Abstract
Exposure to ambient particulate matter (PM) has been associated with adverse health effects, including pulmonary and cardiovascular disease. Studies indicate that ambient PM originated from different sources may cause distinct biological effects. In this study, we sought to investigate the potential of various types of PM to cause epigenetic alterations in the in vitro system. RAW264.7 murine macrophages were exposed for 24 and 72 h to 5- and 50-μg/ml doses of the water soluble extract of 6 types of PM: soil dust, road dust, agricultural dust, traffic exhausts, biomass burning, and pollen, collected in January–April of 2014 in the area of Little Rock, Arkansas. Cytotoxicity, oxidative potential, epigenetic endpoints, and chromosomal aberrations were addressed. Exposure to 6 types of PM resulted in induction of cytotoxicity and oxidative stress in a type-, time-, and dose-dependent manner. Epigenetic alterations were characterized by type-, time-, and dose-dependent decreases of DNA methylation/demethylation machinery, increased DNA methyltransferases enzymatic activity and protein levels, and transcriptional activation and subsequent silencing of transposable elements LINE-1, SINE B1/B2. The most pronounced changes were observed after exposure to soil dust that were also characterized by hypomethylation and reactivation of satellite DNA and structural chromosomal aberrations in the exposed cells. The results of our study indicate that the water-soluble fractions of the various types of PM have differential potential to target the cellular epigenome.
Keywords: chromosomal aberrations, DNA methylation, ambient particulate matter, satellite DNA, transposable elements
Recent epidemiological studies examining the effects of particulate matter (PM2.5) on human health demonstrated that, despite a decrease in PM2.5 concentrations over the period 1998–2000, the magnitude of cardiovascular and respiratory effects persist at current ambient levels in the United States (Crouse et al., 2012; Dominici et al., 2007). The observed effects include respiratory infections, asthma, and ischemic heart disease among others (Dominici et al., 2006; Rückerl et al., 2011). The pathogenesis of PM-associated diseases has been attributed to its potential to cause cytotoxicity, oxidative stress, and inflammatory responses, to name a few (van Berlo et al., 2012).
PM originates from many anthropogenic and natural sources, including formation of secondary inorganic and organic particles with variable physical, chemical, and morphological characteristics. Therefore, PM may elicit various effects and, thus, may affect the degree of proinflammatory response and oxidative potential due to exposure (Kroll et al., 2013; Michael et al., 2013; Shi et al., 2006). Studies performed to evaluate the in vitro toxicity of PM collected at different urban areas (ie, RAPTES and PAMCHAR projects) have clearly demonstrated that chemical composition and oxidative potential are determinants of PM-induced toxicity (Happo et al., 2010; Janssen et al., 2014; Steenhof et al., 2011). Carbonaceous particles (elemental and organic carbon) released primarily from fossil fuel and fresh biomass combustion demonstrated oxidative stress and inflammatory potential (Biswas et al., 2009). Similar effects were also observed for coarse dust particles (Halatek et al., 2011; Happo et al., 2010). The complexity and diversification of particle size, chemical composition, and types, as well as the concurrent activation of multiple biological pathways and synergistic effects, limit the ability to differentiate the components and types that may be better associated with a specific health outcome.
Although coding genes comprise 1%–2% of mammalian genomes, repetitive sequences account for their largest portion, with some estimates suggesting 66%–69% of mammalian genomes to be repetitive or repetitive elements derived (de Koning et al., 2011). The majority of them are the retrotransposons—repeated and mobile DNA sequences capable of moving and invading genomes and immobile centromeric/pericentromeric satellite DNA and tandem repeats. Their activity is regulated by epigenetic mechanisms, where methylation of DNA plays a key role. The loss of epigenetic control over repetitive elements, exhibited as loss of DNA methylation and decondensation of the chromatin structure, may result in their transcriptional activation, insertional mutagenesis, and chromosomal aberrations and has been reported in numerous human diseases, including cancer (reviewed in Miousse et al., 2015).
Accumulating evidence clearly demonstrates that exposure to ambient PM and its contributors may affect the methylation status of transposable elements and satellite DNA in the blood of exposed humans (Baccarelli et al., 2009; Byun et al., 2013; Guo et al., 2014; Hou et al., 2014; Jiang et al., 2014; Madrigano et al., 2011). Recently, we reported dose-dependent decreases in mRNA levels of DNA methyltransferases (DNMTs), redistribution of methylation patterns between the repetitive elements and their transcriptional activation in response to the short-term exposure to ambient PM composed of biomass burning, traffic exhausts in the fine and ultrafine particle size range, and mineral dust and pollen in the coarse size range (Chalbot and Kavouras, 2014; Chalbot et al., 2013a; Miousse et al., 2014a). The purpose of this study was to investigate the potential of various types of PM to cause differential epigenetic alterations in the in vitro system. Specifically, we sought to investigate the effects of the 6 types of particles contributing to atmospheric PM, namely: soil dust, road dust, agricultural dust, traffic exhausts, biomass burning, and pollen on the cellular epigenome and whether these effects were persistent. Using RAW264.7 macrophages, the same experimental system utilized in large-scale international particle toxicological studies (Happo et al., 2010; Steenhof et al., 2011), we report that exposure to various PM types, aside from the effects associated with cytotoxicity and oxidative stress, also targeted the cellular epigenome. The latter effects were exhibited as downregulation of DNA methylation machinery in a dose- and particle type-dependent manner as well as altered methylation and expression of retrotransposons LINE-1 (L1) and SINE B1/B2. Furthermore, we showed that the most pronounced and persistent effects were observed in response to soil dust exposure characterized by hypomethylation and accumulation of satellite DNA transcripts and chromosomal aberrations in RAW264.7 macrophages.
MATERIALS AND METHODS
Sample collection
All dust samples were collected in January–April of 2014. Sampling methods were adopted from Rogge et al. (1993a,b, 1998) previously used to determine the chemical content of organic aerosol. Soil dust (SD) and road dust (RD) samples were collected by sweeping undisturbed soil textures in state parks and several residential streets, respectively, in Little Rock, Arkansas. Agricultural dust (AD) was collected from several farm sites located in rural Arkansas. Pollen (P) from important plant species growing in the South such as pine (Pinus), oak (Quercus), and hickory (Carya) were obtained in state parks during the pollination season and placed in a Teflon bag (March–April, 2014) (Chalbot et al., 2013c). The harvesting effort was done shortly after a rain event to reduce the amount of anthropogenic particles due to their possible deposition. The dust and pollen samples were sieved with a 38-μm brass frame stainless sieve (Humboldt Scientific, Inc, Raleigh, North Carolina) to remove debris and large particles, transferred to clean Teflon bags (Welch Fluorocarbon Inc, Dover, New Hampshire), and agitated (ie, induce continuous disturbance) to resuspend the PM (Rogge et al., 1993a,b). Aerosol samples were collected on prefired (550°C for 4 h) quartz filters using a PM10 Harvard Impactor (HI) sampler (Air Diagnostics and Engineering, Inc, Harrison, Maine) (Marple et al., 1987). Traffic exhausts (TE) were collected at the lowest level of an underground multilevel parking lot at the University of Arkansas for Medical Sciences (Little Rock, Arkansas) from 6 am to 6 pm during weekdays on a 20.3 × 25.4 cm prefired quartz filter mounted on a high volume sampler (Tisch Environmental, Ohio). Biomass burning (BB) aerosol samples were obtained in a traditional brick fireplace using commercially available wood types weighing from 0.5 to 2 kg (Rogge et al., 1998). The collection started shortly after the complete consumption of fire starters. Burning wood logs were stirred and new logs were added as needed. Samples were collected using the PM10 HI. Particle size distribution was measured using an optical particle counter (AeroTrak Model 8220, TSI Inc). The particle number median aerodynamic diameter (NMAD) was computed as previously described (Chalbot and Kavouras, 2014; Van Vaeck and Van Cauwenberghe, 1985).
Morphological characterization of dust and atmospheric particles
The morphology of the generated dust and atmospheric particles was obtained using scanning electron microscopy (SEM, Zeiss SUPRA 55, Mainz, Germany). The samples were all dispersed in deionized water at 10 μg/ml. A drop of solution was placed on the silicon wafer chip (5 × 5 mm, Ted Pella Inc, Redding, California) and left to dry overnight. Then, the chip was mounted on a double-sided sticky carbon tape placed on the Pin stub mounts (Ted Pella Inc). The SEM image of the samples was detected after drying on the silicon wafer chip.
Sample preparation and chemical analysis
The water-soluble fraction of aerosol samples was extracted by sonication of the filters in H2O for 1 h. The aqueous extract was filtered on 0.45-μm polypropylene filter (Target2, Thermo Scientific, Waltham, Massachusetts), transferred to a preweighed vial, dried using the SpeedVac apparatus, and weighed to determine the mass of extracted particles. Aliquots of the extracts were analyzed for elements (Fe, Al, K, Mg, Ca, Cu, Cr, Ni, Co, Ba, P and V) by inductively coupled plasma mass spectrometry (ICP-MS) at the University of Arkansas at Fayetteville Stable Isotope Laboratory. In addition, nuclear magnetic resonance spectroscopy (1H-NMR) spectra were obtained on a Bruker 500 MHz at 298K with 3600 scans, using a spin-lock, acquisition time of 3.2 s, relaxation delay of 1 s, and 1 Hz exponential line broadening and presaturation to the H2O resonance (Chalbot and Kavouras, 2014). The NMR resonance signals were integrated as follows: (1) H-C (δ 0.6–δ 1.8 ppm); (2) H-C-C = (δ 1.8–δ 3.2 ppm); (3) H-C-O (δ 3.2–δ 4.4 ppm); (4) O-CH-O and H-C = (δ 5.0–δ 6.4 ppm); and (5) H-Ar (δ 6.5–δ 8.3 ppm) (Chalbot and Kavouras, 2014; Decesari et al., 2007).
Cell culture and exposures
RAW264.7 murine macrophages were purchased from American Type Culture Collection (ATCC, Rockville, Maryland). Cells were plated at a density of 2 × 106 cells per 60 cm2 plates and allowed to attach. The cells were cultured in high glucose DMEM media (DMEM GlutaMAX, Life Technologies, Carlsbad, California) containing 5% FBS, penicillin, and streptomycin. After 24 h, the media was removed and replaced with 10 ml of fresh media. Cells were treated with 0, 5, or 50 µg/ml of PM water soluble extract. The lower dose corresponds to 2.94 µg/cm2, while the high dose corresponds to 29.9 µg/cm2 exposure levels (Steenhof et al., 2011). In our previous study (Miousse et al., 2014a), the higher dose was shown to cause epigenetic responses without induction of cytotoxicity. The lower dose in this study was twice lower than the lowest dose in the previous study to investigate the exposures to very low doses of PM on cellular epigenome. Cells were harvested after 24 h with trypsin, scraped, and divided in 3 aliquots before freezing. For the 72-h exposure experiment, cells were plated at a density of 0.75 × 106 cells per 60 cm2, and cells were harvested 72 h after the addition of PM. All experiments were performed using the cells between the passage 3 and 5.
Cell viability analysis
RAW264.7 were seeded at 1.25 × 104 per well in a 96-well plate, allowed to fully attach for 24 h and then exposed to soil, road, and agricultural dusts, biomass burning, traffic exhausts, or pollen for 24 h at 5 and 50 μg/ml. The CytoTox-One Homogeneous Membrane Integrity Assay (Promega, Madison, Wisconsin) was used to estimate the number of nonviable cells present after the exposure to these particles by measuring the amount of lactate dehydrogenase (LDH) leaked from the cells based on our previous study (Lu et al., 2015). Briefly, the Lysis Solution was used as a positive control, which was included in CytoTox-One Homogeneous Membrane Integrity Assay to generate maximum LDH release. Fluorescence intensity was detected by SoftMax Pro 6 GxP Microplate Data Acquisition and Analysis System (Molecular Devices, Sunnyvale, California) with an excitation/emission wavelength of 560/590 nm. To ensure that the particles did not interfere with the assay results, particle- and media-only control groups were used. The particle-only controls were particles in media at 5- or 50-μg/ml concentrations (without cells), and media-only controls were only media in the absence of cells. Results indicated that autofluorescence effects were only detected in the particle-only control groups of soil and agricultural dusts and pollen in RAW264.7 media at both 5- and 50-μg/ml doses. Thus, the signals for the aforementioned particle exposures were corrected to take into the account the autofluorescence effects of particle-only controls. Percent cytotoxicity was calculated as following: 100 × (Experimental − Culture Medium Background or Particle-only Background)/(Maximum LDH Release − Culture Medium Background).
Estimation of intracellular superoxide levels using dihydroethidium oxidation
Dihydroethidium (DHE) is a fluorescent probe used to evaluate the increase in steady state levels in cells after exposure to these particles. Fifteen percent hydrogen peroxide (15% H2O2, 4.9 mol/l) was used as a positive control for 30 min at the end of 24-h exposure. Then, cells were incubated with 5 μM DHE at 37°C for 30 min. The fluorescence intensity was detected by SoftMax Pro 6 GxP Microplate Data Acquisition and Analysis System (Molecular Devices) with an excitation/emission wavelength of 518/605 nm. Particle-only control experiments were also included in this assay and compared with a media-only control group. Results indicated that autofluorescence effects were detected in the particle-only control groups of soil and agricultural dusts, biomass burning, and pollen in RAW264.7 media at both 5- and 50-μg/ml doses and road dust at 50-μg/ml dose. Thus, the signals for the aforementioned particle exposures were corrected to take into the account the autofluorescence effects of particle-only controls.
Measurement of mitochondrial function using the XF96-extracellular flux analyzer
Oxygen consumption rate (OCR) was measured at 37°C using an XF96-extracellular analyzer (Seahorse Bioscience, North Billerica, Massachusetts) as previously described (Pathak et al., 2014). RAW264.7 cells were plated in 60-mm dishes at conditions described earlier. The next day, they were treated with soil and road dusts particles at 5- and 50-μg/ml doses and pollen at 50-μg/ml dose for 24 h. The cells were then trypsinized and 50 000 RAW cells/well and plated in XF96 cell culture plates. They were allowed to attach and recover for 4 h at 37°C. The media in the wells were changed to unbuffered DMEM supplemented with 4 mM Glutamate and incubated in a non-CO2 incubator for 1 h at 37°C. Three baseline measurements were acquired before injection of mitochondrial inhibitors or uncouplers. Readings were taken after sequential addition of oligomycin (10 μM), carbonylcyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, 10 μM), and rotenone/antimycin A (10 μM). OCRs were calculated by the Wave 2.2 software for Seahorse XF and represent an average of 3 measurements on 8 different wells. The rate of measured oxygen consumption was reported as pmol O2 consumed per minute per 50 000 cells.
Nucleic acids extraction
RNA and DNA were extracted simultaneously from flash-frozen cells using the AllPrep Mini Kit (Qiagen, Valencia, California) according to the manufacturer’s protocol. DNA concentrations were analyzed by the Nanodrop 2000 (Thermo Scientific), and DNA integrity was evaluated on 1% agarose gel.
Quantitative analysis of gene and transposable elements expression levels
Complementary DNA (cDNA) was synthesized from 1-µg RNA using random primers and a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, California) according to the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) to determine the levels of gene transcripts was performed using 10 ng of cDNA per reaction and the TaqMan Universal PCR Master Mix, no AmpErase UNG (Life Technologies) on a ViiA 7 instrument (Life Technologies). Assay IDs and primers used in the study are provided in Supplementary Table 1. Each plate contained 1 experimental gene and a housekeeping gene. The cycle threshold (Ct) for each sample was determined from the linear region of the amplification plot. The ΔCt values for all genes were determined relative to the control gene Gapdh and to Rps13 for TEs (Life Technologies). The ΔΔCt were calculated using each exposed group means relative to control group means as described previously (Schmittgen and Livak, 2008). The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in duplicate.
Western blot
Nuclear proteins were extracted from cell using the EpiQuik Nuclear Extraction Kit (Epigentek) according to the manufacturer’s protocol. A total of 2 µg of nuclear extract was loaded on a 7.5% SDS-PAGE gel and transferred on a PVDF membrane. The membrane was blotted with primary antibodies against Dnmt1 and the nuclear loading control YY1 (Abcam, Cambridge, Massachusetts), fluorescent secondary antibodies (LI-COR, Lincoln, Nebraska), and visualized with an Odyssey Fc imaging system (LI-COR). Analysis was performed with the Image Studio software (LI-COR)
Analysis of methylation status of retrotransposons
Methylation of LINE-1 (L1) and SINE B1/B2 elements was assessed by methylation-sensitive qRT-PCR. First, 1 µg of genomic DNA was digested with 1 U of SmaI enzyme in 1 × CutSmart buffer at 25°C for 2 h. This was followed by a 16-h digestion at 37°C in the presence of 1 U of the enzymes HpaII, HhaI, and AciI in 1 × CutSmart buffer. The digestion was finalized by adding 0.5 U of BstUI enzyme in 1 × CutSmart buffer for 4 h at 60°C. All enzymes were purchased from New England Biolabs (Ipswich, Massachusetts). Digested DNA was then analyzed by qRT-PCR on a ViiA 7 RT-PCR System (Applied Biosystems). DNA samples not digested with the restriction enzyme mix served as positive control, while samples (1) lacking the specific primers for DNA amplification and/or DNA template and (2) RAW264.7-derived DNA pretreated with 5-azacytidine, a potent demethylating agent, served as negative controls. The threshold cycle (Ct) was defined as the fractional cycle number that passes the fixed threshold. The Ct values were converted into the absolute amount of input DNA using the absolute standard curve method and further normalized toward rDNA readings. Assays for determination of L1 and SINE methylation are provided in Supplementary Table 1.
Methyltransferase activity
Nuclear proteins were extracted from 1 × 106 fresh cells using the EpiQuik Nuclear Extraction Kit (Epigentek, Farmingdale, New York). The nuclear extracts were then analyzed for methyltransferase activity by fluorometry using the EpiQuik DNMT Activity/Inhibition Assay Kit (Epigentek) according to the manufacturer’s protocol.
Analysis of the methylation status of satellite DNA
Methylation status of major and minor satellites was determined by methylation-sensitive McrBC-qRT-PCR assay as previously described (Martens et al., 2005). Primer sequences can be found in the Supplementary Table 1.
Chromosomal aberrations
RAW264.7 cells in log phase were exposed to different concentrations of PM and allowed to incubate for 72 h. Fresh media with colcemid (5 µl/ml of media, stock concentration 10 µg/ml; Invitrogen, Cat No. 15210-040) was added, and cells were further allowed to incubate for 2 h before harvest.
Chromosome preparation was made according to the standard air drying procedure. The cells were harvested by trypsinization, washed with prewarmed PBS twice, hypotonically treated (0.56% KCl, 20 min at 37°C), and subsequently fixed in freshly prepared acetic acid-methanol (1:3). At least 3 changes were given in fixative before the cell suspension was dropped on to a precleaned chilled microscopic glass slide and dried at room temperature at least for 1 day before staining. Giemsa staining method was applied to determine the structural aberrations in different treatment groups. Structural aberrations like chromatid-type break (CTB), chromatid-type exchange (CTE), acentric fragments, dicentric and ring chromosomes, and Robertsonian translocation were scored under × 63 magnification. At least 100 metaphase spreads were scored for each of the treatment group.
Statistical analysis
The significance between experimental treatments was determined by 1-way ANOVA, followed by Dunnett’s test using Graphpad Prism 6.02 software. A P value ≤ 0.05 was considered to be significant. Statistical considerations for the analysis of chromosomal aberrations are provided in the Supplementary Table 2.
RESULTS
Chemical and Morphological Characterization of Particle Types
This section presents the size distribution of resuspended PM10 particles from grab samples for soil, agricultural and road dusts, and pollen as well as airborne particles collected in an underground parking building (ie, traffic exhausts) and over a fireplace (ie, biomass burning).
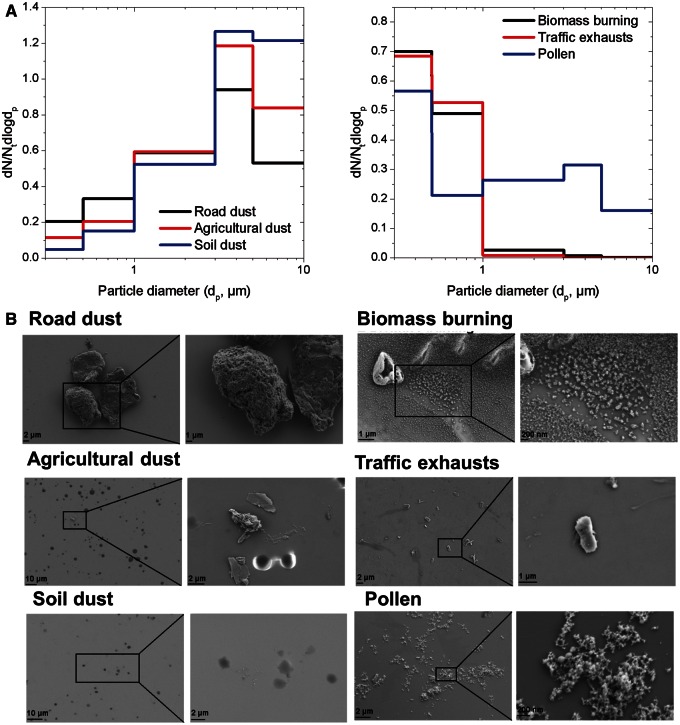
The normalized particle number concentration-based size distributions (dN/Ctdlogdp) and particle morphology by SEM of soil dust, road dust, agricultural dust, traffic exhausts, biomass burning, and pollen are presented in Figure 1 A. Note that particle size distribution refers to the resuspended PM10 samples. Larger than 10-μm particles were not collected in this study (removed by the size selective inlet of the sampler). Comparable mono-modal particle size distributions in the coarse size range were measured for soil, road, and agricultural dust with the highest number concentrations measured for particles with diameter from 3.0 to 10 μm. The NMADs were 2.06, 3.17, and 4.05 μm for road, agricultural, and soil dust, respectively. Biomass burning and traffic exhausts also followed a mono-modal distribution with maxima for particles of 0.3 < dp < 0.5 µm corresponding to a NMAD of 0.42 μm. For pollen, a bimodal distribution was observed with 2 local maxima in the fine (0.3–0.5 μm) and coarse (3.0–5.0 μm) size ranges corresponding to NMAD of 0.45 μm. The morphology of these particles was different from each other and was consistent with the particle number size distribution. Fine and ultrafine particles were observed in biomass burning, traffic exhausts, and pollen and larger particles in dust samples.
FIG. 1.
(A) Particle size distribution and (B) representative scanning electron microscopy images of road dust, agricultural dust, soil dust, traffic exhausts, biomass burning, and pollen particles.
Table 1 shows the elemental and organic composition of the samples. Aluminum (Al), iron (Fe), and potassium (K) were the dominant elements in soil and agricultural dust. Traces of chromium (Cr), nickel (Ni), and magnesium (Mg) were also detected. The road dust sample was composed of the elements found in soil dust (Al, Fe, K, and barium [Ba]) and high quantities of Cr and Ni. The predominant elements in traffic exhausts were Fe, copper (Cu), Cr, and Ni. Comparable elemental composition was measured for biomass burning and pollen with larger quantities of Fe, Cr, Ni, and phosphorous (P). The highest nonexchangeable organic hydrogen concentration measured by proton NMR (1H-NMR) was computed for pollen 128.5 mmol g−1 of particles followed by road dust (35.9 mmol g−1) and biomass burning (21.3 mmol g−1). Supplementary Figure 1 shows the 1D 1H-NMR spectra of the 6 PM types. The aliphatic region (H-C and H-C-C=) of agricultural, road, and soil dust 1H-spectra was dominated by broad convoluted resonances at δΗ 0.88, 1.32, 1.59, 2.18, 2.28, and 3.72 ppm consistent with humic and fulvic acids typically found in soil organic matter. Humic-like substances have also been identified in traffic exhausts (Havers et al., 1998). The resonance at 2.14 ppm was tentatively attributed to the methoxy (O-CH3) in N-acetyl-2-aminoglucosan (Graham et al., 2002). Resonances of amino acids and functionalized carboxylic acids were observed for aerosol of biological origin and wood burning emission (Chalbot and Kavouras, 2014; Chalbot et al., 2013b,c). The carbohydrate region (H-C-O) was dominated by sugars and anhydrides, with glucose, fructose, sucrose, and trehalose present in pollen and biomass burning. Tracers of sucrose were also identified in traffic exhausts, while none of the sugars were observed in dust samples. Levoglucosan was only observed in biomass burning. Well-resolved and intense signals between 3.83 and 3.98 were attributed to Ar-O-CH3 resonances plant-derived material freshly emitted during wood burning (Alves et al., 2012; Graham et al., 2002). The vinylic (H-C=) and aromatic (H-Ar) regions of traffic exhausts and road dust were dominated by phthalic and terephthalic acids (2 broad signals at 7.38 and 7.30 ppm). Tracers of these compounds were also observed in road dust. Acetate (at 1.92 ppm) and formate (at 8.49 ppm) were observed in soil, agricultural, road dust, and biomass burning (Chalbot et al., 2013c). Assuming molar H/C ratios of 2, 2, 1.1, and 0.4 for H-C, H-C-C=, H-C-O, and H-Ar, respectively (Chalbot and Kavouras, 2014), the water soluble organic carbon for the 6 types of particles accounted from 1% of particle mass for agricultural dust and traffic exhausts, 5% for soil dust, 8% for road dust, 13% for wood burning, and 42% for pollen.
TABLE 1.
Elemental and Organic Functional Composition of the Different Types of Particles by ICP-MS and Nuclear Magnetic Resonance, Respectively
| Chemical Component | Particle Type |
|||||
|---|---|---|---|---|---|---|
| Soil Dust | Road Dust | Agricultural Dust | Traffic Exhausts | Biomass Burning | Pollen | |
| Fe (mg g−1) | 36.8 | 118.2 | 47.2 | 143.5 | 43.0 | 51.1 |
| Al (mg g−1) | 21.8 | 29.3 | 24.8 | 1.1 | 1.2 | 0.1 |
| K (mg g−1) | 7.3 | 20.0 | 16.8 | 13.4 | 8.5 | 18.0 |
| Mg (mg g−1) | 1.9 | 3.8 | 2.1 | 3.3 | 1.7 | 2.6 |
| Ca (mg g−1) | 0.5 | 4.7 | 0.7 | 6.4 | 2.7 | 1.6 |
| Cu (mg g−1) | 0.2 | 0.5 | 0.2 | 38.4 | 1.4 | 0.1 |
| Cr (mg g−1) | 4.3 | 20.0 | 7.0 | 20.0 | 8.0 | 7.9 |
| Ni (mg g−1) | 2.7 | 13.5 | 4.7 | 13.5 | 7.0 | 4.9 |
| Co (mg g−1) | > 0.1 | 0.2 | > 0.1 | 0.2 | 0.1 | > 0.1 |
| Ba (mg g−1) | 0.3 | 0.6 | 0.5 | 0.6 | > 0.1 | > 0.1 |
| P (mg g−1) | 0.5 | 1.0 | 0.5 | 1.4 | 1.3 | 2.4 |
| V (mg g−1) | > 0.1 | 0.2 | > 0.1 | 0.2 | > 0.1 | > 0.1 |
| Organic H (mol g−1) | 8.9 | 35.9 | 2.2 | 6.7 | 21.3 | 128.5 |
| % R-H | 22.5 | 36.5 | 31.8 | 34.0 | 17.1 | 17.9 |
| % H-C = C | 33.5 | 22.5 | 26.7 | 22.2 | 21.9 | 19.0 |
| % H-C-O | 38.8 | 36.7 | 36.9 | 40.6 | 47.9 | 59.9 |
| % O-CH-O | 2.2 | 1.5 | 2.3 | 1.6 | 4.9 | 1.0 |
| % Ar-H | 2.9 | 2.8 | 2.3 | 1.8 | 8.1 | 2.1 |
| % H-C = O | n.d. | n.d. | n.d. | n.d. | 0.2 | 0.1 |
n.d.: not detected.
Figure 1B provides indicative SEM images for the PM types used in this study. It is worth noting that the actual size of the PM in these images is in good agreement with the size from the real-time instruments presented earlier.
Selective Cytotoxicity in Response to Various Types of Ambient PM
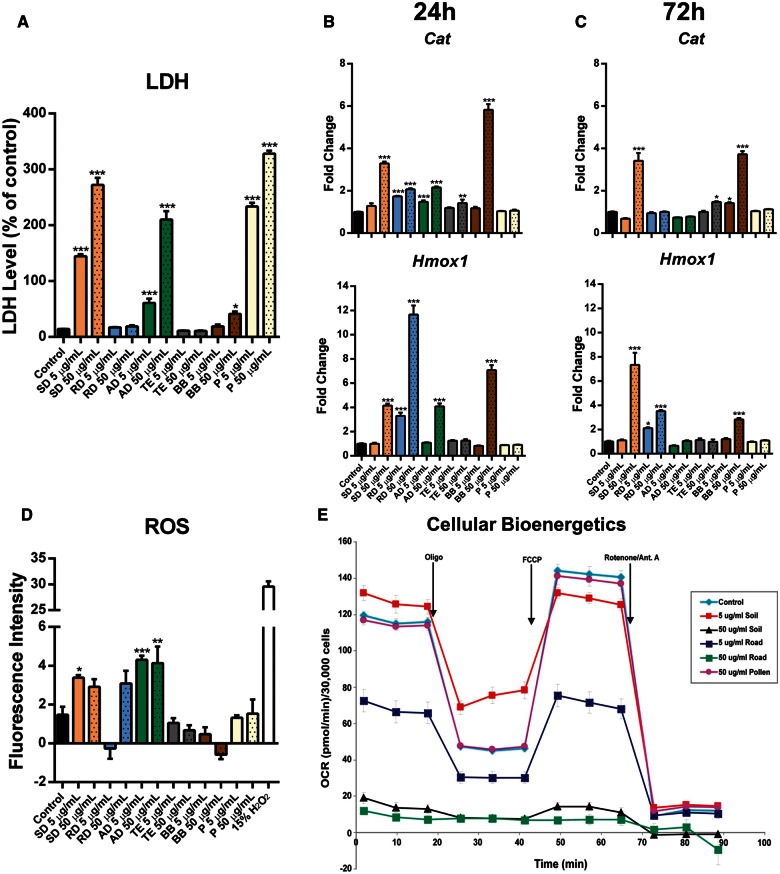
To evaluate the potential impact of various types of PM types on cell viability, RAW 264.7 murine macrophages were exposed to soil, road, and agricultural dusts, traffic exhausts, biomass burning, pollen, and mild steel welding fume (MS-WF) at 5 (low dose) and 50 μg/ml (high dose) for 24 h. LDH analysis provides evidence that soil and agricultural dusts, as well as pollen, had significant effects on cell viability, with the percentage cytotoxicity over 100% at high dose compared with positive control (ie, maximum LDH release). At the same time, the results of road dust, biomass burning, and traffic exhausts exposures indicated that these particles didn’t have a significant effect on LDH release at these 2 doses in RAW264.7 cells, except the high dose of biomass burning (Figure 2 A).
FIG. 2.
Analysis of cytotoxicity and oxidative potential of six types of ambient PM. A, Cytotoxicity was analyzed by calculation of the number of nonviable cells present after the exposure to ambient PM particles measuring the amount of lactate dehydrogenase leaked from the cells. B and C, The differential expression of Cat and Hmox1 was determined by quantitative real-time PCR (qRT-PCR). The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice. D, Induction of reactive oxygen species was analyzed by measuring a superoxide indicator DHE. Fifteen percent hydrogen peroxide (15% H2O2, 4.9 mol/l) was used as a positive control for 30 min at the end of 24-h exposure. E, Cellular bioenergetics was analyzed by measuring the mitochondrial function. Readings were taken after sequential addition of oligomycin (10 μM), carbonylcyanide 4-(trifluoromethoxy)phenylhydrazone (10 μM) and rotenone/antimycin A (10 μM). Oxygen consumption rates were calculated by the Seahorse XF96 software and represent an average of 3 measurements on 8 different wells. Asterisks (*) denotes significant (P < .05), (**) denotes significant (P < .01), and (***) denotes significant (P < .001) difference from control.
Exposure to Road and Soil Dusts Is Characterized by Significant Oxidative Potential
Twenty-four hours exposure to PM led to upregulation of Cat and Hmox1 genes, enzymes responsive to oxidative stress. This upregulation was observed in cells treated with high doses of soil and agricultural dusts and biomass burning, as well as in cells exposed to both doses of road dust (Figure 2, Panel B). The effect was still present after 72 h in cells exposed to high doses of soil dust and biomass burning (for both genes) and both doses of road dust (for Hmox1) (Figure 2, Panel C). Similar trends were observed in the expression patterns of another molecular indicator of alterations in redox homeostasis, Sod2 (Supplementary Figure 2A).
Levels of DHE oxidation were only increased in soil and agricultural dust-treated RAW264.7 cells, suggesting an increase in steady state levels of superoxide (Figure 2D). Although slight increases were observed, no significant difference was evident in total DHE fluorescence between cells exposed to other particles and untreated cells.
To further investigate the oxidative potential of different types of PM, we have performed the analysis of the cellular mitochondrial function in cells exposed to both doses of soil dust (exhibited high cytotoxic and oxidative potentials), road dust (exhibited no cytotoxic but high oxidative potential), as well as pollen (exhibited high cytotoxic potential but no evidence of oxidative stress). Initially, 3 measurements were taken to determine the “basal respiration” of the cells. Treatment with low dose of soil dust slightly but significantly (P < .05) increased the mitochondrial respiration compared with control, whereas there was a significant decrease in the mitochondrial respiration when low-dose road dust exposure was performed (P < .001) (Figure 2, Panel E). Next, by injecting oligomycin, an inhibitor of mitochondrial ATP synthase, a decrease in OCR was obtained. In both low-dose treatments, ATP-linked respiration was significantly decreased (P < .001) compared with control group, suggesting a possible obstruction of electron flow through damaged mitochondrial electron transport chain, decreasing the oxygen consumption. Furthermore, cells exposed to low doses of soil dust were characterized by increased proton leak compared with control (P < .001), supporting the possibility of damaged electron flow. In the road dust low-dose exposure group, the proton leak was significantly less than in the control cells (P < .01).
To determine the maximal respiration potential of the cells, FCCP (an uncoupler) was used. In both treatments, maximum respiration was decreased, and the cells were left with no spare respiratory capacity following treatment with low doses of either soil or road dust, indicating significantly damaged mitochondrial integrity.
The amount of nonmitochondrial oxygen-consumption was determined by inhibiting the respiratory chain activity via an antimycin A and rotenone cocktail. There were no significant decreases in nonmitochondrial OCR in any of the treatment groups with low-dose exposure.
Exposure to soil and road dusts at high doses completely obliterated cellular respiration. No significant alterations in cellular respiration were observed after exposure to high dose of pollen (Figure 2, Panel E).
Exposure to PM Affects DNA Methylation Machinery
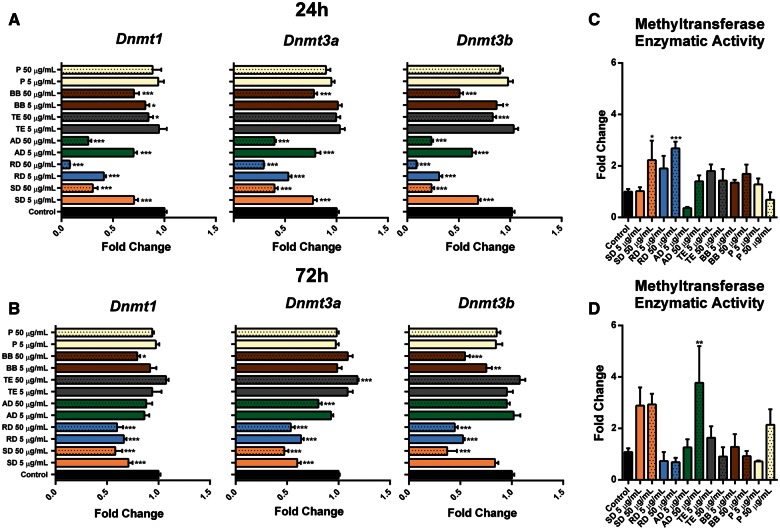
Next, we sought to analyze the effects exerted by PM on DNA methylation machinery. The 24-h exposure to soil, road, and agricultural dusts resulted in substantial and dose-dependent decreases in mRNA levels of all 3 DNMTs—Dnmt1, Dnmt3a, and Dnmt3b (Figure 3 , Panel A). Subtle downregulation of DNMTs was also observed in response to traffic exhaust and biomass burning. At a later time-point (72 h), only exposure to soil and road dusts was associated with the loss of expression of DNA methylation machinery, while the effects of agricultural dust were not persistent. Dnmt1 and Dnmt3b were also slightly downregulated after exposure to the high dose of biomass burning (Figure 3, Panel B). At the same time, opposite to mRNA levels, exposure to soil and road dusts has led to increased nuclear methyltransferase enzymatic activity at 24 h (2- to 2.8-fold), and an increase, albeit insignificant, was still observed in road dust exposure groups at the 72-h time-point (Figs. 3, Panels C and D). These results were confirmed by Western blot analysis, where the elevated protein levels of Dnmt1, although insignificant, were observed (Supplementary Figure 3).
FIG. 3.
Effects of ambient PM exposure on DNA methylation machinery. A and B, The differential expression of Dnmt1, Dnmt3a, and Dnmt3b was determined by qRT-PCR. The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice. C and D, Nuclear DNA methyltransferase (DNMT) enzymatic activity was analyzed by fluorometry using the EpiQuik DNMT Activity/Inhibition Assay Kit (Epigentek). Asterisks (*) denotes significant (P < .05), (**) denotes significant (P < .01), and (***) denotes significant (P < .001) difference from control.
Effects of PM exposure on methylcytosine dioxygenases were less pronounced and consistent. An almost 2-fold downregulation of Tet2 was observed after high-dose agricultural dust exposure at an earlier time-point (P < .001) followed by a 1.5-fold upregulation (P < .001) at a later time-point. Expression of Tet3 was decreased at both 24 and 72 h after exposure to soil and road dusts and after exposure to high-dose agricultural dust at 24 h only (Supplementary Figure 2B).
Effects of PM Exposure on Methylation of Transposable Elements
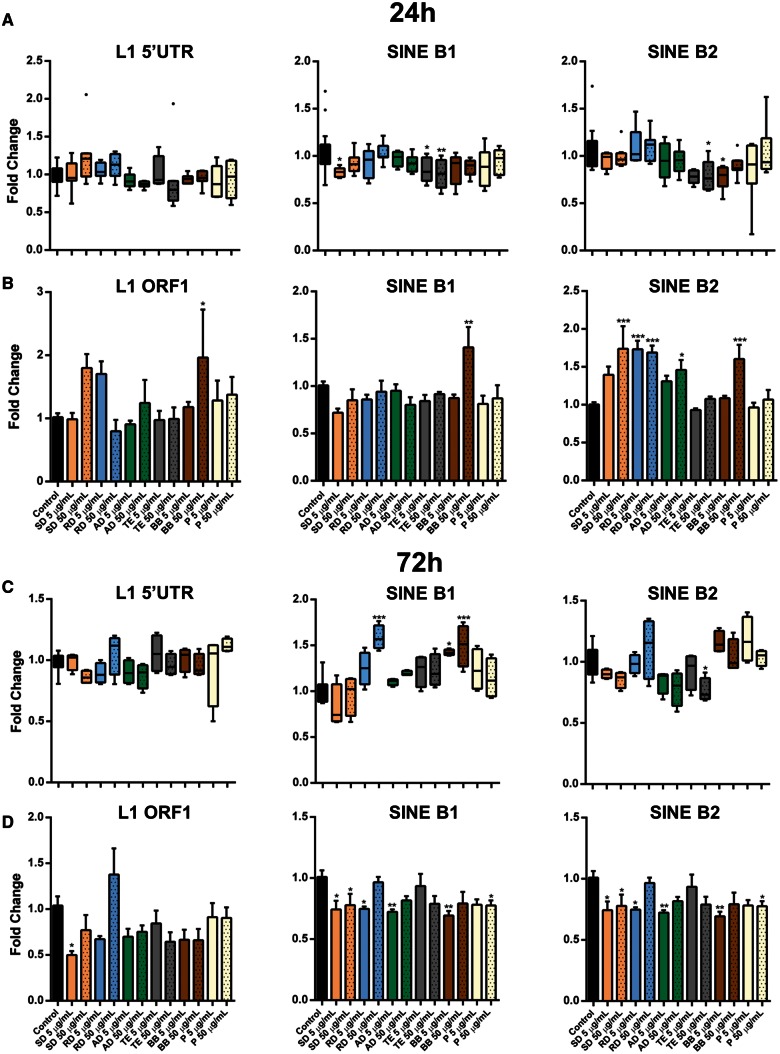
Taking into account the observed alterations in DNA methylation/demethylation machinery and the results of previous studies indicating alterations in the methylation status of transposable elements in response to PM exposure (Baccarelli et al., 2009; Byun et al., 2013; Guo et al., 2014; Hou et al., 2014; Jiang et al., 2014; Madrigano et al., 2011; Miousse et al., 2014a), we hypothesized that exposure to various types of PM may differentially affect the methylation status of the most abundant mammalian transposable elements L1 and SINE B1/B2.
Certain discrepancies exist between studies reporting alterations in DNA methylation within transposable elements in response to environmental stressors, PM in particular. One of the reasons for this is inconsistency in the functional units of L1, in which DNA methylation analysis are performed. Therefore, to systematically analyze the methylation of L1, we addressed its methylation in all 4 functional units—5′- and 3′-untranslated regions (UTRs), as well as in both open reading frames—ORF1 and ORF2. Both short- and long-term exposures did not substantially affect methylation of L1 in any of the evaluated units and only exposure to road dust led to weak hypomethylation at 72 h in 5′-UTR and both ORFs (Figs. 4 , Panels A and C; Supplementary Figs. 2C–D). Somewhat more pronounced alterations in the methylation status of SINE elements were observed, where weak hypomethylation after exposure to soil dust, traffic exhaust, and biomass burning was detected at the 24-h time-point and was followed by hypermethylation at the 72-h time-point in cells exposed to a high-dose road dust and both doses of biomass burning.
FIG. 4.
Transposable elements in response to ambient PM exposure. A and C, DNA methylation in LINE-1 (L1) and SINE B1/B2 transposable elements was measured by methylation-sensitive qPCR assay. B and D, The differential expression of LINE-1 and SINE B1/B2 in murine macrophages was determined by qRT-PCR. The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice. Asterisks (*) denotes significant (P < .05), (**) denotes significant (P < .01), and (***) denotes significant (P < .001) difference from control.
Expression of Transposable Elements Is Affected by Exposure to PM
At 24 h, no major changes in the expression of either L1 ORF1 or ORF2 were observed, except for the 1.95-fold reactivation in the high-dose biomass burning group (P < .01) (Figure 4, Panel B; Supplementary Figs. 2C and 2D). At 72 h, transcriptional silencing of both ORFs was observed in soil, road, and agricultural dusts, which was more evident after exposure to lower doses of PM (Figure 4, Panel D and Supplementary Figs. 2C–D).
More pronounced effects were observed in the expression of SINE elements. A significant upregulation of SINE B2 was observed in cells exposed for 24 h to soil, road, and agricultural dusts and biomass burning (Figure 4, Panel B), and was followed by a downregulation of both SINE B1 and B2 at 72 h (Figure 4, Panel D).
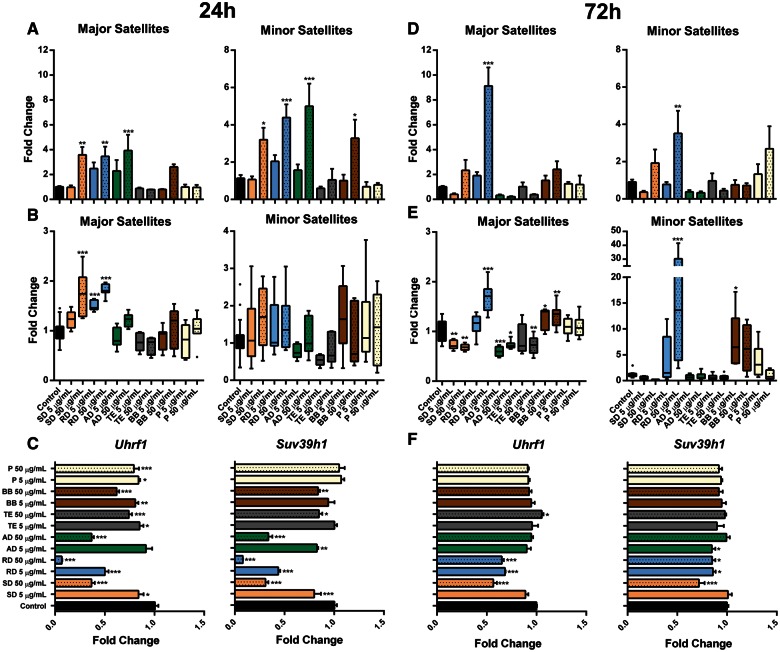
Effects of PM Exposure on Satellite DNA
Expression of major and minor satellites was mostly and significantly affected in response to high doses of soil, road, and agricultural dusts at 24 h after exposure (Figure 5 , Panel A). At the 72-h time-point, elevated levels of satellite DNA were detected after exposure to higher doses of soil (although, insignificantly) and road dust (Figure 5, Panel D). DNA methylation, one of the primary mechanisms in satellite DNA silencing, was increased after 24-h exposure to soil and road dusts in major satellites (Figure 5, Panel B). At 72 h, loss in major satellites methylation was observed after exposure to both doses of soil and agricultural dusts and the high dose of traffic exhaust. At the same time, exposure to the high dose of road dust has led to hypermethylation of both major and minor satellites (Figure 5, Panel E).
FIG. 5.
Satellite DNA is affected by exposure to ambient PM. A and D, The differential expression of major and minor satellites in murine macrophages was determined by qRT-PCR. B and E, DNA methylation major and minor satellites was measured by methylation-sensitive qRT-PCR assay. C and F, The differential expression of Uhrf1 and Suv39h1 was determined by qRT-PCR. The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice. Asterisks (*) denotes significant (P < .05), (**) denotes significant (P < .01), and (***) denotes significant (P < .001) difference from control.
Uhrf1 is a key protein for the maintenance of normal DNA methylation (Ehrlich and Lacey, 2013). However, it is also an important regulator of the pericentromeric heterochromatin replication and silencer of satellite DNA (Papait et al., 2007). Exposure to PM has led to alterations in the Uhrf1 mRNA levels that were inversely correlated with the expression status of major satellites at both time-points (Figs. 5, Panels C and F; Supplementary Figure 2E).
Furthermore, we have identified decreased mRNA levels of Suv39h1, a histone methyltransferase needed for trimethylation of lysine 9 on histone 3—an epigenetic mark responsible for the heterochromatic status of centromeric and pericentromeric regions. Patterns of Suv39h1 downregulation mirrored those of Uhrf1 with a clear dose-dependent response (Figs. 5, Panels C and F).
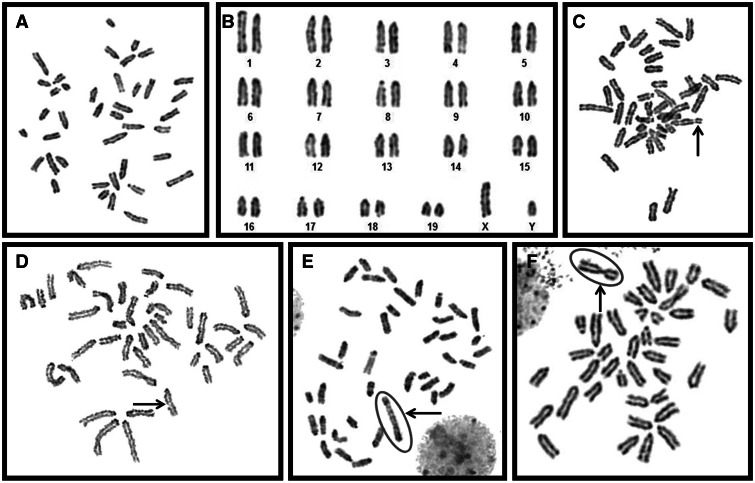
Exposure to Soil Dust Results in Chromosomal Aberrations in RAW264.7 Cells
Increased mRNA transcripts of satellite DNA can disturb the normal centromeric and pericentromeric heterochromatin and lead to chromosomal aberrations (Bouzinba-Segard et al., 2006). Therefore, as a final step of this study, we sought to determine whether or not alterations in DNA methylation and accumulation of satellite DNA mRNA transcripts may lead to structural chromosomal aberrations.
Metaphase spreads with representative structural aberration are shown in Figure 6 . We observed a significant (P < .01; Z value 5.68; Supplementary Table 2) increase in structural chromosomal aberrations after exposure to the high dose of soil dust for 72 h (24.76% vs 3.45% in control, Table 2 ). At the same time, 72 h treatment with the high dose of road dust caused no significant (P > .05; Z value 1.37) increase in structural aberration (7.53% vs 3.45% in control). Increase in structural chromosomal aberrations was also not observed after exposure to the high dose of pollen (5.83% vs 3.45% in control, Table 2). These data clearly indicate that soil dust has a substantial effect on chromosome damage in comparison to other PM investigated in this study. Among different types of structural aberrations scored, CTBs were most prevalent, while CTEs were completely absent in different treatment groups (Table 2).
FIG. 6.
Exposure to soil dust leads to chromosomal aberrations in RAW264.7 cells. Photomicrograph showing different structural aberrations induced in RAW cells after treating with different concentrations of particulate matter. The aberrations were indicated by arrows. A, normal metaphase spread with 40 chromosomes, (B) karyotype of the normal metaphase showing all acrocentric chromosomes, (C) acentric fragment (acentrics), (D) chromatid-type breaks (CTB), (E) dicentric chromosome (Dic), and (F) Robertsonian translocation (RbT).
TABLE 2.
Chromosomal Aberrations in RAW264.7 Cells After Exposure to Ambient Particular Matter
| Treatment | Normal | Aberrant | Total Scored | CTB | CTE | Acentrics | Dic plus r | RbT | Total |
|---|---|---|---|---|---|---|---|---|---|
| Control | 96.55 ± 9.12 | 3.45 ± 1.72 | 116 | 0.86 ± 0.86 | 0.0 ± 0.0 | 1.72 ± 1.22 | 0.0 ± 0.0 | 0.86 ± 0.86 | 3.45 ± 1.72 |
| (112) | (4) | (1) | (0) | (2) | (0) | (1) | (4) | ||
| Soil dust | 95.83 ± 8.94 | 4.17 ± 1.86 | 120 | 3.33 ± 1.67 | 0.0 ± 0.0 | 1.67 ± 1.18 | 0.0 ± 0.0 | 0.83 ± 0.83 | 5.83 ± 2.20 |
| 5 µg/ml | (115) | (5) | (4) | (0) | (2) | (0) | (1) | (7) | |
| Soil dust 50 µg/ml | 75.24 ± 8.46 | 24.76 ± 4.86 | 105 | 28.57 ± 5.22 | 0.0 ± 0.0 | 7.62 ± 2.69 | 0.95 ± 0.95 | 0.0 ± 0.0 | 37.14* ± 5.95 |
| (79) | (26) | (30) | (0) | (7) | (1) | (0) | (38) | ||
| Road dust 5 µg/ml | 93.33 ± 8.31 | 6.67 ± 2.22 | 135 | 2.96 ± 1.48 | 0.0 ± 0.0 | 0.74 ± 0.74 | 1.48 ± 1.05 | 1.48 ± 1.05 | 6.67 ± 2.22 |
| (126) | (9) | (4) | (0) | (1) | (2) | (2) | (9) | ||
| Road dust 50 µg/ml | 92.47 ± 7.96 | 7.53 ± 2.27 | 146 | 4.11 ± 1.68 | 0.0 ± 0.0 | 2.05 ± 1.19 | 0.68 ± 0.68 | 0.68 ± 0.68 | 7.53 ± 2.27 |
| (135) | (11) | (6) | (0) | (3) | (1) | (1) | (11) | ||
| Pollen | 94.17 ± 8.86 | 5.83 ± 2.20 | 120 | 4.17 ± 1.86 | 0.0 ± 0.0 | 0.83 ± 0.83 | 0.83 ± 0.83 | 0.0 ± 0.0 | 5.83 ± 2.20 |
| 50 µg/ml | (113) | (7) | (5) | (0) | (1) | (1) | (0) | (7) |
Aberration percentage ± SE (total number aberration observed) detected in RAW cells after 72 h of exposure to different concentrations of PM. Standard errors on the aberration percentage were calculated by √a/A, where “a” is the number under consideration, and “A” is the total number of cells analyzed (Lee et al., 2005). The abbreviations used for chromosomal aberrations are according to international nomenclature (ISCN, 1978). CTB, chromatid-type break; CTE, chromosome-type exchange; Dic plus r, dicentric chromosomes plus ring chromosomes; RbT, Robertsonian translocation. Asterisks (*) denotes significant (P < .001) difference from control.
DISCUSSION
In this study, we demonstrate that exposure to 6 common types of PM—soil dust, road dust, agricultural dust, traffic exhausts, biomass burning, and pollen—can exhibit differential biological effects in an in vitro experimental system. These effects were characterized by the differences in PM’s abilities to induce cytotoxicity and oxidative damage to DNA, as well as changes in DNA methylation and expression of transposable elements and satellite DNA and DNA methylation machinery.
Fe was a major component in all PM types, and the carbohydrate compounds dominated the organic PM fraction. Al contribution to PM was high in soil, road, and agricultural dust but only traces were measured in the other four PM types. Furthermore, soil dust particles had a higher content in compounds with unsaturated aliphatic functional groups that can be oxidized in the presence of free radicals. Metals, such as Cu, Fe, and Al, present in particles can lead to inflammatory responses, oxidative stress, and may also affect the methylation and expression of repetitive elements (Jalava et al., 2009; Miousse et al., 2015).
Exposure to six types of PM was characterized by different extents of oxidative potential. Soil dust caused the highest effects exhibited as a significant and persistent overexpression of genes, indicative of cellular response to oxidative stress, induction of ROS, and obliterated cellular respiration. Effects of exposure to road dust were less persistent and did not result in the induction of ROS. At the same time, exposure to a high dose of biomass burning has led to persistently increased gene expression but did not result in accumulation of ROS. Pollen did not cause gene expression alterations, induction of reactive oxygen species, or changes in cellular mitochondrial respiration.
Presence of metals and their high oxidative potential may affect DNA methylation machinery (Fragou et al., 2011). We report that exposure to soil and road dust results in persistent dose-dependent decrease in DNMTs expression, in agreement with the recent study reporting dose-dependent loss of DNMTs in RAW264.7 cells after exposure to ambient PM (Miousse et al., 2014a).
At the same time, increased nuclear methyltransferase enzymatic activity was observed in the exposed cells and these results were confirmed by increased protein levels of Dnmt1. One of the possible explanations of this may be the initiation of transcription from alternative transcription start sites which was not detectable using the current assays. This may also explain the lack of substantial alterations in DNA methylation within the retrotransposons. Furthermore, Tet2 and Tet3 methylcytosine deoxygenases were downregulated by exposure to PM, suggesting a possible decline in the 5-methylcytosine to 5-hydroxymethylcytosine conversion rates. The observed alterations in DNA methylation were not uniform, resembling both hypo- and hypermethylation patterns. Similarly, the most recent studies report nonuniform responses to in vitro exposure to PM (Miousse et al., 2014a) as well as in human studies (Jiang et al., 2014).
Transcriptional activation of SINE elements in response to environmental exposure has been documented (Miousse et al., 2014b; Rudin and Thompson, 2001). Similar to these reports, we observed increases in mRNA levels of SINE B2 elements 24 h after exposure. However, analysis of their expression at 72 h revealed the transcriptional silencing of both SINE elements in almost all groups of treatment. Likewise, decreased expression of L1 ORF1 and ORF2 was observed at 72 h. These findings suggest that activation of retrotransposons is rather an early and not necessarily a persistent event that may be followed by their transcriptional silencing.
Satellite DNA, represented as major and minor satellites in mice, are the centromeric- and pericentromeric-specific sequences and the major components of heterochromatin that act as a centromere-building element (Ugarkovic, 2005). Here, we report hypomethylation of major and minor satellites in cells exposed to soil dust at 72 h paralleled by persistent overexpression of both satellite repeats. Furthermore, we identified decreased mRNA levels of Uhrf1, the protein needed for the maintenance of normal patterns of DNA methylation and transcriptionally silent status of major satellites (Ehrlich and Lacey, 2013; Ugarkovic, 2005) as well as decreased levels of Dnmt3b, required for stabilization of pericentromeric satellite repeats (Okano et al., 1999; Xu et al., 1999). Additionally, we report decreased mRNA levels of Suv39h1, a key methyltransferase for trimethylation of lysine 9 on histone H3 associated with silencing of pericentromeric heterochromatin (Guenatri et al., 2004).
Overexpression of pericentromeric satellite repeats has been documented in cancer (Ting et al., 2011) and noncancerous disease (Haider et al., 2012). Accumulation of satellite DNA mRNA transcripts, paralleled by their hypomethylation, was also observed in vitro after exposure to ambient PM (Miousse et al., 2014a) and in murine tissues after inhalational exposure to 1,3-butadiene (Koturbash et al., 2011). As centromeric and pericentromeric RNAs serve as key players for heterochromatin formation, the observed alterations in methylation and expression of satellite DNA may significantly affect normal chromatin assembly and result in chromosomal aberrations (Bouzinba-Segard et al., 2006; Ehrlich et al., 2003; Tsuda et al., 2002). Indeed, we identified that exposure to 50 µg/ml of soil dust had led to an increase in chromosomal aberrations, 9.5-fold higher than the background aberration percentage. Although expression of major and minor satellites after exposure to road dust was higher than after exposure to soil dust, and expression of Uhrf1, Dnmt3b, and Suv39h1 was diminished, both satellite repeats were heavily methylated in response to exposure to road dust, while exposure to soil dust led to satellite DNA hypomethylation. These findings suggest that loss of satellite DNA methylation in concert with loss of Uhrf1, Dnmt3b, and Suv39h1 and subsequent reactivation of the satellite repeats may lead to altered centromeric heterochromatin status and chromosomal aberrations. Exposure to pollen, despite the exhibited high cytotoxicity at both doses, led to subtle epigenetic alterations and did not result in chromosomal aberrations in vitro.
Conceptually, in vitro toxicological assessments are limited by the type of used cell lines, exposure magnitude, and concentrations. We used a well-characterized cell line extensively used in PM in vitro toxicity studies (Jalava et al., 2007, 2008, 2009; Lu et al., 2015; Michael et al., 2013; Steenhof et al., 2011). This allowed us to corroborate the experimental conditions and findings of previously published research as well as perform a detailed evaluation of the effects on the cellular epigenome of different PM types. Exposure concentrations were also consistent with those previously used for this cell line (Loxham et al., 2015; Miousse et al., 2014a; Steenhof et al., 2011). These results, however, have to be taken with caution, because this study was not designed specifically to evaluate the content of each type in the ambient PM and, therefore, do not necessarily represent real-life exposures and their outcomes. Additional limitations are associated with the representativeness of the different types of PM samples. A certain degree of misrepresentation of traffic exhausts and biomass burning emissions may be due to the collection of samples immediately after the release, thus, eliminating atmospheric oxidation and aging. However, the possible effect of atmospheric aging and secondary organic aerosol formation is highly variable and inconsistent because of its relation to the overall atmospheric oxidative burden and distance/time between the source and the receptor site. Also, while the study was performed in murine cells, our recent study on the effects of engineered nanomaterials showed congruent epigenetic responses between the RAW264.7 and human THP-1 cells (Lu et al., 2015).
In this study, we used the water soluble extract, as did previous in vitro PM toxicity studies of different PM types, for which the potential effect of water-insoluble species may not be accounted. Although the effect of particle size was not evaluated, particle size distribution measurements showed distinct differences for the six PM types. Previous studies showed that endotoxins may also contribute to the inflammatory responses due to exposures to PM. We did not directly measure endotoxin levels in this study; thus, their overall effect cannot be determined. However, proton resonances in the carbohydrate region of the NMR spectra may provide information on the presence of functional groups of O-antigen, lipid A and oligosaccharide of endotoxins (Wu et al., 2013). The three types of soil samples had the lowest content in carbohydrate proton as compared with traffic exhausts, biomass burning, and pollen particles (Table 1). The possible effect of other biological material such as (1-3)-β-D-glucan, a glucose polymer associated with endotoxins, may be reduced during the extraction and sample preparation and exposure processes involving sonication. Overall, the limitations of this study are associated with technical limits in particle collection and in vitro toxicity; however, they were comparable for all particle types and to those in previous studies.
In summary, we show that in vitro exposure to various types of PM may cause distinct cellular, molecular, genetic, and epigenetic alterations. The observed epigenetic changes are type- and dose-dependent and may be persistent in nature. Finally, we show that the most pronounced and persistent effects were observed in response to soil dust exposure and were characterized by hypomethylation and accumulation of centromeric and pericentromeric satellite DNA transcripts that led to chromosomal aberrations in exposed cells.
FUNDING
This work was supported by the National Institutes of Health Center of Biological Research Excellence (grant number 1P20GM109005-01A1); National Institutes of Health UAMS Clinical and Translational Science Award (grant numbers UL1TR000039 and KL2TR000063); National Institute for Occupational Safety and Health and Consumer Product Safety Commission (grant number 212-2012-M-51174); Arkansas Space Grant Consortium through National Aeronautics and Space Administration (grant number NNX13AB29A); and the Arkansas Biosciences Institute.
SUPPLEMENTARY DATA
Supplementary data are available online at: http://toxsci.oxfordjournals.org/.
ACKNOWLEDGMENTS
The authors are thankful to Dr Priyanka Chitranshi and Dr Gonçalo Gamboa da Costa for their help with nuclear magnetic resonance spectroscopy (NMR) analysis. They are thankful to Dr Kristy Kutanzi for critical reading and to Dr Rebecca Helm for editing the article.
REFERENCES
- Alves C., Vicente A., Pio C., Kiss G., Hoffer A., Decesari S., Prevôt A. S. H., Minguillon M. C., Querol X., Hillamo R. (2012). Organic compounds in aerosols from selected European sites - Biogenic versus anthropogenic sources. Atmos. Environ. 59, 243–255. [Google Scholar]
- Baccarelli A., Wright R. O., Bollati V., Tarantini L., Litonjua A. A., Suh H. H., Zanobetti A., Sparrow D., Vokonas P. S., Schwartz J. (2009). Rapid DNA methylation changes after exposure to traffic particles. Am. J. Res. Critic. Care Med. 179, 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Verma V., Schauer J. J., Cassee F. R., Cho A. K., Sioutas C. (2009). Oxidative potential of semi-volatile and non volatile particulate matter (PM) from heavy-duty vehicles retrofitted with emission control technologies. Environ. Sci. Technol. 43, 3905–3912. [DOI] [PubMed] [Google Scholar]
- Bouzinba-Segard H., Guais A., Francastel C. (2006). Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. U.S.A. 103, 8709–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun H. M., Motta V., Panni T., Bertazzi P. A., Apostoli P., Hou L., Baccarelli A. A. (2013). Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part. Fibre Toxicol. 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalbot M., McElroy B., Kavouras I. (2013a). Sources, trends and regional impacts of fine particulate matter in southern Mississippi valley: Significance of emissions from sources in the Gulf of Mexico coast. Atmos. Chem. Phys. 13, 3721–3732. [Google Scholar]
- Chalbot M., Nikolich G., Etyemezian V., Dubois D. W., King J., Shafer D., DaCosta G. G., Hinton J. F., Kavouras I. (2013b). Soil humic-like organic compounds in prescribed fire emissions by nuclear magnetic resonance spectroscopy. Environ. Pollut. 181, 167–171. [DOI] [PubMed] [Google Scholar]
- Chalbot M. C., da Costa G. G., Kavouras I. G. (2013c). NMR analysis of the water-soluble fraction of airborne pollen particles. Appl. Magn. Reson. 44, 1347–1358. [Google Scholar]
- Chalbot M. C., Kavouras I. G. (2014). Nuclear magnetic resonance spectroscopy for determining the functional content of organic aerosols: A review. Environ. Pollut. 191, 232–249. [DOI] [PubMed] [Google Scholar]
- Crouse D. L., Peters P. A., van D. A., Goldberg M. S., Villeneuve P. J., Brion O., Khan S., Atari D. O., Jerrett M., Pope C. A., et al. (2012). Risk of nonaccidental and cardiovascular mortality in relation to long-term exposure to low concentrations of fine particulate matter: A Canadian national-level cohort study. Environ. Health Perspect. 120, 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning A. P., Gu W., Castoe T. A., Batzer M. A., Pollock D. D. (2011). Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7, e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decesari S., Mircea M., Cavalli F., Fuzzi S., Moretti F., Tagliavini E., Facchini M. C. (2007). Source attribution of water-soluble organic aerosol by nuclear magnetic resonance spectroscopy. Environ. Sci. Technol. 41, 2479–2484. [DOI] [PubMed] [Google Scholar]
- Dominici F., Peng R. D., Bell M. L., Pham L., McDermott A., Zeger S. L., Samet J. M. (2006). Fine particulate air pollution and hospital admission for cardiovascular and respiratory diseases. JAMA. 295, 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici F., Peng R. D., Zeger S. L., White R. H., Samet J. M. (2007). Particulate air pollution and mortality in the United States: Did the risks change from 1987 to 2000? Am . J. Epidemiol. 166, 880–888. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Hopkins N. E., Jiang G., Dome J. S., Yu M. C., Woods C. B., Tomlinson G. E., Chintagumpala M., Champagne M., Dillerg L., et al. (2003). Satellite DNA hypomethylation in karyotyped Wilms tumors. Cancer Genet. Cytogenet. 141, 97–105. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Lacey M. (2013). DNA hypomethylation and hemimethylation in cancer. Adv. Exp. Med. Biol. 754, 31–56. [DOI] [PubMed] [Google Scholar]
- Fragou D., Fragou A., Kouidou S., Njau S., Kovatsi L. (2011). Epigenetic mechanisms in metal toxicity. Toxicol Mech. Methods 21, 343–352. [DOI] [PubMed] [Google Scholar]
- Graham B., Mayol-Bracero O. L., Guyon P., Roberts G. C., Decesari S., Facchini M. C., Artaxo P., Maenhaut W., KöII P., Andreae M. O. (2002). Water-soluble organic compounds in biomass burning aerosols over Amazonia 1. Characterization by NMR and GC-MS. J. Geophys. Res. 107, LBA–14. [Google Scholar]
- Guenatri M., Bailly D., Maison C., Almouzni G. (2004). Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Byun H. M., Zhong J., Motta V., Barupal J., Zheng Y., Dou C., Zhang F., McCracken J. P., Diaz A., et al. (2014). Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environ. Mol. Mutagen. 55, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S., Cordeddu L., Robinson E., Movassagh M., Siggens L., Vujic A., Choy M. K., Goddard M., Lio P., Foo R. (2012). The landscape of DNA repeat elements in human heart failure. Genome Biol. 13, R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halatek T., Stepnik M., Stetkiewicz J., Krajnow A., Kur B., Szymczak W., Rydzynski K., Dybing E., Cassee F. R. (2011). The inflammatory response in lungs of rats exposed on the airborne particles collected during different seasons in four European cities. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 46, 1469–1481. [DOI] [PubMed] [Google Scholar]
- Happo M. S., Salonen R. O., Halinen A. I., Jalava P. I., Pennanen A. S., Dormans J. A., Gerlofs-Nijland M. E., Cassee F. R., Kosma V. M., Sillanpaa M., et al. (2010). Inflammation and tissue damage in mouse lung by single and repeated dosing of urban air coarse and fine particles collected from six European cities. Inhal. Toxicol. 22, 402–416. [DOI] [PubMed] [Google Scholar]
- Havers N., Burba P., Lambert J., Klockow D. (1998). Spectroscopic characterization of humic-like substances in airborne particulate matter. J. Atmos. Chem. 29, 45–54. [Google Scholar]
- Hou L., Zhang X., Zheng Y., Wang S., Dou C., Guo L., Byun H. M., Motta V., McCracken J., Diaz A., et al. (2014). Altered methylation in tandem repeat element and elemental component levels in inhalable air particles. Environ. Mol. Mutagen. 55, 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISCN (1978). Report of the Standing Commitee on Human Cytogenetic Nomenclature. Cytogenet. Cell. Genet. 21, 309–404. [DOI] [PubMed] [Google Scholar]
- Jalava P. I., Hirvonen M. R., Sillanpaa M., Pennanen A. S., Happo M. S., Hillamo R., Cassee F. R., Gerlofs-Nijland M., Borm P. J., Schins R. P., et al. (2009). Associations of urban air particulate composition with inflammatory and cytotoxic responses in RAW 246.7 cell line. Inhal. Toxicol. 21, 994–1006. [DOI] [PubMed] [Google Scholar]
- Jalava P. I., Salonen R. O., Pennanen A. S., Happo M. S., Penttinen P., Halinen A. I., Sillanpaa M., Hillamo R., Hirvonen M. R. (2008). Effects of solubility of urban air fine and coarse particles on cytotoxic and inflammatory responses in RAW 264.7 macrophage cell line. Toxicol. Appl. Pharmacol. 229, 146–160. [DOI] [PubMed] [Google Scholar]
- Jalava P. I., Salonen R. O., Pennanen A. S., Sillanpaa M., Halinen A. I., Happo M. S., Hillamo R., Brunekreef B., Katsouyanni K., Sunyer J., et al. (2007). Heterogeneities in inflammatory and cytotoxic responses of RAW 264.7 macrophage cell line to urban air coarse, fine, and ultrafine particles from six European sampling campaigns. Inhal. Toxicol. 19, 213–225. [DOI] [PubMed] [Google Scholar]
- Janssen N. A., Yang A., Strak M., Steenhof M., Hellack B., Gerlofs-Nijland M. E., Kuhlbusch T., Kelly F., Harrison R., Brunekreef B., et al. (2014). Oxidative potential of particulate matter collected at sites with different source characteristics. Sci. Total Environ. 472, 572–581. [DOI] [PubMed] [Google Scholar]
- Jiang R., Jones M. J., Sava F., Kobor M. S., Carlsten C. (2014). Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part. Fibre Toxicol. 11, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I., Scherhag A., Sorrentino J., Sexton K., Bodnar W., Tryndyak V., Latendresse J. R., Swenberg J. A., Beland F. A., Pogribny I. P., et al. (2011). Epigenetic alterations in liver of C57BL/6J mice after short-term inhalational exposure to 1,3-butadiene. Environ. Health Perspect. 119, 635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll A., Gietl J. K., Wiesmüller G. A., Günsel A., Wohlleben W., Schnekenburger J., Klemm O. (2013). In vitro toxicology of ambient particulate matter: Correlation of cellular effects with particle size and components. Environ. Toxicol. 28, 76–86. [DOI] [PubMed] [Google Scholar]
- Lee R., Nasonova E., Ritter S. (2005). Chromosome aberration yields and apoptosis in human lymphocytes irradiated with Fe-ions of differing LET. Adv. Space Res. 35, 268–75. [DOI] [PubMed] [Google Scholar]
- Loxham M., Morgan-Walsh R. J., Cooper M. J., Blume C., Swindle E. J., Dennison P. W., Howarth P. H., Cassee F. R., Teagle D. A., Palmer M. R., et al. (2015). The effects on bronchial epithelial mucociliary cultures of coarse, fine, and ultrafine particulate matter from an underground railway station. Toxicol. Sci. 145, 98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Miousse I. R., Pirela S. V., Melnyk S., Koturbash I., Demokritou P. (2015). Short-term exposure to engineered nanomaterials affects cellular epigenome. Nanotoxicology. DOI:10.3109/17435390.2015.1025115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J., Baccarelli A., Mittleman M. A., Wright R. O., Sparrow D., Vokonas P. S., Tarantini L., Schwartz J. (2011). Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ. Health Perspect. 119, 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marple V. A., Rubow K. L., Turner W., Spengler J. D. (1987). Low flow rate sharp cut impactors for indoor air sampling: Design and calibration. JAPCA. 37, 1303–1307. [DOI] [PubMed] [Google Scholar]
- Martens J. H., O'Sullivan R. J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T. (2005). The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael S., Montag M., Dott W. (2013). Pro-inflammatory effects and oxidative stress in lung macrophages and epithelial cells induced by ambient particulate matter. Environ. Pollut. 183, 19–29. [DOI] [PubMed] [Google Scholar]
- Miousse I. R., Chalbot M. C., Aykin-Burns N., Wang X., Basnakian A., Kavouras I. G., Koturbash I. (2014a). Epigenetic alterations induced by ambient particulate matter in mouse macrophages. Environ. Mol. Mutagen. 55, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse I. R., Chalbot M. C., Lumen A., Ferguson A., Kavouras I. G., Koturbash I. (2015). Response of transposable elements to environmental stressors. Mutat. Res. Rev. Mutat. Res. 335, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse I. R., Shao L., Chang J., Feng W., Wang Y., Allen A. R., Turner J., Stewart B., Raber J., Zhou D., et al. (2014b). Exposure to low-dose (56)Fe-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat. Res. 182, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A., Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Papait R., Pistore C., Negri D., Pecoraro D., Cantarini L., Bonapace I. M. (2007). Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol. Biol. Cell 18, 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R., Pawar S. A., Fu Q., Gupta P. K., Berbee M., Garg S., Sridharan V., Wang W., Biju P. G., Krager K. J., et al. (2014). Characterization of transgenic Gfrp knock-in mice: Implications for tetrahydrobiopterin in modulation of normal tissue radiation responses. Antioxid. Redox Signal. 20, 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge W. F., Hildemann L. M., Mazurek M. A., Cass G. R., Simoneit B. R. T. (1993a). Sources of fine organic aerosol. 3. Road dust, tire debris, and organometallic brake lining dust: Roads as sources and sinks. Environ. Sci. Technol. 27, 1892–1904. [Google Scholar]
- Rogge W. F., Hildemann L. M., Mazurek M. A., Cass G. R., Simoneit B. R. T. (1993b). Sources of fine organic aerosol. 4. Particulate abrasion products from leaf surfaces of urban plants. Environ. Sci. Technol. 27, 2700–2711. [Google Scholar]
- Rogge W. F., Hildemann L. M., Mazurek M. A., Cass G. R., Simoneit B. R. T. (1998). Sources of fine organic aerosol. 9. Pine, oak, and synthetic log combustion in residential fireplaces. Environ. Sci. Technol. 32, 13–22. [Google Scholar]
- Rückerl R., Schneider A., Breitner S., Cyrys J., Peters A. (2011). Health effects of particulate air pollution: A review of epidemiological evidence. Inhal. Toxicol. 23, 555–592. [DOI] [PubMed] [Google Scholar]
- Rudin C. M., Thompson C. B. (2001). Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 30, 64–71. [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shi T., Duffin R., Borm P. J. A., Li H., Weishaupt C., Schins R. P. F. (2006). Hydroxyl-radical-dependent DNA damage by ambient particulate matter from contrasting sampling locations. Environ. Res. 101, 18–24. [DOI] [PubMed] [Google Scholar]
- Steenhof M., Gosens I., Strak M., Godri K. J., Hoek G., Cassee F. R., Mudway I. S., Kelly F. J., Harrison R. M., Lebret E., et al. (2011). In vitro toxicity of particulate matter (PM) collected at different sites in the Netherlands is associated with PM composition, size fraction and oxidative potential–the RAPTES project. Part. Fibre Toxicol. 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting D. T., Lipson D., Paul S., Brannigan B. W., Akhavanfard S., Coffman E. J., Contino G., Deshpande V., Iafrate A. J., Letovsky S., et al. (2011). Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H., Takarabe T., Kanai Y., Fukutomi T., Hirohashi S. (2002). Correlation of DNA hypomethylation at pericentromeric heterochromatin regions of chromosomes 16 and 1 with histological features and chromosomal abnormalities of human breast carcinomas. Am. J. Pathol. 161, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic D. (2005). Functional elements residing within satellite DNAs. EMBO Rep. 6, 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo D., Hullmann M., Schins R. P. (2012). Toxicology of ambient particulate matter. EXS. 101, 165–217. [DOI] [PubMed] [Google Scholar]
- Van Vaeck L., Van Cauwenberghe K. A. (1985). Characteristic parameters of particle size distributions of primary organic constituents of ambient aerosols. Environ. Sci. Technol. 19, 707–716. [DOI] [PubMed] [Google Scholar]
- Wu E. L., Engström O., Jo S., Stuhlsatz D., Yeom M. S., Klauda J. B., Im W. (2013). Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J, 105, 1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. L., Bestor T. H., Bourc'his D., Hsieh C. L., Tommerup N., Bugge M., Hulten M., Qu X., Russo J. J., Viegas-Pequignot E. (1999). Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.