Abstract
Compared with the sequence and expression similarity, miRNA functional similarity is so important for biology researches and many applications such as miRNA clustering, miRNA function prediction, miRNA synergism identification and disease miRNA prioritization. However, the existing methods always utilized the predicted miRNA target which has high false positive and false negative to calculate the miRNA functional similarity. Meanwhile, it is difficult to achieve high reliability of miRNA functional similarity with miRNA-disease associations. Therefore, it is increasingly needed to improve the measurement of miRNA functional similarity. In this study, we develop a novel path-based calculation method of miRNA functional similarity based on miRNA-disease associations, called MFSP. Compared with other methods, our method obtains higher average functional similarity of intra-family and intra-cluster selected groups. Meanwhile, the lower average functional similarity of inter-family and inter-cluster miRNA pair is obtained. In addition, the smaller p-value is achieved, while applying Wilcoxon rank-sum test and Kruskal-Wallis test to different miRNA groups. The relationship between miRNA functional similarity and other information sources is exhibited. Furthermore, the constructed miRNA functional network based on MFSP is a scale-free and small-world network. Moreover, the higher AUC for miRNA-disease prediction indicates the ability of MFSP uncovering miRNA functional similarity.
In recent years, more and more researchers pay attention to measure miRNA functional similarity in biology research, since it is very important for many applications such as miRNA clustering1, miRNA function prediction2, miRNA synergism identification3,4,5,6, miRNA-mRNA interaction inference7 and disease miRNA prioritization8,9,10,11,12,13. Although miRNA similarity could be calculated based on miRNA sequence or expression data6,14, functional similarity is more beneficial to fully understand the functions and biological roles of miRNA15.
In previous studies, some computational approaches based on miRNA-target associations were developed for studying miRNA functional similarity. Shalgi et al.16 utilized Jaccard similarity method to quantify the functional similarity of two miRNAs based on their common target genes. However, most of miRNA functional similarities using the Jaccard similarity measurement are zeros, because there is no intersection among the target gene sets of most human miRNAs17,18. Yu et al.19 systematically measured the functional similarity of miRNA pair using GO annotation of their target genes. Sun et al.20 calculated the miRNA functional similarity based on miRNAs targeting propensity and proteins connectivity in the integrated protein-protein interaction network. Xu et al.21 combined the site accessibility and the interactive context of target genes in functional gene networks, which was constructed with semantic similarity weights generated from the GO terms of the target genes, to infer the functional similarity of miRNA pair. Meng et al.22 proposed a method, called PPImiRFS, for calculating the functional similarity of plant miRNAs inferred from similarity of their target gene sets. PPImiRFS firstly constructed a protein-protein interaction network using the gene semantic similarity by GOSemSim23 and then quantified the functional similarity of target pair based on the shortest paths. A modified best-match average method was employed to calculate the functional similarity of miRNA pair using the predicted miRNA gene24. In addition, Xu et al.3 defined miRNA synergistic pair which significantly related to at least one co-regulating functional module established by a protein-protein interaction network25,26. Nevertheless, this method could not measure the level of miRNA similarity, since the calculated similarities are only 0 or 1. Moreover, the above methods always make use of the predicted miRNA target which has high false positives and false negatives. It makes these methods difficult to obtain high reliability for miRNA functional similarity. So, Liu et al.27 integrated the miRNA-target association, the lncRNA-disease association and the miRNA-lncRNA association to quantify the miRNA similarity. However, there is informative subtype information of disease verified by expertise in the US National Library of Medicine28. Furthermore, lncRNA-disease associations are always utilized to calculate lncRNA functional similarity in many methods29,30,31 and the density of miRNA-disease associations validated by biology experiment is greater than that of miRNA target in human. Therefore, Wang et al.32 utilized human miRNA-disease associations to compute the functional similarity scores based on the supposition that similar miRNAs tend to be associated with similar diseases, named MISIM. In this method, an inferring GO term similarity algorithm was applied to measure the semantic similarity of diseases structured as directed acyclic33, and then the miRNA functional similarity was inferred by best-match average (BMA) method. Moreover, Xuan et al.34 improved the calculation of information content (IC) of diseases based on the intuition that the more general the disease term is and the less semantic contribution it has, which ensure higher reliability of semantic similarity of disease. Similarly, BMA method was employed to quantify the miRNA functional similarity based on disease similarity. However, these methods using miRNA-disease associations, namely based on BMA method, did’nt consider the topology of disease semantic network. Furthermore, path-based similarity measurements have been successfully applied on various types of relationship data35,36. Therefore, an efficiently path-based method is required to measure miRNA functional similarity using miRNA-disease associations.
In this study, we designed a method, called MFSP (MiRNA Functional Similarity based on Path), to infer the functional similarity of miRNA pair using miRNA-disease associations. To validate MFSP, we compared it with two other state-of-the-art methods based on disease-related miRNAs, namely Wang’s method32 and Xuan’s method34, with functional similarity scores of intra-family, inter-family, intra-cluster and inter-cluster miRNA pairs. Meanwhile, the lower p-value was obtained while applying Wilcoxon rank-sum test and Kruskal-Wallis test on different miRNA groups. Furthermore, it was verified that the positive correlation is between the expression similarity and the functional similarity. The negative effect of distance of genome coordinate was exhibited for functional similarity. In addition, the effect of varying parameters was analyzed. For formed miRNA network based on miRNA functional similarity, it is scale-free network and small world network. The higher AUC was achieved while MIDP9 was applied to the miRNA network constructed by MFSP. Moreover, a Cosine Similarity of Disease, named CSD, was developed to improve the reliability of dieasese semantic similarity. To validate CSD, the different similarity calculation methods of disease and BMA method for miRNA functional similarity calculation were combined to compare performance.
Results
Design of experiment
MFSP was developed to infer miRNA functional similarity by combining subtype information of disease obtained from MeSH (http:www.ncbi.nlm.nih.gov/)28 and the known miRNA-disease associations provided by HMDD37. In the MFSP method, first, the semantic similarity of disease is calculated based on subtype information of disease. Then, unlike the Wang’s method32 and Xuan’s method34 which are only consider the direct neighbor diseases of miRNA, MFSP measures the miRNA functional similarity based on the topology of disease network constructed by semantic similarity of disease. Furthermore, functional similarities of all miRNA pairs are provided in Supplementary Material 1. The performance of MFSP was evaluated by average similarity of different miRNA groups and p-value obtained by Wilcoxon rank-sum test and Kruskal-Wallis test. The relationships between other biology informations and miRNA functional similarity were exhibited. The results of predicting disease-related miRNAs were compared. In addition, we analyzed the effect of parameters and constructed miRNA network based on miRNA functional similarity calculated by MFSP.
Performance
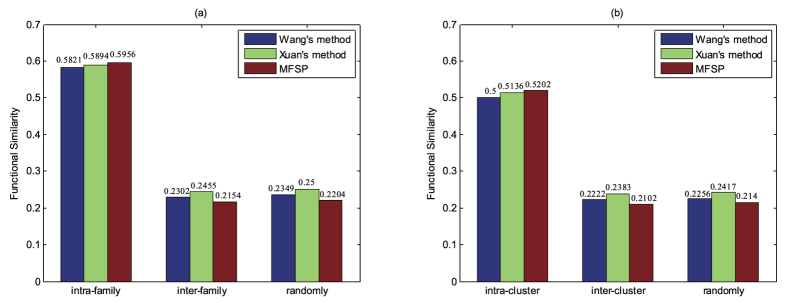
The miRNA-disease associations dataset of human used in the experiment can be downloaded from HMDD37. The latest version of HMDD (updated in 2015) includes 330 diseases which contained in the US National Library of Medicine (MeSH) and their associated 574 miRNAs (Supplementary Material 2). After a comprehensive exploration, the performances of MFSP and the existing methods are compared intuitively while parameters a = 0.6 and b = 5. A family of miRNAs exhibit sequence similarity and has completed identical seed regions. Therefore, miRNAs in the same family are likely to show high functional similarity. MiRNAs belonging to the same family are provided by RFam38. In order to measure the performance of MFSP, the human miRNAs are divided three classes: intra-family, inter-family and randomly selected miRNA group which included intra-family pairs and inter-family pairs (76 families that contain 534 miRNAs). The computed functional similarity scores are shown in Fig. 1(a). It can be seen that, in the intra-family selected miRNA groups, the functional similarity score calculated by MFSP is higher than that of Wang’s method32 and Xuan’s method34, which implemented with the same version of database. Meanwhile, the functional similarity score calculated by MFSP is lower than that of other methods in terms of inter-family groups.
Figure 1. Similarity comparison for MFSP and other methods using miRNA family and miRNA cluster.
In addition, we applied Wilcoxon rank-sum test and Kruskal-Wallis test22 implemented by Matlab to demonstrate significant differences (Table 1). As shown in the Table 1, the functional similarity of intra-family group was significantly greater than that of inter-family (Wilcoxon rank-sum test; intra-inter family p-value = 0.00E-00). The result of Kruskal-Wallis test further clarified that our method is effective (p-value = 0.00E-00). Furthermore, the smaller p-value calculated by Wilcoxon rank-sum test between inter-family and randomly miRNA pairs confirmed that our method performed better than other methods.
Table 1. P-values obtained by Wilcoxon rank-sum testing and Kruskal-Wallis testing functional similarity of the intra-family, inter-family and randomly selected miRNAs.
| Wilcoxon rank-sum test |
Kruskal-Wallis test | |||
|---|---|---|---|---|
| intra-inter | intra-random | inter-random | Intra-inter-random | |
| Wang’s method | 0.00E-00 | 0.00E-00 | 1.30E-03 | 0.00E-00 |
| Xuan’s method | 0.00E-00 | 0.00E-00 | 1.10E-03 | 0.00E-00 |
| CSD+BMA | 0.00E-00 | 0.00E-00 | 9.02E-04 | 0.00E-00 |
| MFSP | 0.00E-00 | 0.00E-00 | 1.94E-04 | 0.00E-00 |
In addition, to validate the advantage of CSD compared with other methods using disease DAG, we integrated CSD with BMA to infer the miRNA similarity since the difference between Wang’s method and Xuan’s method is disease similarity calculation and BMA was employed in the two methods for miRNA functional similarity. From Table 1, the lower p-value is obtained by CSD+BMA, which demonstrates that CSD is more effective than two other methods on calculating the disease semantic similarity.
Mature miRNAs in the same cluster tend to transcribe and express synchronously. Therefore, miRNAs with the higher functional similarity tend in the same cluster. Similarly, we selected 50 kb for genome coordinate data, which are downloaded from miRBase39, as a distance cutoff separated into three classes: intra-cluster, inter-cluster and randomly selected miRNA group which contained intra-cluster and inter-cluster pairs (77 clusters that include 574 miRNAs). The computed functional similarity of intra-cluster, inter-cluster and randomly miRNAs are shown in Fig. 1(b). Compared with other methods, the functional similarity of intra-cluster group obtained by MFSP is higher. The functional similarity of inter-cluster achieved by MFSP is lower than that of other methods simultaneously. Similarly, Wilcoxon rank-sum test and Kruskal-Wallis test were applied for intra-cluster group, inter-cluster group and randomly group to demonstrate significant differences (Table 2). The smaller p-values were obtained by MFSP based on Wilcoxon rank-sum test and Kruskal-Wallis test. It further demonstrated that the performance of MFSP is better than the calculated results of other methods. In addition, CSD+BMA method also could receive lower p-value compared with Wang’s method and Xuan’s method for intra-cluster, inter-cluster and randomly groups, which indicated that CSD may be a better method for disease semantic similarity calculation using DAG of disease.
Table 2. P-values obtained by Wilcoxon rank-sum testing and Kruskal-Wallis testing functional similarity of the intra-cluster, inter-cluster and ran-domly selected miRNAs.
| Wilcoxon rank-sum test |
Kruskal-Wallis test | |||
|---|---|---|---|---|
| intra-inter | intra-random | inter-random | Intra-inter-random | |
| Wang’s method | 3.02E-211 | 3.23E-206 | 1.53E-02 | 1.17E-209 |
| Xuan’s method | 5.58E-223 | 1.15E-217 | 1.27E-02 | 2.23E-221 |
| CSD+BMA | 4.46E-226 | 1.09E-220 | 1.20E-02 | 1.79E-224 |
| MFSP | 0.00E-00 | 0.00E-00 | 2.50E-03 | 0.00E-00 |
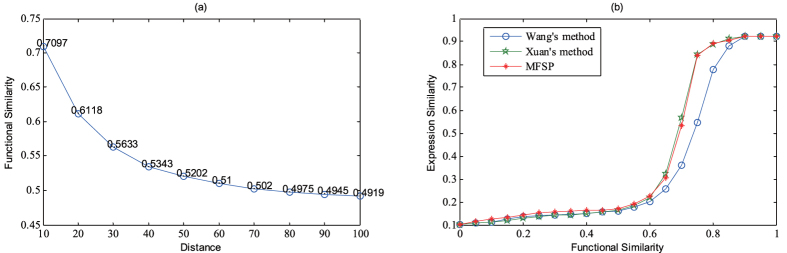
MiRNAs with similar functions tend to be located in the nearby genome coordinate. We grouped miRNAs into different clusters using distance cutoffs from 10 kb to 100 kb by a step of 10 kb. Then, the average functional similarity of intra-cluster miRNA pairs were calculated as shown in the Fig. 2(a). It can be seen that the functional similarity calculated by MFSP is negatively correlated with the distance of genome coordinate.
Figure 2. The relationship between miRNA functional similarity and other information sources of miRNA.
(a) The relationship between distance cutoff for identifying miRNA clusters and miRNA MFSP functional similarity. (b) The relationship between expression similarity and miRNA MFSP functional similarity.
MiRNAs with similar functions are likely to act on the similar cellular components and relate to similar biological processes. Therefore, miRNA with similar functions tend to have similar expression profiles. The expression profiles of 345 miRNAs across 40 normal human tissues were obtained from the supplementary files of the paper by Liang et al.40. PCC (Pearson Correlation Coefficient) was used as the measurement for expression similarity of miRNAs. MiRNAs were grouped into different groups according the threshold value t. It ranges in [0, 1] and the step is 0.05. Then, we calculated the average expression similarity of miRNA pairs whose functional similarity is higher than the threshold value t. As shown in Fig. 2(b), the functional similarity calculated by MFSP is positively correlated with expression similarity of miRNA. In addition, Pearson correlation coefficient (PCC) is employed to numerically demonstrate the relationship between the functional similarity and the miRNA expression similarity (R = 0.8792, P = 0.1551E-06 for MFSP; R = 0.866, P = 0.3919E-06 for Wang’s method; R = 0.8835, P = 0.1112E-06 for Xuan’s method). In respect of correlation coefficient, MFSP is similar to Xuan’s method and better than Wang’s method.
Parameter analysis
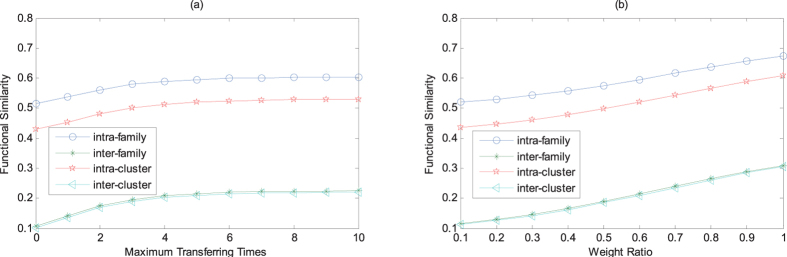
Two issues about the maximum transferring times b and the weight ratio a are analyzed in the experiment for MFSP. MFSP was implemented varying the maximum transferring times b while the weight ratio a is 0.6. As can be seen in Fig. 3(a), the first few transfer times can effectively boost the similarity of miRNA pairs containing intra-family, inter-family, intra-cluster and inter-cluster pairs. In addition, the similarity of miRNA pair is stabilized as the maximum transferring times increase to 5. In Supplementary Table 1, the p-value keeps decreasing expect b ≤ 1 as the maximum transferring times gradually increase Table 3.
Figure 3. MFSP functional similarity with different maximum transferring times and weight ratio.
Table 3. AUC of MIDP applied on miRNA functional similarity network constructed by different methods.
| Disease | MFSP | Wang’s method | Xuan’s method |
|---|---|---|---|
| Melanoma | 0.948 | 0.9467 | 0.9469 |
| Hepatocellular Carcinoma | 0.9674 | 0.9678 | 0.9671 |
| Breast Neoplasms | 0.9592 | 0.9589 | 0.9588 |
| Colorectal Neoplasms | 0.9608 | 0.9583 | 0.9578 |
| Stomach Neoplasms | 0.9435 | 0.9414 | 0.9402 |
| Average AUC | 0.95578 | 0.95462 | 0.95416 |
To test the impact of the weight ratio a, we vary the value of the weight ratio and set b = 5. Results were shown in Fig. 3(b), where it plotted the average similarity of miRNA pairs with different weight ratio. It can be seen that the average similarity of miRNA pairs included intra-family, inter-family, intra-cluster and inter-cluster pairs keeps getting growth as the weight ratio increase. Furthermore, these functional similarity scores for varying weight ratio demonstrate significant differences based on Kruskal-Wallis test and Wilcoxon rank-sum test (Supplementary Table 2).
Construction of miRNA network
We construct the miRNA functional network based on the criterion that an edge is added to link two miRNAs whose functional similarity is greater than 0.7. The threshold is set as 0.7 based on three reasons. Firstly, the average functional similarity of miRNA pairs within 10 kb for genome coordinate is about 0.7. Secondly, while functional similarity of miRNA pair is greater than 0.75, its expression similarity raises slowly. Thirdly, while the threshold is 0.7 rather than 0.75, 85 more miRNAs are contained in the constructed network. The constructed miRNA network includes 422 nodes and 1794 edges and is visualized by Cytoscape (Fig. 4). As shown in Supplementary Figure 1, there are a large number of miRNA partners for a few miRNAs, whereas many miRNAs interact with few miRNA partners. A power law fitting is performed to demonstrate that the node degree follows a power law with a slope of −1.165 and R-squared is 0.676, as expected for a scale-free network (Supplementary Figure 1). Furthermore, the “Random Networks” plugin of Cytoscape41 is executed to achieve the topological measurements and random networks. We find that the characteristic path length of 3.358 for the constructed miRNA functional network is similar to that of random graphs generated by the ER model (3.35035 ± 0.0231). Meanwhile, the higher average clustering coefficient of 0.595 is obtained compared the constructed network with random networks (0.1318098 ± 0.0060293), which demonstrate that the miRNA functional network is a small-world network42. In addition, the CFinder is implemented43 to infer cliques which are all of complete sub-graphs in the miRNA functional network. As shown in Supplementary Figure 2, with an increase in the value of k, the number of cliques is on rise peaking at 29598 while the k value is 7. However, the number of cliques decreases while the k value continues to grow. This phenomenon is according with a principle that the specific regulation is implemented by small clusters rather than individual or big modules.
Figure 4. MiRNA functional network constructed by miRNA functional similarity.
Application
In order to verify the effectiveness of MFSP comparing with other methods, MIDP9 which walks on the miRNA similarity network predicts miRNA-disease associations on miRNA network constructed by different methods (rQ = 0.4, rU = 0.1 for MIDP). We performed experiments using leave-one-out cross validation (LOOCV) scheme which is always used on prediction of miRNA-disease association13. In previous studies8,9,10,11,12,13, the area under the ROC44 curve (AUC) was employed as the main metric for performance evaluation. Five diseases with more than 160 related miRNAs were performed in the experiment (Melanoma, Hepatocellular Carcinoma, Breast Neoplasms, Colorectal Neoplasms, Stomach Neoplasms), since disease with a few miRNAs was not sufficient to evaluate the prediction performance. As a result, our method achieved the average AUC of 0.95578 which is higher than that of other methods (Table 3). The performance of MFSP for all five diseases is superior to other methods except AUC of hepatocellular carcinoma calculated by Wang’s method. It indicated that our method is preferable for predicting miRNA-disease association based on miRNA similarity network. In addition, using MFSP rather than MISIM to calculate the miRNA functional similarity, results of other prediction methods (such as WBSMDA45 and KATZ46) may be improved.
Discussion
In this paper, we presented a path-based calculation method of the miRNA-miRNA functional similarity, called MFSP. It measured the miRNA functional similarity based on the paths among miRNA-related disease sets. The similarities of intra-family, inter-family, intra-cluster and inter-cluster miRNA groups were compared for MFSP and other state-of-the-art methods. The superior effectiveness of MFSP was verified by Wilcoxon rank-sum test and Kruskal-Wallis test. Furthermore, we demonstrated the negative correlation between distance of genome coordinate and miRNA functional similarity and the positive correlation between expression similarity and miRNA functional similarity. The constructed miRNA network based on functional similarity is a scale-free and small-world network. For application of miRNA-disease association prediction, MIDP could obtain higher AUC based on miRNA functional similarity calculated by MFSP. In addition, MFSP could be applied to calculate lncRNA functional similarity using lncRNA-disease associations29,30,31. It also could infer gene functional similarity based on GO terms33, which may contribute to the performance of predicting disease-related target47.
Moreover, we proposed a calculation for disease semantic similarity, called CSD, which transformed all structural disease data into attribute feature vector of disease and then calculated the semantic similarity by traditional cosine similarity. For experimental results, when CSD is employed rather than other calculation methods, results of BMA could be improved.
As future work, other biological information may contribute to further improve the reliability of miRNA functional similarity. For example, instead of using just miRNA-disease association, multiple associations including miRNA-target association and miRNA-disease association may be used for miRNA functional similarity calculation.
Methods
Method Overview
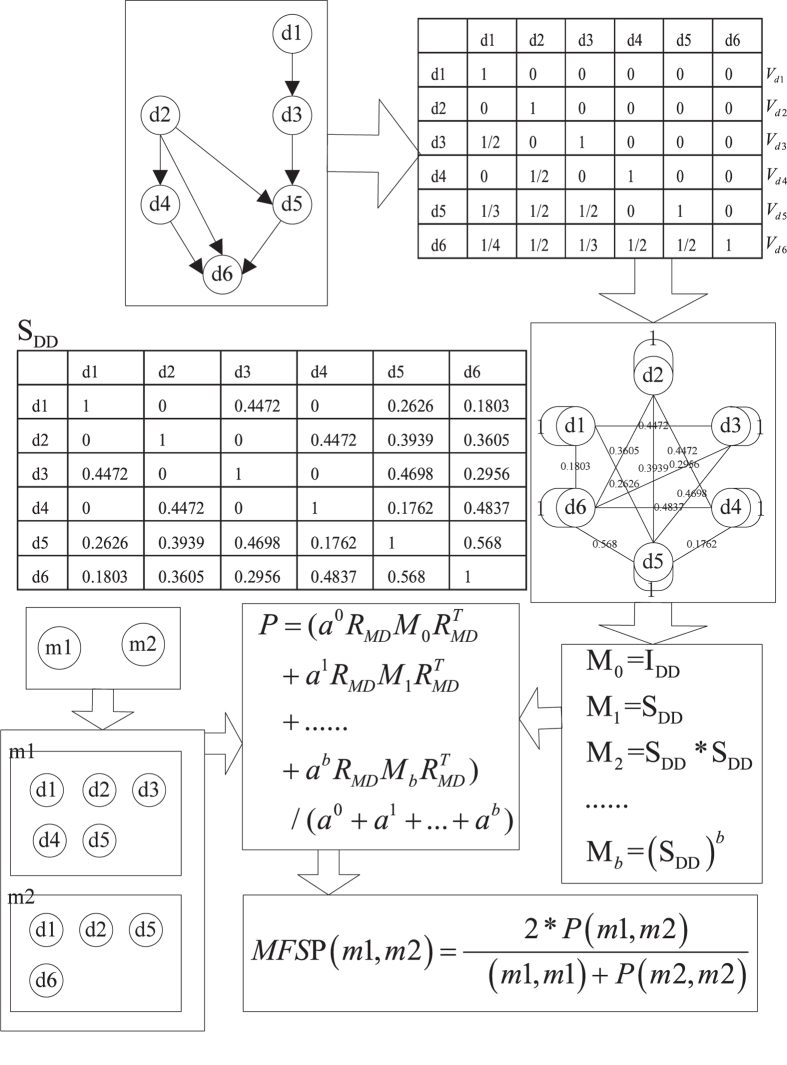
In this study, we presented a method, MFSP, to measure the functional similarity of miRNA pair. The flow chart of MFSP is shown in Fig. 5. First, the hierarchical structure about disease obtained from MeSH descriptor was transferred into features of diseases. Second, the semantic similarity of disease pair was calculated by cosine similarity and the disease similarity network was constructed. Third, the weight sum of paths among diseases was achieved based on the different transferring times respectively. Forth, the miRNA-miRNA path matrix was formed based on the weight sum of pathes among disease sets. Finally, we measured the functional similarity by miRNA-miRNA path matrix. Details of the procedures are given in the following sections.
Figure 5. The flow chart of MFSP.
Cosine Similarity of Disease (CSD)
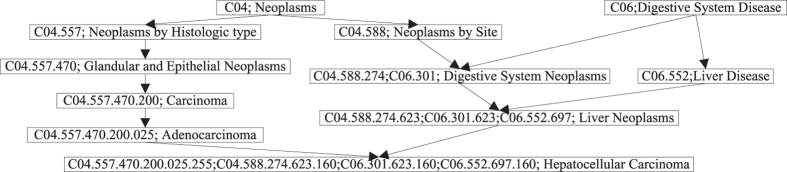
The procedures of CSD are mainly composed of two steps: 1) attribute feature of disease is obtained; 2) cosine similarity is employed. Hierarchical DAG, in which the nodes represent diseases while the links represent relationship between nodes, is provided by the MeSH database (http:www.ncbi.nlm.nih.gov/). Only one type of relationship which represents a child node connecting to a parent node is contained in the DAG. Each disease corresponds to one or more MeSH ID which numerically defines its location in MeSH graph. The codes of a child node are composed of the codes of its parent nodes and the child’s addresses. For instance as shown in Fig. 6, MeSH IDs: C06.301 and C04.588.274 are all corresponding to Digestive System Neoplasms. The entries on its parent nodes (Digestive System Diseases and Neoplasms by Site) have only MeSH ID: C06 and C04.588, respectively.
Figure 6. Hierarchical DAG of Hepatocellular Carcinoma.
In order to apply the traditional methods (similarity measurement, classification and link prediction, and so on) in relational database, the relational structure are always transferred into attribute features of object48,49. In this study, we represent a disease d as a vector Vd of size 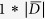 , where
, where 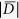 is the number of diseases in the MeSH database. The i-th element of Vd is defined as:
is the number of diseases in the MeSH database. The i-th element of Vd is defined as:
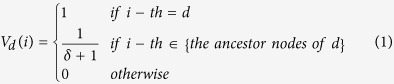 |
where δ is the shortest distance between i-th node, an ancestor node of d, and the disease d. Therefore, the shorter distance of the ancestor node from node d is, namely the more specific denomination of the ancestor node is, the higher value corresponding to the ancestor node will be. For example as shown in Fig. 6, Neoplasms (C04) is an ancestor node of Hepatocellular Carcinoma (C04.588.274.623.160, C04.557.470.200.025.255). The length of two paths from Neoplasms to Hepatocellular Carcinoma is 4 and 5, respectively. Therefore, the element of feature vector for Hepatocellular Carcinoma corresponding to Neoplasms is 1/(4 + 1) = 1/5. Based on the feature vector of each disease formed by equation (1), we then calculated the semantic similarity of disease pair di − dj by cosine similarity:
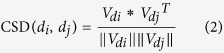 |
where Vdi and Vdj are the feature vector of disease di and dj, respectively. The disease similarity network can be constructed via the semantic similarity of disease pair.
MiRNA functional similarity
Many path-based similarity measurements are utilized in the relationship data and verified effectively35,36. To obtain the more accurate similarity of miRNA pair using paths between disease sets, we need to consider the topological structure of disease network and diseases related to these two miRNAs. Therefore, human miRNA-disease associations are downloaded from HMDD database updated in 201537, which contained 6197 distinct human miRNA-disease associations among 330 diseases, which are included in MeSH, and their related 574 miRNAs. Assumed that |M| and |D| denote the number of miRNAs and diseases in the HMDD respectively, matrix RMD of size  represents the adjacency matrix of miRNA-disease association, where the entry RMD(i, j) in the row i column j is 1 if miRNA i is related to disease j, 0 otherwise. Matrix SDD of size
represents the adjacency matrix of miRNA-disease association, where the entry RMD(i, j) in the row i column j is 1 if miRNA i is related to disease j, 0 otherwise. Matrix SDD of size  is the similarity matrix of disease and its element SDD(di, dj) represents the semantic similarity CSD(di, dj) of di − dj pair. In this study, we defined transferring matrix to describe the topological structure of the disease similarity network. Given the maximum transferring times b, transferring matrix Mi for each transferring times i is defined as follows:
is the similarity matrix of disease and its element SDD(di, dj) represents the semantic similarity CSD(di, dj) of di − dj pair. In this study, we defined transferring matrix to describe the topological structure of the disease similarity network. Given the maximum transferring times b, transferring matrix Mi for each transferring times i is defined as follows:
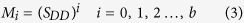 |
Assume that the weight of path is the product of weights of links on it, transferring matrix Mi (i denotes transferring times) is a symmetric matrix, whose each element representing the sum of the i-length path weight for the disease pair.
Here, we utilized the miRNA-miRNA path matrix P to describe the sum of weights of paths between disease sets related to miRNA pair. Since longer path utilized more remote relationships, it was assigned a smaller weight. The different transferring times on the disease similarity networks are combined with a weight ratio a to dampen the contributions from longer paths (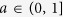 ). Consequently, the miRNA-miRNA path matrix P is defined as follows:
). Consequently, the miRNA-miRNA path matrix P is defined as follows:
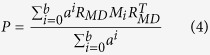 |
Based on the formula (4), we can see that a longer path namely larger transferring times may contribute less than a shorter path namely smaller transferring times. The element P(mi, mj) denotes the sum of the weight of the b maximum-length paths from mi-related diseases to mj-related diseases. Then, MFSP between miRNA mi and mj can be calculated as:
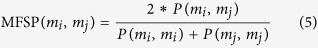 |
The formula (5) shows that MFSP(mi, mj) is composed of two parts: 1) the numerator denotes the connectivity of two miRNA-related disease sets defined by the sum of the path weights between them; and 2) the denominator is used for suppressing miRNA related with many diseases. In addition, MFSP has the symmetric property as the miRNA-miRNA path matrix P is a symmetric matrix, which makes it useful in many applications50.
The symmetric property shows that MFSP has the more general symmetric property for different maximum transferring times. This property is useful for many applications. For instance, if the functional similarity of miRNA pair can be measured and the similarity is symmetric, MIDP algorithm9 can be applied in the similarity matrix directly. Moreover, Cluster One51 method can be performed on the miRNA functional network, a weighted undirected graph which is constructed based on the functional similarity of miRNA pair.
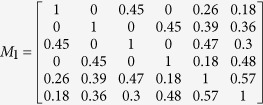
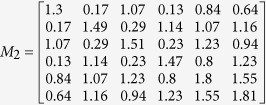
Assume that a directed acyclic graph and miRNA-disease associations are shown in Fig. 5, we simply showed the procedure for calculating the MFSP between m1 and m2. The maximum transferring times b is set as 2 and the weight ratio a is set as 0.6. We can obtain the feature vector Vd1, Vd2, Vd3, Vd4, Vd5 and Vd6 to calculate the disease semantic similarity matrix SDD using cosine similarity (shown in Fig. 5). Then, transferring matrixes for different transferring times are achieved by formula (3) as follows:
 |
 |
 |
We can obtain the adjacency matrix RMD of miRNA-disease based on miRNA-disease association is:
 |
After combined the different transferring matrix and the adjacency matrix of miRNA-disease, the miRNA-miRNA path matrix P is:
 |
As a result, the functional similarity of m1 and m2 is calculated to be (2*7.377)/(9.3741 + 7.5149) = 0.8736, and the functional similarity of m1 − m1 pair is (2*9.3741)/(9.3741 + 9.3741) = 1.
MFSP is implemented in C++ and can be downloaded at https://github.com/KDDing/MFSP.
Additional Information
How to cite this article: Ding, P. et al. A path-based measurement for human miRNA functional similarities using miRNA-disease associations. Sci. Rep. 6, 32533; doi: 10.1038/srep32533 (2016).
Supplementary Material
Acknowledgments
This work has been supported by the National Natural Science Foundation of China (Grant no. 61572180) and Hunan Provincial Natural Science Foundation of China (Grant no. 13JJ2017, no. 2015JJ2032).
Footnotes
Author Contributions P.D. and J.L. conceived the experiments, P.D. and J.L. conducted the experiments, P.D., J.L., Q.X. and X.C. analysed the results. All authors reviewed the manuscript.
References
- Yu J. et al. Human microrna clusters: genomic organization and expression profile in leukemia cell lines. Biochem Bioph Res Co 349, 59–68 (2006). [DOI] [PubMed] [Google Scholar]
- Meng J., Shi G.-L. & Luan Y.-S. Plant mirna function prediction based on functional similarity network and transductive multi-label classification algorithm. Neurocomputing 179, 283–289 (2016). [Google Scholar]
- Xu J. et al. Mirna-mirna synergistic network: construction via co-regulating functional modules and disease mirna topological features. Nucleic Acids Res 39, 825–836 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boross G., Orosz K. & Farkas I.-J. Human micrornas co-silence in well-separated groups and have different predicted essentialities. Bioinformatics 25, 1063–1069 (2009). [DOI] [PubMed] [Google Scholar]
- Zhou Y., Ferguson J., Chang J.-T. & Kluger Y. Inter- and intra- combinatorial regulation by transcription factors and micrornas. Bmc Genomics 8, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liang C., Wong K.-C., Luo J. & Zhang Z. Mirsynergy: detecting synergistic mirna regulatory modules by overlapping neighbourhood expansion. Bioinformatics 30, btu373 (2014). [DOI] [PubMed] [Google Scholar]
- Li Y., Liang C., Wong K.-C., Jin K. & Zhang Z. Inferring probabilistic mirna-mrna interaction signatures in cancers: a role-switch approach. Nucleic Acids Res 42, e76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Xuan Z. & Quan Z. Integrative approaches for predicting microrna function and prioritizing disease-related microrna using biological interaction networks. Briefings in bioinformatics 8, bbv033 (2015). [DOI] [PubMed] [Google Scholar]
- Xuan P. et al. Prediction of potential disease-associated micrornas based on random walk. Bioinformatics 31, btv039 (2015). [DOI] [PubMed] [Google Scholar]
- Chen X., Liu M.-X. & Yan G.-Y. Rwrmda: predicting novel human microrna-disease associations. Mol Biosyst 8, 2792–2798 (2012). [DOI] [PubMed] [Google Scholar]
- Chen H.-L. & Zhang Z.-P. Similarity-based methods for potential human microrna-disease association prediction. Bmc Med Genomics 6, Artn 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Li J., Song L., Zeng X. & Wang G. Similarity computation strategies in the microrna-disease network: a survey. Brief Funct Genomics 15, 55–64 (2016). [DOI] [PubMed] [Google Scholar]
- Chen X. & Yan G.-Y. Semi-supervised learning for potential human microrna-disease associations inference. Sci Rep-Uk 4, 5501, 10.1038/srep05501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. mirbase: the microrna sequence database. Methods in molecular biology 342, 129–138 (2006). [DOI] [PubMed] [Google Scholar]
- Teng Z. et al. Measuring gene functional similarity based on group-wise comparison of go terms. Bioinformatics 29, btt160 (2013). [DOI] [PubMed] [Google Scholar]
- Shalgi R., Lieber D., Oren M. & Pilpel Y. Global and local architecture of the mammalian microrna-transcription factor regulatory network. Plos Comput Biol 3, e131 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.-H. et al. Mirtarbase 2016: updates to the experimentally validated mirna-target interactions database. Nucleic Acids Res 44, D239–D247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos G.-L., Reczko M., Simossis V.-A., Sethupathy P. & Hatzigeorgiou A.-G. The database of experimentally supported targets: a functional update of tarbase. Nucleic Acids Res 37 (suppl 1), D155–D158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. et al. A new method for measuring functional similarity of micrornas. Journal of integrated omics 1, 49–54 (2010). [Google Scholar]
- Sun J. et al. Inferring potential microrna-microrna associations based on targeting propensity and connectivity in the context of protein interaction network. Plos One 8, e69719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Guo M., Liu X., Wang C. & Liu Y. Inferring the soybean (glycine max) microrna functional network based on target gene network. Bioinformatics 30, 94–103 (2014). [DOI] [PubMed] [Google Scholar]
- Meng J., Liu D. & Luan Y.-S. Inferring plant microrna functional similarity using a weighted protein-protein interaction network. Bmc Bioinformatics 16, Artn 360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. et al. Gosemsim: an r package for measuring semantic similarity among go terms and gene products. Bioinformatics 26, 976–978 (2010). [DOI] [PubMed] [Google Scholar]
- Meng J., Shi L. & Luan Y.-S. Plant microrna-target interaction identification model based on the integration of prediction tools and support vector machine. Plos One 9, e103181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Luo J., Liang C., Wang S. & Song D. Moepga: A novel method to detect protein complexes in yeast protein-protein interaction networks based on multi objective evolutionary programming genetic algorithm. Comput Biol Chem 58, 173–181 (2015). [DOI] [PubMed] [Google Scholar]
- Luo J. & Liang S. Prioritization of potential candidate disease genes by topological similarity of protein-protein interaction network and phenotype data. J Biomed Inform 53, 229–236 (2015). [DOI] [PubMed] [Google Scholar]
- Liu Y., Zeng X., He Z. & Zou Q. Inferring microrna-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Transactions on Computational Biology and Bioinformatics 2550432, 10.1109/TCBB.2016.2550432 (2016). [DOI] [PubMed] [Google Scholar]
- Lipscomb C.-E. Medical subject headings (mesh). Bulletin of the Medical Library Association 88, 265 (2000). [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-A., Chen X., You Z.-H., Huang D.-S. & Chan K.-C. Ilncsim: improved lncrna functional similarity calculation model. Oncotarget 8296, 10.18632/oncotarget.8296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Predicting lncrna-disease associations and constructing lncrna functional similarity network based on the information of mirna. Scientific reports 5, 13186, 10.1038/srep13186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. Constructing lncrna functional similarity network based on lncrna-disease associations and disease semantic similarity. Scientific reports 5, 11338, 10.1038/srep11338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Wang J., Lu M., Song F. & Cui Q. Inferring the human microrna functional similarity and functional network based on microrna-associated diseases. Bioinformatics 26, 1644–1650 (2010). [DOI] [PubMed] [Google Scholar]
- Wang J.-Z., Du Z., Payattakool R., Philip S.-Y. & Chen C.-F. A new method to measure the semantic similarity of go terms. Bioinformatics 23, 1274–1281 (2007). [DOI] [PubMed] [Google Scholar]
- Xuan P. et al. Prediction of micrornas associated with human diseases based on weighted k most similar neighbors. Plos One 8, e70204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han J., Yan X., Yu P.-S. & Wu T. Pathsim: Meta path-based top-k similarity search in heterogeneous information networks. Proceedings of the VLDB Endowment 4, 992–1003 (2011). [Google Scholar]
- Shi C., Kong X., Huang Y., Philip S.-Y. & Wu B. Hetesim: A general framework for relevance measure in heterogeneous networks. IEEE Transactions on Knowledge and Data Engineering 26, 2479–2492 (2014). [Google Scholar]
- Li Y. et al. Hmdd v2.0: a database for experimentally supported human microrna and disease associations. Nucleic Acids Res 42, gkt1023 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E.-P. et al. Rfam 12.0: updates to the rna families database. Nucleic Acids Res 43, gku1063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S. & Bartel D.-P. Microarray profiling of micrornas reveals frequent coexpression with neighboring mirnas and host genes. Rna 11, 241–247 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L. & Chen C. Characterization of microrna expression profiles in normal human tissues. Bmc Genomics 8, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A.-L. & Albert R. Emergence of scaling in random networks. Science 286, 509–512 (1999). [DOI] [PubMed] [Google Scholar]
- Adamcsek B., Palla G., Farkas I.-J., Dere’nyi I. & Vicsek T. Cfinder: locating cliques and overlapping modules in biological networks. Bioinformatics 22, 1021–1023 (2006). [DOI] [PubMed] [Google Scholar]
- Davis J. & Goadrich M. The relationship between precision-recall and roc curves. ICML 23, 233–240 (2006). [Google Scholar]
- Chen X. et al. Wbsmda: within and between score for mirna-disease association prediction. Scientific reports 6, 21106, 10.1038/srep21106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q. et al. Prediction of microrna-disease associations based on social network analysis methods. BioMed research international 810514, 10.1155/2015/810514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Liao Y. & Zou Q. Prediction and validation of disease genes using hetesim scores. IEEE/ACM Transactions on Computational Biology and Bioinformatics 2520947, 10.1109/TCBB.2016.2520947 (2016). [DOI] [PubMed] [Google Scholar]
- Kong X., Yu P.-S., Ding Y. & Wild D.-J. Meta path-based collective classification in heterogeneous information networks. CIKM 21, 1567–1571 (2012). [Google Scholar]
- Zhang J., Yu P.-S. & Zhou Z.-H. Meta-path based multi-network collective link prediction. SIGKDD 20, 1286–1295 (2014). [Google Scholar]
- Xia Q. The geodesic problem in quasimetric spaces. J Geom Anal 19, 452–479 (2009). [Google Scholar]
- Nepusz T., Yu H. & Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 9, 471–472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.