Abstract
Transcriptional regulation of the murine immunoglobulin (Ig) heavy chain gene (Igh) involves several regulatory elements including the 3′Igh regulatory region (3′IghRR), which is composed of at least 4 enhancers (hs3A, hs1.2, hs3B, and hs4). The hs1.2 and hs4 enhancers exhibit the greatest transcriptional activity and contain binding sites for several transcription factors including nuclear factor kappaB/Rel (NF-κB/Rel) proteins and the aryl hydrocarbon receptor (AhR). Interestingly, the environmental immunosuppressant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), which potently inhibits antibody secretion, also profoundly inhibits 3′IghRR and hs1.2 enhancer activation induced by the B-lymphocyte activator lipopolysaccharide (LPS), but enhances LPS-induced activation of the hs4 enhancer. Within the hs1.2 and hs4 enhancers, the AhR binding site is in close proximity or overlaps an NF-κB/Rel binding site suggesting a potential reciprocal modulation of the 3′IghRR by AhR and NF-κB/Rel. The objective of the current study was to evaluate the role of NF-κB/Rel and the AhR on the 3′IghRR and its enhancers using the AhR ligand TCDD, the AhR antagonist CH223191, and toll-like receptor agonists LPS, Resiquimod (R848), or cytosine-phosphate-guanine-oligodeoxynucleotides (CpG). Utilizing the CH12.LX B-lymphocyte cell line and variants expressing either a 3′IghRR-regulated transgene reporter or an inducible IκBα (inhibitor kappa B-alpha protein) superrepressor (IκBαAA), we demonstrate an AhR- and NF-κB/Rel-dependent modulation of 3′IghRR and hs4 activity. Additionally, in mouse splenocytes or CH12.LX cells, binding within the hs1.2 and hs4 enhancer of the AhR and the NF-κB/Rel proteins RelA and RelB was differentially altered by the cotreatment of LPS and TCDD. These results suggest that the AhR and NF-κB/Rel protein binding profile within the 3′IghRR mediates the inhibitory effects of TCDD on Ig expression and therefore antibody levels.
Keywords: aryl hydrocarbon receptor, NF-κB/Rel, gene regulation, 3′Igh regulatory region, immunoglobulin, immunosuppression, TCDD
The immune system is complex and multifaceted, requiring the temporal and spatial regulation and interaction of many cell types and cellular mediators to mediate specific immune responses. Altered immune function by environmental chemicals could lead to serious pathophysiological effects such as immune suppression or hypersensitivity and autoimmunity. 2,3,7,8-Tetracholordibenzo-p-dioxin (TCDD), a potent and persistent environmental contaminant, elicits a variety of biological effects in both animal and cellular models including a marked suppression of immune function (reviewed in Birnbaum and Tuomisto, 2000; White and Birnbaum, 2009). TCDD targets, either directly or indirectly, many cell types and cellular functions of the immune response (Esser et al., 2009). Our work and others have demonstrated a direct effect of TCDD on B lymphocytes, resulting in the inhibition of B-lymphocyte stimulation and differentiation into antibody-secreting cells (reviewed in Sulentic and Kaminski, 2011). Several studies support an involvement of the aryl hydrocarbon receptor (AhR) signaling pathway in these effects; however, the specific mechanism remains unclear (Holsapple et al., 1991; Sulentic and Kaminski, 2011; Sulentic et al., 1998, 2000; Vorderstrasse et al., 2001). The AhR and its dimerization partner AhR nuclear translocator (ARNT) are classically believed to regulate transcription by binding dioxin-responsive elements (DREs) in regulatory regions of dioxin-sensitive genes (Okey, 2007). In addition to the direct binding of the AhR to DREs, the AhR has been shown to associate with other cellular proteins including transcription factors such as nuclear factor kappaB/Rel (NF-κB/Rel). NF-κB/Rel proteins play a significant physiological and pathophysiological role in many cellular processes and are prominent regulatory proteins of immune cell function (Vallabhapurapu and Karin, 2009). Interestingly, associations between the AhR and NF-κB/Rel have been shown to mediate TCDD-induced biological effects, such as cytokine expression (Beischlag et al., 2008; Kim et al., 2000; Tian, 2009; Tian et al., 1999; Vogel et al., 2007). As NF-κB/Rel is an essential regulator of B-lymphocyte activation and differentiation (Gerondakis and Siebenlist, 2009), interactions between the AhR and NF-κB/Rel may also mediate the inhibitory effects of TCDD on immunoglobulin (Ig) expression and therefore antibody levels.
Our previous work has identified a novel transcriptional target of TCDD within the Ig heavy chain (Igh) gene locus: A large (approximately 30 kb) transcriptional regulatory region located downstream of the Igh constant regions (referred to as 3′IghRR). The 3′IghRR mediates upregulation of Igh expression and class switch recombination (CSR), processes central to B-lymphocyte differentiation and mounting an effective antibody response (Manis et al., 1998; Pinaud et al., 2001, 2011; Vincent-Fabert et al., 2010). In a murine B-lymphocyte cell line model (CH12.LX), we demonstrated a profound inhibition by TCDD of 3′IghRR activation in cells stimulated with lipopolysaccharide (LPS), a toll-like receptor 4 (TLR4) ligand, which mirrored the effect on Igh gene expression and antibody secretion (Henseler et al., 2009; Sulentic et al., 2000, 2004b). This effect of TCDD on 3′IghRR activation may be mediated through the AhR-DRE signaling pathway as 2 DRE-like sites were identified within the 3′IghRR that were capable of binding the AhR/ARNT complex (Sulentic et al., 2000). However, the 3′IghRR, which is most often associated with 4 enhancers (hs3A, hs1.2, hs3B, and hs4), contains DNA binding sites for several transcription factors, including NF-κB/Rel, that appear to be important regulators of individual enhancer and overall 3′IghRR activity (Khamlichi et al., 2000; Pinaud et al., 2011). Supporting a potential interaction between the AhR and NF-κB/Rel proteins and/or signaling pathways, an NF-κB/Rel binding site (κB site) is in close proximity to the DRE site within the hs1.2 enhancer and a κB site overlaps the DRE site within the hs4 enhancer (Sulentic et al., 2000). Additionally, TCDD increased NF-κB/Rel protein binding to the hs4 κB site, which appeared to be at least partially independent of the AhR (Sulentic et al., 2000). However, binding to both the DRE and κB sites cooperatively influenced hs4 luciferase reporter activity (Sulentic et al., 2004a,b). Therefore, the objective of the present study was to determine the role of the AhR and NF-κB/Rel proteins and potential interactions between these proteins in mediating the inhibitory effects of TCDD on 3′IghRR activation. Using an AhR antagonist (CH223191), the well-characterized CH12.LX B-lymphocyte cell line, and 2 variants of this line that either stably expressed a transgene regulated by the 3′IghRR (CH12.γ2 b-3′IghRR) or an inducible IκBα superrepressor to inhibit NF-κB/Rel activity (CH12.IκBαAA [CH12.LX B-lymphocyte cell line expressing an IPTG {isopropyl β-d-1-thiogalactopyranoside}-inducible IκBα superrepressor]), we determined that both the AhR and NF-κB/Rel proteins are essential for mediating the effects of TCDD on 3′IghRR activity and that these effects appear to be mediated by an altered NF-κB/Rel binding profile within the hs1.2 and hs4 enhancer. These results suggest that interactions between the AhR and NF-κB/Rel within the 3′IghRR mediate the inhibitory effects of TCDD on Ig expression and therefore antibody levels.
MATERIALS AND METHODS
Chemicals and reagents
TCDD in 100% dimethyl sulfoxide (DMSO) was purchased from AccuStandard Inc (New Haven, Connecticut). The certificates of product analysis stated the purity of TCDD to be 99.1%. IPTG, LPS (Escherichia coli), and DMSO were purchased from Sigma Aldrich (Milwaukee, Wisconsin). IPTG and LPS were dissolved in water and 1× PBS, respectively. The AhR antagonist CH223191 was purchased from Calbiochem (San Diego, California) and dissolved in 100% DMSO. Resiquimod (R848) was purchased from Enzo Life Sciences (Farmingdale, New York) and dissolved in 100% DMSO. Cytosine-phosphate-guanine (CpG) oligodeoxynucleotides (ODN) (5′-TCCATGACGTTCCTGACGTT-3′) was purchased from Eurofins MWG Operon (Huntsville, Alabama) and dissolved in RNase and DNase free water.
Cell lines
The CH12.IκBαAA B-lymphocyte cell line (IgM+) was developed and generously provided by Dr Gail Bishop (Hsing and Bishop, 1999) and is a variant of the parental CH12.LX cell line (Bishop and Haughton, 1986), which was derived from the murine CH12 B-cell lymphoma in B10.H-2aH-4 bp/Wts mice (B10.A × B10.129) (Arnold et al., 1983). The CH12.IκBαAA cell line stably expresses an IPTG-inducible, degradation resistant IκBα superrepressor protein (IκBαAA), which sequesters NFκB/Rel proteins in the cytoplasm (Hsing and Bishop, 1999). The CH12.γ2b-3′IghRR cell line (IgA+) was generated from CH12.LX cells and is a subclone that stably expresses a transgene (γ2b Igh gene) regulated by the 3′IghRR (Henseler et al., 2009; Shi and Eckhardt, 2001). Cells were grown in RPMI 1640 media (MediaTech, Herndon, Virginia) supplemented with 10% bovine calf serum (Hyclone, Logan, Utah), 13.5 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 100 units/ml penicillin, 100 µg/ml streptomycin, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 50 µM β-mercaptoethanol. Cells were maintained at 37°C in an atmosphere of 5% CO2. Cell viability was determined by assaying 1.0 ml of cell suspension for Trypan Blue exclusion with a ViCell instrument (Beckman Coulter, Brea, California).
Vertebrate animals
To validate the results from the cell line studies, splenocytes from 6-week-old female B6C3F1 mice were also utilized. No in vivo treatments were performed and all animals were sacrificed by decapitation in accordance with university policy and approved under our animal use protocol (AUP 685). After decapitation, the spleens were aseptically removed and placed in sterile ice-cold 1× PBS. The spleens were combined then homogenized with frosted microscope slides, and the debris cleared by passing the homogenate through a screen filter. After filtration, the cells were centrifuged for 5 min at 250 × g at 4°C, the supernatant was discarded, and the cells were washed with ice-cold PBS with inversion. The cells were then subjected once more to centrifugation for 5 min at 250 × g at 4°C. The supernatant was again discarded and the cell pellet was resuspended in 1 ml of red blood cell lysis buffer (10 mM KHCO3, 150 mM NH4CL, and 0.1 mM EDTA-pH 8.0) for 5 min, then diluted 1:10 with ice-cold PBS. The cells were subjected to a final centrifugation for 5 min at 250 × g at 4°C. The supernatant was discarded and the pellet was resuspended in culture media. The cells were allowed to incubate at 37°C in an atmosphere of 5% CO2 for 2 h. After the incubation, the cells were slowly decanted out of the culture flask, counted with a ViCell instrument (Beckman Coulter), and resuspended in culture media for use in chromatin immunoprecipitation (ChIP) experiments.
Protein isolation and enzyme-linked immunosorbent assay for γ2b analysis
CH12.γ2b-3′IghRR cells were stimulated with either 1 µg/ml LPS (TLR4 ligand), 1 µg/ml R848 (TLR7 and 8 ligand) or 1 µM CpG (TLR9 ligand) and cotreated with either the vehicle control (0.01% DMSO) or increasing concentrations of TCDD (0.003–30 nM). Treated and naïve control CH12.γ2b-3′IghRR cells were plated in triplicate into 12-well plates at a concentration of 2.5 × 104 cells/well and incubated for 48 h. Following the incubation period, cells were centrifuged at 3000 rpm, lysed with mild lysis buffer (150 mM NaCl, 10 mM sodium phosphate pH 7.2, 2 mM EDTA, and 1% Igepal) then centrifuged at 14 000 rpm. Supernatants were collected and stored at −80°C until analysis. To measure γ2b, cell lysates were thawed on ice and protein concentrations were determined by a Bradford assay (Bio-Rad Laboratories, Hercules, California) according to manufacturer specifications. Samples were then diluted to the lowest sample concentration and 2 μg of total protein was analyzed for γ2b by sandwich enzyme-linked immunosorbent assay as described by Henseler et al. (2009). Colorimetric detection was performed every minute over a 1-h period using a Spectramax plus 384 automated microplate reader with a 405-nm filter (Molecular Devices, Sunnyvale, California). The SOFTmax PRO analysis software (Molecular Devices) calculated the concentration of γ2b in each sample from a standard curve generated from the kinetic rate of absorption for known γ2b concentrations. Results are represented as percent effect relative to the DMSO control (set to 100% effect).
Western blot analysis
Following the appropriate treatment concentration and incubation period (see Figure 2 and Supplementary Figure S1), CH12.IκBαAA or CH12.LX cells were harvested using centrifugation (3000 rpm for 5 min at 4°C) and washed once with 1× PBS. The cells were resuspended in 150 µl of mild lysis buffer containing freshly added protease inhibitors (Complete Mini Protease Inhibitor Cocktail; Roche Diagnostics, Indianapolis, Indiana) and frozen at −80°C. For protein quantification, the lysate was thawed on ice and resuspended briefly then centrifuged at 14 000 rpm for 5 min at 4°C. The whole-cell lysate was removed from the pelleted cell debris, quantified by a Bradford assay, and frozen at −80°C until Western blot analysis. Briefly, whole-cell lysates were thawed on ice and 50 µg of protein from each extract was run on a 10% polyacrylamide gel at 200 V for 30–40 min. The protein was transferred from the gel to a polyvinylidene fluoride membrane (Millipore, Bedford, Massachusetts) using an electric current of 100 V for 75 min. The membrane was then immediately immersed in 3% BSA (bovine serum albumin)/TTBS (tris-buffered saline with 0.05% tween-20) and rocked overnight at 4°C. The membranes were incubated overnight at room temperature with either anti-IκBα (sc-371 (C-21), Santa Cruz, Santa Cruz, California) at a 1:1000 dilution, anti-AhR (ab2770 Abcam, Cambridge, Massachusetts) at a 1:1000 dilution, or anti-β-actin (Sigma Aldrich) at a 1:10 000 dilution in 3% BSA/TTBS. The membrane was then washed 4 times in TTBS at 10-min intervals, and the blot was incubated with the appropriate horse-radish-peroxidase-conjugated secondary antibody (goat anti-mouse at 1:8000 or goat anti-rabbit at 1:2500) for 1 h. The blot was washed again 4 times in TTBS, exposed to ECL substrate (ThermoScientific, Waltham, Massachusetts) and analyzed on a Fuji LAS-3000 Bioimager (Tokyo, Japan).
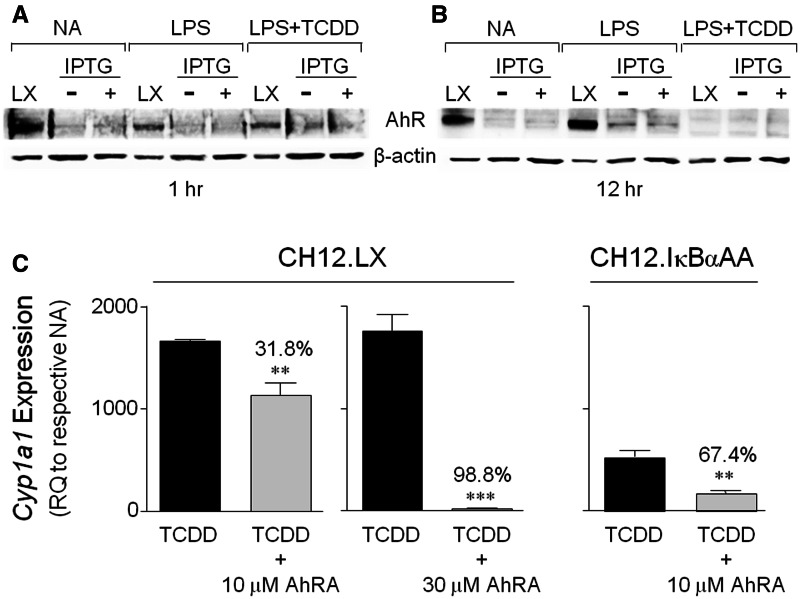
FIG. 2.
AhR expression and function in the CH12.IκBαAA cells. CH12.LX (denoted LX) and CH12.IκBαAA cells (pretreated with [+] or without [−] 100 μM IPTG for 2 h) were incubated for 1 h (A) or 12 h (B) in the absence of further treatment or in the presence of 1 μg/ml LPS with or without a 10 nM TCDD cotreatment. “NA” denotes the unstimulated control. Whole cell protein was isolated and analyzed by Western blot analysis. An anti-AhR antibody identified the AhR protein (approximately 95 kDa) and β-actin served as a loading control. Results are representative of at least 3 separate experiments. C, CH12.LX or CH12.IκBαAA cells were pretreated for 1 h with media alone, dimethyl sulfoxide (DMSO) or the AhR antagonist CH223191 (AhRA, 10 or 30 µM). The cells were then treated with DMSO or 10 nM TCDD and incubated for 8 h. Total RNA was isolated, converted to cDNA, and analyzed by real-time PCR for Cyp1a1 transcripts. Results from 3 to 4 separate RNA isolations per treatment are represented as the relative quantitation (RQ) compared with the respective NA set to 1. The DMSO (0.11% final concentration) vehicle control ranged from less than 1 to 30 RQ and the AhRA alone control ranged from less than 1 to 1.5 RQ (data not shown). Significance between the TCDD+AhRA treatment and the TCDD alone treatment was determined by an unpaired, 2-tailed t test. “**” and “***” denote significance at P < .01 and P < .001, respectively, from the appropriate TCDD treatment. Numbers above bars indicate the percent antagonism induced by the AhRA. AhR, aryl hydrocarbon receptor; CH12.IκBαAA, CH12.LX B-lymphocyte cell line expressing an IPTG-inducible IκBα superrepressor; Cyp1a1, cytochrome P4501a1 gene; IPTG, isopropyl β-d-1-thiogalactopyranoside; LX, CH12.LX parental cells.
RNA isolation, cDNA synthesis, and real-time PCR
CH12.LX or CH12.IκBαAA cells at a concentration of 5 × 105 cells/ml were pretreated for 1 h with media alone, DMSO or the AhR antagonist CH223191 (AhRA, 10 or 30 µM). The cells were then treated with DMSO or 10 nM TCDD and incubated for 8 h. The final DMSO vehicle concentration was 0.11%. Total RNA was isolated using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s protocol. The RNA concentration was determined using a NanoDrop (ThermoScientific, Wilmington, Delaware) and 200 ng total RNA was reverse transcribed to cDNA using the Taqman Reverse Transcription Reagents kit (Applied Biosystems, Foster City, California). The expression of β-actin (endogenous control to normalize cDNA concentrations) and cytochrome P4501a1 (Cyp1a1) genes was quantified by real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems) as previously described (Fernando et al., 2012; Romer and Sulentic, 2011). Primers for Cyp1a1 and β-actin span an intron and were as follows: Cyp1a1 Forward Primer—AAGTGCAGATGCGGTCTTCT, Cyp1a1 Reverse Primer—AAAGTAGGAGGCAGGCACAA, β-actin Forward Primer—GCTACAGCTTCACCACCACA, and β-actin Reverse Primer—TCTCCAGGGAGGAAGAGGAT. The results of the PCR amplification were analyzed using the 7500 system SDS software to determine relative quantification values (ie, fold-change) using the 2−ΔΔCT equation.
Transient transfection and luciferase assay
The Igh luciferase reporter plasmids were generously provided by Dr Robert Roeder (Rockefeller University, New York, New York). The VH-Luc-hs4 and VH-Luc-3′IghRR plasmids consist of an upstream variable heavy chain (VH) promoter, a luciferase gene and the hs4 enhancer or the 3′IghRR, respectively, located downstream of the luciferase gene. Plasmids were constructed using a pGL3 basic luciferase reporter construct (Promega, Madison, Wisconsin) as described previously (Ong et al., 1998). Transient transfections were performed as previously described (Henseler et al., 2009; Sulentic et al., 2004b). Briefly, CH12.LX or CH12.IκBαAA cells (1.0 × 107) were resuspended in 200 µl of culture media with 10 µg of plasmid (VH-Luc-hs4, or VH-Luc-3′IghRR) and transferred into a 2-mm gap electroporation cuvette (Molecular BioProducts, San Diego, California). Cells were electroporated using an electro cell manipulator (ECM 630; BTX, San Diego, California) with the voltage at 250 V, the capacitance at 150 µF, and the resistance at 75 Ω. For each plasmid, multiple transfections were pooled in fresh media at 2.0 × 105 cells/ml then immediately treated with the following treatment conditions and aliquoted in quadruplicate into a 12-well plate and cultured for a 24-h (VH-Luc-hs4) or 48-h (VH-Luc-3′IghRR) incubation period in 5% CO2 at 37°C. For the CH12.LX cells, they were pretreated for 1 h with DMSO or 30 µM CH223191 (AhRA) then treated with DMSO or 10 nM TCDD in the presence of 1 µg/ml LPS stimulation. The final DMSO concentration was 0.11%. For the CH12.IκBαAA cells, they were divided into 2 equal portions and 1 portion was treated with 100 μM IPTG for 2 h to activate the IPTG-inducible IκBαAA transgene whereas the other portion was cultured in the absence of IPTG to provide a control that lacked IκBαAA transgene expression. An initial concentration response and time course were conducted to determine the optimum concentration and time of addition for IPTG-induced IκBαAA expression (Supplementary Figure S1). After 2 h, the CH12.IκBαAA cells were treated with DMSO (0.01%) or varying concentrations of TCDD (0.001–10 nM) with or without LPS stimulation (0.001–1 µg/ml). Following the appropriate incubation period, cells were lysed with a 1 × reporter lysis buffer (Promega) and samples were immediately frozen at −80°C. To measure luciferase enzyme activity, samples were thawed at room temperature and 20 µl of sample lysate was mixed with 100 µl of luciferase assay reagent (Promega). Luciferase activity or luminescence was measured with a luminometer (Berthold Detection Systems, Oak Ridge, Tennessee) and represented as relative light units or percent effect relative to the appropriate DMSO control (set to 100% effect).
ChIP assay
CH12.LX cells or splenocytes (1.0 × 107 per treatment condition) were treated with 0.01% DMSO or 30 nM TCDD in the absence or presence of 1 µg/ml LPS stimulation and incubated for 90 min at 37°C with 5% CO2. After incubation, proteins were cross-linked to chromatin by incubating the cells with 1% formaldehyde for 10 min at room temperature with agitation. Glycine for a 125 mM final concentration was then added to each treatment condition to quench the crosslinking and samples were agitated for an additional 10 min. Samples were then centrifuged at 1800 × g for 5 min at 4°C. The supernatant was discarded and the pellet resuspended in ice-cold 1× PBS and centrifuged at 1800 × g for 5 min at 4°C. The supernatant was again discarded and the cells were subjected to ice-cold lysis buffer-1 (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% Igepal, 0.25% Triton X-100, and Protease Inhibitor Cocktail) for 20 min on ice. The samples were then centrifuged at 1800 × g for 5 min at 4°C. The supernatant was discarded and the pellet resuspended in lysis buffer-2 (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA (ethylene glycol tetraacetic acid), and Protease Inhibitor Cocktail) for 10 min at room temperature. The samples were then centrifuged at 1800 × g for 5 min at 4°C. The supernatant was discarded and the pellet resuspended in 350 µl ice-cold low salt sonication buffer (40 mM Tris-HCL pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, and Protease Inhibitor Cocktail). DNA was sheared by sonication (W-225 Sonicator Cell Distrupter, Heat Systems-Ultrasound, Inc) with a microtip horn at 40% power for eight 20 s, constant pulses and resting for 20 s in an ice/water mix between pulses. After sonication, samples were heated at 37°C for 5 min then supplemented with CaCl2 (5 mM final concentration) and treated with 120 units of micrococcal nuclease (MNase) for 15 min at 37°C. EDTA and EGTA for a 20 mM final concentration of each were added to the samples after heating to quench the MNase activity. Additionally, the final concentration of NaCl was adjusted to 300 mM, and the samples placed on ice for 10 min. Samples were then centrifuged at 20 000× g for 10 min at 4°C, and the supernatant transferred to 1.5-ml Eppendorf tubes. The samples were then precleared of free antibody and proteins that nonspecifically bind the immunoprecipitation (IP) antibodies by mixing with 25 µl of a protein-G coated magnetic bead slurry (MagnaBind Protein G magnetic beads, Thermal Scientific, Rockford, Illinois) and rotated at 4°C for 1 h. The samples were then cleared of the magnetic beads with a magnetic Eppendorf rack. A 10 -µl aliquot of each sample was then evaluated for total DNA by phenol-chloroform extraction and DNA quantification with a NanoDrop Spectrophotometer (ThermoScientific). All cleared pre-IP samples were then diluted to the same amount of DNA/sample and stored at −80°C until prepared for use in the ChIP assay. The IP antibodies (anti-AhR [ab2770 Abcam], anti-RelA [A301-823A Bethyl Laboratories, Inc, Montgomery, Texas], or anti-RelB [A302-183A Bethyl Laboratories, Inc]) were incubated at a concentration of 3 µg antibody to every 25 µl of protein G-coated magnetic bead slurry in 1 ml of 1× NET buffer (300 mM NaCl, 2 mM EDTA, 20 mM Tris-HCL, 1% Triton X-100, and Protease Inhibitor Cocktail) for a minimum of 4 h at 4°C with rotation. After the first incubation, 2 mg of sheared salmon sperm DNA (E213-5 ml; Amresco, Scion, Ohio) was added to the IP/bead complex and rotated an additional 1 h. The magnetic beads were then cleared of excess/unbound antibody and salmon sperm with a magnetic Eppendorf rack. The IP/bead complex was then resuspended in 200 µl of 1× NET buffer and 25 µl added to 50 µl of each precleared sample. Each sample was then adjusted to a final volume of 1 ml using 1× NET buffer. Samples were incubated overnight with rotation at 4°C. Using the magnetic Eppendorf rack, the samples were sequentially washed with 1 ml of 150 mM NaCl, 500 mM NaCl, 250 mM LiCl, and finally 10 mM TE. The 2 NaCl washes included 2 mM EDTA, 20 mM Tris-HCL, 1% Triton X-100, 0.1% SDS, and Protease Inhibitor Cocktail. The LiCl wash also had 1 mM EDTA, 10 mM Tris-HCL, 1% Igepal, 1% sodium deoxycholate, and Protease Inhibitor Cocktail. The samples (IP/bead complex with the protein cross-linked to the DNA) were treated 2 times with 100 µl elution buffer (100 mM NaHCO3, 1% SDS) for 15 min, and the eluent collected each time. To reverse the cross-linking, the eluent was placed in a heat block set to 65°C then spiked with NaCl (195 mM final concentration) and 1 µl of 10 mg/ml RNase was added to each sample. The samples were incubated for a minimum of 5 h, cooled to 45°C, and treated with 1 µl of 10 mg/ml proteinase K for 2 h. The DNA from the samples was extracted with phenol/chloroform extraction, and the presence of DNA was validated with a NanoDrop Spectrophotometer. SYBRGreen real-time PCR was utilized to analyze the DNA samples (2 µl) as previously described (Romer and Sulentic, 2011) with the exception of using the CFX96 Touch Real-Time PCR System (Bio-Rad). PCR primers were as follows: hs4 Forward Primer—CACACCCCACCTGTAGCAC, hs4 Reverse Primer—TGAGGAGGTTGACATGATGG, hs1.2 Forward Primer—CTGATATCTGAGCCCCCAAC, hs1.2 Reverse Primer—GTGGTCTGGGTAATGGATGG, β-actin Forward Primer—GCTACAGCTTCACCACCACA, and β-actin Reverse Primer—TCTCCAGGGAGGAGGAGGAT. β-actin was measured to determine whether the initial DNA (pre-IP samples) was equally diluted so that changes in threshold cycle (CT) could be attributed to treatment conditions within a sample set. ChIP data are reported as % input (100/2−ΔCT [normalized ChIP]), where ΔCT[normalized ChIP] = (CTChIP − (CTinput − log2 (Dilution Input Factor))) (Livak and Schmittgen, 2001). The Dilution Input Factor was 100. The data are reported as the combined average of at least 3 separate experiments.
Statistical analysis of data
Mean ± SE was determined for each treatment group (n = 3–4) within a given experiment. A statistical difference between treatment groups and the controls was determined by an unpaired, 2-tailed t test for comparisons of 2 groups or by a 1-way ANOVA with a Dunnett’s or Bonferroni’s Multiple Comparison post hoc test when comparing more than 2 groups. Statistical differences between and within treatments in the non-IκBαAA and IκBαAA-expressing cells were determined by a 2-way ANOVA with a Bonferroni’s post hoc test. The results are either representative of or the combined average of at least 3 separate experiments.
RESULTS
3′IghRR activation by different Toll-like receptor signaling pathways is equally sensitive to TCDD-induced inhibition
Toll-like receptors (TLRs) play an important role in the first line of host defense by recognition of microbial products. Mouse B lymphocytes express several TLR receptors including TLR4, 7, 8, and 9. Activation of these TLRs results in polyclonal B-lymphocyte activation and increased Ig expression and antibody secretion. Interestingly, human B lymphocytes express very low levels of TLR4 but express higher levels of TLR7 and 9. Correspondingly, human B lymphocytes are refractory to LPS stimulation but can be stimulated by ligands for TLR7 or 9 (reviewed in Bekeredjian-Ding and Jego, 2009). Because our previous results evaluating the effect of TCDD on 3′IghRR activation were limited to TLR4 stimulation via LPS (Sulentic et al., 2004a,b), which is an ineffective stimuli for human B lymphocytes, we evaluated the effect of TLR7/8 or 9 activation on the 3′IghRR and the corresponding effect of TCDD. The reason for this was 2-fold: (1) to determine whether TCDD uniquely targets signaling through the extracellular membrane-bound TLR4 or more likely a downstream effector of all TLRs such as NF-κB/Rel proteins; and (2) to utilize stimulation that is more representative of human B-lymphocyte activation via TLRs.
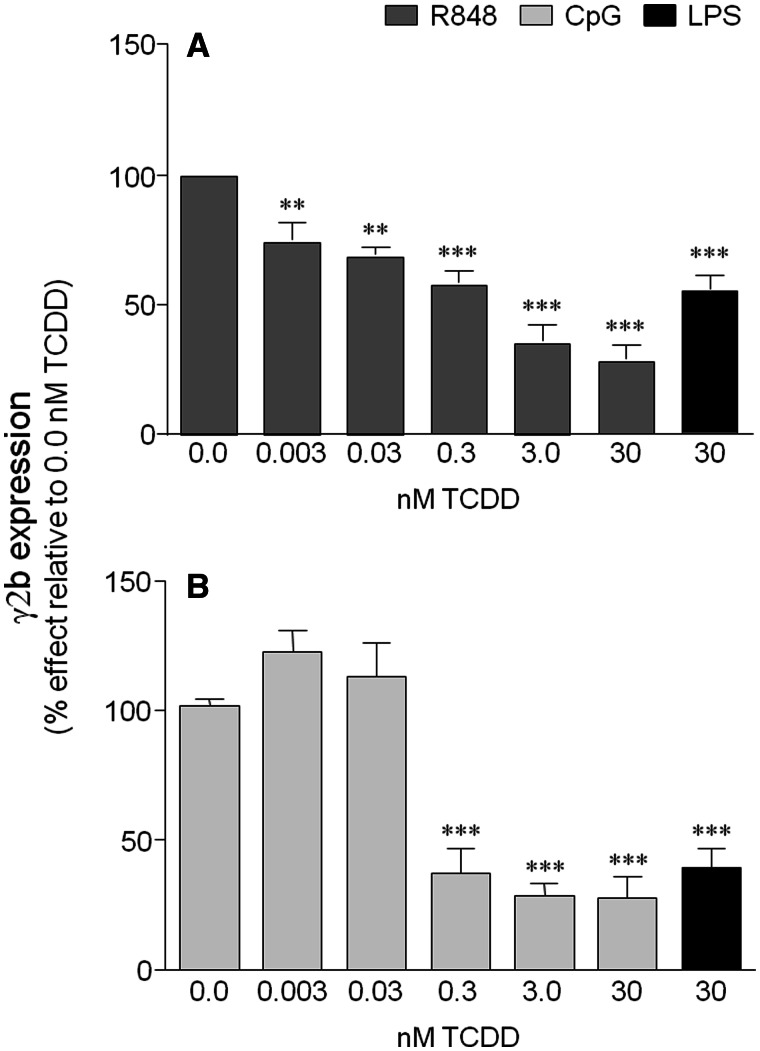
Our previous studies have utilized the well-characterized CH12.LX mouse B-lymphocyte cell line model, which has been extensively utilized in studying the effects of TCDD on B-lymphocyte function and Ig expression (Fernando et al., 2012; Sulentic et al., 1998, 2000, 2004a,b). Utilizing a variant of the CH12.LX cell line (ie, CH12.γ2b-3′IghRR [CH12.LX B-lymphocyte cell line stably expressing a 3′IghRR {mouse 3′Igh regulatory region}-regulated γ2b transgene]) that endogenously expresses IgA and stably expresses a transgene (γ2b Igh gene) regulated by the 3′IghRR (Henseler et al., 2009; Shi and Eckhardt, 2001), we have previously demonstrated an LPS-induced expression of the γ2b transgene and endogenous IgA that was markedly inhibited by TCDD (Henseler et al., 2009). Similar to the activation induced by a TLR4 ligand (ie, LPS), activation of TLR7/8 by Resiquimod (R848) and of TLR9 by hypomethylated/unmethylated CpG ODN resulted in activation of the 3′IghRR-regulated γ2b transgene (Figure 1). Additionally, TCDD significantly inhibited 3′IghRR activation (and endogenous IgA) in a concentration-dependent manner regardless of the specific TLR that was stimulated (Figure 1 and data not shown). As activation of the NF-κB/Rel signaling pathway is a common result of TLR activation and our previous results demonstrated a potential cooperative interaction between the AhR and NF-κB/Rel proteins in the activity of the hs4 enhancer of the 3′IghRR (Sulentic et al., 2004a,b), we further investigated the role of NF-κB/Rel proteins and the AhR in the inhibitory effect of TCDD on the 3′IghRR.
FIG. 1.
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a general inhibitor of the 3′IghRR. CH12.γ2b-3′IghRR cells were treated with increasing concentrations of TCDD (0–30 nM) and cotreated with the following toll-like receptor ligands: A, R848 (1 μg/ml); B, CpG (1 μM); or A and B, LPS (1 μg/ml). The LPS and TCDD cotreatment served as a positive control for TCDD-induced inhibition of the 3′IghRR. 3′IghRR-regulated γ2b transgene expression (n = 3 per treatment group) normalized to 2 μg total protein was determined by enzyme-linked immunosorbent assay. Results were normalized to the appropriate vehicle control set to 100%, ie, cotreatment of 0.01% dimethyl sulfoxide (vehicle control denoted as 0.0 nM TCDD) and stimulation. The means from 3 separate experiments (overall mean ± SE) are represented in the bar graph. The stimulation index for LPS, R848, and CpG did not differ significantly and was approximately 3-fold above the unstimulated, naive control. Statistical significance was determined by a 1-way ANOVA followed by Dunnett’s Multiple Comparison test. “**” and “***” denote significance from the vehicle control (0.0 nM TCDD) at P < .01 and P < .001, respectively. R848, Resiquimod; CpG, cytosine-phosphate-guanine (TLR9 agonist); LPS, lipopolysaccharide (TLR4 agonist); 3′IghRR, mouse 3′Igh regulatory region.
AhR expression and function in the CH12.IκBαAA cells
To initiate studies that directly evaluate the role of NF-κB/Rel proteins in the effects of LPS and TCDD on 3′IghRR activity, we utilized another variant of the CH12.LX cell line (ie, CH12.IκBαAA). The CH12.IκBαAA cell line stably expresses an IPTG-inducible IκBα superrepressor protein (IκBαAA), which is resistant to negative feedback regulation by NF-κB/Rel proteins (Hsing and Bishop, 1999; Romer and Sulentic, 2011). We verified the inducibility of IκBαAA expression, by treating the CH12.IκBαAA cells with varying concentrations of IPTG overnight (approximately 17 h) or with 100 μM IPTG from 0 to 5 h followed by whole-cell protein isolation and Western blot analysis. Concentration-response and time course studies demonstrated maximal expression of IκBαAA following a 2-h treatment with 100 µM IPTG (Supplementary Figure S1).
Additionally, a functioning AhR signaling pathway is an important characteristic of a suitable model to study the effects of TCDD on Igh transcriptional regulation. Therefore AhR expression and function were evaluated in the CH12.IκBαAA cells and compared with the parental CH12.LX cell line, which has been well-characterized in terms of the AhR signaling pathway (De Abrew et al., 2010; Suh et al., 2002; Sulentic et al., 1998, 2000). AhR protein levels were analyzed by Western blot analysis and demonstrated much lower basal levels in CH12.IκBαAA cells compared with CH12.LX cells (Figure 2A). However, LPS stimulation for 12 h induced AhR protein levels in both cell lines (Figure 2) as previously seen in the CH12.LX cells (Sulentic et al., 1998) and in stimulated primary mouse B lymphocytes (Marcus et al., 1998; Tanaka et al., 2005) as well as other cell types (Vogel et al., 2013). Alternatively, treatment with 10 nM TCDD for 12 h decreased AhR levels in both cell lines in agreement with previous studies (Pollenz, 2002) (Figure 2B). IκBαAA expression had no effect on basal AhR expression or the expression profile induced by LPS or TCDD (Figure 2). Moreover, confirming the expression of a functional AhR, TCDD induced Cyp1a1 expression in the CH12.IκBαAA cells, which was significantly reversed by pretreatment with 10 μM of the AhR antagonist CH223191 (AhRA) (Figure 2C). However, the level of Cyp1a1 induction by TCDD was significantly less in the CH12.IκBαAA cells (approximately 3-fold less) and more sensitive to the AhR antagonist as compared with the CH12.LX cells, which was consistent with the difference in AhR protein levels. Additionally, antagonism of TCDD-induced Cyp1a1 induction in the CH12.LX cells was much more effective with a 30 μM rather than 10 μM concentration of the AhR antagonist, ie, 99% inhibition compared with 32% inhibition, respectively (Figure 2C). Despite the differences in AhR expression levels between the CH12.IκBαAA and CH12.LX cells, the AhR expressed in the CH12.IκBαAA cell line is functional and regulated as expected.
IκBαAA expression or AhR antagonism reverse the effects of TCDD on 3′IghRR and hs4 enhancer activity
Interestingly, TLR activation and TCDD treatment produce a dichotomous effect on 3′IghRR and hs1.2 enhancer activity versus hs4 enhancer activity. TLR4 activation significantly increases 3′IghRR and hs1.2 activity but has little effect on hs4 activity, whereas a TCDD cotreatment profoundly inhibits LPS-induced 3′IghRR and hs1.2 activation but synergistically increases hs4 activity (Fernando et al., 2012; Sulentic et al., 2004b). These effects may be at least partially mediated by NF-κB/Rel proteins as supported by (1) protein-protein interactions between the AhR and NF-κB/Rel (Kim et al., 2000; Tian, 2009; Tian et al., 1999; Vogel et al., 2007); (2) the presence of an NF-κB/Rel binding site (κB site) either in close proximity or overlapping a DRE-like site within the hs1.2 or hs4 enhancers, respectively (Sulentic et al., 2000); and (3) TCDD-induced NF-κB/Rel protein binding to the hs4 κB site as well as a cooperative influence of protein binding to both the DRE and κB sites on hs4 luciferase reporter activity (Sulentic et al., 2000, 2004a,b).
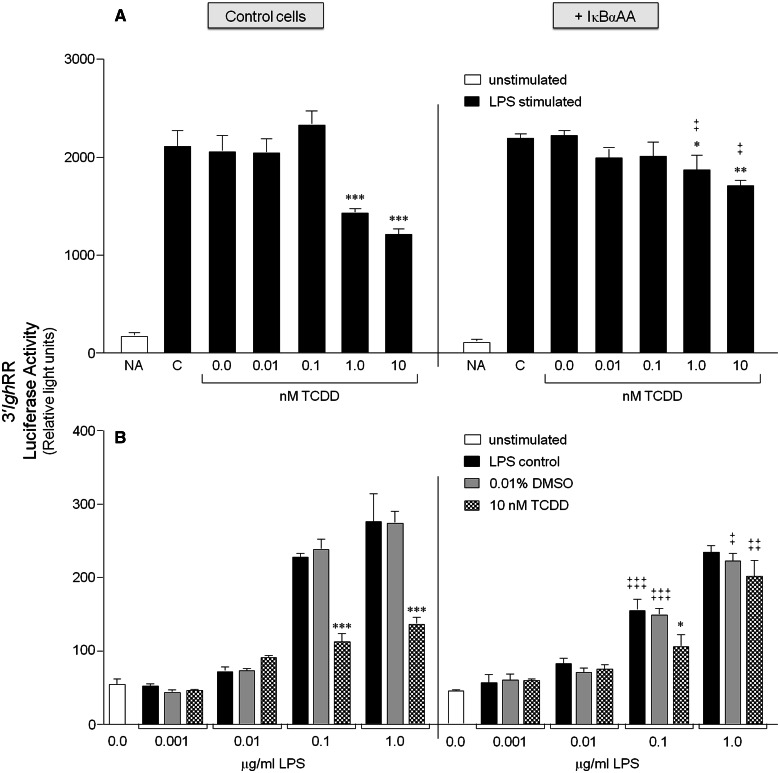
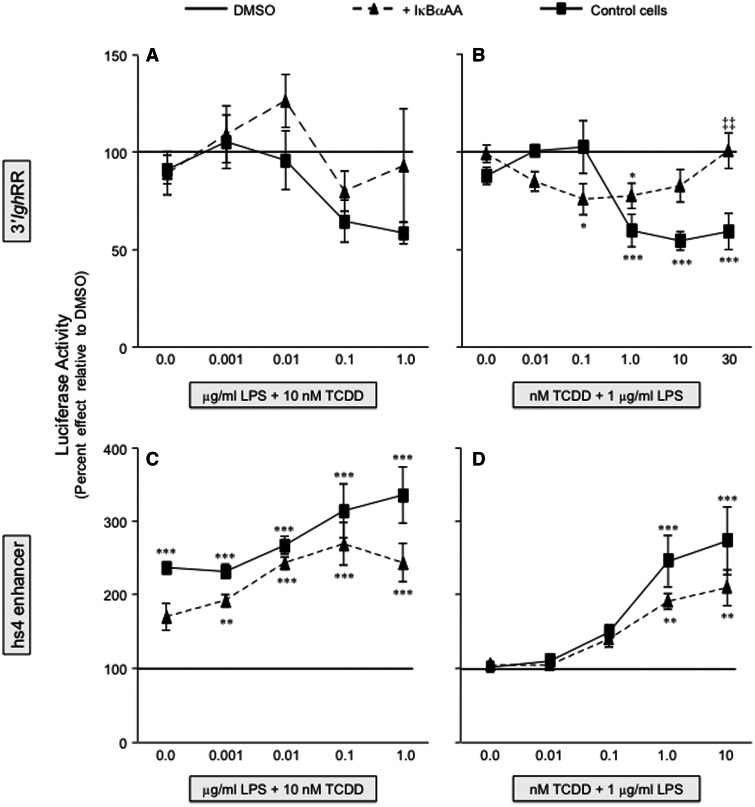
To explore the role of NF-κB/Rel proteins in the divergent effects of TCDD on 3′IghRR versus hs4 enhancer activity, we utilized the CH12.IκBαAA cells to modulate the activity of NF-κB/Rel proteins regulated by IκBα (primarily RelA and to a lesser extent c-Rel). In the absence of IκBαAA expression, LPS and TCDD induced a concentration-dependent profile of effects on the 3′IghRR and hs4 luciferase reporters (Figs. 3 and 4) that corresponded with previous results using the parental CH12.LX cells (Henseler et al., 2009; Sulentic et al., 2004b), with the exception of a modest decrease in sensitivity to TCDD, likely due to the lower levels of AhR expressed in CH12.IκBαAA as compared with CH12.LX cells (Figure 2). The analysis of 3′IghRR activation by LPS demonstrated a significant and concentration-dependent increase in overall reporter activity that was significantly inhibited by 10 nM TCDD (Figure 3B). However, TCDD had no inhibitory effect on basal 3′IghRR reporter activity. TCDD also demonstrated a concentration-dependent inhibition of LPS-induced 3′IghRR activation (Figure 3A), though the overall magnitude of inhibition was greater in the CH12.LX cells as compared with the CH12.IκBαAA cells, ie, close to 100% compared with 50%, respectively (Figs. 5A and 5B; Henseler et al., 2009; Sulentic et al., 2004b). With the induction of IκBαAA expression, we have previously demonstrated a suppression of LPS-induced 3′IghRR reporter activity (Romer and Sulentic, 2011). In the current study, expression of IκBαAA also suppressed LPS-induced 3′IghRR reporter activity but this effect was concentration-dependent and was overcome by a greater concentration of LPS (Figure 3B, compare 0.1 with 1.0 μg/ml LPS). Regardless, the most dramatic effect was seen with a cotreatment of LPS and TCDD in that IκBαAA expression significantly reversed the inhibitory effect of TCDD (Figure 3). The average of 3 separate experiments expressed as percent inhibition demonstrated a complete reversal of TCDD-induced inhibition with IκBαAA expression (Figs. 5A and 5B).
FIG. 3.
IκBαAA expression abrogates the inhibitory effect of TCDD on 3′IghRR activation. CH12.IκBαAA cells transiently transfected with the VH (variable Ig heavy chain promoter)-Luc-3′IghRR luciferase reporter plasmid (3′IghRR) were either cultured for 2 h in media alone or with IPTG to activate the IκBαAA superrepressor. The cells were then cultured in the absence or presence of increasing concentrations of TCDD with 1 μg/ml LPS (A) or increasing concentrations of LPS with 10 nM TCDD (B). Luciferase enzyme activity is represented on the y-axis as relative light units (mean ± SE, n = 4 per treatment group). For graph A, “NA” denotes the unstimulated control; “C”, the LPS control; and “0.0 nM TCDD”, the 0.01% DMSO control. For graph B, “0.0 μg/ml LPS” denotes the unstimulated control; gray bars are treated with 0.01% DMSO and increasing concentrations of LPS; and checkered bars are treated with 10 nM TCDD and increasing concentrations of LPS. Statistical significance was determined by a 2-way ANOVA followed by a Bonferroni’s post hoc test. “*”, “**”, “***” denote significance at P < .05, P < .01, and P < .001, respectively, from the appropriate vehicle control (0.0 nM TCDD for A or 0.01% DMSO for B). “‡”, “‡‡”, “‡‡‡” denote significance for a specific treatment at P < .05, P < .01, and P < .001, respectively, between the control cells (no IκBαAA) and the cells induced to express the IκBαAA superrepressor (+ IκBαAA). Results are representative of 3 separate experiments. IκBαAA, IκBα superrepressor.
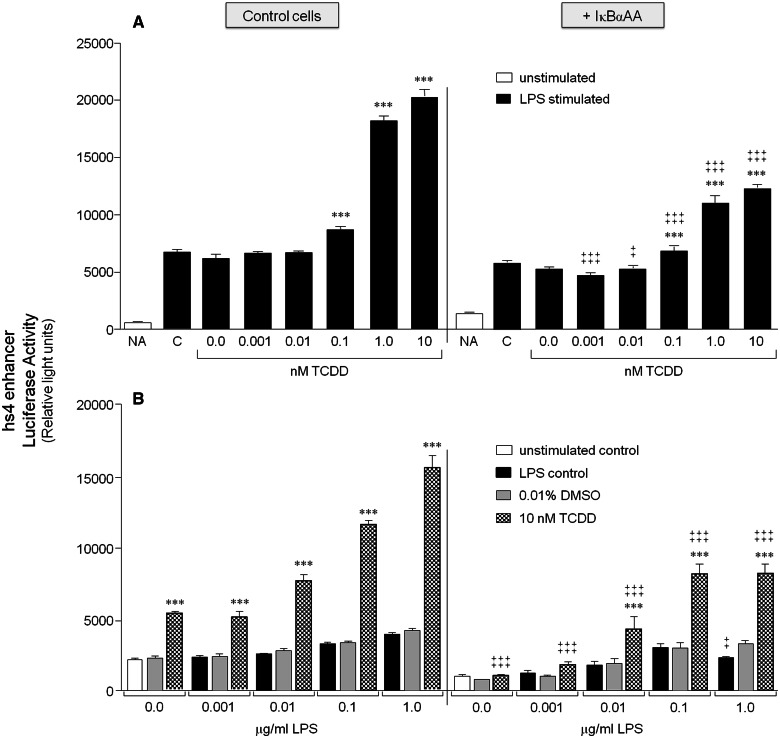
FIG. 4.
IκBαAA expression blunts the synergistic activation of hs4 by TCDD and LPS stimulation. CH12.IκBαAA cells transiently transfected with the VH-Luc-hs4 luciferase reporter plasmid (hs4 enhancer) were either cultured for 2 h in media alone or with IPTG to activate the IκBαAA superrepressor. The cells were then cultured in the absence or presence of increasing concentrations of TCDD with 1 μg/ml LPS (A) or increasing concentrations of LPS with 10 nM TCDD (B). Luciferase enzyme activity is represented on the y-axis as relative light units (mean ± SE, n = 4 per treatment group). For graph (A), “NA” denotes the unstimulated control; “C,” the LPS control; and “0.0 nM TCDD,” the 0.01% DMSO control. For graph (B), “0.0 μg/ml LPS” denotes the unstimulated control; gray bars are treated with 0.01% DMSO and increasing concentrations of LPS; and checkered bars are treated with 10 nM TCDD and increasing concentrations of LPS. Statistical significance was determined by a 2-way ANOVA followed by a Bonferroni’s post hoc test. “***” denote significance at P < .001 from the appropriate vehicle control (0.0 nM TCDD for [A] or 0.01% DMSO for [B]). “‡” and “‡‡‡” denote significance for a specific treatment at P < .05 and P < .001, respectively, between the control cells (no IκBαAA) and the cells induced to express the IκBαAA superrepressor (+ IκBαAA). Results are representative of at least 3 separate experiments.
FIG. 5.
IκBαAA expression abrogates the inhibitory effect of TCDD on 3′IghRR activation and reduces the synergistic activation of hs4. The CH12.IκBαAA cells were transiently transfected with VH-Luc-3′IghRR (A and B) or VH-Luc-hs4 (C and D) and treated with either increasing concentrations of LPS (A and C) or increasing concentrations of TCDD (B and D). Luciferase enzyme activity (a single representative experiment is shown in Figs. 3 and 4) was normalized to percent effect relative to the appropriate DMSO vehicle control (represented by the line at 100%) and the mean from 3 separate experiments (n = 3–4 per treatment group) was averaged and represented on the y-axis as the overall mean ± SE. Statistical significance was determined by a 2-way ANOVA followed by a Bonferroni’s post hoc test. “*,” “**,” “***” denote significance at P < .05, P < .01, and P < .001, respectively, from the appropriate vehicle control (0.01% DMSO for [A] and [C]; 0.01% DMSO + 1 μg/ml LPS for [B] and [D]), which is represented by the line at 100%. “‡‡” denotes significance for a specific treatment at P < .01, respectively, between the control cells (no IκBαAA) and the cells induced to express the IκBαAA superrepressor (+ IκBαAA). Results are the overall average ± SE of 3 separate experiments.
As demonstrated previously, analysis of hs4 activation confirmed a variable activation by LPS alone, a significant activation by TCDD alone, and a synergistic increase in overall reporter activity by an LPS and TCDD cotreatment that was dependent on both the concentration of LPS and of TCDD (Figs. 4A and 4B; Sulentic et al., 2004b). Expression of IκBαAA significantly blunted the synergistic increase in hs4 activity (Figure 4). However, unlike the complete reversal of TCDD-induced 3′IghRR inhibition by IκBαAA, TCDD and a cotreatment of TCDD and LPS still induced an approximately 200% increase in hs4 activity in the presence of the IκBαAA superrepressor (Figs. 5C and 5D). Taken together, these data support a prominent role of IκBα-regulated NF-κB/Rel proteins in the regulation of both the 3′IghRR and the hs4 enhancer. Surprisingly, these NF-κB/Rel proteins not only appear to partially mediate LPS-induced activation of the 3′IghRR but also mediate the inhibitory effect of TCDD on LPS-induced 3′IghRR activation (Figs. 3 and 5; Romer and Sulentic, 2011). However, consistent with previous mutational analysis (Sulentic et al., 2004b), IκBα-regulated NF-κB/Rel proteins significantly, but partially, mediated TCDD-induced activation of the hs4 enhancer as well as the synergistic activation of the hs4 enhancer by a TCDD and LPS cotreatment (Figs. 4 and 5).
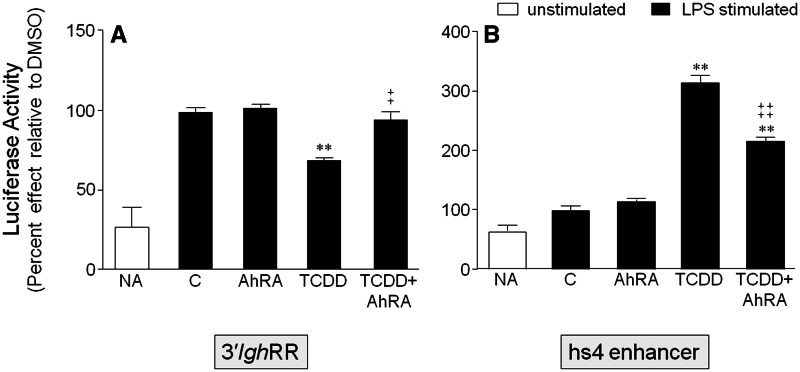
As the effects of TCDD are presumed to be AhR-dependent and we have previously demonstrated a cooperative interaction between proteins binding to the overlapping κB and DRE binding site in the hs4 enhancer (Sulentic et al., 2004a,b), we examined the effect of AhR antagonism on the effects of LPS and TCDD on the 3′IghRR and hs4 enhancer. The AhR antagonist, CH223191, had no effect on LPS-induced activation of the 3′IghRR but completely reversed the inhibitory effect of TCDD (Figure 6A) as previously demonstrated (Wourms and Sulentic, 2015). AhR antagonism also partially inhibited the synergistic activation of the hs4 enhancer by LPS and TCDD (Figure 6B). Taken together, these results suggest an interaction between the AhR and specific NF-κB/Rel proteins, which leads to significant modulation of 3′IghRR and hs4 enhancer activity.
FIG. 6.
Antagonism of the AhR reverses the inhibitory effect of TCDD on 3′IghRR activation and partially reverses the synergistic activation of hs4. CH12.LX cells were transiently transfected with VH-Luc-3′IghRR (A) or VH-Luc-hs4 (B) and pretreated for 1 h with DMSO or 30 µM CH223191 (AhR antagonist, AhRA) then treated with DMSO or 10 nM TCDD in the presence of 1 µg/ml LPS stimulation. Results were normalized to the appropriate vehicle control set to 100%, ie, cotreatment of 0.11% DMSO (vehicle control, represented as “C”) and stimulation; and the means from 3 to 4 separate experiments (overall mean ± SE) are represented in the bar graph. The vehicle control did not significantly differ from the LPS alone control (data not shown). Statistical significance was determined by a 1-way ANOVA followed by Dunnett’s Multiple Comparison test. “**” denotes significance from the vehicle control (C) at P < .01. “‡” and “‡‡” denote significance of the TCDD+AhRA treatment group from the TCDD treatment group at P < .05 and P < .01, respectively. Results are the overall average± SE of 3–4 separate experiments.
TCDD alters the binding profile of NF-κB/Rel proteins within the hs1.2 and hs4 enhancer
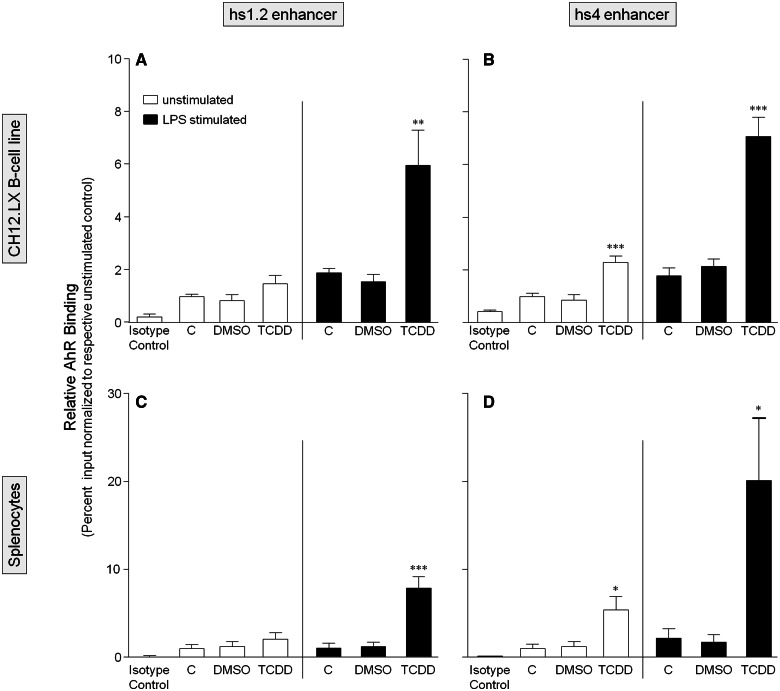
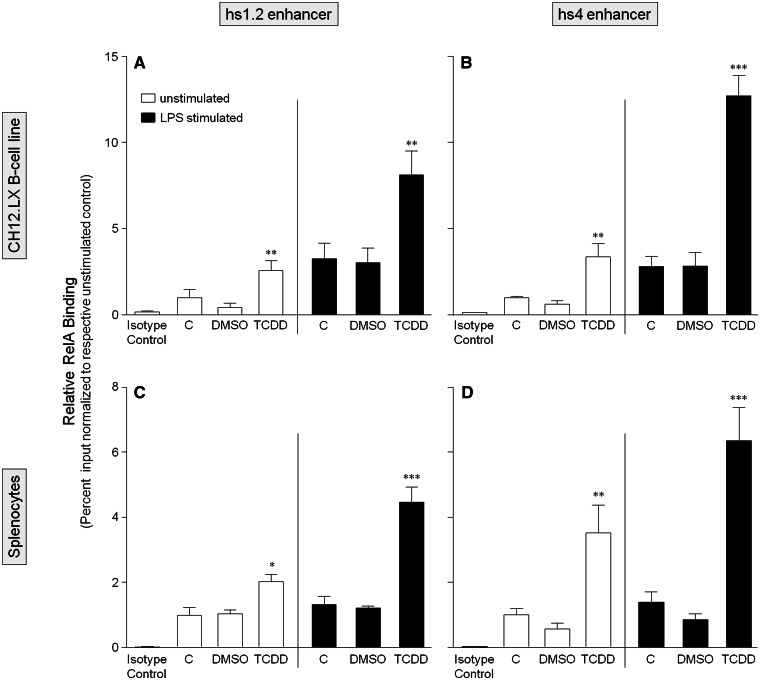
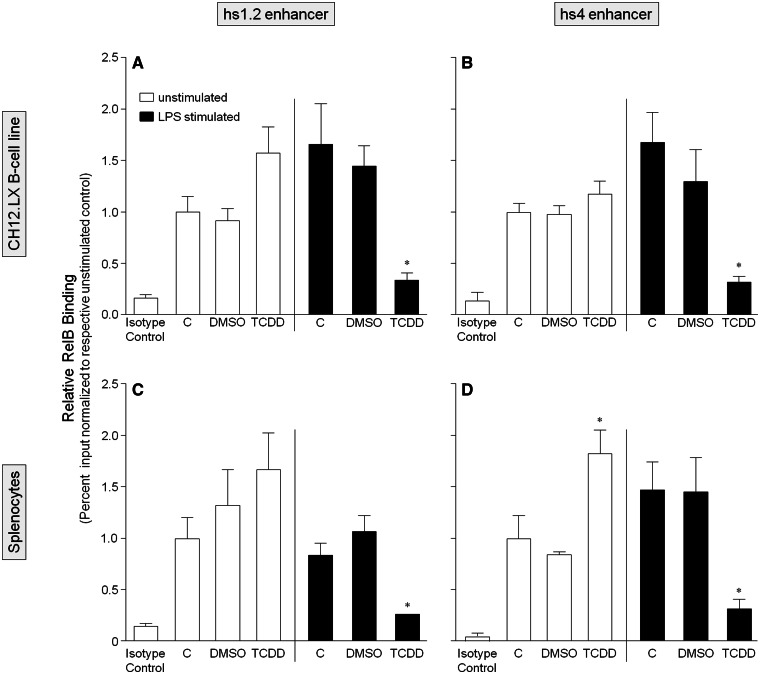
To further characterize the role of the AhR and NF-κB/Rel proteins in the transcriptional activity of the 3′IghRR, we evaluated by ChIP analysis the binding profile within the hs1.2 and hs4 enhancers of the AhR and the NF-κB/Rel subunits RelA and RelB, which have been previously shown to physically interact with the AhR (Kim et al., 2000; Tian, 2009; Tian et al., 1999; Vogel et al., 2007). For these studies, we utilized the parental CH12.LX cell line as it expresses high levels of the AhR. To correlate these studies with primary cells, we also utilized primary splenocytes isolated from female B6C3F1 mice, which express a high affinity AhR and a functional signaling pathway. ChIP analysis of the CH12.LX cells or mouse splenocytes treated with DMSO (0.01%), TCDD (30 nM), and/or LPS (1 µg/ml) for 90 min demonstrated similar binding profiles of the AhR, RelA, or RelB in both cellular models (Figs. 7–9). Consistent with our previous electrophoretic mobility shift assay (EMSA)-Western and ChIP analysis (Sulentic et al., 2000, 2004b), TCDD in the absence of cellular stimulation induced AhR binding within the hs4 enhancer (Figs. 7B and 7D). However, contrasting with previous EMSA-Western results (Sulentic et al., 2000), TCDD did not induce significant AhR binding within the hs1.2 enhancer (Figs. 7A and 7C). Previous studies did not evaluate the effect of cellular stimulation on AhR binding to the hs1.2 and hs4 enhancers but previous transfection studies with a hs4 luciferase reporter demonstrated a synergistic activation of the hs4 reporter with a TCDD and LPS cotreatment (Sulentic et al., 2000, 2004a,b). Correspondingly, LPS-induced stimulation and TCDD cotreatment synergistically increased AhR binding within the hs4 enhancer as well as the hs1.2 enhancer (Figure 7). For RelA and RelB binding, previous EMSA-Western analysis demonstrated TCDD-induced RelA and RelB binding within the hs4 enhancer; cellular stimulation was not evaluated, nor was NF-κB/Rel binding to the hs1.2 enhancer (Sulentic et al., 2000). The current ChIP analysis demonstrated TCDD-induced RelA binding to both the hs4 and hs1.2 enhancers; and similar to AhR binding, a TCDD and LPS-cotreatment synergistically increased RelA binding within both the hs4 and hs1.2 enhancers (Figure 8). However, RelB exhibited a very different binding profile from RelA in that TCDD did not significantly increase binding within either the hs1.2 or hs4 enhancers (except for an increased binding to hs4 only in splenocytes) and the cotreatment of LPS and TCDD resulted in a marked decrease in RelB binding to either enhancer (Figure 9). Taken together these results suggest a significantly altered NF-κB/Rel binding profile (ie, RelA>>RelB) with TCDD treatment that is greatly enhanced under cellular stimulation. Additionally, in both the CH12.LX cells and mouse splenocytes, the LPS and TCDD cotreatment induces a greater RelA binding within the hs4 enhancer compared with the hs1.2 enhancer (Figure 8).
FIG. 7.
TCDD alone only increases AhR binding within the hs4 enhancer but TCDD and LPS stimulation synergistically increases AhR binding within both the hs1.2 and hs4 enhancers of the 3′IghRR. CH12.LX cells (A and B) or mouse splenocytes (C and D) were treated with 0.01% DMSO or 30 nM TCDD in the absence or presence of 1 µg/ml LPS stimulation and incubated for 90 min. The cells were then cross-linked with formaldehyde and the chromatin was immunoprecipitated with an anti-AhR antibody. Immunoprecipitated chromatin was analyzed by PCR and represented as % input as described in the Materials and Methods. Each bar represents the overall mean ± SE of 3–4 separate experiments. “C” denotes the naïve (white bar) or LPS (black bar) control. The isotype control represents chromatin immunoprecipitation with polyclonal IgG. Statistical significance was determined by a 1-way ANOVA followed by Bonferroni’s Multiple Comparison test to determine significance within unstimulated or stimulated samples. There was no significant difference between the LPS-stimulated and naïve controls. “*,” “**,” and “***” denote significance from the appropriate DMSO control at P < .05, P < .01, and P < .001, respectively. Results are the overall average ± SE of 3–4 separate experiments.
FIG. 8.
TCDD alone increases RelA binding within both the hs1.2 and hs4 enhancers of the 3′IghRR but TCDD and LPS stimulation synergistically increases RelA binding within both enhancers. CH12.LX cells (A and B) or mouse splenocytes (C and D) were treated with 0.01% DMSO or 30 nM TCDD in the absence or presence of 1 µg/ml LPS stimulation and incubated for 90 min. The cells were then cross-linked with formaldehyde and the chromatin was immunoprecipitated with an anti-RelA antibody. Immunoprecipitated chromatin was analyzed by PCR and represented as % input as described in the Materials and Methods. Each bar represents the overall mean ± SE of 3–4 separate experiments. “C” denotes the naïve (white bar) or LPS (black bar) control. The isotype control represents chromatin immunoprecipitation with polyclonal IgG. Statistical significance was determined by a 1-way ANOVA followed by Bonferroni’s Multiple Comparison test to determine significance within unstimulated or stimulated samples. There was no significant difference between the LPS-stimulated and naïve controls. “*,” “**,” and “***” denote significance from the appropriate DMSO control at P < .05, P < .01, and P < .001, respectively. Results are the overall average ± SE of 3–4 separate experiments.
FIG. 9.
TCDD and LPS stimulation markedly decreases RelB binding within both the hs1.2 and hs4 enhancers of the 3′IghRR. CH12.LX cells (A and B) or mouse splenocytes (C and D) were treated with 0.01% DMSO or 30 nM TCDD in the absence or presence of 1 µg/ml LPS stimulation and incubated for 90 min. The cells were then cross-linked with formaldehyde and the chromatin was immunoprecipitated with an anti-RelB antibody. Immunoprecipitated chromatin was analyzed by PCR and represented as % input as described in the Materials and Methods. Each bar represents the overall mean ± SE of 3–4 separate experiments. “C” denotes the naïve (white bar) or LPS (black bar) control. The isotype control represents chromatin immunoprecipitation with polyclonal IgG. Statistical significance was determined by a 1-way ANOVA followed by Bonferroni’s Multiple Comparison test to determine significance within unstimulated or stimulated samples. There was no significant difference between the LPS-stimulated and naïve controls. “*” denotes significance from the LPS-stimulated DMSO control at P < .05. Results are the overall average ± SE of 3–4 separate experiments.
DISCUSSION
The 3′IghRR mediates upregulation of Igh expression and CSR, processes central to B-lymphocyte differentiation and to mounting an effective antibody response (Manis et al., 1998; Pinaud et al., 2001; Vincent-Fabert et al., 2010). The 3′IghRR is also a sensitive target of exogenous chemicals, such as TCDD (Henseler et al., 2009). The current study suggests that the AhR and the NF-κB/Rel pathway, specifically the NF-κB/Rel proteins regulated through IκBα (inhibitor kappa B-alpha protein) degradation (ie, RelA), play an integral role in the overall suppressive effects of TCDD on the 3′IghRR (Figure 10). This may seem counterintuitive to the stimulatory effect on B-lymphocyte activation and differentiation usually associated with NF-κB/Rel activation (Hsing and Bishop, 1999). However, there are 5 NF-κB/Rel subunits (RelA, RelB, c-Rel, p50, and p52) that can form homo- or heterodimers and these dimers may have inhibitory or stimulatory roles depending on the specific transcriptional target and potential interactions with other transcription factors. For example, previous studies have demonstrated a stimuli-specific binding profile of NF-κB/Rel dimers that correlated with Igh germline transcript expression. Specifically, CD40 ligand (CD40L) stimulation induced binding within the γ1 Igh germline promoter of more p50/RelB and p50/RelA heterodimers whereas LPS induced more p50/cRel and p50/p50 dimers, which correlated with a significantly greater induction of γ1 transcripts by CD40L as compared with LPS stimulation (Lin et al., 1998). Additionally, overexpression of NF-κB/Rel fusion proteins demonstrated a stimulatory effect of p50-RelB or p50-RelA on the germline γ1 promoter but coexpression of p50-cRel inhibited this activation. However, p50-cRel was not a general inhibitor of germline Igh promoters in that overexpression of p50-cRel induced the germline ε promoter (Lin et al., 1998).
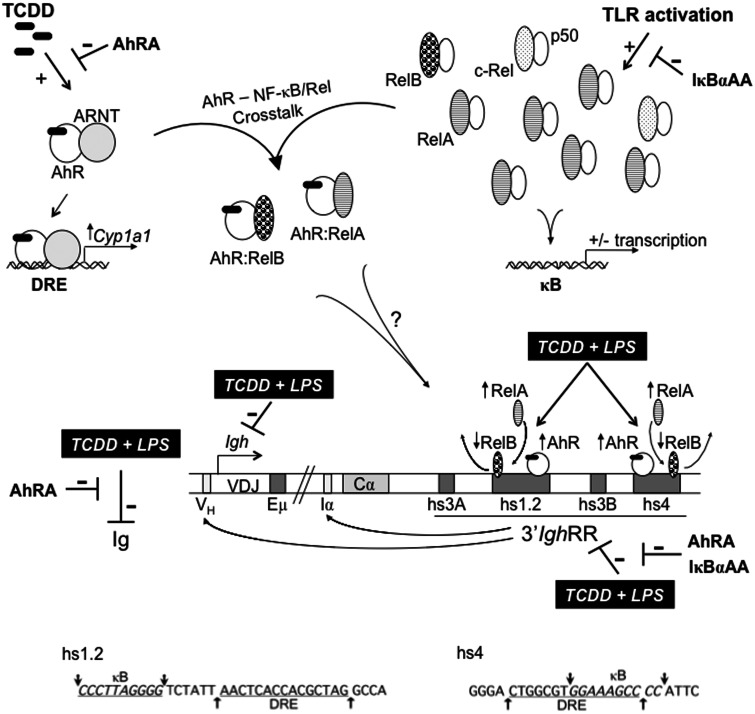
FIG. 10.
Schematic representation of mouse Igh regulation by the AhR and NF-κB/Rel proteins. A schematic depicting a portion of the mouse Igh gene locus including the VDJ antigen recognition region, the 3′ most Igh constant region Cα, which will encode for the heavy chain of IgA, and the regulatory elements: variable Igh promoter (VH), µ or intronic enhancer (Eµ), intronic promoter for Cα (Iα), and the 3′Igh regulatory region (3′IghRR) with its 4 enhancer regions (ie, hypersensitive sites [hs] hs3A, hs1.2, hs3B, and hs4). Long-range interactions between the 3′IghRR and the VH promoter and the intronic promoters just upstream of each constant region are noted with arrows from the 3′IghRR to these sites (Birshtein, 2014). The AhR and NF-κB/Rel nuclear pathways are depicted and show (1) TCDD-induced AhR activation and the prototypical response of Cyp1a1 induction; (2) toll-like receptor activation of NF-κB/Rel proteins, which can form various homo- or heterodimers and upregulate or downregulate the expression of various genes; and (3) crosstalk between the AhR and NF-κB/Rel proteins. The typical NF-κB/Rel heterodimers following IκBα degradation are depicted and will largely be RelA-p50 and less so of c-Rel-p50. Excess RelB not bound by inactivated, unprocessed p100 can be sequestered by IκBs, including IκBα (inhibitor kappa B-alpha protein) (Millet et al., 2013). The potential interactions between the AhR and the NF-κB/Rel proteins RelA and RelB are shown, which may account for the changes in RelA versus RelB binding within the hs1.2 and hs4 enhancers following TCDD and LPS cotreatment. The inhibitory effects of TCDD on LPS-induced 3′IghRR activation, Igh expression, and Ig secretion are depicted by( ). The inhibitory effects of the AhR antagonist (AhRA) and the IκBαAA superrepressor are also illustrated. Nucleotide sequences for the κB (NF-κB/Rel DNA binding motif) (italicized, top arrows) and DRE (dioxin-responsive element) (bottom arrows) motifs are shown for the hs1.2 and hs4 enhancers; arrows indicate the boundary for each binding site.
). The inhibitory effects of the AhR antagonist (AhRA) and the IκBαAA superrepressor are also illustrated. Nucleotide sequences for the κB (NF-κB/Rel DNA binding motif) (italicized, top arrows) and DRE (dioxin-responsive element) (bottom arrows) motifs are shown for the hs1.2 and hs4 enhancers; arrows indicate the boundary for each binding site.
In the current study, the 3′IghRR rather than intronic promoters was evaluated and based on our earlier mutational studies demonstrating a cooperative interaction between the proteins (presumably NF-κB/Rel and AhR) binding to the overlapping κB and DRE motifs in the hs4 enhancer and the close proximity of the κB and DRE in the hs1.2 enhancer (Sulentic et al., 2000, 2004a,b), we hypothesized that TCDD inhibited 3′IghRR by inducing a shift in the NF-κB/Rel binding profile to κB sites within the 3′IghRR enhancers (Figure 10). Correspondingly, our ChIP analysis demonstrated a marked increase in RelA binding and a significant decrease in RelB binding within both the hs1.2 and hs4 enhancers following a cotreatment with TCDD and LPS stimulation that was not as pronounced or was not seen in the individual treatments. This altered NF-κB/Rel binding profile may be responsible for the inhibitory effect of TCDD on LPS-induced 3′IghRR activation (Figure 10). These effects on NF-κB/Rel binding were not only demonstrated in the well-characterized CH12.LX B-lymphocyte cell line model but were also replicated in primary splenocytes.
Notably, the similarity in the treatment-induced AhR and NF-κB/Rel binding profiles within the hs1.2 and hs4 enhancers does not appear to explain the dichotomous effect of TCDD cotreatment on LPS-induced hs1.2 versus hs4 reporter activity. However, the TCDD and LPS cotreatment did induce greater binding of RelA to the hs4 enhancer as compared with hs1.2, which may influence the overall transcriptional effect perhaps through differential interactions with other transcription factors or transcriptional machinery. Alternatively, previous mutational analysis studies by Michaelson et al. (1996) have demonstrated a cooperative transcriptional activation of the hs4 enhancer by the same transcription factors (ie, NF-κB/Rel, Oct, and Pax5) that induced a cooperative transcriptional repression of the hs1.2 enhancer. These authors also demonstrated a lack of RelA binding within the hs1.2 enhancer using an EMSA and competition with an anti-RelA antibody; however, they did not evaluate RelA binding within the hs4 enhancer (Michaelson et al., 1996). Our previous EMSA-Western analysis, on the other hand, identified strong TCDD-inducible binding of all NF-κB/Rel subunits including RelA to the hs4 enhancer; however, we did not evaluate the effect of stimulation or NF-κB/Rel binding within the hs1.2 enhancer in those studies (Sulentic et al., 2000). Taken together, these results are consistent with the potential for differential transcriptional effects of RelA on hs1.2 versus hs4 activity.
Moreover, the AhR has been shown to physically interact with RelA and RelB resulting in altered transactivation (Beischlag et al., 2008; Kim et al., 2000; Tian, 2009; Tian et al., 1999; Vogel et al., 2007). Correspondingly, our lab has identified an interaction by co-IP between the AhR and the NF-κB/Rel proteins, RelA and RelB, in the CH12.LX murine B-lymphocyte cell line. However, we were unable to establish a treatment-dependent association, which may be partly due to the protein-protein interactions masking the antibody-specific epitopes (data not shown). Several studies have explored the potential functional roles of an AhR-NF-κB/Rel interaction in various cellular models and support a significant impact on a variety of intracellular processes (reviewed in Tian, 2009; Vogel and Matsumura, 2009). Furthermore, as alluded to above, previous studies demonstrated an inhibitory effect of protein binding to a κB site within the hs1.2 enhancer in a mature, unstimulated B-lymphocyte cell line, but binding to this same site was stimulatory in a plasma B-cell line (Michaelson et al., 1996). This dichotomy in regulation was attributed to the presence or absence of Pax5, an inhibitor of B-lymphocyte differentiation, which is degraded in activated B lymphocytes and absent in plasma cells (Michaelson et al., 1996). Therefore, Pax5 may directly inhibit transcription and/or possibly facilitate an inhibitory NF-κB/Rel binding profile (ie, perhaps RelA) in the hs1.2 enhancer in unstimulated B lymphocytes. Additionally, previous studies have demonstrated an AhR-dependent increase in Pax5 expression in stimulated B lymphocytes following treatment with TCDD (Yoo et al., 2004). Therefore, AhR activation in stimulated B lymphocytes may lead to altered protein binding within the hs1.2 and hs4 enhancers, and altered transactivation, by a combination of directly interacting with NF-κB/Rel proteins and sustaining Pax5 expression, which may also influence the protein binding profile within the 3′IghRR enhancers—ultimately leading to altered Igh gene expression.
The significant reversal of TCDD-induced inhibition of the 3′IghRR with either the AhR antagonist or expression of the IκBαAA superrepressor further supports an interaction between RelA and AhR in the inhibitory effect of TCDD (Figure 10). Qualitative comparisons via EMSA-analysis of RelA and RelB binding to the hs4 κB site that overlaps the DRE in a cell line with a functional AhR signaling pathway (ie, CH12.LX) versus a cell line with a nonfunctional AhR signaling pathway (ie, BCL-1) suggested a greater induction of RelA compared with RelB following TCDD treatment (without cellular stimulation). However, in the absence of the AhR, RelB binding, as compared with RelA, appeared to be induced to a greater extent following TCDD treatment (without cellular stimulation) (Sulentic et al., 2000). A caveat to this interpretation is the likely differences in affinity of each primary antibody. However, further supporting a distinct AhR-NF-κB/Rel interaction, previous mutational analysis identified a transcriptional role of proteins binding to both the overlapping κB and DRE sites in the synergistic activation of the hs4 enhancer following a TCDD and LPS cotreatment (Sulentic et al., 2004b). Interestingly, an AhR-deficient cell line (ie, BCL-1) only demonstrated LPS-induced VH promoter activity and was completely refractory to TCDD and to the transcriptional activity of the overlapping κB and DRE sites. In contrast, addition of these binding motifs greatly enhanced reporter activity in the CH12.LX cells and this activation was dependent on both the κB and DRE binding sites (Sulentic et al., 2004a), suggesting an essential role of the AhR in TCDD inducibility. This partly corresponds with the current results utilizing the AhR antagonist; however, the antagonist did not result in a complete loss of hs4 activation, which may be due to an incomplete antagonism of the AhR or differences in cellular models (ie, BCL-1 vs CH12.LX). Taken together, these results suggest an interaction between the AhR and specific NF-κB/Rel proteins causing an altered NF-κB/Rel binding profile within the hs1.2 and hs4 enhancers that mediates the effects of TCDD and LPS on the 3′IghRR.
It is difficult to put the stimulatory effect of TCDD and LPS on hs4 enhancer activity in the context of the inhibitory effect of TCDD on LPS-induced activation of the entire 3′IghRR. However, this dichotomy in activation has been seen previously and was thought to be due to the maturation state of the B lymphocyte and the involvement of inhibitory transcription factors such as Pax-5, which maintains the hs1.2 enhancer but not the hs4 enhancer in an inhibited state until B-lymphocyte activation when the levels of Pax-5 are downregulated (Michaelson et al., 1996). In conjunction with this, TCDD maintains Pax-5 expression, which as mentioned above may be partially responsible for the overall inhibitory effect of TCDD on the 3′IghRR (Schneider et al., 2008; Yoo et al., 2004). Various in vivo knockout animal models have suggested that deletion of any single enhancer within the 3′IghRR has modest to no phenotypic effects but deletion of the hs3b and hs4 enhancer resulted in partially impaired CSR and Ig secretion and deletion of the entire 3′IghRR resulted in impaired CSR and Ig secretion of all isotypes (reviewed by Pinaud et al., 2011). Therefore, the apparent dichotomous regulation of the individual enhancer elements appears to resolve into an overall cooperative regulation of CSR and Igh expression by the intact 3′IghRR.
Although less is known regarding its regulatory influence on the human IGH gene, the human 3′IGHRR has been associated with multiple human disease states such as celiac disease, IgA nephropathy, systemic sclerosis, plaque psoriasis, psoriatic arthritis, dermatitis herpetiformis, rheumatoid arthritis, systemic lupus erythematosus, and Burkitt’s lymphoma. These diseases have been associated with a polymorphism within the hs1.2 enhancer excepting Burkitt’s lymphoma, which has not been evaluated for the hs1.2 polymorphism but does exhibit a translocation between the c-myc gene and the regulatory elements of IGH (ie, µ enhancer and/or the 3′IGHRR) (Aupetit et al., 2000; Cianci et al., 2008; Frezza et al., 2004, 2007, 2012; Madisen and Groudine, 1994; Tolusso et al., 2009). Interestingly, the polymorphism has been characterized as an approximately 53-bp invariant sequence within the hs1.2 enhancer that is repeated up to 4 times and this invariant sequence contains putative binding sites for NF-κB/Rel, AP-1, SP-1, and NF-1, as well as a DRE core motif (Chen and Birshtein, 1997; Denizot et al., 2001; Fernando et al., 2012; Giambra et al., 2005). Consistent with the current results, our previous studies have identified an AhR-dependent influence of TCDD on the polymorphic hs1.2 enhancer; however, we also demonstrated a species difference in that TCDD activated the human hs1.2 enhancer but inhibited the mouse hs1.2 enhancer (Fernando et al., 2012). It is unclear what effect TCDD would have on the complete human 3′IGHRR or the actual influence of the human 3′IGHRR on CSR and IGH expression; however, previous luciferase reporter studies have demonstrated the ability of the human 3′IGHRR enhancers to activate intronic promoters for different IGH constant regions, therefore supporting a role of the human 3′IGHRR enhancers in CSR (Chen and Birshtein, 1997; Hu et al., 2000; Kim et al., 2004). Interestingly, NF-κB/Rel dysregulation and activation have been implicated in the same autoimmune disease states associated with the hs1.2 polymorphism (Abdou and Hanout, 2008; Ashizawa et al., 2003; Bell et al., 2003; Dozmorov et al., 2014; Maiuri et al., 2003; Trynka et al., 2009) and given that the hs1.2 polymorphism contains putative binding sites for both the AhR and NF-κB/Rel proteins, AhR ligands may influence NF-κB/Rel binding in the human 3′IGHRR as seen in the current study and therefore may influence various disease states. The convergence of these 2 transcriptional pathways may provide novel therapeutic strategies for autoimmune disease and specific B-cell lymphomas.
FUNDING
Boonshoft School of Medicine at WSU and the National Institute of Environmental Health Sciences (NIEHS) (R01ES014676).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Geoffrey Haughton (in memoriam) for the CH12.LX cells, Dr Gail Bishop for the CH12.IκBαAA cells, Dr Robert Roeder for the Igh luciferase reporter plasmids, and Dr Laurel Eckhardt for the γ2b mini-locus plasmid. The CH12.γ2b-3′IghRR cell line was generated and characterized by Dilini Ranatunga and Eric Romer. We also greatly appreciate the technical support and feedback from Michael Wourms and Eric Romer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organization acknowledged in the Funding section.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Abdou A. G., Hanout H. M. (2008). Evaluation of survivin and NF-kappaB in psoriasis, an immunohistochemical study. J. Cutan. Pathol. 35, 445–451. [DOI] [PubMed] [Google Scholar]
- Arnold L. W., LoCascio N. J., Lutz P. M., Pennell C. A., Klapper D., Haughton G. (1983). Antigen-induced lymphomagenesis: Identification of a murine B cell lymphoma with known antigen specificity. J. Immunol. 131, 2064–2068. [PubMed] [Google Scholar]
- Ashizawa M., Miyazaki M., Abe K., Furusu A., Isomoto H., Harada T., Ozono Y., Sakai H., Koji T., Kohno S. (2003). Detection of nuclear factor-kappaB in IgA nephropathy using Southwestern histochemistry. Am. J. Kidney Dis. 42, 76–86. [DOI] [PubMed] [Google Scholar]
- Aupetit C., Drouet M., Pinaud E., Denizot Y., Aldigier J. C., Bridoux F., Cogne M. (2000). Alleles of the alpha1 immunoglobulin gene 3′ enhancer control evolution of IgA nephropathy toward renal failure. Kidney Int. 58, 966–971. [DOI] [PubMed] [Google Scholar]
- Beischlag T. V., Luis Morales J., Hollingshead B. D., Perdew G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18, 207–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekeredjian-Ding I., Jego G. (2009). Toll-like receptors–sentries in the B-cell response. Immunology 128, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S., Degitz K., Quirling M., Jilg N., Page S., Brand K. (2003). Involvement of NF-kappaB signalling in skin physiology and disease. Cell. Signal. 15, 1–7. [DOI] [PubMed] [Google Scholar]
- Birnbaum L. S., Tuomisto J. (2000). Non-carcinogenic effects of TCDD in animals. Food Addit. Contam. 17, 275–288. [DOI] [PubMed] [Google Scholar]
- Birshtein B. K. (2014). Epigenetic Regulation of Individual Modules of the immunoglobulin heavy chain locus 3′ Regulatory Region. Front. Immunol. 5, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop G. A., Haughton G. (1986). Induced differentiation of a transformed clone of Ly-1+ B cells by clonal T cells and antigen. Proc. Natl. Acad. Sci. U.S.A. 83, 7410–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Birshtein B. K. (1997). Virtually identical enhancers containing a segment of homology to murine 3′IgH-E(hs1,2) lie downstream of human Ig C alpha 1 and C alpha 2 genes. J. Immunol. 159, 1310–1318. [PubMed] [Google Scholar]
- Cianci R., Giambra V., Mattioli C., Esposito M., Cammarota G., Scibilia G., Magazzu G., Orlando A., Sandri G., Bianchi L., et al. (2008). Increased frequency of Ig heavy-chain HS1,2-A enhancer *2 allele in dermatitis herpetiformis, plaque psoriasis, and psoriatic arthritis. J. Investig. Dermatol. 128, 1920–1924. [DOI] [PubMed] [Google Scholar]
- De Abrew K. N., Kaminski N. E., Thomas R. S. (2010). An integrated genomic analysis of aryl hydrocarbon receptor-mediated inhibition of B-cell differentiation. Toxicol. Sci. 118, 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot Y., Pinaud E., Aupetit C., Le Morvan C., Magnoux E., Aldigier J. C., Cogne M. (2001). Polymorphism of the human alpha1 immunoglobulin gene 3′ enhancer hs1,2 and its relation to gene expression. Immunology 103, 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov M. G., Wren J. D., Alarcon-Riquelme M. E. (2014). Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes. Epigenetics 9, 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C., Rannug A., Stockinger B. (2009). The aryl hydrocarbon receptor in immunity. Trends Immunol. 30, 447–454. [DOI] [PubMed] [Google Scholar]
- Fernando T. M., Ochs S. D., Liu J., Chambers-Turner R. C., Sulentic C. E. W. (2012). 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces transcriptional activity of the human polymorphic hs1,2 enhancer of the 3′Igh regulatory region. J. Immunol. 188, 3294–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza D., Giambra V., Cianci R., Fruscalzo A., Giufre M., Cammarota G., Martinez-Labarga C., Rickards O., Scibilia G., Sferlazzas C., et al. (2004). Increased frequency of the immunoglobulin enhancer HS1,2 allele 2 in coeliac disease. Scand. J. Gastroenterol. 39, 1083–1087. [DOI] [PubMed] [Google Scholar]
- Frezza D., Giambra V., Tolusso B., De Santis M., Bosello S., Vettori S., Triolo G., Valentini G., Ferraccioli G. (2007). Polymorphism of immunoglobulin enhancer element HS1,2A: Allele *2 associates with systemic sclerosis. Comparison with HLA-DR and DQ allele frequency. Ann. Rheum. Dis. 66, 1210–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza D., Tolusso B., Giambra V., Gremese E., Marchini M., Nowik M., Serone E., D’Addabbo P., Mattioli C., Canestri S., et al. (2012). Polymorphisms of the IgH enhancer HS1.2 and risk of systemic lupus erythematosus. Ann. Rheum. Dis. 71, 1309–1315. [DOI] [PubMed] [Google Scholar]
- Gerondakis S., Siebenlist U. (2009). Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb. Perspect. Biol. 2, a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambra V., Fruscalzo A., Giufre M., Martinez-Labarga C., Favaro M., Rocchi M., Frezza D. (2005). Evolution of human IgH3′EC duplicated structures: Both enhancers HS1,2 are polymorphic with variation of transcription factor’s consensus sites. Gene 346, 105–114. [DOI] [PubMed] [Google Scholar]
- Henseler R. A., Romer E. J., Sulentic C. E. W. (2009). Diverse chemicals including aryl hydrocarbon receptor ligands modulate transcriptional activity of the 3′immunoglobulin heavy chain regulatory region. Toxicology 261, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsapple M. P., Morris D. L., Wood S. C., Snyder N. K. (1991). 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced changes in immunocompetence: Possible mechanisms. Annu. Rev. Pharmacol. Toxicol. 31, 73–100. [DOI] [PubMed] [Google Scholar]
- Hsing Y., Bishop G. A. (1999). Requirement for nuclear factor-kappaB activation by a distinct subset of CD40-mediated effector functions in B lymphocytes. J. Immunol. 162, 2804–2811. [PubMed] [Google Scholar]
- Hu Y., Pan Q., Pardali E., Mills F. C., Bernstein R. M., Max E. E., Sideras P., Hammarstrom L. (2000). Regulation of germline promoters by the two human Ig heavy chain 3′ alpha enhancers. J. Immunol. 164, 6380–6386. [DOI] [PubMed] [Google Scholar]
- Khamlichi A. A., Pinaud E., Decourt C., Chauveau C., Cogne M. (2000). The 3′ IgH regulatory region: A complex structure in a search for a function. Adv. Immunol. 75, 317–345. [DOI] [PubMed] [Google Scholar]
- Kim D. W., Gazourian L., Quadri S. A., Romieu-Mourez R., Sherr D. H., Sonenshein G. E. (2000). The RelA NF-kappaB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene 19, 5498–5506. [DOI] [PubMed] [Google Scholar]
- Kim E. C., Edmonston C. R., Wu X., Schaffer A., Casali P. (2004). The HoxC4 homeodomain protein mediates activation of the immunoglobulin heavy chain 3′ hs1,2 enhancer in human B cells. Relevance to class switch DNA recombination. J. Biol. Chem. 279, 42258–42269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. C., Wortis H. H., Stavnezer J. (1998). The ability of CD40L, but not lipopolysaccharide, to initiate immunoglobulin switching to immunoglobulin G1 is explained by differential induction of NF-kappaB/Rel proteins. Mol. Cell Biol. 18, 5523–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Madisen L., Groudine M. (1994). Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt’s lymphoma cells. Genes Dev. 8, 2212–2226. [DOI] [PubMed] [Google Scholar]
- Maiuri M. C., De Stefano D., Mele G., Fecarotta S., Greco L., Troncone R., Carnuccio R. (2003). Nuclear factor kappa B is activated in small intestinal mucosa of celiac patients. J. Mol. Med. (Berl.) 81, 373–379. [DOI] [PubMed] [Google Scholar]
- Manis J. P., van der Stoep N., Tian M., Ferrini R., Davidson L., Bottaro A., Alt F. W. (1998). Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 188, 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus R. S., Holsapple M. P., Kaminski N. E. (1998). Lipopolysaccharide activation of murine splenocytes and splenic B cells increased the expression of aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator. J. Pharmacol. Exp. Ther. 287, 1113–1118. [PubMed] [Google Scholar]
- Michaelson J. S., Singh M., Snapper C. M., Sha W. C., Baltimore D., Birshtein B. K. (1996). Regulation of 3′ IgH enhancers by a common set of factors, including kappa B-binding proteins. J. Immunol. 156, 2828–2839. [PubMed] [Google Scholar]
- Millet P., McCall C., Yoza B. (2013). RelB: An outlier in leukocyte biology. J. Leukoc. Biol. 94, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okey A. B. (2007). An aryl hydrocarbon receptor odyssey to the shores of toxicology: The Deichmann Lecture, International Congress of Toxicology-XI. Toxicol. Sci. 98, 5–38. [DOI] [PubMed] [Google Scholar]
- Ong J., Stevens S., Roeder R. G., Eckhardt L. A. (1998). 3′ IgH enhancer elements shift synergistic interactions during B cell development. J. Immunol. 160, 4896–4903. [PubMed] [Google Scholar]
- Pinaud E., Khamlichi A. A., Le Morvan C., Drouet M., Nalesso V., Le Bert M., Cogne M. (2001). Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity 15, 187–199. [DOI] [PubMed] [Google Scholar]
- Pinaud E., Marquet M., Fiancette R., Peron S., Vincent-Fabert C., Denizot Y., Cogne M. (2011). The IgH locus 3′ regulatory region: Pulling the strings from behind. Adv. Immunol. 110, 27–70. [DOI] [PubMed] [Google Scholar]
- Pollenz R. S. (2002). The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem. Biol. Interact. 141, 41–61. [DOI] [PubMed] [Google Scholar]
- Romer E. J., Sulentic C. E. W. (2011). Hydrogen peroxide modulates immunoglobulin expression by targeting the 3′Igh regulatory region through an NFkappaB-dependent mechanism. Free Radic. Res. 45, 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D., Manzan M. A., Crawford R. B., Chen W., Kaminski N. E. (2008). 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated impairment of B cell differentiation involves dysregulation of paired box 5 (Pax5) isoform, Pax5a. J. Pharmacol. Exp. Ther. 326, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Eckhardt L. A. (2001). Deletional analyses reveal an essential role for the hs3b/hs4 IgH 3′ enhancer pair in an Ig-secreting but not an earlier-stage B cell line. Int. Immunol. 13, 1003–1012. [DOI] [PubMed] [Google Scholar]
- Suh J., Jeon Y. J., Kim H. M., Kang J. S., Kaminski N. E., Yang K. H. (2002). Aryl hydrocarbon receptor-dependent inhibition of AP-1 activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin in activated B cells. Toxicol. Appl. Pharmacol. 181, 116–123. [DOI] [PubMed] [Google Scholar]
- Sulentic C. E., Kaminski N. E. (2011). The long winding road toward understanding the molecular mechanisms for B-cell suppression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 120(Suppl. 1), S171–S191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulentic C. E., Holsapple M. P., Kaminski N. E. (1998). Aryl hydrocarbon receptor-dependent suppression by 2,3,7, 8-tetrachlorodibenzo-p-dioxin of IgM secretion in activated B cells. Mol. Pharmacol. 53, 623–629. [PubMed] [Google Scholar]
- Sulentic C. E., Holsapple M. P., Kaminski N. E. (2000). Putative link between transcriptional regulation of IgM expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin and the aryl hydrocarbon receptor/dioxin-responsive enhancer signaling pathway. J. Pharmacol. Exp. Ther. 295, 705–716. [PubMed] [Google Scholar]
- Sulentic C. E., Kang J. S., Na Y. J., Kaminski N. E. (2004a). Interactions at a dioxin responsive element (DRE) and an overlapping kappaB site within the hs4 domain of the 3′alpha immunoglobulin heavy chain enhancer. Toxicology 200, 235–246. [DOI] [PubMed] [Google Scholar]
- Sulentic C. E., Zhang W., Na Y. J., Kaminski N. E. (2004b). 2,3,7,8-tetrachlorodibenzo-p-dioxin, an exogenous modulator of the 3′alpha immunoglobulin heavy chain enhancer in the CH12.LX mouse cell line. J. Pharmacol. Exp. Ther. 309, 71–78. [DOI] [PubMed] [Google Scholar]
- Tanaka G., Kanaji S., Hirano A., Arima K., Shinagawa A., Goda C., Yasunaga S., Ikizawa K., Yanagihara Y., Kubo M., et al. (2005). Induction and activation of the aryl hydrocarbon receptor by IL-4 in B cells. Int. Immunol. 17, 797–805. [DOI] [PubMed] [Google Scholar]
- Tian Y. (2009). Ah receptor and NF-kappaB interplay on the stage of epigenome. Biochem. Pharmacol. 77, 670–680. [DOI] [PubMed] [Google Scholar]
- Tian Y., Ke S., Denison M. S., Rabson A. B., Gallo M. A. (1999). Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J. Biol. Chem. 274, 510–515. [DOI] [PubMed] [Google Scholar]
- Tolusso B., Frezza D., Mattioli C., Fedele A. L., Bosello S., Faustini F., Peluso G., Giambra V., Pietrapertosa D., Morelli A., et al. (2009). Allele *2 of the HS1,2A enhancer of the Ig regulatory region associates with rheumatoid arthritis. Ann. Rheum. Dis. 68, 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G., Zhernakova A., Romanos J., Franke L., Hunt K. A., Turner G., Bruinenberg M., Heap G. A., Platteel M., Ryan A. W., et al. (2009). Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut 58, 1078–1083. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S., Karin M. (2009). Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733. [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C., Fiancette R., Pinaud E., Truffinet V., Cogne N., Cogne M., Denizot Y. (2010). Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood 116, 1895–1898. [DOI] [PubMed] [Google Scholar]
- Vogel C. F., Matsumura F. (2009). A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem. Pharmacol. 77, 734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C. F., Khan E. M., Leung P. S., Gershwin M. E., Chang W. L., Wu D., Haarmann-Stemmann T., Hoffmann A., Denison M. S. (2013). Cross-talk between aryl hydrocarbon receptor and the inflammatory response: A role for nuclear factor-kappaB. J. Biol. Chem. 289, 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C. F., Sciullo E., Li W., Wong P., Lazennec G., Matsumura F. (2007). RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol. Endocrinol. 21, 2941–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorderstrasse B. A., Steppan L. B., Silverstone A. E., Kerkvliet N. I. (2001). Aryl hydrocarbon receptor-deficient mice generate normal immune responses to model antigens and are resistant to TCDD-induced immune suppression. Toxicol. Appl. Pharmacol. 171, 157–164. [DOI] [PubMed] [Google Scholar]
- White S. S., Birnbaum L. S. (2009). An overview of the effects of dioxins and dioxin-like compounds on vertebrates, as documented in human and ecological epidemiology. J. Environ. Sci. Health 27, 197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wourms M. J., Sulentic C. E. (2015). The aryl hydrocarbon receptor regulates an essential transcriptional element in the immunoglobulin heavy chain gene. Cell. Immunol. 295, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo B. S., Boverhof D. R., Shnaider D., Crawford R. B., Zacharewski T. R., Kaminski N. E. (2004). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the regulation of Pax5 in lipopolysaccharide-activated B cells. Toxicol. Sci. 77, 272–279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.