Abstract
HNF1A-antisense 1 (HNF1A-AS1), a long non-coding RNA (lncRNA), is associated with metastasis and is an independent prognostic factor for lung cancer. Recent studies demonstrated that HNF1A-AS1 play important roles in cacinogenesis. However, the exact effects and molecular mechanisms of HNF1A-AS1 in osteosarcoma (OS) progression is still unclear. In the present study, we found that HNF1A-AS1 was markedly up-regulated in OS tissues compared to their adjacent non-tumor tissues. HNF1A-AS1 expression levels were positively associated with the clinical stage, distant metastasis, and reduced overall survival of OS patients. In addition, knockdown HNF1A-AS1 expression inhibited cell proliferation, metastasis and influences the activity of the Wnt/β-catenin pathway in OS cells. Wnt/β-catenin pathway activator (LiCl) rescued the anticancer effect of knockdown HNF1A-AS1 expression in OS cells. In conclusion, our study demonstrated that HNF1A-AS1 promoted the progression of OS via regulating the activity of the Wnt/β-catenin pathway, indicating that HNF1A-AS1 might be a potential target for the treatment of OS.
Keywords: Osteosarcoma, long non-coding RNA, HNF1A-AS1, Wnt/β-catenin pathway
Introduction
Osteosarcoma (OS) is a primary bone malignancy that mainly affects the rapidly growing bones of children and adolescents and is associated with high morbidity [1]. The 5-year survival rate of OS patients has significantly improved over the past decades since the introduction of combinatorial chemotherapy [2]. However, there are a large number of OS patients who respond poorly to chemotherapy and have a high risk of local relapse or distant metastasis even after curative resection of the primary tumor and intensive chemotherapy [3,4]. Thus, there is an urgent need to explore the molecular mechanisms underlying in OS and identify the specific biomarkers that cause tumor progression.
The human genome sequencing project revealed that 70% of the genome is transcribed, but only up to 2% of the human genome serves as blueprints for proteins [5]. Long non-coding RNAs (lncRNAs) are defined as endogenous cellular RNAs more than 200 nucleotides in length that lack an open reading frame of significant length [6]. In recent years, several lncRNAs have been shown to be involved in the cancer progression [7]. For example, Zhang et al showed that upregulation of lncRNA MALAT1 correlated with tumor progression and poor prognosis in clear cell renal cell carcinoma [8]. Lin et al indicated that increased expression of the lncRNA ANRIL promoted lung cancer cell metastasis and correlates with poor prognosis [9]. Yan et al suggested that upregulation of the lncRNA HOTAIR predicted recurrence in stage Ta/T1 bladder cancer [10].
The long non-coding RNA HNF1A antisense RNA 1 (lncRNA HNF1A-AS1) located on chromosome 12 is transcribed as a 2.455 kb lncRNA in the opposite direction of HNF1A gene transcription [11]. Yang et al showed that that dysregulation of HNF1A-AS1 participated in oesophageal tumorigenesis, and that this participation may be mediated, at least in part, by modulation of chromatin and nucleosome assembly as well as by H19 induction [12]. Wu et al found that increased HNF1A-AS1 expression could regulate cell proliferation and metastasis and identified it as a poor prognostic biomarker in lung adenocarcinoma [13]. Liu et al showed that lncRNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p [14]. These results suggested that the dysregulation of HNF1A-AS1 play a critical role in carcinogenesis. However, little is known about the role of HNF1A-AS1 in OS progression. Thus, the aim of this study was to discover whether HNF1A-AS1 contributed to OS progression and identify the underlying regulatory mechanisms.
Materials and methods
Tissue specimens
A total of 43 pairs of OS tissues and their matched adjacent normal bone were obtained from patients who underwent surgery at The First Affiliated Hospital of Xinxiang Medical University from June 2006 to July 2011. Tissue specimens were immediately frozen in liquid nitrogen after surgery and stored at -80°C until the extraction of total RNA. No patients had received radiotherapy or chemotherapy before surgery. Tumor stage was classified according to the sixth edition of the TNM classification of the International Union against Cancer (UICC). This study was approved by the Research Ethics Committee of Shanghai Jiao Tong University. Written informed consent was obtained from all of the patients. The clinicopathological information of the patients is shown in Table 1.
Table 1.
Association of HNF1A-AS1 expression with clinicopathologic features in OS patients
| Parameter | Total | HNF1A-AS1 | P | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age (years) | 0.455 | |||
| <25 | 25 | 11 | 14 | |
| ≥25 | 18 | 10 | 8 | |
| Gender | 0.172 | |||
| Female | 20 | 12 | 8 | |
| Male | 23 | 9 | 14 | |
| Tumor size | 0.454 | |||
| <8 cm | 16 | 9 | 7 | |
| ≥8 cm | 27 | 12 | 15 | |
| Location | 0.159 | |||
| Tibia/Femur | 29 | 12 | 17 | |
| Elsewhere | 14 | 9 | 5 | |
| Clinical stage | 0.004 | |||
| I/II | 19 | 14 | 5 | |
| III | 24 | 7 | 17 | |
| Distant metastasis | 0.018 | |||
| Absent | 32 | 19 | 13 | |
| Present | 11 | 2 | 9 | |
Cell culture
Osteosarcoma cell lines (HOS, SaOS2, MG63 and U2OS), and osteoblastic cell line (hFOB1.19) were purchased from American Type Culture Collection (ATCC). Human OS cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin and 100 mg/mL streptomycin (Invitrogen). hFOB1.19 cells were cultured in Ham’s F12/Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% FBS, 100 U/mL penicillin and 100 mg/mL streptomycin (Invitrogen). Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2.
Cell transfection
The siRNA specifically targeting HNF1A-AS1 (si-HNF1A-AS1) was commercially constructed by Shanghai GenePharma. The sequences were as follows: HNF1A-AS1 siRNA1 (5’-UUAAAUUGCAGAGUUGCACUU-3’); HNF1A-AS1 siRNA2 (5’-GGGUGAGCAGCUGUUUGCAAGACUA-3’) [13]. The scrambled nucleotide was used as the negative control (si-NC). Cells were transfected with siRNAs using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Each experiment was performed in triplicate. The interfering effciency was determined by qRT-PCR after transfection 48 hours.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the tissue samples or cultured cells with Trizol reagent (Takara), and first-strand cDNA was synthesized from 2 μg of total RNA using random primers and the M-MLV Reverse Transcriptase (Invitrogen). The following primers were used to detect the expression of HNF1A-AS1 and GAPDH: HNF1-AS1 sense, 5’-TCAAGAAATGGTGGCTAT-3’, reverse 5’-GCTCTGAGACTGGCTG-AA-3’; GAPDH sense, 5’-AGAAGGCTGGGGCTCATTTG-3’; reverse 5’-AGGGGCCATCCACAGTCTTC-3’. RNA expression was measured by qRT-PCR using the SYBR-Green method (Takara) according to the manufacturer’s instructions. All experiments were performed using the 2-ΔΔCt method. The results were normalized to the expression of GAPDH.
MTT assay
The affects of HNF1A-AS1 on cell proliferation were determined by the MTT assay. In brief, 2×103 cells/well were seeded into 96-well plates and routinely cultured. At the indicated time points, 10 μl MTT solution (5 mg/ml; Sigma-Aldrich) was added into each well, and the reaction was terminated by 200 μl DMSO 2 hours later. The absorbance was measured on a microplate reader at 570 nm. This experiment was repeated in triplicate and at least three times.
Cell cycle analysis
Cells were harvested by trypsinization, washed twice in cold PBS, and fixed in 70% ethanol at 4°C overnight. After fixation, the cells were washed and re-suspended in cold PBS and incubated in a solution of 10 mg/mL RNase and 1 mg/mL propidium iodide (Sigma) at 37°C for 30 minutes in the dark. Finally, the DNA content was determined by flow cytometry (BD Biosciences).
Cell apoptosis analysis
Cells were harvested 48 hours after transfection for apoptosis analysis. Floating and adherent cells were collected using 0.1% trypsin, washed twice with cold PBS, and suspended in 1000 ul binding buffer. The cells were then treated with FITC-Annexin V and propidium iodide (PI) in the dark at room temperature, according to the manufacturer’s protocol. The cells were then examined by flow cytometry (BD Biosciences) on in instrument equipped with CellQuest software (BD Biosciences).
Cell migration and invasion assays
Cell migration ability was assessed using 6.5-mm transwell chambers with a pore size of 8 μm. Cell invasion ability was assessed using the chamber with 100 µg Matrigel (BD Biosciences). The assays were performed according to the manufacturer’s instructions. Briefly, 2×104 cells from each group were suspended in serum-free medium and were seeded into the upper chamber. The lower chamber was filled with medium containing 10% FBS. After incubation for 24 h, the migrated/invaded cells in the lower chamber (below the filter surface) were fixed in 4% paraformaldehyde, stained with 0.1 mg/ml crystal violet solution, and counted under a microscope. The cells on the lower surface were photographed and five random fields were counted. Three independent experiments were performed.
Western blot
For analysis of protein expression, the proteins extracted from cultured cells were separated on 10% SDS-polyacrylamide gels. The separated proteins were then electro-transferred to a PVDF membrane (Bio-Rad). The membrane was then blocked by blocking buffer and incubated with primary antibody at 4°C overnight. Then, the membrane was incubated with horseradish peroxidase (HRP) labeled secondary antibody. The protein bands were visualized using an enhanced chemiluminescence system (Amersham). Primary antibodies, β-actin and HRP labeled secondary antibody were purchased from Santa Cruz Biotechnology (Santa Cruz).
Statistical analysis
Statistical analyses were performed using SPSS 18.0 software package. All the experiments were repeated in triplicate, and data was expressed as mean ± SD. Differences were assessed by two-tailed Student’s t test, and P<0.05 was considered statistically significant.
Results
HNF1A-AS1 levels were up-regulated in OS
To explore the role of HNF1A-AS1 in the progression of osteosarcoma. We first performed qRT-PCR analysis to detected HNF1A-AS1 expression in 43 paired OS tissues. Our data showed that the expression levels of HNF1A-AS1 was significant up-regulated in OS tissues compared with adjacent non-tumor tissues (P<0.05, Figure 1A). Furthermore, OS cells were also detected, compared with osteoblastic cells hFOB1.19, HNF1A-AS1 expression was significantly upregulated in all four OS cell lines (HOS, SaOS2, MG63 and U2OS) (P<0.05, Figure 1B). Then, 43 OS patients were divided into two groups based on the median value of relative HNF1A-AS1 expression. As shown in Table 1, HNF1A-AS1 expression was correlated with clinical stage and distant metastasis (P<0.05). However, there were no significant correlations between HNF1A-AS1 expression and other clinicopathologic factors including age, gender, tumor size and tumor location (P>0.05).
Figure 1.
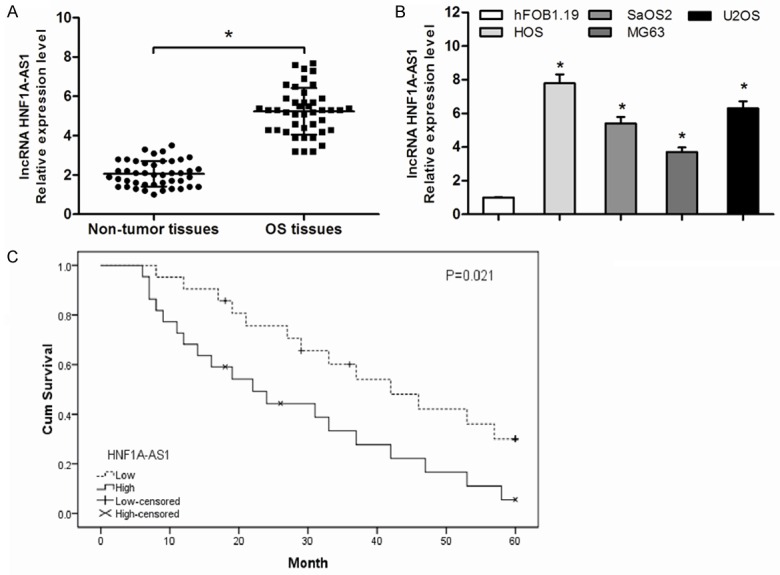
Relative HNF1A-AS1 expression in OS and its clinical significance. A: The expression levels of HNF1A-AS1 in 43 pairs of OS tissues and matched adjacent non-tumor tissues were detected by qRT-PCR. B: The relative expression levels of HNF1A-AS1 in four OS cell lines (HOS, SaOS2, MG63 and U2OS) and osteoblastic cell line hFOB1.19 were confirmed via qRT-PCR. C: The Kaplan-Meier curve indicated that patients with high expression of HNF1A-AS1 has a worse overall survival compared to patient with low expression of HNF1A-AS1. *P<0.05.
High HNF1A-AS1 expression was associated with poor prognosis of OS patients
Considering that the level of HNF1A-AS1 expression was correlated with clinical stage and distant metastasis. We further investigate the prognosis of HNF1A-AS1 in OS patients. Kaplan-Meier analysis showed that the overall survival rate of high HNF1A-AS1 expression group was significantly poor than that of low HNF1A-AS1 expression group (P<0.05, Figure 1C). The univariate analysis showed that clinical stage, distant metastasis and HNF1A-AS1 expression were significantly related to overall survival of OS patients (P<0.05, Table 2). Moreover, the multivariate Cox regression analysis suggested that clinical stage, distant metastasis and HNF1A-AS1 expression were independent prognostic factors for overall survival of OS patients (P<0.05, Table 2). Thus, these observations suggested that increased expression of HNF1A-AS1 was associated with the progression and development of OS.
Table 2.
Univariate and multivariate analyses of prognostic factors in OS patients
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Hazard ratio | 95% CI | P | Hazard ratio | 95% CI | P | |
| Age (years) | 1.174 | 0.628-4.073 | 0.417 | |||
| ≥25 vs <25 | ||||||
| Gender | 0.739 | 0.492-1.947 | 0.591 | |||
| Male vs Female | ||||||
| Tumor size | 1.635 | 0.528-5.874 | 0.171 | |||
| ≥8 cm vs <8 cm | ||||||
| Location | 0.894 | 0.651-6.178 | 0.363 | |||
| Tibia/Femur vs Elsewhere | ||||||
| Clinical stage | 2.739 | 1.215-8.936 | 0.007 | 2.318 | 1.095-8.163 | 0.009 |
| III vs I/II | ||||||
| Distant metastasis | 3.917 | 1.781-12.117 | 0.004 | 3.285 | 1.536-11.741 | 0.008 |
| Present vs Absent | ||||||
| HNF1A-AS1 | 2.976 | 1.487-7.945 | 0.003 | 2.637 | 1.391-7.417 | 0.005 |
| High vs Low | ||||||
Suppressing HNF1A-AS1 expression inhibited OS cell proliferation
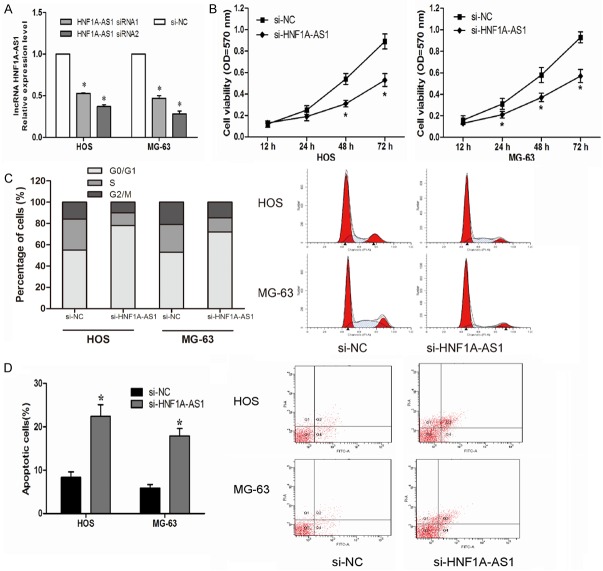
To determine the role of HNF1A-AS1 in OS tumorigenesis, we knocked-down HNF1A-AS1 expression in HOS and MG-63 cells by transfecting si-HNF1A-AS1 (P<0.05, Figure 2A). MTT assay indicated that decreased expression of HNF1A-AS1 significantly suppressed HOS and MG-63 cells proliferation compared with control group (si-NC) (P<0.05, Figure 2B). To investigated the potential mechanisms underlying cell proliferation, flow cytometry was used. Cell cycle analysis showed that reduced expression of HNF1A-AS1 increased the number of HOS and MG-63 cells in G0/G1 phase and decreased the number of HOS and MG-63 cells in S phase compared with cells transfected with si-NC (P<0.05, Figure 2C). Apoptosis analysis indicated that knockdown HNF1A-AS1 expression induced HOS and MG-63 cells apoptosis compared with control group (P>0.05, Figure 2D). These results demonstrated that HNF1A-AS1 promoted OS cells proliferation by promoting cell cycle progression and inhibiting cell apoptosis.
Figure 2.
Suppressing HNF1A-AS1 inhibited cell proliferation in the OS cell lines. A: The expression level of HNF1A-AS1 in HOS and MG-63 cells was significantly decreased by si-HNF1A-AS1 compared with the si-NC group. B: MTT assay showed that inhibition of the HNF1A-AS1 suppressed proliferation of HOS and MG-63 cells. C: Cell cycle analysis showed that inhibition of the HNF1A-AS1 increased the percentage of G0/G1 phase cells and decreased the percentage of S phase cells. D: Cell apoptosis assay showed that inhibition of the HNF1A-AS1 promoted HOS and MG-63 cells apoptosis. *P<0.05.
Suppressing HNF1A-AS1 expression inhibited OS cell migration and invasion
To further assess the effect of HNF1A-AS1 on the invasiveness of OS cells, Transwell migration assay and transwell invasion assay were used. Transwell migration assay indicated that reduced expression of HNF1A-AS1 inhibited HOS and MG-63 cells migration capacity than si-NC group (P<0.05, Figure 3A). Similarly, transwell invasion assay showed that decreased expression of HNF1A-AS1 significantly inhibited HOS and MG-63 cells invasion capacity compared to si-NC group (P<0.05, Figure 3B). These results indicated that HNF1A-AS1 promoted OS cells migration and invasion ability.
Figure 3.
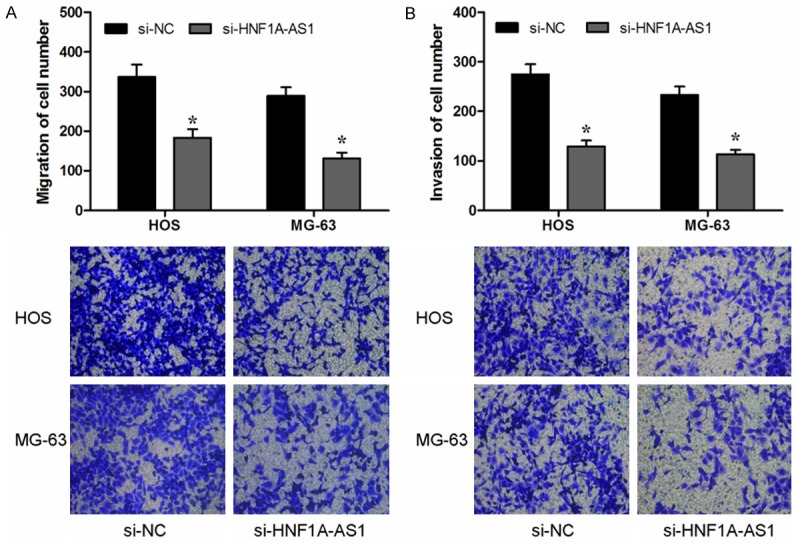
Suppressing HNF1A-AS1 inhibited cell migration and invasion in the OS cell lines. A: Transwell migration assay showed that inhibition of the HNF1A-AS1 suppressed migration ability of HOS and MG-63 cells. B: Transwell invasion assay showed that inhibition of the HNF1A-AS1 suppressed invasion ability of HOS and MG-63 cells. *P<0.05.
Suppressing HNF1A-AS1 expression affected the Wnt/β-catenin signaling pathway
To better understand the detailed regulation mechanism of HNF1A-AS1 in OS progression, we performed western blot analysis to investigated whether suppressing HNF1A-AS1 affected the Wnt/β-catenin signaling pathway, which play important roles in tumor development and progression [15,16]. Western blot showed that knockdown HNF1A-AS1 expression significantly inhibited β-catenin expression compared with control groups. In addition, we found that reduced expression of HNF1A-AS1 significantly decreased expression of cyclin D1 and c-myc expression (classic downstream genes of the Wnt/β-catenin signaling pathway) (P<0.05, Figure 4). Indicating that HNF1A-AS1 could affect the activity of the Wnt/β-catenin pathway. To further confirm the role of HNF1A-AS1 in regulating the activity of the Wnt/β-catenin pathway, LiCl (Sigma) was used to activate the Wnt/β-catenin pathway. Our data showed that the anticancer ability of knockdown HNF1A-AS1 expression on OS cells was rescued by OS cells treat with LiCl (P<0.05, Figure 5A-C). Therefore, the studies indicated that HNF1A-AS1 might serve as an oncogenic lncRNA that promoted the proliferation and metastasis of OS and activated the Wnt/β-catenin signaling pathway.
Figure 4.
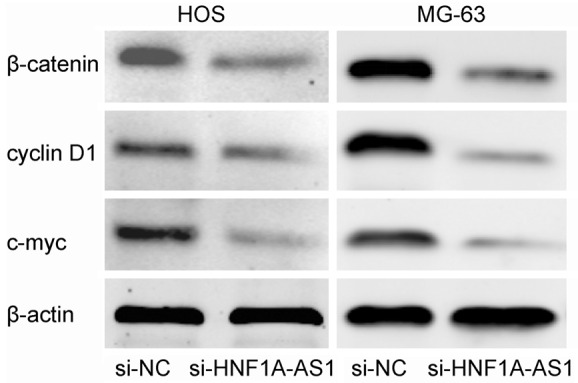
Suppressing HNF1A-AS1 affected the Wnt/β-catenin signaling pathway. Western blot analysis showed that si-HNF1A-AS1 inhibited the β-catenin, cyclin D1 and c-myc expression in the HOS and MG-63 cells.
Figure 5.
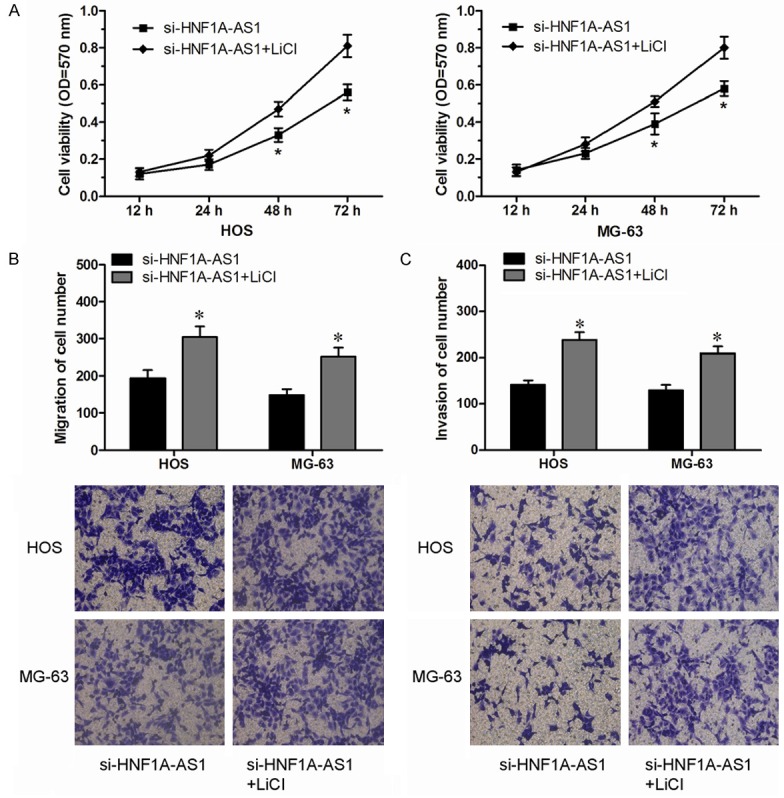
Wnt/β-catenin pathway was involved in the tumor oncogenic effect of HNF1A-AS1 in OS cells. A: MTT assay evaluated the effect of LiCl in HNF1A-AS1 knockdown HOS and MG-63 cells. B: Transwell migration assay evaluated the effect of LiCl in HNF1A-AS1 knockdown HOS and MG-63 cells. C: Transwell invasion assay evaluated the effect of LiCl in HNF1A-AS1 knockdown HOS and MG-63 cells.
Discussion
Osteosarcoma is the most common primary malignancy in the world [1]. Despite of the recent rapid advances in the diagnosis and treatment, the survival of OS patients with recurrence or metastasis remains poor [3]. Thus, to improve the OS survival rate requires a clear understanding of pivotal molecular mechanisms from the initiation and progression of OS.
Accumulating evidence showed that lncRNAs play critical roles in the regulation of cell proliferation, metastasis, infiltration and apoptosis [17]. Aberrant lncRNA expression is involved in various cancers, including OS. For example, Sun et al showed that increased expression of lncRNA HULC could indicate a poor prognosis and promoted cell metastasis in osteosarcoma [18]. Wang et al showed that overexpression of lncRNA HOTAIR could promote tumor growth and metastasis in human osteosarcoma [19]. Dong et al suggested that lncRNA MALAT1 promoted the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway [20]. However, the possible role and associated molecular mechanisms of HNF1A-AS1 in OS are still unclear.
In this study, we found that the expression level of HNF1A-AS1 was upregulated in OS tissues than those in adjacent non-tumor tissues. HNF1A-AS1 expression were associated with clinical stage and distant metastasis of OS patients. Furthermore, high HNF1A-AS1 expression was correlated with lower overall survival rates and could be an independent prognostic factor in OS patients. To understand its biological role in OS. HOS and MG-63 cells were chosen to investigate the effect of HNF1A-AS1 knockdown on OS cell phenotype. Our data revealed that decreased expression of HNF1A-AS1 significantly suppressed proliferation and metastasis capability of HOS and MG-63 cells in vitro. These results suggested that HNF1A-AS1 might function as a tumor oncogene in OS progression.
Wnt/β-catenin signaling is known to regulate a broad range of cellular processes, such as proliferation, invasion, differentiation, and other signaling pathways, through regulating the ability of the multifunctional β-catenin protein, which is a crucial signaling molecule in the Wnt/β-catenin pathway [21]. Recent studies suggested that Wnt/β-catenin signaling activation were involved in various human cancers. For example. Mohammadi-Yeganeh et al suggested that miRNA-340 could inhibit the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway [22]. Zhang et al found that lncRNA CASC11 interacted with hnRNP-K and activated the Wnt/β-catenin pathway to promote growth and metastasis in colorectal cancer [23]. Yuan et al showed that lncRNA-CTD903 inhibited colorectal cancer invasion and migration by repressing Wnt/β-catenin signaling and predicted favorable prognosis [24]. In the present study, we found that knockdown HNF1A-AS1 expression significantly inhibited β-catenin, cyclin D1 and c-myc expression in OS cells. Furthermore, we found that LiCl (Wnt/β-catenin pathway activator) could rescue the anticancer effect of knockdown HNF1A-AS1 expression in OS cells. These results suggested that tumor oncogenic effect of HNF1A-AS1 was at least partly mediated by Wnt/β-catenin pathway in OS cells.
In conclusion, our study demonstrated that lncRNA HNF1A-AS1 was increased expression in OS and HNF1A-AS1 was likely to be a useful biomarker for this disease. Moreover, knockdown of HNF1A-AS1 could inhibit OS cell proliferation, migration and invasion by repressing Wnt/β-catenin signaling in vitro. Taken together, our study suggested that lncRNA HNF1A-AS1 could be a new potential target and prognostic factor for the treatment of OS.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. doi: 10.1155/2011/548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrle D, Bielack SS. Current strategies of chemotherapy in osteosarcoma. Int Orthop. 2006;30:445–451. doi: 10.1007/s00264-006-0192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 9.Lin L, Gu ZT, Chen WH, Cao KJ. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagn Pathol. 2015;10:14. doi: 10.1186/s13000-015-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan TH, Lu SW, Huang YQ, Que GB, Chen JH, Chen YP, Zhang HB, Liang XL, Jiang JH. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol. 2014;35:10249–57. doi: 10.1007/s13277-014-2344-8. [DOI] [PubMed] [Google Scholar]
- 11.Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, Coin LJ, Deng G, Gieger C, Heard-Costa NL, Hottenga JJ, Kühnel B, Kumar V, Lagou V, Liang L, Luan J, Vidal PM, Mateo Leach I, O’Reilly PF, Peden JF, Rahmioglu N, Soininen P, Speliotes EK, Yuan X, Thorleifsson G, Alizadeh BZ, Atwood LD, Borecki IB, Brown MJ, Charoen P, Cucca F, Das D, de Geus EJ, Dixon AL, Döring A, Ehret G, Eyjolfsson GI, Farrall M, Forouhi NG, Friedrich N, Goessling W, Gudbjartsson DF, Harris TB, Hartikainen AL, Heath S, Hirschfield GM, Hofman A, Homuth G, Hyppönen E, Janssen HL, Johnson T, Kangas AJ, Kema IP, Kühn JP, Lai S, Lathrop M, Lerch MM, Li Y, Liang TJ, Lin JP, Loos RJ, Martin NG, Moffatt MF, Montgomery GW, Munroe PB, Musunuru K, Nakamura Y, O'Donnell CJ, Olafsson I, Penninx BW, Pouta A, Prins BP, Prokopenko I, Puls R, Ruokonen A, Savolainen MJ, Schlessinger D, Schouten JN, Seedorf U, Sen-Chowdhry S, Siminovitch KA, Smit JH, Spector TD, Tan W, Teslovich TM, Tukiainen T, Uitterlinden AG, Van der Klauw MM, Vasan RS, Wallace C, Wallaschofski H, Wichmann HE, Willemsen G, Würtz P, Xu C, Yerges-Armstrong LM Alcohol Genome-wide Association (AlcGen) Consortium; Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study; Genetic Investigation of Anthropometric Traits (GIANT) Consortium; Global Lipids Genetics Consortium; Genetics of Liver Disease (GOLD) Consortium; International Consortium for Blood Pressure (ICBP-GWAS); Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) Abecasis GR, Ahmadi KR, Boomsma DI, Caulfield M, Cookson WO, van Duijn CM, Froguel P, Matsuda K, McCarthy MI, Meisinger C, Mooser V, Pietiläinen KH, Schumann G, Snieder H, Sternberg MJ, Stolk RP, Thomas HC, Thorsteinsdottir U, Uda M, Waeber G, Wareham NJ, Waterworth DM, Watkins H, Whitfield JB, Witteman JC, Wolffenbuttel BH, Fox CS, Ala-Korpela M, Stefansson K, Vollenweider P, Völzke H, Schadt EE, Scott J, Järvelin MR, Elliott P, Kooner JS. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, Khashab M, Singh VK, Shin EJ, Verma AK, Meltzer SJ, Mori Y. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Wei X, Zhang A, Li C, Bai J, Dong J. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473:1268–1275. doi: 10.1016/j.bbrc.2016.04.054. [DOI] [PubMed] [Google Scholar]
- 15.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8:2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, Wang H, Zhang R, Liu Y. Overexpression of Long Non-Coding RNA HOTAIR Promotes Tumor Growth and Metastasis in Human Osteosarcoma. Mol Cells. 2015;38:432–440. doi: 10.14348/molcells.2015.2327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 21.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Mohammadi-Yeganeh S, Paryan M, Arefian E, Vasei M, Ghanbarian H, Mahdian R, Karimipoor M, Soleimani M. MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway. Tumour Biol. 2016 doi: 10.1007/s13277-015-4513-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Zhou C, Chang Y, Hu Y, Zhang F, Lu Y, Zheng L, Zhang W, Li X. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT beta-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376:62–73. doi: 10.1016/j.canlet.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Z, Yu X, Ni B, Chen D, Yang Z, Huang J, Wang J, Wang L. Overexpression of long non-coding RNA-CTD903 inhibits colorectal cancer invasion and migration by repressing Wnt/beta-catenin signaling and predicts favorable prognosis. Int J Oncol. 2016;48:2675–2685. doi: 10.3892/ijo.2016.3447. [DOI] [PubMed] [Google Scholar]