Abstract
Hepatocellular carcinoma (HCC) is the leading cause for cancer death worldwide, new prognostic factors and targets are critical for HCC treatment. Here, we found GRAMD1A was upregulated in HCC tissues, patients with high GRAMD1A levels had poor outcome, statistical analyses found GRAMD1A expression was positively correlated with pathologic differentiation and survival or mortality. It was an unfavorable prognostic factor for HCC patients. Functional analyses revealed GRAMD1A contributed to the self-renewal of HCC stem cells, resistance to chemotherapy and tumor growth of HCC determined by hepatosphere formation assay, side population (SP) analysis, TUNEL assay, soft agar growth ability assay and tumor growth model in vivo. Mechanism analyses found signal transducer and activator of transcription 5 (STAT5) was the target of GRAMD1A, GRAMD1A regulated the target genes of STAT5 and the transcriptional activity of STAT5. Inhibition of STAT5 in indicated HCC cells overexpressing GRAMD1A suppressed the effects of GRAMD1A on the self-renewal of HCC stem cell, resistance to chemotherapy and tumor growth, suggesting GRAMD1A promoted the self-renewal of HCC stem cells and the development of HCC by increasing STAT5 level. GRAMD1A might be a useful biomarker and target for HCC.
According to the data of GLOBOCAN 2012 provided by the IARC, about 782,5002 new HCC cases and 745,500 deaths occurred during 2014 worldwide. Almost 50% of new cases and deaths were Chinese, so the development of new prognostic factors and therapy targets is a big challenge for Chinese scientists1. Despite of the advances in diagnosis and treatment, the recurrence rate of patients after resection, chemotherapy and radiotherapy is still high due to high metastasis ability and treatment resistance. Our understanding of regulatory mechanisms of HCC initiation and progression is limited so far.
Recent years, a growing number evidences suggest cancer stem cells (CSCs) play a critical role in tumor relapse, CSCs are a small fraction of a tumor, and have the ability of initiation tumorigenecity, they can maintain the CSCs number or generate more CSCs by self-renewal, and differentiate into non-tumorigenic tumor cells to maintain tumor heterogeneity2,3. Chemotherapy and radiotherapy only kill non-CSCs, but CSCs still exist, so the tumors often relapse. Combining targeting CSCs drugs with chemotherapy or radiotherapy could kill all cancer cells4. HCC stem cells can be enriched using specific surface markers, like CD1335, CD906, CD247 and so on.
Many signaling pathways regulate the self-renewal of HCC stem cell and development of HCC, like annexin A3/ c-Jun N-terminal kinase (ANXA3/JNK) signaling promotes tumorigenesis, chemoresistance, metastasis, angiogenesis and self-renewal of HCC stem cells8. IL-6/Jak2/Stat3 pathway promotes HCC growth, chemoresistance and the self-renewal of HCC stem cells9,10,11. GRAMD1A first finds in human embryonic stem cell, and expresses in ectoderm, mesoderm and endoderm tissues and many tumor cells12, its function hasn’t been explored. Here, we studied the role of GRAMD1A in the prognosis, initiation and progression of HCC, we analyzed the correlation between GRAMD1A levels and many clinicopathological features, and examined whether GRAMD1A could function as a prognostic factor for HCC patients. Then we determined the role of GRAM1DA in the self-renewal of HCC stem cell and resistance to chemotherapy by overexpressing GRAMD1A, and the role of GRAMD1A in tumor growth by overexpressing or downregulating GRAMD1A. In the end, we studied its regulatory target, we found GRAMD1A was a prognostic factor for HCC, it promoted the self-renewal of HCC stem cell, resistance to chemotherapy and tumor growth through promoting STAT5, Inhibition of STAT5 in indicated cells with GRAMD1A overexpression inhibited the self-renewal of HCC stem cell, resistance to chemotherapy and tumor growth.
Materials and Methods
Ethics statement
All the clinical specimens were obtained with informed consent and approved by the First Affiliated Hospital of Sun Yat-sen University Ethics Committee, Informed consent was obtained from all patients involved in this study. Animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. All the experiments were performed in accordance with the approved guidelines of the Institutional Research Ethics Committee of Sun Yat-Sen University.
Tissue specimens and cell culture
A total of 96 human advanced HCC specimens were obtained from Department of Medical Oncology, the First Affiliated Hospital of Sun Yat-sen University. The detailed information was shown in Supplemental Table 1. Human HCC cells Huh7 and HepG2 were obtained from American Type Culture Collection (ATCC), and growth in DMEM medium supplanted with 10% FBS (GIBCO) and 1% penicillin-streptomycin (Hyclone).
Vectors, oligonucleotides and transfection
The CDS of GRAMD1A was amplified form cDNA of human embryonic stem cell, and cloned into the pMSCV-Puro vector (indicated as GRAMD1A), the empty vector was used as negative control (indicated as Vector). small inference RNAs (siRNA) of GRAMD1A (indicated as siGRAMD1A) and their Scramble control were purchased from RiboBio Co. Ltd. The siRNAs were modified with 2′-OMe. The Vectors and oligonucleotides was transfected into Hun-7 and HepG2 using lipofectamine 2000 (Invitrogen).
Quantitative real-time PCR
Total RNA was isolated using Trizol reagent (Life Technologies), cDNA was synthesized using PrimeScript RT Master Mix (Takara). Quantitative real-time PCR was performed using SYBR Green PCR Master Mix (Takara) on a ABI Prism 7500 Sequence Detection system (Applied Biosystems). The quantitative real-time PCR reactions were carried out in triplicate. GAPDH was used as an endogenous control to normalize the amount of mRNA in each sample. The relative expression was calculated with the 2−ΔΔCt method. The primers for these genes were shown in Supplemental Table 2.
Western blot
Tumor cells were homogenized with PIPA buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP40 and 1 mM EDTA) supplemented with cocktail protease inhibitor (Roche). Western blot was performed according to the standard methods. The following antibodies were used: GRAMD1A (Sigma, HPA008852), caspase-3 (Cell Signaling Technology, #9662), poly(ADP-ribose) polymerase (PARP) (Cell Signaling Technology, #9542), BCL-XL (Cell Signaling Technology, #2762), CD133 (Miltenyi Biotec, 130-092-395), CD90 (Abcam, ab133350), STAT5 (Cell Signaling Technology, #9363), β-actin (Abcam, ab8226) and GAPDH (Abcam, ab8245).
Luciferase reporter assay
The promoter sequence of STAT5 was amplified from genomic DNA prepared from 293T cells using PCR, and cloned into psi-CHECK2 vector (Promega). The vector was cotransfected into indicated cells with GRAMD1A overexpression vector or siRNAs of GRAMD1A using lipofectamine 2000 (Invitrogen). The luciferase activity was measured after 48 h of transfection using Dual Luciferase Assay (Promega) according to the manufacturer’s protocol.
Soft agar growth ability assay
Soft agar growth ability assay was performed according to previously reported method13.
Apoptosis assay
Cells were treated with various concentrations of Doxorubicin (0.5 uM and 1.0 uM, respectively) for 48 h. Following treatment, cells were harvested and stained with 0.1% Trypan blue, cell number was counted with a microscope. For TUNEL assay, slides were plated in six-well plates, then cells were seeded in the plates, and treated with 1.0 uM Doxorubicin for 48 h, after treatment, The TUMEL assay was performed using In situ Direct DNA Fragmentation Assay Kit (Abcam, ab66108) according to the instructions of manufacturer.
Immunohistochemistry (IHC)
IHC was performed according to previous methods14,15. Anti-GRAMD1A antibody (Sigma, HPA008852) was used. The tissue sections were scored using two-blinded method. The proportion of tumor cells was scored as follows: Score 0, no positive cells; Score 1, 1–10% positive cells; Score 2, 11~50% positive cells; Score 3, 51–80% positive cells; Score 4, mane than 80% positive cells. The intensity of protein expression was shown as follows: 0 (no staining); 1 (weak staining, light yellow); 2 (moderate staining, yellowish brown) and 3 (strong staining, brown).
The staining index (SI) was calculated as the product of the staining intensity and the proportion of positive cell scores (scored as 0, 1, 2, 3, 4, 6, 8, 9 or 12). Cut-off values for GRAMD1A expression were chosen based on a measurement of heterogeneity using the log-rank test with respect to overall survival.
Hepatosphere formation assay
200 Huh-7 or HepG2 cells were seeded in Ultra Low Attachment 6-well plates (Corning) and maintained with DMEM/F12 medium (Life Technologies) supplemented with 20 ng/ml human recombinant epidermal growth factor (Sigma), 10 ng/ml human recombinant basic fibroblast growth factor (bFGF, Millipore), 4 ug/ml insulin (Sigma), B27 (Life Technologies), 500 U/ml penicillin, 500 ug/ml streptomycin and 1% methylcellulose. Spheres were incubated in suspension for 2 weeks and counted under a microscope.
Side population (SP) assay
Cells were resuspended at the density of 1 × 106/ml in DMEM (Life Technologies), supplementing with 2% Fetal calf serum (FCS) (Life Technologies) and HEPES buffer (Life Technologies), and incubated with 5 ug/ml Hoechst 33342 dye in the presence or absence of Verapamil for 90 min at 37 °C with intermittent shaking. Then cells were washed using cold HBSS with 2% FCS and 10 mmol/L HEPES following centrifugation at 4 °C, and resuspended in cold HBSS with 2% FCS and 10 mmol/L HEPES. PI (propidium iodide) was added to gate viable cells. Cells were analyzed using a FACS Vantage-SE (BD).
Animal studies
BALB/c-nu mice were purchased from the Experimental Animal Center of the Guangzhou University of Chinese Medicine. Xenograft tumors were established by subcutaneous injection of different number (1 × 105, 1 × 104 and 1 × 103) Huh-7 cells into the flank of female BALB/C nude mice about 4-to-5 week old. Tumor sizes were measured every 6 days by calipers, tumor volumes were calculated according to the formula V = L × W2 × 0.5 (L: tumor length, W: tumor width). On day 31, animals were euthanized and tumors were excised. For orthotopic transplantation mouse model, 5 × 106 Hub-7 cells with GRAMD1A knockdown or negative control were transplanted into the liver of mouse (n = 8) respectively, the mouse was fed for 40 days, the survival of mice was observed. The blood of mouse was extracted to investigate the concentration of ALT and AST.
Statistical analysis
All statistical analyses were performed with SPSS 19.0 software (SPSS) or Excel (Microsoft). GRAMD1A expression data was downloaded from The Cancer Genome Atlas (TCGA) (https://gdc-portal.nci.nih.gov/projects/TCGA-LIHC). The Chi-square test and Fisher’s Exact test were performed to analyze the correlation between GRAMD1A levels and HCC clinical features. The Spearman correlation analysis was used to confirm the correlation between GRAMD1A levels and clinical features. Independent prognostic factors were examined by the Cox proportional hazards stepwise regression model. Survival curve was plotted by Kaplan-Meier survival analysis and compared by the log-rank test. Gene set enrichment analysis (GSEA) analysis was performed using online website (http://software.broadinstitute.org/gsea/index.jsp)16. Results were showed as the Mean ± SEM. A two-tailed paired student’s t test was used to assess the significant difference of two groups of data. A p value of less than 0.05 was considered statistical significance.
Results
GRAMD1A overexpression is positively associated with HCC progression
To determine the role of GRAMD1A in HCC progression, we used TCGA dataset to investigate GRAMD1A expression in HCC tissues and normal live tissues, and found GRAMD1A was significantly upregulated in HCC tissues (Fig. 1a). To examine the association between GRAMD1A expression and advanced HCC, we chose 78 patients with advanced HCC (Pathologic Stage III–IV) to analyze the association between GRAMD1A expression and survival time, the log-rank test suggested patients with high GRAMD1A levels had poor outcome (p = 0.000, Fig. 1b). GSEA was used to confirm this results, and found high GRAMD1A expression was positively correlated with low HCC survival and inversely correlated with high HCC survival (Fig. 1c). These results suggested GRAMD1A was associated with HCC progression.
Figure 1. HCC patients with high GRAMD1A levels have poor outcome.
(a) Analysis GRAMD1A mRNA levels in HCC tissues and normal liver tissues using TCGA data set. (b) Kaplan-Meier curves with log rank test based on the expression of GRAMD1A using TCGA data set. (c) Analysis of the correlation between GRAMD1A levels and survival state using GSEA, data was downloaded from TCGA data set (Pathologic stage III∽IV). (d) IHC assay of GRAMD1A levels in HCC tissues (top, 100X; bottom, 400X). Representative figure (left), statistical analysis of GRAMD1A levels in patients with good prognosis and poor prognosis (right). (e) Kaplan-Meier plots of overall survival based on the expression of GRAMD1A.
To further demonstrate above results, a cohort of 96 advanced HCC tissues was used to study the role of GRAMD1A in clinical progression, IHC analysis suggested GRAMD1A expression was positive in 96.9% (93/96) of examined tissues, only 3% (3/96) was negative. GRAMD1A is upregulated in 60.4% (58/96) tissues, only 39.6% was low expression (Supplemental Table 3). We next analyzed the association between GRAMD1A levels and the clinicopathological features of HCC tissues, as shown in Table 1, high GRAMD1A was significantly correlated with pathological differentiation (p = 0.036 measured by Chi-square test, p = 0.043 measured by Fisher’s Exact test) and Survive or Mortality (p = 0.002, measured by both Chi-square test and Fisher’s Exact test) (Table 1). In contrast, GRAMD1A levels wasn’t associated with gender, age, clinical stage, TNM classification, cirrhosis or HbsAg. We used Spearman correlation analysis to confirm this result, and found high GRAMD1A levels was significantly correlated with pathologic differentiation (rs = 0.210, p = 0.040) and Survival or Mortality (rs = 0.329, p = 0.001), suggesting GRAMD1A was associated with HCC progression. IHC analysis also found GRAMD1A was overexpressed in HCC tissues of patients with poor prognosis compared to HCC tissues of patients with good prognosis (p < 0.001). The detail data was shown as follows: GRAMD1A was overexpressed in 26.3% (5/19) tissues of patients with good prognosis, but it’s overexpressed in 68.8% (53/77) tissues of patients with poor prognosis (Fig. 1d). The log-rank test also revealed patients with high GRAMD1A levels had poor outcome (p = 0.000, Fig. 1e), suggesting GRAMD1A might be a prognostic factor for HCC patient. Univariate Cox regression analyses revealed T classification (p = 0.007) and GRAMD1A level (p = 0.000) were poor prognostic factors for HCC patients, Multivariate Cox regression analyses further revealed they are independent prognostic factors for HCC patients (Table 2). Together, these findings revealed GRAMD1A is associated with HCC progression and is an independent prognostic factor for HCC patients.
Table 1. Correlation between GRAMD1A expression and clinicopathologic characteristics of HCC.
| Characteristics | GRAMD1A | Chi-square test P-value | Fisher’s Exact test P-value | ||
|---|---|---|---|---|---|
| Low No. cases | High No. cases | ||||
| Gender | Male | 30 | 46 | 1.000 | 1.000 |
| Female | 8 | 12 | |||
| Age (years) | >45 | 24 | 44 | 0.251 | 0.251 |
| ≤45 | 14 | 14 | |||
| Clinical Stage | IIIa | 21 | 37 | 0.296 | 0.296 |
| IIIb | 14 | 13 | |||
| IIV | 3 | 8 | |||
| T classification | T1 | 15 | 20 | 0.973 | 0.973 |
| T2 | 7 | 12 | |||
| T3 | 9 | 15 | |||
| T4 | 7 | 11 | |||
| N classification | N0 | 23 | 30 | 0.411 | 0.411 |
| N1 | 15 | 28 | |||
| M classification | No | 35 | 50 | 0.518 | 0.518 |
| Yes | 3 | 8 | |||
| Pathologic Differentiation | Well | 2 | 0 | 0.036 | 0.043 |
| Moderate | 27 | 34 | |||
| poor | 9 | 24 | |||
| Cirrhosis | Yes | 21 | 26 | 0.404 | 0.404 |
| No | 17 | 32 | |||
| HBsAg | Yes | 28 | 40 | 0.654 | 0.654 |
| No | 10 | 18 | |||
| Survive or Mortality | Yes | 11 | 3 | 0.002 | 0.002 |
| No | 27 | 55 | |||
Table 2. Univariate and multivariate analyses of various prognostic parameters in patients with HCC Cox-regression analysis.
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| No. patients | P | Regression coefficient (SE) | P | Relative risk | 95% confidence interval | |
| T classification | ||||||
| T1 | 35 | 0.007 | 0.273 (0.102) | 0.005 | 1.333 | 1.090–1.630 |
| T2 | 19 | |||||
| T3 | 24 | |||||
| T4 | 18 | |||||
| Expression of GRAMD1A | ||||||
| Low expression | 38 | 0.000 | 1.041 (0.248) | 0.000 | 2.901 | 1.776–4.739 |
| High expression | 58 | |||||
GRAMD1A promotes the self-renewal of HCC stem cells and resistance to chemotherapy
We used GSEA to analyze the association between GRAMD1A levels and tumor recurrence, and found patients with high GRAMD1A levels had high recurrence rate (Fig. 2a). Meanwhile, we found low GRAMD1A levels correlated with repressed self-renewal of HCC stem cell, suggesting GRAMD1A might regulate the tumor growth, drug resistance and the expansion of HCC stem cells. To determine GRAMD1A’s role in the initiation and progression of HCC, we overexpressed GRAMD1A in HCC cells Huh7 and HepG2. Hepatosphere formation assay suggested overexpression of GRAMD1A increased the number and volume of hepatosphere (Fig. 2b). SP assay found overexpression of GRAMD1A increased the number of SP population (Fig. 2c). The markers of HCC stem cells, CD90 and CD133, were used to demonstrate GRAMD1A regulates HCC stem cells, western blot assay found overexpression of GRAMD1A increased CD90 and CD133 levels, while knockdown of GRAMD1A decreased CD90 and CD133 levels (Fig. 2d). These results suggested GRAMD1A promoted the expansion of HCC stem cells. In order to investigate the effect of GRAMD1A on drug resistance, we use chemotherapy drug Doxorubicin (Sigma) to treat indicated HCC cells. Cell survival assay found after treatment with Doxorubicin (dissolved with DMSO, 0.5 uM and 1.0 uM, respectively), the number of survival cells with GRAMD1A was significantly more than negative control cells (Fig. 2e). Apoptosis assay was used to confirm this result, TUNEL assay found after treatment with Doxorubicin (1.0 uM), the number of TUNEL positive cells was significantly decreased compared to negative control (Fig. 2f). Cleavages of pro-caspase and PARP are the markers of apoptosis17,18, western blot assay found GRAMD1A overexpression inhibited the activation of caspase 3 and the cleavage of PARP, but the expression of an anti-apoptotic protein BCL-XL was increased (Fig. 2g), suggesting GRAMD1A increased the drug resistance of HCC by inhibiting apoptosis. Together, these results suggested GRAMD1A promotes the expansion of HCC stem cells and resistance to chemotherapy.
Figure 2. GRAMD1A promotes the self-renewal of HCC stem cell and the resistance to chemotherapy.
(a) GSEA analysis of the correlation between GRAMD1A levels and tumor recurrence and HCC stem cells gene signature, data set was downloaded from TCGA data set. (b) Hepatosphere formation assay for the effect of GRAMD1A overexpression on self-renewal of HCC stem cells in Huh-7 and HepG2 cells. Representative micrographs (left); quantification of hepatosphere number (right). (c) SP analysis demonstrated the role of GRAMD1A in self-renewal of HCC stem cells by GRAMD1A overexpression. (d) Western blot assay of the expression of HCC stem cells markers CD130 and CD90 by overexpressing or downregulating GRAMD1A in HepG2. (e) Cell viability assay demonstrated the role of GRAMD1A in resistance to Doxorubicin by GRAMD1A overexpression. (f) TUNEL assay for the effect of GRAMD1A overexpression on resistance to Doxorubicin. Representative micrographs (left); quantification of TUNEL positive cell number (right). DAPI was used to stain nucleus. (g) Western blot assay for caspase 3 activity, PARP cleavage and the BCL-XL levels after GRAMD1A overexpression in Huh-7 and HepG2. Data are expressed as mean ± SEM. *p < 0.05.
GRAMD1A promotes HCC growth
We further determined the role of GRAMD1A in tumor growth. Soft agar growth ability assay suggested overexpression of GRAMD1A significantly promoted anchorage-independent growth, while knockdown of GRAMD1A significantly suppressed anchorage-independent growth (Fig. 3a). We injected subcutaneously different number of cells into the flank of nude mice, and found overexpression of GRAMD1A promoted tumor growth, the tumor volume was larger than negative control. While knockdown of GRAMD1A suppressed tumor growth, the tumor volume was smaller than Scramble control (Fig. 3b). We used orthotopic transplantation mouse model to conform this findings, knockdown of GRAMD1A inhibited HCC growth (Fig. 3c), survival cure analysis revealed the survival of mouse with GRAMD1A was significantly longer than Scramble group (Fig. 3d). We also analyzed the markers of liver injure, including ALT (Alanine aminotransferase) and AST (Aspartate aminotransferase), and found GRAMD1A knockdown significantly reduced the concentration of AST and ALT, confirming GRAMD1A promotes HCC growth (Fig. 3e).
Figure 3. GRAMD1A promotes HCC growth.
(a) Soft agar growth ability assay for the role of GRAMD1A in the anchorage-independent cell growth. Representative micrographs (left); quantification of colony number with diameter greater than 0.1 mm (right). (b) Xenograft tumors model for effect of GRAMD1A overexpression or knockdown on the growth of Huh-7 xenografts. Representative tumor images (left); the tumor volume (right). (c) Representative tumor images of orthotopic transplantation mouse model using Huh-7 with GRAMD1A knockdown and negative control. (d) Kaplan-Meier curves with log rank test for orthotopic transplantation mouse model. (e) the concentration of AST and ALT of orthotopic transplantation mouse model. Data are expressed as mean ± SEM. *p < 0.05.
STAT5 is a downstream target of GRAMD1A
STATs play a key role in cell proliferation, differentiation, migration ad survival. STAT3 promotes HCC initiation and development, but the role of STAT5 in the initiation and development of human HCC has been reported rarely, we used TCGA data to analyze the relationship between GRAMD1A levels and the levels of STAT5 targeting genes by GSEA, and found a positive correlation between GRAMD1A levels and the levels of STAT5 targeting genes, suggesting GRAMD1A might regulate STAT5 (Fig. 4a). We also found overexpression of GRAMD1A increased STAT5 levels in both Huh-7 and HepG2 cells, while knockdown of GRAMD1A reduced STAT5 levels (Fig. 4b). Cyclin D1, Bcl-2, c-Jun and c-Myc are the target of STAT519, GRAMD1A overexpression promoted these genes expression, while GRAMD1A knockdown inhibited these genes expression (Fig. 4b), suggesting STAT5 is a downstream of GRAMD1A. We cloned the promoter sequence of STAT5 into luciferase reporter vector, and transfected into indicated cells. Overexpression of GRAMD1A significantly increased luciferase activity, knockdown of GRAMD1A significantly decreased luciferase activity (Fig. 4c). These findings revealed GRAMD1A could regulate STAT5.
Figure 4. STAT5 is a downstream target of GRAMD1A.
(a) Analysis of the correlation between GRAMD1A levels and STAT5 levels using GSEA. (b) Quantitative real-time PCR assay for the mRNA levels of STAT5, CyclinD1, Bcl-2, c-jun and c-Myc after GRAMD1A overexpression or knockdown. (c) Luciferase activity assay was used to determine whether GRAMD1A could regulate the transcriptional activity of STAT5. Data are expressed as mean ± SEM. *p < 0.05.
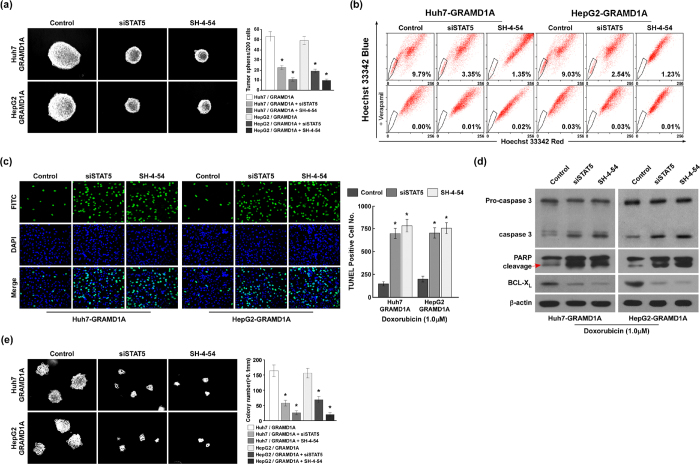
GRAMD1A regulates the expansion of HCC stem cells, resistance to chemotherapy and tumor growth through regulating STAT5
To investigate whether GRAMD1A regulates HCC initiation and development through regulating STAT5, we downregulated STAT5 in indicated HCC cells with GRAMD1A overexpression by siRNA for STAT5 and STAT inhibitor SH-4-54 (Selleckchem)20. Hepatosphere formation assay revealed downregulation of STAT5 significantly inhibited the number and volume of hepatosphere (Fig. 5a). SP analysis found downregulation of STAT5 reduced the percentage of the cell number of SP (Fig. 5b). These findings suggested GRAMD1A regulates the expansion of HCC stem cells through regulating STAT5. TUNEL assay revealed that TUNEL positive cells were significantly increased when STAT5 was downregulated in indicated cells with GRAMD1A overexpression (Fig. 5c). Western blot assay found downregulation of STAT5 promoted caspase 3 activation and PARP cleavage, and inhibited BCL-XL levels (Fig. 5d). These findings suggested GRAMD1A regulated resistance to chemotherapy through regulating STAT5. Soft agar growth ability assay found downregulation of STAT5 significantly inhibited anchorage-independent growth (Fig. 5e), suggesting GRAMD1A regulates tumor growth through regulating STAT5.
Figure 5. GRAMD1A promotes the self-renewal of HCC stem cells, resistance to chemotherapy and tumor growth.
(a) Hepatosphere formation assay for the effect of STAT5 downregulation on self-renewal of HCC stem cells in Huh-7 and HepG2 cells with GRAMD1A overexpression. Representative micrographs (left); quantification of hepatosphere number (right). (b) SP analysis demonstrated the role of STAT5 downregulation in self-renewal of HCC stem cells in Huh7 and HepG2 with GRAMD1A overexpression. (c) TUNEL assay for the effect of STAT5 downregulation on resistance to Doxorubicin in Huh7 and HepG2 with overexpression. Representative micrographs (left); quantification of TUNEL positive cell number (right). DAPI was used to stain nucleus. (d) Western blot assay for caspase 3 activity, PARP cleavage and BCL-XL levels after STAT5 downregulation in Huh-7 and HepG2 with GRAMD1A overexpression. (e) Soft agar growth ability assay for the role of STAT5 downregulation in the anchorage-independent cell growth of Huh7 and HepG2 with GRAMD1A overexpression. Representative micrographs (left); quantification of colony number with diameter greater than 0.1 mm (right). Data are expressed as mean ± SEM. *p < 0.05.
Discussion
GRAMD1A is a newly discovered gene, it’s function never been reported in the development of various diseases. In present study, we analyzed the role of GRAMD1A in HCC prognosis and progression, and found GRAMD1A was upregulated in HCC tissues, GRAMD1A expression was significantly positively correlated with pathologic differentiation and survival state, patients with high GRAMD1A had poor outcome, Further analysis revealed GRAMD1A was an independent prognostic factor for HCC patients.
We found GRAMD1A overexpression promoted the self-renewal of HCC stem cells, the resistance to chemotherapy and tumor growth. Here, we found GRAMD1A regulated the target genes of STAT5, such as CyclinD1, Bcl-2, c-jun and c-Myc. GRAMD1A also regulated the transcriptional activity of STAT5, overexpression of STAT5 promoted the STAT5 activity, while knockdown of STAT5 inhibited the STAT5 activity. This finding suggested GRAMD1A regulated STAT5 expression, but we couldn’t study whether GRAMD1A regulates the phosphorylation of STAT5 and the location of STAT5. The molecular mechanisms of GRAMD1A are still to be elucidated, the direct interaction proteins of GRAMD1A must be analyzed using co-immunoprecipitation coupled with tandem mass spectrometry.
Previous results have demonstrated that STAT5 which has two isoforms, STA5A and STAT5B, plays critical role in normal and malignant cells, it can be phosphorylated by Jak2, and causes it translocate to nuclear to regulate the transcription of STAT5′ target genes21,22. For example, STAT5 promotes the proliferation, survival and self-renewal of hematopoietic stem cell23,24. STAT5 increases the AR stability, promotes castration-resistant prostate cancer growth, tumor metastasis and the self-renewal of prostate cancer stem cells25,26,27.
The role of STAT5 in HCC progression is studied by several labs, but their results are contradictory. Lother Hennighausen, et al. find CCl4 induces liver tumorigenesis in STAT5 knockout mice by increasing TGF-β stability and STAT3 activity, the N-terminal of STAT5 interacts with TGF-β to decrease TGF-β levels, overexpression of N-terminal STAT5 inhibits HCC formation. TGF-β upregulation abrogates growth hormone-induced STAT5 activation. Instead, TGF-β upregulation promotes growth hormone-induced STAT3 activation28. Meanwhile, they find STAT5 is essential for growth hormone induced NOX4 expression, in turn induces ROS generation and proapototic proteins PUMA and BIM expression29. Other labs also find in growth hormone signaling hyperactivated mice, deletion STAT5 protects liver tumorigenesis30. Knockdown of STAT5 in HCC cell SMMC7721 inhibits cell proliferation and induce apoptosis31. These findings suggest STAT5 is a tumor suppressor for HCC. Other labs also find STAT5B level is positively correlated with HCC progression, overexpression of STAT5B promotes epithelial-mesenchymal transition (EMT) and invasion in HCC induced by HBV32. We found knockdown of STAT5 in Huh-7 and HepG2 with GRAMD1A overexpression suppressed the self-renewal of HCC stem cells, resistance to chemotherapy and tumor growth, suggesting STAT5 was an oncogene. The role of STAT5 in HCC might be context-dependent.
In summary, we provide evidences that GRAMD1A is a prognostic factor, it promotes the self-renewal of HCC stem cells, resistance to chemotherapy and tumor growth through regulating STAT5.
Additional Information
How to cite this article: Fu, B. et al. GRAM domain-containing protein 1A (GRAMD1A) promotes the expansion of hepatocellular carcinoma stem cell and hepatocellular carcinoma growth through STAT5. Sci. Rep. 6, 31963; doi: 10.1038/srep31963 (2016).
Supplementary Material
Acknowledgments
This work is supported by the Natural Science Foundation of Guangdong Province (No. 2014A030313090, No. 2014A030313190 and No. 2016A030313195), the Science and Technology Projects Foundation of Guangdong Province (No. 2015A070710006 and No. 2016A020215053) and the Science and Technology Projects Foundation of Guangzhou City (No. 201507020037 and No. 201607010260).
Footnotes
Author Contributions Conceived and designed the experiments: T.Z., H.L., B.F., W.M. and H.Z. Performed the experiments: B.F., W.M., H.Z., B.Z., Y.Z., H.T. and J.Y. Analyzed the data: B.F., T.Z., H.L., W.M., H.Z. and B.Z. Contributed reagents/materials: Y.Z., H.T. and J.Y. Contributed clinical specimens: H.L. and Y.Z. Wrote the paper: T.Z. and H.L. Conceived and supervised the project: H.L. and T.Z.
References
- Torre L. A. et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- Zhao Z., Li S., Song E. & Liu S. The roles of ncRNAs and histone-modifiers in regulating breast cancer stem cells. Protein & cell 7, 89–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Cho R. W. & Clarke M. F. Cancer stem cells: models and concepts. Annual review of medicine 58, 267–284 (2007). [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison S. J., Clarke M. F. & Weissman I. L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001). [DOI] [PubMed] [Google Scholar]
- Ma S. et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 132, 2542–2556 (2007). [DOI] [PubMed] [Google Scholar]
- Yang Z. F. et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer cell 13, 153–166 (2008). [DOI] [PubMed] [Google Scholar]
- Lee T. K. et al. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell stem cell 9, 50–63 (2011). [DOI] [PubMed] [Google Scholar]
- Tong M. et al. ANXA3/JNK Signaling Promotes Self-Renewal and Tumor Growth, and Its Blockade Provides a Therapeutic Target for Hepatocellular Carcinoma. Stem cell reports 5, 45–59 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Long Non-coding RNA DILC Represses Self-renewal of Liver Cancer Stem Cells via Inhibiting Autocrine IL-6/STAT3 Axis. Journal of hepatology(2016). [DOI] [PubMed] [Google Scholar]
- Zhao H. et al. A novel anti-cancer agent Icaritin suppresses hepatocellular carcinoma initiation and malignant growth through the IL-6/Jak2/Stat3 pathway. Oncotarget 6, 31927–31943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Fernandez P. P., Rajendran P., Hui K. M. & Sethi G. Diosgenin, a steroidal saponin, inhibits STAT3 signaling pathway leading to suppression of proliferation and chemosensitization of human hepatocellular carcinoma cells. Cancer letters 292, 197–207 (2010). [DOI] [PubMed] [Google Scholar]
- Song X. et al. Production and characterization of a monoclonal antibody against GRAM domain-containing protein 1A. Monoclonal antibodies in immunodiagnosis and immunotherapy 33, 246–253 (2014). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Mir-208 promotes cell proliferation by repressing SOX6 expression in human esophageal squamous cell carcinoma. Journal of translational medicine 12, 196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. et al. TPX2 as a novel prognostic biomarker for hepatocellular carcinoma. Hepatology research : the official journal of the Japan Society of Hepatology 45, 906–918 (2015). [DOI] [PubMed] [Google Scholar]
- Cheang T. Y. et al. C14orf166 overexpression correlates with tumor progression and poor prognosis of breast cancer. Journal of translational medicine 14, 54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary G. S., Al-Harbi S. & Almasan A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods in molecular biology 1219, 1–9 (2015). [DOI] [PubMed] [Google Scholar]
- Heeres J. T. & Hergenrother P. J. Poly(ADP-ribose) makes a date with death. Current opinion in chemical biology 11, 644–653 (2007). [DOI] [PubMed] [Google Scholar]
- Ferbeyre G. & Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochimica et biophysica acta 1815, 104–114 (2011). [DOI] [PubMed] [Google Scholar]
- Haftchenary S. et al. Potent Targeting of the STAT3 Protein in Brain Cancer Stem Cells: A Promising Route for Treating Glioblastoma. ACS medicinal chemistry letters 4, 1102–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltemes-Harris L. M. & Farrar M. A. The role of STAT5 in lymphocyte development and transformation. Current opinion in immunology 24, 146–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T. et al. Identification of STAT5A and STAT5B target genes in human T cells. PloS one 9, e86790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. & Bunting K. D. STAT5 in hematopoietic stem cell biology and transplantation. Jak-Stat 2, e27159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haetscher N. et al. STAT5-regulated microRNA-193b controls haematopoietic stem and progenitor cell expansion by modulating cytokine receptor signalling. Nature communications 6, 8928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. et al. Transcription factor Stat5 knockdown enhances androgen receptor degradation and delays castration-resistant prostate cancer progression in vivo. Molecular cancer therapeutics 10, 347–359 (2011). [DOI] [PubMed] [Google Scholar]
- Talati P. G. et al. Jak2-Stat5a/b Signaling Induces Epithelial-to-Mesenchymal Transition and Stem-Like Cell Properties in Prostate Cancer. The American journal of pathology 185, 2505–2522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koptyra M., Gupta S., Talati P. & Nevalainen M. T. Signal transducer and activator of transcription 5a/b: biomarker and therapeutic target in prostate and breast cancer. The international journal of biochemistry & cell biology 43, 1417–1421 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosui A. et al. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-{beta} and STAT3 activation. The Journal of experimental medicine 206, 819–831 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Zhu B. M., Riedlinger G., Kang K. & Hennighausen L. The liver-specific tumor suppressor STAT5 controls expression of the reactive oxygen species-generating enzyme NOX4 and the proapoptotic proteins PUMA and BIM in mice. Hepatology 56, 2375–2386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedbichler K. et al. Growth-hormone-induced signal transducer and activator of transcription 5 signaling causes gigantism, inflammation, and premature death but protects mice from aggressive liver cancer. Hepatology 55, 941–952 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. RNA interference-mediated silencing of Stat5 induces apoptosis and growth suppression of hepatocellular carcinoma cells. Neoplasma 59, 302–309 (2012). [DOI] [PubMed] [Google Scholar]
- Lee T. K. et al. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer research 66, 9948–9956 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.