Abstract
Laboratory rodents are available as either genetically defined inbred strains or genetically undefined outbred stocks. As outbred rodents are generally thought to display a higher level of phenotypic variation compared to inbred strains, it has been argued that experimental studies should preferentially be performed by using inbred rodents. However, very few studies with adequate sample sizes have in fact compared phenotypic variation between inbred strains and outbred stocks of rodents and moreover, these studies have not reached consistent conclusions. The aim of the present study was to compare the phenotypic variation in commonly used experimental readouts within obesity and diabetes research, for four of the most frequently used mouse strains: inbred C57BL/6 and BALB/c and outbred NMRI and CD-1 mice. The variation for all readouts was examined by calculating the coefficient of variation (CV), i.e., the relative variation, including a 95% confidence interval for the CV. We observed that for the majority of the selected readouts, inbred and outbred mice showed comparable phenotypic variation. The observed variation appeared highly influenced by strain choice and type of readout, which suggests that these collectively would serve as more predictive of the phenotypic variation than the more general classification of mice as inbred or outbred based on genetic heterogeneity.
Keywords: Phenotypic variability, inbred, outbred, coefficient of variation, diabetes and obesity research
Introduction
Experimental animals are a valuable resource in preclinical development of new pharmaceutical compounds [1,2]. Thus, in vivo experiments using in particular rodents have contributed to our understanding of the pathophysiology of e.g. common lifestyle-associated conditions such as type 2 diabetes and obesity but also provided the necessary experimental model systems to test novel therapeutics [3].
Inbred strains and outbred stocks constitute the two major classes of available laboratory rodents [4]. Inbred strains are generally characterized by genetic uniformity, which is expected to result in less phenotypic variation in responses to toxic or pharmacologic effects compared to outbred stocks. Arguments favoring the use of inbred strains include increased statistical power, improved experimental reproducibility and thus fewer animals needed to test a specific hypothesis. Additionally, extensive genetic information resources are available allowing the selection of the exact genetic makeup necessary for a particular experiment [5]. In contrast, outbred stocks are characterized by being genetically variable. Arguments favoring the use of outbred stocks emphasize particularly the importance of genetic heterogeneity when modelling the “outbred” human population, higher fecundity, and lower cost [6].
For the last six decades, arguments for the increased utilization of inbred strains in research have been put forward, but controlled and targeted studies comparing the within-strain variation of commonly used inbred and outbred animals are few and contradictory [7] and to our knowledge, no studies have previously been conducted with particular focus on variation in experimental endpoints relevant to diabetes and obesity research. The use of outbred animals continues to be widespread within many areas of in vivo research and scientists generally agree that there is a need for experiments comparing variation between commonly used inbred and outbred rodents [7,8]. In order to investigate if inbred rodents are less variable than outbred in the context of preclinical diabetes and obesity research, the within-strain variation of four commonly used mouse strains (Inbred: C57BL/6 and BALB/c; Outbred: NMRI and CD-1) was compared. In addition to their popularity within the field of lifestyle associated conditions, these mouse strains were recently reported to be among the most frequently used in biomedical research in general [8]. Here, we report from two studies: an acute study of insulin tolerance and a six week dietary intervention trial on an energy-dense diet commonly used in studies of diet-induced obesity (Western Diet, D12079B, Research Diets). Using the data obtained from these studies, the coefficients of variation (CV) were calculated and compared between the four strains for parameters related to pharmacodynamic effects of insulin and metabolic effects of a high-energy diet.
Materials and methods
Animals
Outbred NMRI and inbred C57BL/6 and BALB/c male mice were purchased from Taconic Biosciences (Lille Skensved, Denmark). Outbred CD-1 male mice were purchased from Charles River Laboratories (Sulzfeld, Germany). Animals were acclimatized for two weeks upon arrival and were on average seven weeks old at the beginning of each experiment, weighing approximately 25-30 g. They were housed in Scantainers in groups of ten (with exception of one group/strain of five, due to sample size), and had access to water and standard rodent chow (RC; Altromin 1324) ad libitum until beginning of experiments. Temperature in the animal rooms was maintained at 20-25°C with a light/dark cycle of 12/12 h. The study was approved by the Danish Animal Experiments Inspectorate under the Ministry of Food, Agriculture and Fisheries and in accordance with European Union directive 2010/63/EU.
Experimental design
To obtain reliable estimates of the coefficient of variation (CV) for each strain, we wanted to calculate point-estimates of the CV with associated 95% confidence intervals (CI). As the range of such CIs is dependent only on sample size [9], ranges corresponding to different sample sizes were calculated in R studio (Rstudio Inc., Boston, MA, USA). We used the ratio of the upper and lower limit of a 95% CI as an expression of the range of the CI and calculated that about 40-50 animals per strain would result in estimation of a sufficiently narrow 95% CI for CV comparison (corresponding to ratios of 1.49-1.57 of upper vs. lower limit of a 95% CI). To be able to detect an effect of the two treatments, the sample sizes of the control/vehicle groups were calculated based on previous data from similar experiments conducted at Novo Nordisk. This resulted in n=5 and n=15 for the acute and chronic study, respectively, and a final total sample size per strain of 55 mice. Two standard preclinical experiments were conducted to examine the variability within each strain. First, an acute insulin tolerance test was performed to compare within-strain variation of selected pharmacodynamic parameters in response to subcutaneous dosing with human insulin (HI, Novo Nordisk A/S, Måløv, DK). The experiment was performed on four experimental days and mice were randomly allocated to each of the four experimental days, ensuring that all strains were represented on each day. On the morning of each experimental day, the mice were acclimatized to the procedure room for 30 min prior to blood sampling. Animals were non-fasted and food but not water was removed for the duration of the procedure. Blood samples from a tail vein (t=0) were collected prior to treatment with HI, after which animals assigned to treatment groups (n=50/strain) were dosed subcutaneously in the flank with HI (0.76 µM formulated in vehicle containing 5 mM phosphate; 140 mM sodium chloride and 70 ppm polysorbate 20 at pH=7.4) at a dose of 3 nmol/kg. Subsequently, blood samples were collected from a tail vein at 20, 40, 60, 90, 120 and 180 min. Following the acute insulin tolerance test, 40 randomly selected mice from each strain were switched to an energy-dense diet (Western Diet (WD), 41 kcal% fat, 43 kcal% carbohydrates, 17 kcal% protein, D12079B, Research Diets), while the remaining 15 mice were maintained on the RC control diet. The mice were kept on the respective diets for six weeks and weighed once weekly. On day 42, an MRI scan was performed on all animals to determine lean body mass and fat mass. Subsequently, all mice were euthanized by cervical dislocation and the epididymal fat depots and liver were collected and weighed.
Blood glucose measurements
Samples for blood glucose measurements (5 µL) were collected in capillary tubes and transferred to 250 µL system solution. Blood glucose levels were analysed using the glucose oxidase method at a Biosenapparatur (EKF Diagnostics, Barleben, Germany) according to manufacturer’s instructions.
Echo magnetic resonance imaging
To determine lean body mass and total fat mass an EchoMRI scan was performed on all animals using an EchoMRI Body Composition Analyser (EchoMRI, Houston, TX, USA). Mass measurements of fat, lean tissue, free water and total water were performed according to manufacturer’s instructions and as previously described [10].
Calculation of the coefficient of variation and statistical analysis
The CV was chosen as the relative measure of variability. Data were assumed to follow a normal distribution when log-transformed, which was confirmed by visual inspection of qq-plots. When transforming a series of data points using the natural logarithm, the standard deviation (SD) of the log-transformed data set will be approximately equal to the CV of the original data set [11], and this concept was utilized in the present statistical analyses. For all parameters, CVs were calculated with 95% CIs. Statistical analysis was performed using SAS software version 10.0.2 (SAS institute Inc., Cary, NC, USA) and graphs were drawn using GraphPad Prism software version 6.05 (GraphPad Software Inc., La Jolla, CA, USA). After completion of the insulin tolerance test, the area over the Δblood glucose vs. time curve (AOC), describing blood glucose after treatment with insulin normalized to t=0, was calculated for each mouse using the trapezoidal method. After completion of the chronic experiment, individual bodyweights over time were plotted, as were the data for fat mass, lean mass, liver weight and epididymal fat mass. Significant differences in CVs between strains were analyzed by comparing the ratios of the sampling variances with critical values of an F-distribution. Comparisons between each inbred and each outbred strain were performed for all parameters, resulting in a total of 32 direct comparisons of CVs. Differences in means between strains for each parameter were analyzed using a one-way ANOVA and Bonferroni corrections were used as adjustment for multiple comparisons. P-values < 0.05 were considered significant.
Results
Acute effects of human insulin on blood glucose in inbred and outbred mice
Figure 1 shows mean changes in blood glucose within each strain following subcutaneous dosing with HI. HI lowered blood glucose significantly in all strains compared to age-matched un-treated controls (Table 1). Blood glucose returned to the same levels as controls after ~120-180 min in all strains (Figure 1). At 40 min, the inbred C57BL/6 strain revealed the most substantial effect on blood glucose (AOCglucose0-40), an effect which was significantly larger compared to the other mouse strains (Table 2). These differences were also present in AOC calculated from 0-60 and 0-90 min.However for AOC calculated from 0-180 min, there were no significant between-strain differences in mean AOC-values (data not shown). Mean maximal blood glucose response to HI was observed in all four strains ~40 min after treatment and was significantly greater in C57BL/6 mice compared to BALB/c, NMRI and CD-1. The mean maximal blood glucose response was significantly lower in the BALB/c strain compared to outbred NMRI and CD-1 mice (Table 2), i.e., BALB/c mice appeared to be the least insulin-sensitive strain.
Figure 1.
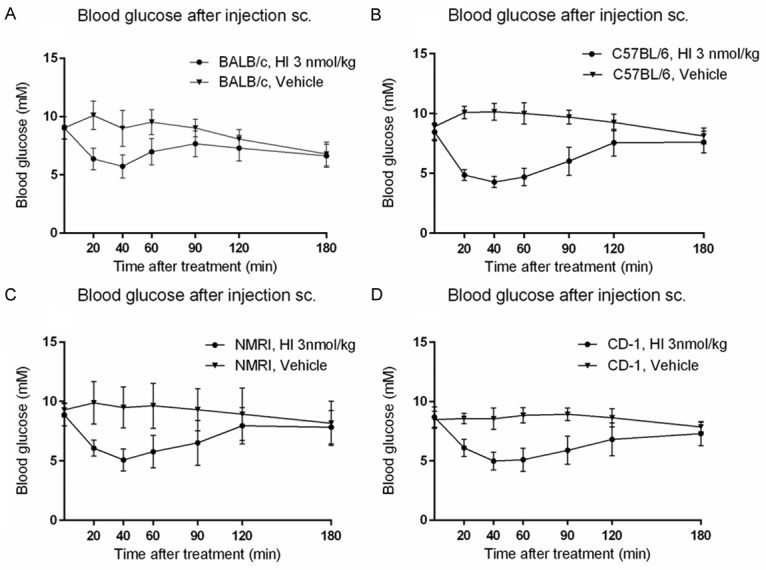
Blood glucose changes after subcutaneous flank dosing with human insulin in inbred (BALB/c, C57BL/6) and outbred (NMRI, CD-1) male mice. Circular and triangular symbols and curves designate treatment groups (BALB/c: n=48; C57BL/6: n=48; NMRI: n=49; CD-1: n=50), and vehicle-treated mice (n=5/strain), respectively. Data points represent the mean value in each group, error bars represent the standard deviation. Maximal effect was observed in all four strains 40 min after treatment. Blood glucose returned to near basal levels after ≈120-180 min. A: BALB/c mice. B: C57BL/6 mice. C: NMRI mice. D: CD-1 mice.
Table 1.
Blood glucose at t=40 min after subcutaneous flank dosing with human insulin compared to age-matched controls (mean ± SD)
| Strain/stock | Treatment | Mean ± SD (mM) | n |
|---|---|---|---|
| BALB/c | HI | 5.7±1.0**** | 48 |
| Vehicle | 9.0±1.5 | 5 | |
| C57BL/6 | HI | 4.3±0.5**** | 48 |
| Vehicle | 10.1±0.7 | 5 | |
| NMRI | HI | 5.1±0.9**** | 49 |
| Vehicle | 9.5±1.7 | 5 | |
| CD-1 | HI | 5.0±0.8**** | 50 |
| Vehicle | 8.5±0.9 | 5 |
indicates: P<0.0001 when HI and vehicle treated mice were compared for each strain.
HI=Human Insulin.
Table 2.
Phenotypic variation of selected parameters in inbred and outbred mice before and after subcutaneous dosing with human insulin
| BALB/c (n=48) | C57BL/6 (n=48) | NMRI (n=49) | CD-1 (n=50) | |||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Mean ± SD | CV (%) | Mean ± SD | CV (%) | Mean ± SD | CV (%) | Mean ± SD | CV (%) | |
| Baseline blood glucose (mM) | 9.0±1.0†† | 10.8 [9.0; 13.5] | 8.5±0.7† | 8.6 [7.2; 10.8] | 8.9±0.9††,† | 10.2 [8.5; 12.8] | 8.7±0. 9††,† | 9.5 [8.0; 11.9%] |
| AOCglucose0-40 (mM×min) | 85.5±22.4†† | 27.8 [23.2; 34.8]a | 114.0±19.3† | 17.0 [14.2; 21.3]b | 94.3±23.5†† | 25.9 [21.6; 32.3]a | 89.2±24.3†† | 24.7 [20.7; 30.8]a |
| Maximal blood glucose effect of HI (mM) | 5.6±0.9††† | 16.4 [13.7; 20.6]d | 4.2±0.5† | 11.0 [9.1; 13.7]c | 4.9±1.0†† | 20.8 [17.4; 26.0]d | 4.8±0.8†† | 17.3 [14.5; 21.6]d |
Numbers given in percentage are coefficients of variations. Brackets designate 95% confidence intervals for coefficients of variation. Superscripts indicate significant (P<0.05) differences in CVs (a,b,c,d) and effect (†, ††, †††) between strains. CV=Coefficient of variation, AOC=Area over the Δblood glucose vs. time curve.
Within-strain variation in acute effects of subcutaneous dosing with human insulin
Baseline blood glucose was the least variable of the investigated parameters with CVs ranging from 8.6 to 10.8%, all of which were comparable between strains (Table 2). The inbred strain C57BL/6 exhibited the lowest level of variation within parameters “AOCglucose0-40” and “Mean maximal blood glucose effect” and was significantly less variable than both outbred stocks and the inbred BALB/c mice. Within-strain variation of inbred strain BALB/c was comparable to that of the outbred stocks for all parameters.
Metabolic effects of Western diet intervention in inbred and outbred mice
To investigate whether exposure to an energy-dense diet resulted in changes of selected metabolic parameters, mice were fed a WD for six weeks and compared with age-matched controls fed a standard RC. Figure 2 shows the weight curve during the six week diet trial. Exposure to WD significantly increased bodyweight in the C57BL/6 strain and the CD-1 strain compared to animals on RC. Surprisingly, BALB/c mice fed WD gained significantly less bodyweight compared to BALB/c mice fed RC during the six weeks (Table 3).
Figure 2.
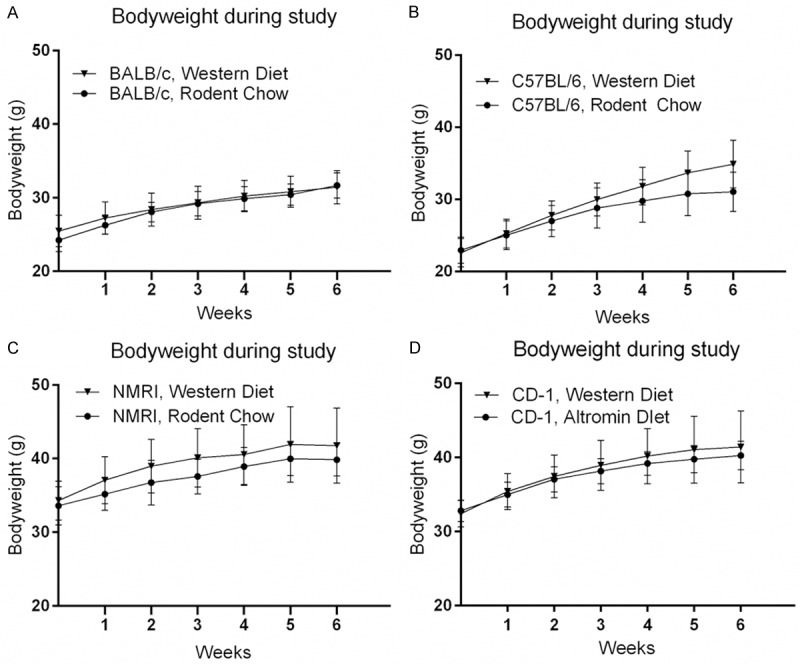
Development in bodyweight over time in inbred (BALB/c, C57BL/6) and outbred (NMRI, CD-1) strains of mice on Western Diet compared to control mice fed standard rodent chow. Mice were maintained on either WD or RC for 6 weeks, and weighed on a weekly basis. Circular and triangular symbols and curves designate treatment groups (BALB/c: n=40; C57BL/6: n=39; NMRI: n=40; CD-1: n=40), and age-matched RC-fed control mice (n=15, all strains except NMRI: n=14), respectively. Data points represent the mean value in each group, error bars represent the standard deviation. Significant effect of WD on bodyweight gain was only observed in the C57BL/6 strain (P<0.0001). Conversely, significant difference was found between the BALB/c WD and RC group, with the RC group demonstrating a larger gain in bodyweight compared to the WD group. WD=Western Diet, RC=Rodent chow. A: BALB/c mice. B: C57BL/6 mice. C: NMRI mice. D: CD-1 mice.
Table 3.
Selected metabolic parameters after 6 weeks exposure to an energy-dense diet (WD) compared with age-matched controls fed standard rodent chow (RC)
| Strain/Stock | Diet | Bodyweight change (g) | Fat mass (g) | Lean mass (g) | Liver weight (g) | Epididymal fat depot (g) |
|---|---|---|---|---|---|---|
| BALB/c | WD | 6.0±1.4*** | 2.5±1.1** | 23.2±1.9 | 2.0±0.3** | 0.6±0.1*** |
| RC | 7.4±1.0 | 1.7±0.5 | 23.5±1.5 | 1.8±0.2 | 0.4±0.1 | |
| C57BL/6 | WD | 12.3±2.7**** | 5.1±2.5**** | 24.8±2.3 | 1.6±0.2* | 1.2±0.5**** |
| RC | 8.1±2.1 | 1.3±0.8 | 24.5±3.1 | 1.4±0.2 | 0.3±0.1 | |
| NMRI | WD | 7.7±4.5 | 5.0±3.7* | 30.6±2.3 | 2.0±0.3** | 1.4±0.9** |
| RC | 6.2±2.2 | 2.6±1.4 | 30.5±1.6 | 1.7±0.03 | 0.7±0.2 | |
| CD-1 | WD | 9.4±4.0* | 5.8±3.6*** | 29.7±2.2** | 2.0±0.3 | 1.6±0.9*** |
| RC | 7.4±1.5 | 2.3±1.0 | 31.5±1.7 | 1.9±0.2 | 0.7±0.2 |
RC=Rodent Chow, WD=Western Diet.
indicates p<0.05;
indicates p<0.01;
indicates p<0.001;
indicates p<0.0001.
Comparisons are between WD groups and RC groups within each strain. WD groups: (BALB/c: n=40; C57BL/6: n=39; NMRI: n=40; CD-1: n=40); RC groups: (BALB/c: n=15; C57BL/6: n=15; NMRI: n=14; CD-1: n=15).
Total fat mass and epididymal fat mass weights were significantly increased in all four strains in mice fed WD compared to controls. Lean mass was decreased in the outbred CD-1 strain, with no significant effects in the remaining strains, and liver weights were significantly increased in BALB/c, C57BL/6 and NMRI mice compared to controls (Table 3). Compared to standard rodent chow, exposure to an energy-dense diet significantly increased a minimum of three out of five metabolic parameters in all strains.
The effects of WD between strains were also assessed. The WD-fed mice of the C57BL/6 strain displayed a significantly higher mean bodyweight gain compared to WD-fed BALB/c, NMRI and CD-1 mice. Total fat mass and epididymal fat mass were significantly higher in C57BL/6, NMRI and CD-1 WD-fed mice compared to the BALB/c, while liver weights were significantly lower in C57BL/6 mice compared to the other strains.
Within-strain variation of metabolic parameters
Point-estimates of within-strain variation in bodyweight gain ranged from 22.9-53.4% (Table 4). The lowest level of variation was observed in the two inbred strains C57BL/6 and BALB/c, both of which were significantly less variable compared to the outbred stocks. Additionally, mean epididymal fat mass was significantly less variable in inbred BALB/c mice compared to all other strains. No difference in variability between strains was observed when comparing total fat mass, lean mass and liver weight.
Table 4.
Phenotypic variation of selected metabolic parameters in inbred and outbred mice after 6 weeks exposure to an energy-dense diet (Western Diet)
| BALB/c (n=40) | C57BL/6 (n=39) | NMRI (n=40) | CD-1 (n=40) | |||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Mean ± SD | CV (%) | Mean ± SD | CV (%) | Mean ± SD | CV (%) | Mean ± SD | CV (%) | |
| Bodyweight change (g) | 6.0±1.4† | 25.8 [21.1; 33.1]a | 12.3±2.7†† | 22.9 [18.7; 29.4]a | 7.7±4.5†,††† | 53.4 [43.7; 68.9]b | 9.4±4.0††† | 43.7 [35.6; 56.5]b |
| Fat mass (EchoMRI) (g) | 2.5±1.1† | 63.1 [51.7; 81.0] | 5.1±2.5†† | 71.0 [58.0; 91.5] | 5.0±3.7†† | 69.0 [56.5; 88.6] | 5.8±3.6†† | 73.0 [59.8; 93.8] |
| Lean mass (EchoMRI) (g) | 23.2±1.9† | 8.3 [6.8; 10.6] | 24.8±2.3†† | 9.7 [7.9; 12.5] | 30.6±2.3††† | 7.9 [6.4; 10.1] | 29.7±2.2††† | 7.4 [6.0; 9.5] |
| Liver weight (g) | 2.0±0.3† | 12.0 [9.9; 15.5] | 1.6±0.2†† | 15.8 [12.9; 20.4] | 2.0±0.3† | 14.8 [12.1; 19.0] | 2.0±0.3† | 13.4 [11.0; 17.2] |
| Epididymal fat weight (g) | 0.6±0.1† | 25.0 [20.5; 32.1]c | 1.2±0.5†† | 46.9 [38.3; 60.4]d | 1.4±0.9†† | 58.4 [47.8; 75.0]d | 1.6±0.9†† | 56.2 [46.1; 72.2]d |
Numbers given in percentage are coefficients of variations. Brackets designate 95% confidence intervals for coefficients of variation. Superscripts indicate significant (P<0.05) differences in CV’s (a,b,c,d) and effect (†, ††, †††) between strains. CV=Coefficient of variation.
Discussion
To our knowledge, this is the first study to estimate and evaluate the relative within-strain variation of preclinical parameters in response to insulin dosing and metabolic effects of an energy-dense diet in inbred and outbred mouse strains commonly used in diabetes and obesity research. Here, we show that phenotypic variability in most of the typical readouts was comparable between inbred and outbred strains of mice. Thus, our findings are not in agreement with the common notion that inbred strains will display lower levels of phenotypic variation due to their genetic homogeneity [12].
Overall, comparable phenotypic variation was observed in 23/32 (~72%) of direct comparisons between inbred and outbred mice. However, the inbred strain C57BL/6 was significantly less variable in response to HI in 2/3 of parameters compared to the two outbred stocks. When examining the variability of the metabolic response to WD, inbred BALB/c mice were significantly less variable than the outbred stocks in 2/5 parameters, while within-strain variation in the C57BL/6 mice was significantly lower in only 1/5 parameters.
Besides genetic background, a wide range of factors arising from experimental, analytical and environmental conditions have the potential to contribute to the phenotypic variation observed within a strain [13]. Estimating the relative contribution of each of these variables to the total variation within a strain was not attempted in this study. However, sources of experimental, analytical and environmental variation were limited by careful planning of the study and are expected to have been comparable between experiments and strains. However, since the variability of the majority of the readouts examined in this study was comparable between inbred and outbred mice, our data suggest that the potential contribution from genetic heterogeneity to the total phenotypic variation is relatively small, and that the phenotypic variation is likely primarily influenced by experimental, analytical and environmental variation.
Previously, it has been suggested that phenotypic variation in inbred strains is relatively more influenced by environmental factors than outbred stocks, because the environmental component of variability will increase relatively as the genetic component is reduced [6,14]. Also, inadequate ongoing quality control among outbred stocks may introduce the possibility of outbred stocks being more or less inbred, depending on breeding facility. Although breeders try to monitor genetic variation and drift within their stocks by use of microsatellite markers, the screening procedures are often only performed on a few animals and may be separated by years [15,16]. Genetic drift and bottlenecks within outbred colonies are well-known problems capable of interfering with population structure over a span of only a few generations, introducing less variability by decreasing the level of heterozygosity [17-19]. Thus, we cannot exclude the possibility that the outbred stocks used in this study may have been subject to a decline in genetic variation brought on by genetic drift or non-random breeding, which would likely result in the phenotypic variation being more comparable to inbred strains of mice.
Breeder identity has previously been identified as a possible confounder in relation to altered genotypic and phenotypic variability [20-22]. Sprague-Dawley rats from three different breeders showed genetically and phenotypically distinguishable variation, both between breeders and between individual colonies within facilities [15]. The same has been shown in outbred ICR-mice from separated colonies within the same breeding facility; however, similar studies in other stocks demonstrated only a negligible loss of variation [22].
As expected, the sensitivity to the experimental treatment varied between strains in the present study. When the mice were fed a WD, the least sensitive strain (BALB/c) was also the least variable for several of the chosen readouts, which agrees with a previous study showing that in strains prone to diet-induced obesity, high fat feeding induces highly variable phenotypes (even in genetically uniform C57BL/6 mice) [23]. In agreement with our observations, NMRI, CD-1 and C57BL/6 mice have previously been shown to be prone to diet-induced obesity, whereas BALB/c mice are relatively resistant [24-26]. However, by using the CV as measure of variability, the difference in effect between strains has only little impact on the variation as the CV represents a relative measure of variability.
In the present study, the within-strain variation seemed more closely associated with the sensitivity of the individual strain to the applied treatment and the endpoint than to the classification of inbred or outbred. This suggest that choice of animal model should be based on pilot-studies determining the relative efficacy and variation of individual strains to the experimental procedure or intervention, rather than on the assumption that outbred stock variation surpasses that of inbred strains.
Previous studies comparing the within-strain variation of inbred and outbred rodents have yielded contradicting results, but the endpoints and compared strains also vary between studies [7,14,27-36]. Some studies used low sample sizes resulting in a questionable reliability of the estimate of phenotypic variation [7,27,29,33-35]. Larger sample sizes provide better estimates of the true phenotypic variation per se, and the sample sizes used in this study were calculated based on a predefined range of a 95% confidence interval for the CV, thus predicting the reliability of the estimate of the variation. In the majority of previous studies referenced in this text, it has not been reported if formal sample size calculations were performed. Only very few previous studies have employed sample sizes as large as those used here and they too have reached conflicting conclusions [30,31,36]. Hence, one study examined the variation in skeletal muscle fiber number in inbred and outbred mice and rats (n=97-113/strain) and found inbred to be more phenotypically variable than outbred [31]. Another study found the variability in neurobehavioral response to acute trimethyltin neurotoxicity to be comparable between inbred and outbred rats (n=40/strain) [30], whereas a third study, observing the sleeping time after administration of hexobarbital in mice, showed outbred to be more variable than inbred (n=25-64/strain)[36]. However, the contradicting results may at least partly be explained by the variation not being inbred/outbred-related but rather strain sensitivity and endpoint-related. Thus, as the studies are separated by decades and as breeder effects and genetic drift is likely to occur within such a time frame, it is questionable if the results can be directly compared or used to predict the variability of other phenotypic traits.
It is important to note, that in cases where no significant difference in variation was observed between inbred and outbred strains, significant differences in within-strain variation could exist if tested using larger sample sizes. However, detection of such differences would potentially require thousands of animals in each group suggesting that the relative contribution from difference in true phenotypic variability may be so small, that it would have no practical impact on the number of animals needed to perform an experiment. When retrospectively calculating the sample sizes for a theoretical experiment based on the CVs obtained in the present study, in only relatively few cases did the difference in CV between inbred and outbred result in the requirement of a considerably larger number of animals (Table 5). In relation to this, the use of a multi-strain assay has previously been suggested [37]. In an attempt to model the genetic heterogeneity of the human population, this type of experimental design employs a panel of 8-10 different inbred strains. This is in contrast to the traditional approach of testing against a genetically diverse background in a single outbred stock. One argument for the use of this design is that, despite the employment of more strains of inbred mice, the lower phenotypic variation within these animals will ultimately lead to a lower sample size compared to if the same experiment was done in outbred animals. However, with the comparable within-strain variation for inbred and outbred mice observed in this study, the multi-strain approach would paradoxically lead to the use of a far higher number of animals.
Table 5.
Sample size calculation in a hypothetical experiment based on the coefficients of variation estimated in this study. Power was set at 0.80, α=0.05 and the minimal detectable difference to a fold-change of 1.5
| Parameter | Number of mice per group to be able to detect a difference >1.5-fold between two groups (e.g., treatment versus control) | |||
|---|---|---|---|---|
|
|
||||
| BALB/c (inbred) | C57BL/6 (inbred) | NMRI (outbred) | CD-1 (outbred) | |
| AOCglucose0-40 | 9 | 4 | 8 | 7 |
| Maximal blood glucose effect of HI | 4 | 3 | 6 | 5 |
| Body weight change | 7 | 8 | 23 | 19 |
| Fat mass | 40 | 50 | 47 | 52 |
| Lean mass | 3 | 3 | 3 | 3 |
| Liver weight | 3 | 4 | 4 | 4 |
| Epididymal fat mass | 8 | 22 | 34 | 32 |
Sample size calculations were performed in SAS JMP. HI=Human insulin. AOC=Area over the Δblood glucose vs. time curve.
The main objective of this study was to provide reliable estimates of within-strain variation for selected pharmacological and metabolic parameters in inbred strains and outbred stocks of mice. In conclusion, we found that the variability of the majority of tested readouts was comparable between inbred and outbred mice. The results suggest that within-strain variation is highly strain- and endpoint dependent and highlight the impact of choice of animal model (whether inbred or outbred) on the outcome of pharmacological studies within diabetes and obesity.
Acknowledgements
We are indebted to professor Ib Skovgaard for his invaluable advice with the experimental design and statistical approach of the study. We furthermore wish to thank Anne Bowman, Mikkel Schrøder Jørgensen, Sara Louise Riisberg, Marianne BojsenJappe, Emilie Due Jensen and Trine Rørmose Løjmand for excellent technical assistance. This study was funded by Novo Nordisk A/S and partly supported by the Lifepharm Centre for In Vivo Pharmacology.
Disclosure of conflict of interest
None.
Authors’ contribution
The study was designed by VSJ, TP, JL and HH. The experiments were carried out by VSJ. The initial data analysis was performed by VSJ and HH followed by data interpretation by all authors. The draft manuscript was written by VSJ and subsequently edited by all authors.
References
- 1.Proetzel G, Wiles MV. Mouse Models for Drug Discovery [Google Scholar]
- 2.Singh SS. Preclinical pharmacokinetics: an approach towards safer and efficacious drugs. Curr Drug Metab. 2006;7:165–182. doi: 10.2174/138920006775541552. [DOI] [PubMed] [Google Scholar]
- 3.Chatzigeorgiou A, Halapas A, Kalafatakis K, Kamper E. The use of animal models in the study of diabetes mellitus. In Vivo. 2009;23:245–258. [PubMed] [Google Scholar]
- 4.Festing MF. Variation and its implications for the design of experiments in toxicological research. Comp Haematol Int. 1997;7:202–207. [Google Scholar]
- 5.Festing MF. Inbred strains should replace outbred stocks in toxicology, safety testing, and drug development. Toxicol Pathol. 2010;38:681–690. doi: 10.1177/0192623310373776. [DOI] [PubMed] [Google Scholar]
- 6.Gill TJ. The use of randomly bred and genetically defined animals in biomedical research. Am J Pathol. 1980;101:S21–32. [PMC free article] [PubMed] [Google Scholar]
- 7.Vaickus LJ, Bouchard J, Kim J, Natarajan S, Remick DG. Inbred and outbred mice have equivalent variability in a cockroach allergen-induced model of asthma. Comparative Med. 2010;60:420–426. [PMC free article] [PubMed] [Google Scholar]
- 8.Festing MF. Evidence should trump intuition by preferring inbred strains to outbred stocks in preclinical research. ILAR J. 2014;55:399–404. doi: 10.1093/ilar/ilu036. [DOI] [PubMed] [Google Scholar]
- 9.Kelley K. Sample size planning for the coefficient of variation from the accuracy in parameter estimation approach. Behav Res Methods. 2007;39:755–766. doi: 10.3758/bf03192966. [DOI] [PubMed] [Google Scholar]
- 10.Metzinger MN, Miramontes B, Zhou P, Liu Y, Chapman S, Sun L, Sasser TA, Duffield GE, Stack MS, Leevy WM. Correlation of X-Ray computed tomography with quantitative nuclear magnetic resonance methods for pre-clinical measurement of adipose and lean tissues in living mice. Sensors. 2014;14:18526–18542. doi: 10.3390/s141018526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diletti E, Hauschke D, Steinijans V. Sample size determination for bioequivalence assessment by means of confidence intervals. Int J Clin Pharmacol Ther Toxicol. 1991;29:1–8. [PubMed] [Google Scholar]
- 12.Festing MFW. Warning: the use of heterogeneous mice may seriously damage your research. Neurobiol Aging. 1999;20:237–244. doi: 10.1016/s0197-4580(99)00040-8. [DOI] [PubMed] [Google Scholar]
- 13.Svendsen O, Hansen AK. Biological variation, reproducibility and predictability of in vivo drug testing. Scand J Lab Anim Sci. 1998;25:86–98. [Google Scholar]
- 14.Bader RS. Variability in wild and inbred mammalian populations. Quart Jour Florida Acad Sci. 1956;19:14–34. [Google Scholar]
- 15.Fitzpatrick CJ, Gopalakrishnan S, Cogan ES, Yager LM, Meyer PJ, Lovic V, Saunders BT, Parker CC, Gonzales NM, Aryee E, Flagel SB, Palmer AA, Robinson TE, Morrow JD. Variation in the form of pavlovian conditioned approach behavior among outbred male Sprague-Dawley rats from different vendors and colonies: sign-tracking vs. goal-tracking. PLoS One. 2013;8:e75042. doi: 10.1371/journal.pone.0075042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldinger KA, Sokoloff G, Rosenberg DM, Palmer AA, Millen KJ. Genetic variation and population substructure in outbred CD-1 mice: implications for genome-wide association studies. PLoS One. 2009;4:e4729. doi: 10.1371/journal.pone.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoger H. Genetic drift in an outbred stock of mice. Experimental Anim Tokyo. 1992;41:215–220. [PubMed] [Google Scholar]
- 18.Papaioannou VE, Festing MFW. Genetic drift in a stock of laboratory mice. Lab Anim. 1980;14:11–13. doi: 10.1258/002367780780943015. [DOI] [PubMed] [Google Scholar]
- 19.Kloting I, Nitschke C, van den Brandt J. Impact of genetic profiles on experimental studies: outbred versus wild rats. Toxicol Appl Pharm. 2003;189:68–71. doi: 10.1016/s0041-008x(03)00099-1. [DOI] [PubMed] [Google Scholar]
- 20.Turnbull AV, Rivier CL. Sprague-Dawley rats obtained from different vendors exhibit distinct adrenocorticotropin responses to inflammatory stimuli. Neuroendocrinology. 1999;70:186–195. doi: 10.1159/000054475. [DOI] [PubMed] [Google Scholar]
- 21.Portelli J, Aourz N, De Bundel D, Meurs A, Smolders I, Michotte Y, Clinckers R. Intrastrain differences in seizure susceptibility, pharmacological response and basal neurochemistry of Wistar rats. Epilepsy Res. 2009;87:234–246. doi: 10.1016/j.eplepsyres.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Hayakawa J, Koizumi T, Natsuumesakai S. Constancy of genetic variability in mice from non-inbred closed colonies. Lab Anim. 1980;14:233–236. doi: 10.1258/002367780780937625. [DOI] [PubMed] [Google Scholar]
- 23.Burcelin R, Crivelli V, Dacosta A, Roy-Tirelli A, Thorens B. Heterogeneous metabolic adaptation of C57BL/6J mice to high-fat diet. Am J Physiol. 2002;282:E834–E842. doi: 10.1152/ajpendo.00332.2001. [DOI] [PubMed] [Google Scholar]
- 24.Montgomery MK, Hallahan NL, Brown SH, Liu M, Mitchell TW, Cooney GJ, Turner N. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia. 2013;56:1129–1139. doi: 10.1007/s00125-013-2846-8. [DOI] [PubMed] [Google Scholar]
- 25.Herberg L, Doppen W, Major E, Gries FA. Dietary induced hypertrophic hyperplastic obesity in mice. J Lipid Res. 1974;15:580–585. [PubMed] [Google Scholar]
- 26.Breslin WL, Strohacker K, Carpenter KC, Esposito L, McFarlin BK. Weight gain in response to high-fat feeding in CD-1 male mice. Lab Anim. 2010;44:231–237. doi: 10.1258/la.2010.009114. [DOI] [PubMed] [Google Scholar]
- 27.McLaren A, Michie D. Variability of response in experimental animals. J Genet. 1956;54:440–455. [Google Scholar]
- 28.Biggers JD, McLaren A, Michie D. Variance control in the animal house. Nature. 1958;182:77–80. doi: 10.1038/182077a0. [DOI] [PubMed] [Google Scholar]
- 29.Buttner D, Hackbarth H, Wollnik F, Borggreve H. Blood pressure in rats-a comparison of a multifactorial experimental design to measurements in an outbred stock. Lab Anim. 1984;18:110–114. doi: 10.1258/002367784780891334. [DOI] [PubMed] [Google Scholar]
- 30.Moser VC. Rat strain- and gender-related differences in neurobehavioral screening: acute trimethyltin neurotoxicity. J Toxicol Environ Health. 1996;47:567–586. doi: 10.1080/009841096161546. [DOI] [PubMed] [Google Scholar]
- 31.Dudenhoeffer GA, Bowlin BK, Timson BF. A brief study of within litter and within strain variation in skeletal muscle fiber number in 3 lines of laboratory rodents. Growth. 1985;49:450–454. [PubMed] [Google Scholar]
- 32.Festing MF, Diamanti P, Turton JA. Strain differences in haematological response to chloroamphenicol succinate in mice: implications for toxicological research. Food Chem Toxicol. 2001;39:375–383. doi: 10.1016/s0278-6915(00)00149-6. [DOI] [PubMed] [Google Scholar]
- 33.Ward JA, Robinson J, Morris ID. Strain-dependency of procarbazine-induced testicular toxicity. Reprod Toxicol. 1989;3:43–50. doi: 10.1016/0890-6238(89)90037-3. [DOI] [PubMed] [Google Scholar]
- 34.Tordoff MG, Alarcon LK, Lawler MP. Preferences of 14 rat strains for 17 taste compounds. Physiol Behav. 2008;95:308–332. doi: 10.1016/j.physbeh.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Festing MF. Phenotypic variability of inbred and outbred mice. Nature. 1976;263:230–232. doi: 10.1038/263230a0. [DOI] [PubMed] [Google Scholar]
- 36.Jay GE. Variation in response of various mouse strains to hexobarbital (Evipal) P Soc Exp Biol Med. 1955;90:378–380. doi: 10.3181/00379727-90-22039. [DOI] [PubMed] [Google Scholar]
- 37.Festing MF. Use of a multistrain assay could improve the NTP carcinogenesis bioassay. Environ Health Persp. 1995;103:44–52. doi: 10.1289/ehp.9510344. [DOI] [PMC free article] [PubMed] [Google Scholar]


