) | <0.0001 |
| Current smoking | 130 (43%) | 239 (51%) | 90 (69%) | <0.0001 |
| Diabetes mellitus | 28 (9%) | 42 (9%) | 16 (12%) | 0.52 |
| Hypertension | 54 (18%) | 125 (27%) | 46 (35%) | 0.0004 |
| LDL (mg/dL) | 106 (83–132) | 104 (80–134) | 93 (69–130) | 0.067 |
| HDL (mg/dL) | 45 (36–58) | 43 (35–54) | 44 (32–56) | 0.079 |
| Triglycerides (mg/dL) | 130 (87–188) | 132 (94–196) | 139 (97–200) | 0.54 |
| BMI (kg/m2) | 26 (23–31) | 27 (24–32) | 25 (22–29) | 0.0005 |
| Waist circumference (cm) | 87 (80–96) | 89 (80–101) | 86 (77–97) | 0.0060 |
| Current HAART use | 186 (62%) | 280 (60%) | 62 (47%) | 0.016 |
| Current CD4 | 443 (274–622) | 414 (254–581) | 284 (161–441) | <0.0001 |
| Nadir CD4 | 230 (117–340) | 220 (120–330) | 163 (77–281) | 0.0012 |
| History of AIDS | 125 (42%) | 228 (49%) | 88 (67%) | <0.0001 |
| Detectable HIV RNA | 204 (68%) | 315 (67%) | 109 (83%) | <0.0001 |
| Hepatitis C | 72 (24%) | 147 (32%) | 58 (45%) | <0.0001 |
| eGFRCys | 96 (85–110) | 87 (75–105) | 71 (58–85) | <0.0001 |
| Albuminuria | 46 (15%) | 106 (23%) | 60 (46%) | <0.0001 |
| Urine biomarkers | ||||
| NAG (mU/mL)a | 0.9 (0.5–1.3) | 3.2 (2.2–4.5) | 7.0 (4.6–11.0) | <0.0001 |
| α1m (ng/mL)a | 0.6 (0.6–0.6) | 0.8 (0.6–1.5) | 4.4 (3.2–7.2) | <0.0001 |
| KIM-1 (pg/mL)a | 199 (109–291) | 678 (439–1042) | 1036 (648–1669) | <0.0001 |
| IL-18 (pg/mL) | 59 (34–108) | 148 (84–262) | 249 (152–411) | <0.0001 |
| NGAL (ng/mL) | 20 (10–45) | 41 (21–85) | 68 (36–129) | <0.0001 |
| ACR (mg/g) | 10 (6–17) | 9 (5–19) | 24 (10–85) | <0.0001 |
| L-FABP (ng/mL) | 1.9 (0.5–4.1) | 5.6 (2.9–9.9) | 15.0 (6.8–30.2) | <0.0001 |
| AAG (µg/mL) | 1.2 (0.6–2.8) | 3.2 (1.2–6.6) | 22.4 (7.4–38.6) | <0.0001 |
Data are presented as median (IQR) or numbers (percent).
LDL, low-density lipoprotein; HDL, high-density lipoprotein; NAG, N-acetyl-β-d-glucosaminidase; α1m, α-1-microglobulin; KIM-1, kidney injury molecule-1; IL-18, interleukin 18; NGAL, neutrophil gelatinase-associated lipocalin; ACR, albumin-to-creatinine ratio; L-FABP, liver fatty acid-binding protein; AAG, α-1-acid-glycoprotein.
aClusters were derived using NAG, α1m and KIM-1, independent of study outcomes.
There was a progressive increase in all biomarker levels by cluster, although only three (NAG, α1m and KIM-1) were used to derive the clusters (Table 1). Cluster 2 had intermediate levels for six of the eight biomarkers; however, α1m and ACR levels were similar in Clusters 1 and 2.
We then used our biomarker-derived clusters to examine associations with incident CKD. Median follow-up time for incident CKD was 7.9 years, during which 177 cases of CKD occurred. Rates of incident CKD were lowest in Cluster 1, intermediate in Cluster 2 and highest in Cluster 3 (Table 2). In unadjusted analysis, we found that Cluster 2 was associated with a 67% higher risk of CKD, whereas being in Cluster 3 was associated with a 4-fold higher risk of developing CKD compared with being in Cluster 1. After multivariable adjustment for traditional kidney risk factors and HIV-related factors, we found that both clusters remained associated with a higher risk of CKD compared with being in Cluster 1.
Table 2.
Association of biomarker-derived clustera with incident CKD and all-cause mortality among HIV-infected women
| Outcome | Cluster 1 (n = 289) | Cluster 2 (n = 435) | Cluster 3 (n = 94) |
|---|---|---|---|
| Incident CKDb | |||
| Event rate | 13% | 21% | 50% |
| Unadjusted risk ratio (95% CI) | Reference | 1.67 (1.18, 2.37) P = 0.0042 | 3.91 (2.72, 5.61) P < 0.0001 |
| Adjusted risk ratio (95% CI) | Reference | 1.59 (1.13, 2.25) P = 0.0076 | 2.89 (1.97, 4.25) P < 0.0001 |
| Cluster 1 (n = 301) | Cluster 2 (n = 470) | Cluster 3 (n = 131) | |
| All-cause mortality | |||
| Event rate | 7.3% | 13% | 34% |
| Unadjusted hazard ratio (95% CI) | Reference | 1.85 (1.14, 3.01) P = 0.013 | 5.38 (3.22, 8.97) P < 0.0001 |
| Adjusted hazard ratio (95% CI) | Reference | 1.51 (0.92, 2.47) P = 0.10 | 2.79 (1.62, 4.81) P = 0.0002 |
Adjusted models control for age, race, hypertension, diabetes mellitus, hepatitis C virus infection, HIV viral load and CD4 lymphocyte count. Mortality models adjust additionally for eGFR calculated using cystatin C.
CI, confidence interval; CKD, chronic kidney disease.
aClusters were derived using NAG, α1m and KIM-1.
bEighty-four of the 902 participants had CKD at baseline, leaving 818 available for analysis of incident CKD.
Next, we used biomarker-derived clusters to examine associations with all-cause mortality. A total of 128 deaths occurred over a median of 7.9 years of follow-up. Rates of all-cause mortality were also lowest in Cluster 1, intermediate in Cluster 2 and highest in Cluster 3. After multivariable adjustment for traditional kidney risk factors and HIV-related factors, we found that Cluster 2 was associated with a marginally higher risk of mortality, whereas being in Cluster 3 was associated with a >2-fold risk of mortality compared with being in Cluster 1. Results for both CKD and mortality were similar in alternative models in which we constructed clusters using all eight biomarkers instead of just the most distinguishing three biomarkers (Supplementary data, Table S1).
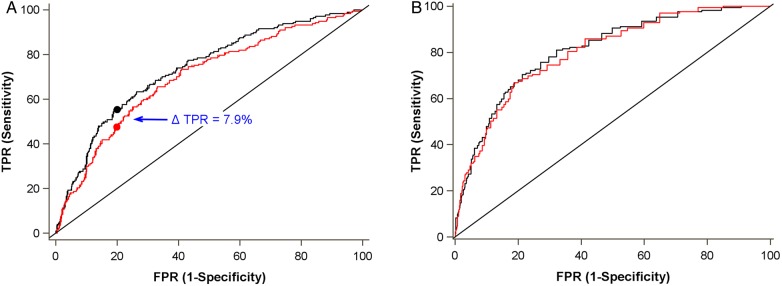
We next assessed measures of overall fit, discrimination, calibration and reclassification for multivariable models with and without the novel urine biomarkers and cluster variable (Table 3). For incident CKD, model discrimination was significantly improved by adding the cluster variable to the traditional model (c = 0.72–0.76, P = 0.0029). At a false-positive rate of 20%, the true-positive rate improved by 7.9% with the addition of the clusters (Figure 3). Overall model fit was also increased (R2 = 0.15–0.21). Models incorporating individual biomarkers (analyzed continuously) instead of the categorical cluster variable had performances that were similar to or slightly weaker than the cluster variable.
Table 3.
Comparison of model performance for incident CKD and all-cause mortality among HIV-infected women
| Outcome | Overall fit (R2) | Discrimination (c) |
|
|---|---|---|---|
| Incident CKD | |||
| Traditional model (without biomarkers) | 0.15 | 0.72 | P = 0.0029 |
| Traditional model + clustera | 0.21 | 0.76 | |
| Traditional model + ACR | 0.18 | 0.73 | |
| Traditional model + NAG | 0.20 | 0.75 | |
| Traditional model + α1m | 0.19 | 0.75 | |
| Traditional model + KIM-1 | 0.18 | 0.74 | |
| Traditional model + eight-marker cluster | 0.21 | 0.75 | |
| Traditional model + all eight markers | 0.24 | 0.77 | |
| All-cause mortality | |||
| Traditional model (without biomarkers) | 0.25 | 0.78 | P = 0.18 |
| Traditional model + clustera | 0.26 | 0.79 | |
| Traditional model + ACR | 0.25 | 0.78 | |
| Traditional model + NAG | 0.26 | 0.78 | |
| Traditional model + α1m | 0.26 | 0.78 | |
| Traditional model + KIM-1 | 0.26 | 0.79 | |
| Traditional model + eight-marker cluster | 0.25 | 0.78 | |
| Traditional model + all eight markers | 0.28 | 0.79 | |
Traditional models control for age, race, hypertension, diabetes mellitus, hepatitis C virus infection, HIV viral load and CD4 lymphocyte count. Mortality models adjust additionally for eGFR calculated using cystatin C.
CKD, chronic kidney disease; R2, Nagelkerke R-squared coefficient of determination is useful for comparing non-nested models; c, c-statistic.
aClusters were derived using NAG, α1m and KIM-1.
FIGURE 3:
Comparison of receiver operator characteristic curves for (A) incident CKD and (B) all-cause mortality. Red line denotes traditional model and black line denotes traditional model plus biomarker cluster. Traditional models control for age, race, hypertension, diabetes mellitus, hepatitis C virus infection, HIV viral load and CD4 lymphocyte count. Mortality model adjusts additionally for eGFR calculated using cystatin C. (A) At a false positive rate (FPR) of 20%, the true positive rate (TPR) is 55.4% with the biomarker cluster versus 47.5% without the biomarker cluster. Overall c-statistic is 0.76 with the biomarker cluster and 0.72 without the biomarker cluster. (B) Overall c-statistic is 0.80 with the biomarker cluster and 0.79 without the biomarker cluster.
Net reclassification improvement for CKD was 11.2% (P = 0.006, Table 4). Of the 177 CKD cases in this study, we found that addition of the categorical cluster variable to the model allowed us to identify an additional 27 patients (15%) as high risk who would otherwise have been missed by the traditional model. However, 14 of the 177 cases (7.9%) were inappropriately reclassified as lower risk when the biomarker cluster was included in the model. Although the Hosmer–Lemeshow goodness-of-fit test was indicative of good calibration (P = 0.80), a plot of predicted versus actual probability of CKD suggested that calibration was somewhat worse at higher probabilities (Supplementary data, Figure S1).
Table 4.
Use of biomarker-derived clustersa to improve net reclassification of study outcomes
| Incident CKDb | All-cause mortality | |
|---|---|---|
| Participants with event | 177 | 169 |
| Appropriately reclassified (as higher risk) | 27 (15.3%) | 10 (5.9%) |
| No change | 136 (76.8%) | 154 (91%) |
| Inappropriately reclassified (as lower risk) | 14 (7.9%) | 5 (3.0%) |
| NRI (95% CI), cases | 7.3% (0.3%, 14.4%), P = 0.042 | 3.0% (−1.5%, 7.5%), P = 0.20 |
| Participants without event | 641 | 733 |
| Inappropriately reclassified (as higher risk) | 69 (10.6%) | 36 (4.9%) |
| No change | 480 (74.9%) | 656 (90%) |
| Appropriately reclassified (as lower risk) | 93 (14.5%) | 41 (5.6%) |
| NRI (95% CI), non-cases | −3.9% (−7.8%, 0.0%), P = 0.049 | −0.7% (−3.0%, 1.7%), P = 0.57 |
| Overall NRIc (95% CI) | 11.2% (3.2%, 19.3%), P = 0.0064 | 3.6% (−1.4%, 8.7%), P = 0.16 |
Traditional models control for age, race, hypertension, diabetes mellitus, hepatitis C virus infection, HIV viral load and CD4 lymphocyte count. Mortality model adjusts additionally for eGFR calculated using cystatin C.
CKD, chronic kidney disease; NRI, net reclassification improvement.
aClusters were derived using NAG, α1m and KIM-1.
bEighty-four of the 902 participants had CKD at baseline, leaving 818 available for the analysis of incident CKD.
cThree-category NRI calculated using tertiles of predicted risk to compare changes in risk categories. Models compared are traditional model versus traditional model plus biomarker cluster.
For all-cause mortality, model discrimination was only marginally improved by adding the cluster variable to the traditional model (c = 0.79–0.80, P = 0.099). Overall model fit increased marginally from R2 = 0.27–0.28, and net reclassification improvement was 3.6%. Models incorporating individual biomarkers (analyzed continuously) instead of the cluster variable (which was categorical) had performances that were similar to the cluster variable. We also examined associations of individual biomarkers with both CKD and mortality. Biomarkers used to derive the cluster variable are shown in Supplementary data, Table S2, along with ACR, an established marker of glomerular injury. Each biomarker was individually associated with an increased risk of CKD and mortality, even after controlling for traditional and HIV-related risk factors.
In a separate analysis, we defined CKD using serum creatinine instead of cystatin C to calculate eGFR (Supplementary data, Table S3). Although the overall event rate was lower using this alternative definition (11.5% instead of 21.6%), we still found that rates of incident CKD were lowest in Cluster 1, intermediate in Cluster 2 and highest in Cluster 3. In unadjusted analysis, we found that Cluster 3 was associated with a 3.6-fold higher risk of developing CKD compared with being in Cluster 1. After multivariable adjustment for traditional kidney risk factors and HIV-related factors, Cluster 3 remained associated with a 3.3-fold higher risk of CKD compared with being in Cluster 1.
DISCUSSION
We investigated the use of urine biomarker-derived clusters for predicting incident CKD and all-cause mortality in 902 HIV+ women. We used the three most distinguishing markers (NAG, KIM-1 and α1m) to classify participants into separate clusters, without incorporating any clinical information. We then used these clusters to predict CKD and mortality, and found that event rates were highest in Cluster 3 and lowest in Cluster 1. Participants classified into Cluster 3 had more comorbidities and worse HIV-related characteristics at baseline. Relative to those who were in Cluster 1, those in Cluster 3 had 4-fold higher rates of incident CKD (50 versus 13%) and all-cause mortality (41 versus 10%). Although the new cluster variable only moderately improved model discrimination for CKD risk compared with a model containing traditional risk factors, the cluster was independently associated with each study outcome.
We previously reported that urine α1m is independently associated with kidney function decline and mortality risk [5], even after controlling for other urine biomarkers including ACR, IL-18, KIM-1 and NGAL. In this analysis, we found that our cluster variable had strong, independent associations with both incident CKD and all-cause mortality. While individual biomarkers also showed statistically significant associations with CKD and mortality, an advantage of our cluster-based analysis is that it allows partitioning of subjects into discrete categories of risk. To our knowledge, this is the first study to use biomarker-based cluster analysis to predict CKD and mortality in the setting of HIV infection. The Framingham Offspring Study found that a seven-biomarker panel that included homocysteine and aldosterone was associated with incident CKD and microalbuminuria; rather than using clusters, this study created scores with the median value of each biomarker to define the cutpoint for high risk [24].
A recent study of heart failure patients used phenotype data (67 variables including electrocardiogram, echocardiography, clinical and laboratory measures) to cluster patients into three distinct risk categories, and the authors found strong associations with clinical outcomes [25]. In contrast, our study used biomarkers alone to define the clusters, so that we could distinguish and evaluate the biomarkers' ability to stratify patients into distinct risk categories. Although the clusters' prediction ability would certainly improve if they were comprised of biomarkers combined with clinical risk factors, our goal was to evaluate urine biomarkers in isolation to measure their incremental contribution. An advantage of using biomarkers from just one sample type (i.e. urine) is that the selected panel can be used to develop a multiplex assay, such as the Meso Scale Discovery (MSD) array, which has advantages of both efficiency and enhanced precision over individual ELISAs. Multiplex assays could be used to facilitate clinical applicability, providing a simple, clinically relevant message to clinicians and patients about the risk of disease.
Our results illustrate the strengths and limitations of the use of urine biomarkers for the prediction of kidney disease and mortality in the setting of HIV infection. Our novel cluster method allowed us to select a relevant trio from eight candidates, independent of study outcomes. We found that our three-marker cluster, which was a categorical variable, was as good as or better than any individual biomarker, and as strong as the totality of eight. A strength of this approach is that clusters can define useful groups of patients, and can mitigate the problems of multicollinearity [26] that may arise with the inclusion of multiple correlated measures in a multivariable regression model. Although our results have not yet been validated in an external cohort, our work serves as a prototype for future biomarker studies. We envision that clinicians will be able to use such a biomarker panel to determine the level of risk (low, moderate or high) of an individual HIV-infected patient for chronic kidney disease and early mortality. This biomarker panel could be repeated to provide updated information on a patient's kidney health.
Our study includes several limitations. We were only able to consider markers of proximal tubular injury and function, with the exception of ACR (glomerular injury) and NGAL (maybe distal or mixed proximal/distal). Markers that identify other aspects of nephron function and injury will need to be incorporated to allow better discrimination of risk [27]. Additional work is needed to understand the role of nephrotoxic drugs, such as tenofovir, and to validate in other cohorts. Our study included only women, and little is known about associations of urine biomarkers with kidney outcomes in HIV-infected men. Urine biomarker measurements were made using samples collected over a decade ago, and sample degradation due to the length of freezer storage [28] may have increased measurement error. We were unable to confirm diagnosis of CKD by two consecutive measures because only one serum sample was available at each visit per participant: this may have weakened the specificity of our CKD diagnosis. However, we repeated our analysis using serum creatinine, and found that our urine biomarker cluster was strongly associated with CKD. Finally, there may have been incomplete or inadequate control for factors that may confound or mediate the association of elevations in urine biomarkers with CKD and mortality.
In summary, we have shown that unsupervised cluster analysis (i.e. uninformed by the outcome variable) of urine biomarkers can identify distinct categories of risk and thereby differentiate the risk of CKD and mortality. Future studies of HIV-infected persons (including men) are needed to validate these results. A broader array of candidate biomarkers (interstitial fibrosis, distal tubule and collecting duct) may be needed to improve discrimination potential. In the future, the use of biomarker panels could help inform the diagnosis and staging of CKD, and may be used to identify patients who are at risk of drug toxicity.
SUPPLEMENTARY DATA
Supplementary data are available online at http://ndt.oxfordjournals.org.
CONFLICT OF INTEREST STATEMENT
C.R.P. is a co-inventor on the IL-18 patent issued to the University of Colorado. R.S. received an honorarium from Merck for participating in a Renal Expert Input Forum; this honorarium was donated to NCIRE to support kidney research.
(See related article by Pontillo and Mischak. Urinary biomarkers to predict CKD: is the future in multi-marker panels? Nephrol Dial Transplant 2016; 31: 1373–1375)
Supplementary Material
ACKNOWLEDGEMENTS
The WIHS Kidney Aging Study is funded by grant 1 R01 AG034853-01A2 (PI, M.G.S.), which was administered by the Northern California Institute for Research and Education, and with resources of the Veterans Affairs Medical Center, San Francisco, California. M.G.S. and R.S. were also supported by the Connie Wofsy Women's HIV Study (U01-AI-034989). Data in this manuscript were collected by the WIHS. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH). WIHS (Principal Investigators): UAB-MS WIHS (Michael Saag, Mirjam-Colette Kempf and Deborah Konkle-Parker), U01-AI-103401; Atlanta WIHS (Ighovwerha Ofotokun and Gina Wingood), U01-AI-103408; Bronx WIHS (Kathryn Anastos), U01-AI-035004; Brooklyn WIHS (Howard Minkoff and Deborah Gustafson), U01-AI-031834; Chicago WIHS (Mardge Cohen and Audrey French), U01-AI-034993; Metropolitan Washington WIHS (Mary Young), U01-AI-034994; Miami WIHS (Margaret Fischl and Lisa Metsch), U01-AI-103397; UNC WIHS (Adaora Adimora), U01-AI-103390; Connie Wofsy Women's HIV Study, Northern California (Ruth Greenblatt, Bradley Aouizerat and Phyllis Tien), U01-AI-034989; WIHS Data Management and Analysis Center (Stephen Gange and Elizabeth Golub), U01-AI-042590; Southern California WIHS (Joel Milam), U01-HD-032632 (WIHS I–WIHS IV). The WIHS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the National Cancer Institute (NCI), the National Institute on Drug Abuse (NIDA) and the National Institute on Mental Health (NIMH). Targeted supplemental funding for specific projects is also provided by the National Institute of Dental and Craniofacial Research (NIDCR), the National Institute on Alcohol Abuse and Alcoholism (NIAAA), the National Institute on Deafness and other Communication Disorders (NIDCD) and the NIH Office of Research on Women's Health. WIHS data collection is also supported by UL1-TR000004 (UCSF CTSA) and UL1-TR000454 (Atlanta CTSA).
REFERENCES
- 1.Nadkarni GN, Konstantinidis I, Wyatt CM. HIV and the aging kidney. Curr Opin HIV AIDS 2014; 9: 340–345 [DOI] [PubMed] [Google Scholar]
- 2.Bostom AG, Kronenberg F, Ritz E. Predictive performance of renal function equations for patients with chronic kidney disease and normal serum creatinine levels. J Am Soc Nephrol 2002; 13: 2140–2144 [DOI] [PubMed] [Google Scholar]
- 3.Coresh J, Turin TC, Matsushita K et al. . Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. JAMA 2014; 311: 2518–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peralta C, Scherzer R, Grunfeld C et al. . Urinary biomarkers of kidney injury are associated with all-cause mortality in the Women's Interagency HIV Study (WIHS). HIV Med 2014; 15: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jotwani V, Scherzer R, Abraham A et al. . Association of urine alpha1-microglobulin with kidney function decline and mortality in HIV-infected women. Clin J Am Soc Nephrol 2015; 10: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shlipak MG, Scherzer R, Abraham A et al. . Urinary markers of kidney injury and kidney function decline in HIV-infected women. J Acquir Immune Defic Syndr 2012; 61: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parikh CR, Devarajan P. New biomarkers of acute kidney injury. Crit Care Med 2008; 36 (4 Suppl): S159–S165 [DOI] [PubMed] [Google Scholar]
- 8.Barkan SE, Melnick SL, Preston-Martin S et al. . The Women's Interagency HIV Study. WIHS Collaborative Study Group. Epidemiology 1998; 9: 117–125 [PubMed] [Google Scholar]
- 9.Bacon MC, von Wyl V, Alden C et al. . The Women's Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol 2005; 12: 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hessol NA, Weber KM, Holman S et al. . Retention and attendance of women enrolled in a large prospective study of HIV-1 in the United States. J Womens Health (Larchmt) 2009; 18: 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundaram N, Bennett M, Wilhelm J et al. . Biomarkers for early detection of sickle nephropathy. Am J Hematol 2011; 86: 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaturvedi S, Farmer T, Kapke GF. Assay validation for KIM-1: human urinary renal dysfunction biomarker. Int J Biol Sci 2009; 5: 128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett M, Dent CL, Ma Q et al. . Urine NGAL predicts severity of acute kidney injury after cardiac surgery: a prospective study. Clin J Am Soc Nephrol 2008; 3: 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inker LA, Eckfeldt J, Levey AS et al. . Expressing the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) cystatin C equations for estimating GFR with standardized serum cystatin C values. Am J Kidney Dis 2011; 58: 682–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens LA, Coresh J, Schmid CH et al. . Estimating GFR using serum cystatin C alone and in combination with serum creatinine: a pooled analysis of 3,418 individuals with CKD. Am J Kidney Dis 2008; 51: 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odden MC, Scherzer R, Bacchetti P et al. . Cystatin C level as a marker of kidney function in human immunodeficiency virus infection: the FRAM study. Arch Intern Med 2007; 167: 2213–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levey AS, Stevens LA, Schmid CH et al. . A new equation to estimate glomerular filtration rate. Ann Intern Med 2009; 150: 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyatt CM, Hoover DR, Shi Q et al. . Pre-existing albuminuria predicts AIDS and non-AIDS mortality in women initiating antiretroviral therapy. Antivir Ther 2011; 16: 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen MH, French AL, Benning L et al. . Causes of death among women with human immunodeficiency virus infection in the era of combination antiretroviral therapy. Am J Med 2002; 113: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French AL, Gawel SH, Hershow R et al. . Trends in mortality and causes of death among women with HIV in the United States: a 10-year study. J Acquir Immune Defic Syndr 2009; 51: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones BL, Nagin DS. Advances in group-based trajectory modeling and a SAS procedure for estimating them. Sociol Methods Res 2007; 35: 542–572 [Google Scholar]
- 22.Steyerberg EW, Vickers AJ, Cook NR et al. . Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010; 21: 128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996; 15: 361–387 [DOI] [PubMed] [Google Scholar]
- 24.Fox CS, Gona P, Larson MG et al. . A multi-marker approach to predict incident CKD and microalbuminuria. J Am Soc Nephrol 2010; 21: 2143–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah SJ, Katz DH, Selvaraj S et al. . Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 2015; 131: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belsley DA, Kuh E, Welsch RE. Regression Diagnostics: Identifying Influential Data and Sources of Collinearity. New York: John Wiley & Sons, 2005 [Google Scholar]
- 27.Bonventre JV, Vaidya VS, Schmouder R et al. . Next-generation biomarkers for detecting kidney toxicity. Nat Biotechnol 2010; 28: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tworoger SS, Hankinson SE. Collection, processing, and storage of biological samples in epidemiologic studies: sex hormones, carotenoids, inflammatory markers, and proteomics as examples. Cancer Epidemiol Biomarkers Prev 2006; 15: 1578–1581 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.