Abstract
Nanoparticle-mediated gene and siRNA delivery has been an appealing area to gene therapists when they attempt to treat the diseases by manipulating the genetic information in the target cells. However, the advances in materials science could not keep up with the demand for multifunctional nanomaterials to achieve desired delivery efficiency. Researchers have thus taken an alternative approach to incorporate various materials into single composite nanoparticle using different fabrication methods. This approach allows nanoparticles to possess defined nanostructures as well as multiple functionalities to overcome the critical extracellular and intracellular barriers to successful gene delivery. This chapter will highlight the advances of fabrication methods that have the most potential to translate nanoparticles from bench to bedside. Furthermore, a major class of composite nanoparticle–lipid-based composite nanoparticles will be classified based on the components and reviewed in details.
1. NANOMEDICINE AND GENE THERAPY
Nanomedicine generally refers to the medical application of nanotechnology. It is an interdisciplinary field that exploits the distinguishing features of nanomaterials to fulfill the demanding needs of future research and clinical purposes. Due to the fact that the nanomaterials fall in the same size range as biological molecules and vesicles, researchers are seeking to integrate nanomaterials with biology to develop novel diagnostic devices, contrast agents, analytic tools, and drug delivery carriers.
Nanoparticle-based drug delivery systems are gradually shifting the paradigm of the traditional pharmaceutical industry through targeted delivery and releasing therapeutics to specific cells in order to minimize undesired adverse effects. Moreover, some highly potent drugs with low bioavailability due to pharmaceutically unfavorable physical or chemical properties can now be formulated into nanoparticles, manifesting their real therapeutic efficacy. One such example is nucleic acid-based therapeutics, which encompass a large class of highly potent drugs.
The development of recombinant DNA technology has provided a tool to manipulate DNA and RNA sequences at will. This has led to the emergence of Gene Therapy, a promising technology that treats inherited or acquired diseases by introducing exogenous genetic information into specific cells of the patients (Mulligan, 1993). Later, the discovery of RNA interference expanded the field of gene therapy by administrating regulatory RNA molecules, specifically and effectively silencing the targeted gene expression (Fire et al., 1998). Both strategies are appealing to researchers due to the simplicity of the drug development. As long as the therapeutic targets are identified, a new drug can be instantly generated with high specificity and potency based on the genetic code. This cost-effective drug development strategy circumvents the high-throughput screening process for the development of chemically based drugs, which is costly and time consuming.
As promising as it looks, the clinical translation of gene therapy has been successful only in limited indications using viral vectors due to the physicochemical properties of nucleic acid drugs, such as vulnerability to nuclease degradation, high molecular weight, and anionic charge. All of these significantly reduce the bioavailability of the drugs after systemic administration. Therefore, advances in gene therapy demand the development of carriers to deliver the therapeutics to the target cells with high efficiency. Among all the viral or nonviral approaches, the development of nanoparticle-mediated gene delivery has been put on center stage due to the progress of materials science.
2. COMPOSITE NANOPARTICLES
The emergence of novel nanomaterials with outstanding physicochemical properties and biological performances has fueled the application of nanotechnology in gene delivery. The carriers fabricated with these materials have to go through a variety of physiological conditions, such as pHs, ionic and osmotic strengths after systemic administration. Meanwhile, the carriers need to keep their integrity during blood circulation and respond to particular stimuli for intracellular cargo release. However, it is difficult for single component-based nanoparticles to satisfy the complicated needs to achieve a sophisticated controlled-release platform for gene delivery.
Instead of developing a single novel material, it may be advantageous to fabricate carriers with multiple materials that are equipped with diverse functionalities. These classes of nanoparticles are referred to as composite nanoparticles. The fabrication of composite nanoparticles is an area of materials science, which has gained an increasing attention due to its scientific and technological importance. The most important step in developing these nanoparticles is the preparation of tailored composite nanostructures. To achieve structurally defined composite nanoparticles, virtually all physicochemical properties of the novel materials have been exploited to set up reproducible and well-controlled fabrication protocols, each one with its specific advantages and shortcomings.
In this chapter, we will review the three well-established and popular fabrication methods and analyze their pros and cons. Later, we will review the most extensively studied lipid-based core–shell-structured composite nanoparticles reported in recent years and categorize these nanoparticles based on the materials used. The specific features of the nanostructures as well as the properties of the materials will be discussed in light of their contributions to gene delivery.
3. FABRICATION METHODS OF COMPOSITE NANOPARTICLES
Composite nanoparticles are fabricated with various methods and usually with multiple steps due to the complicated composition of the nanoparticles. The synthesis methods of inorganic nanoparticles such as gold nanoparticles (Bao, Mitragotri, & Tong, 2013), magnetic nanoparticles (Cohen & Shoushan, 2013), quantum dots (Probst, Zrazhevskiy, Bagalkot, & Gao, 2013), and silica nanoparticles (Fine et al., 2013), as well as polymeric nanoparticles (Feng et al., 2013) for drug delivery have been extensively reviewed. The composite nanoparticles, however, require further steps to incorporate other materials into the nanosystem. The fabrication of such nanoparticles primarily involves bulk mixing, which takes advantage of the physicochemical properties of the nanomaterials to achieve a defined nanostructure via self-assembly mechanism. As is recognized by the field of nanomedicine, bulk mixing often results in nanoparticles with large polydispersity as well as batch-to-batch variation. These issues represent critical challenges to clinical translation of the nanoparticles. To date, the applications of customized mixing devices and lithography technology to microfabrication have shown great potentials in solving the issues. Therefore, we will review the advances of the technologies in the fabrication of composite nanoparticles.
3.1 Self-assembly: Microfluidic Mixing
One of the major hurdles to the clinical translation of nanomedicine is the difficulty in reproducing batches of nanoparticles with identical properties in large-scale manufacturing for clinical use (Murday, Siegel, Stein, & Wright, 2009). Microfluidics is an interdisciplinary technology, which incorporates engineering, physics, chemistry, nanotechnology, and biotechnology, with extensive applications to systems in which small volumes of fluids are handled (10−9–10−18 l). Microfluidics has expanded from chemical separations and its original semiconductor technology to the processing of ultralow sample volumes as well as accessing biological length scales. This expansion is also supported by the development of soft lithography, which allows rapid prototyping of microfluidic devices (McDonald et al., 2000), as well as the development of a simple method for fabricating pneumatically activated valves, mixers, and pumps (Thorsen, Maerkl, & Quake, 2002). These innovations significantly shorten the time needed to fabricate prototype devices for testing new ideas (Whitesides, 2006). In the drug delivery field, the applications of microfluidics are focused on the synthesis of nanoparticles. So far, this technology is anticipated to be the very solution to the reproducibility and large-scale issues for clinical evaluation (Valencia, Farokhzad, Karnik, & Langer, 2012).
Amphiphilic molecules such as lipids and copolymers will self-assemble into aggregates when the polarity of the solvent changes. The conventional way to cause this solvent change is to mix the molecularly favorable solvent with an unfavorable solvent. This forces the molecules to form nanoparticles. The mixing timescale (τmix) is usually a few seconds, which is longer than the characteristic 10–100 ms timescale (τagg) for chains to aggregate (Valencia et al., 2012). The long mixing time causes the aggregates to be exposed to the heterogeneous solvent environment, preventing the effective stabilization of the nanoparticles by the hydrophilic portion of the molecules. This will lead to further aggregation of the molecules and result in larger, polydisperse nanoparticles. Microfluidics technology shortens the τmix from milliseconds to microseconds (Johnson & Prud’homme, 2003; Karnik et al., 2008) by mixing the two solvents in an ultrasmall volume. When τmix < τagg, the molecular aggregates are exposed to the homogenous solvent and the hydrophilic portion will stabilize the nanoparticle more effectively. In this way, smaller and more homogenous nanoparticles are produced.
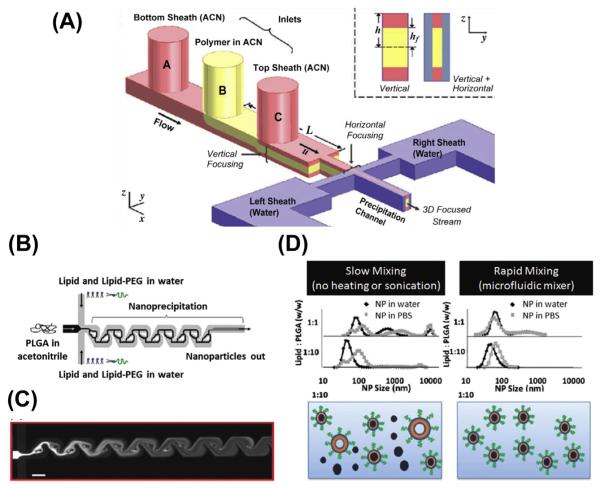
Compared with macroscale mixing, which is achieved by a turbulent flow, the microfluidic system does not generate turbulence due to the hydrodynamic stability. In order to overcome this issue, microfluidic devices are designed to dramatically increase the effect of diffusion and advection by exploiting the small length of the system (Capretto, Cheng, Hill, & Zhang, 2011). Microfluidic mixing devices are generally classified as either passive or active mixing according to the design principle. Active micromixers use external energy input such as a pressure field, acoustics, or temperature to introduce perturbations, which result in efficient mixing. However, these devices require the integration of an energy source referred to as an actuator and is inconvenient to researchers working on chemical and biological applications. On the other hand, passive mixing relies entirely on pumping energy. The devices restructure the flow via channel design to maximize the contact surface area between flows. These devices are relatively inexpensive, convenient, and popular in nanoparticle engineering. Liu et al. (2010) have reported a digital droplet generator for the fabrication of nanoparticles with multiple building blocks including 1-adamantanamine (Ad)–polyamidoamine dendrimer conjugate, Ad-PEG (polyethylene glycol) conjugate, β-cyclodextrin–polyethylenimine (PEI) conjugate, and Ad-Arg-Gly-Asp (RGD)-PEG conjugate. The building blocks are introduced into the mixing device in a sequential pattern with digitally controlled processing parameters on a single chip. They have demonstrated successful fabrication of uniform, RGD-functionalized nanoparticles with defined sizes ranging from 30 to 350 nm. In addition, they are also capable of adjusting the density of targeting ligands on the nanoparticles (0–10% based on feeding) and correlating this with the cellular uptake efficiency (Liu et al., 2010). Rhee et al. have demonstrated preparation of (polylactic-co-glycolic acid) PLGA-PEG nanoparticles using a simplified 3D hydrodynamic focusing technique in microfluidic channel design (Figure 5.1) (Rhee et al., 2011). This device is composed of a monolithic single layer with three sequential vertical inlets followed by horizontal focusing. This method avoids the clogging of channels due to the aggregation of high molecular weight polymers in the channel walls. Valencia et al. (2010) have demonstrated the fabrication of a core–shell-structured nanoparticle composed of PLGA in the core, lipid in the core–shell interface, and PEG on the surface (Figure 5.1 B, C and D). Their microfluidic device has passive mixing Tesla structures built into the mixing channels to facilitate the formation of nanoprecipitation. They have shown that a single, rapid mixing of PLGA in acetonitrile and lipid/lipid-PEG micelles in water using hydrodynamic flow focusing can fabricate homogenous nanoparticles with narrow size distribution. Similarly, they were able to control the physicochemical properties of the nanoparticles such as zeta potential, size, and surface functionalization. It is also noteworthy that the nanoparticles fabricated in one-step mixing showed no difference from the nanoparticles formed by mixing lipid with preformed PLGA cores. The explanation by the author was that the formation of the PLGA core was not affected by the presence of lipids. It is unclear if this one-step mixing could be applied to core–shell-structured nanoparticles made of other materials (Valencia et al., 2010).
Figure 5.1.
(A) Schematic illustration of 3D hydrodynamic focusing composed of three inlets for vertical focusing and separate inlet for side sheath flows. (Reprinted with permission from Rhee et al. (2011).) (B) Schematic illustration of the microfluidic device with three inlets that allows the formation of lipid-coated polymeric nanoparticles in the microchannels with Tesla structures. (C) Solvent mixing in the Tesla channels using fluorescent dye and water respectively. The mixing is complete at the fourth turn in the channel. (D) Comparison of slow and rapid mixing of lipid and PLGA solutions. Aggregation forms under slow mixing conditions without the input of any energy, but not under rapid mixing conditions. (See the color plate.) (Reprinted with permission from Valencia et al. (2010).)
Although nanoparticles prepared with microfluidic devices are often in micro- to milligram scale, the stackability and reproducibility of this preparation allows gram to kilogram scale manufacturing of nanoparticles for clinical evaluation. This undoubtedly is the most feasible approach to commercialize self-assembled nanomedicine to comply with good manufacturing practices.
3.2 Self-assembly: Layer-by-Layer
Layer-by-layer (LbL) self-assembly is a method that can be used to construct nanoparticles with multilayer structures. This method involves alternative and repetitive adsorption of materials with opposite charges on the surface of the core materials (Deshmukh et al., 2013). In 2001, Qiu et al. applied the LbL method to the fabrication of ibuprofen microparticles. Biocompatible polyelectrolytes such as chitosan, dextran sulfate, carboxymethyl cellulose, and alginate were used as coating materials to fabricate polyelectrolyte microcapsules with a shell as thick as 20–60 nm. The capsule thickness affects the release rate of the drugs. This represented the first LbL-based self-assembly system in drug delivery (Qiu, Leporatti, Donath, & Mohwald, 2001). Electrostatic interaction between oppositely charged polyelectrolytes is considered to be the major stabilizing force. However, hydrogen bonding, hydrophobic interactions, and van der Waals forces contribute to LbL formation as well (de Villiers, Otto, Strydom, & Lvov, 2011). The coating stability, morphology, thickness, drug depositions, and permeation of the film are primarily affected by these forces (Hammond, 1999; Lvov, Ariga, Onda, Ichinose, & Kunitake, 1999).
LbL-based multilayers offer several distinct advantages compared to other fabrication methods. (1) The thickness of the walls can be tailored to control the particle size; (2) The selection of polyelectrolytes for coating can be engineered to control the stability of the nanoparticle; (3) the location and sequence of the layers can be controlled to manipulate the drug release kinetics.
3.3 Imprint Lithography: PRINT (Particle Replication in Nonwetting Templates) Technology
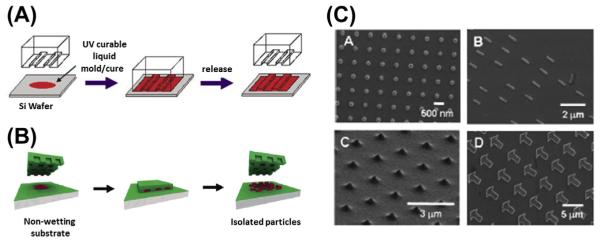
The imprint lithography-based method is considered to be a promising technique for scalable preparation of colloidal particles with specific shape and size (Merkel et al., 2010). This technology involves the use of a rigid template for casting an elastomeric mold, which will be used to replicate the shape of the original template (Qin, Xia, & Whitesides, 2010). It features high resolution, high fidelity, and low cost for large-scale manufacturing of particles. PRINT is a top-down fabrication method that is capable of producing uniform, micro- and nanoparticles with absolute control over size, shape, and composition. This versatile technology can be applied to fabricate particles with a variety of chemical structures. In 2004, DeSimone et al. reported the successfully generation of nanoparticles using photocurable perfluoropolyether (PFPE)-based materials with high-resolution imprint lithography (Figure 5.2) (Rolland, Hagberg, Denison, Carter, & De Simone, 2004). They have developed a chemically robust and durable PFPE-based mold that is also solvent resistant. More importantly, PFPE solved the swelling (Lee, Park, & Whitesides, 2003) and surface energy issues of polydimethylsiloxane-based soft lithography and allowed the fabrication of nanomaterials with high fidelity and quality (Rolland et al., 2004). Another impressive feature of PRINT compared with traditional imprint lithography is that PFPE-based molds are nonwetting to both inorganic and organic materials (Rolland, Van Dam, Schorzman, Quake, & DeSimone, 2004). This unique feature allows the production of isolated nanoparticles instead of embossed films. With this PFPE-based mold, they were able to fabricate monodispersed particles with a variety of materials including PEG, poly-(d-lactic acid), poly-(pyrrole), and triacrylate resin. Also, they were able to incorporate fragile biological molecules such as DNA or protein into these particles without sacrificing their activity (Rolland et al., 2005). Later, Kelly et al. applied this technology to protein-based materials. They molded insulin,albumin,and albumin mixtures with siRNA or paclitaxel using PRINT technology and generated uniform nano- and microparticles that have potential applications in drug delivery (Kelly & DeSimone, 2008).
Figure 5.2.
(A) Schematic illustration of the imprint lithography process. (Reprinted with permission from Rolland et al. (2004).) (B) Schematic illustration of the PRINT process. In PRINT, the nonwetting feature allows the generation of isolated particles. (C) Manipulation of PRINT nanoparticles with different shape and size. (See the color plate.) (Reprinted with permission from Rolland et al. (2005).)
The unique feature of imprint lithography-based fabrication compared to self-assembled nanoparticles is the ability to control the shape of the particles. Therefore a variety of shapes including cylinders, spheres, prolate ellipsoids, and toroidal particles can be fabricated using this technology. As potential drug delivery carriers, the cellular uptake efficiency is usually affected by size, surface charge, targeting ligands, and shape. The first three parameters can be manipulated by adjusting the input of building blocks during fabrication. However, shape can only be controlled by using a mold. Therefore, imprint lithography offers a useful tool to study the correlation between the particle shape and cellular uptake efficiency. It was demonstrated that HeLa cells can readily internalize both cubic and cylindrical particles with dimensions as large as 3 μm. However, the cylindrical particles were more efficiently taken up. A higher aspect ratio also contributes to the uptake rate (Gratton et al., 2008). Therefore, the shape of the particle is taken into consideration for rational design of drug delivery carriers.
4. COMPOSITE NANOPARTICLES FOR TARGETED GENE DELIVERY
The pursuit of successful delivery of targeted nucleic acid-based therapeutics will never cease. Numerous talented researchers are committed to the engineering and development of ideal nanoparticles to achieve this goal. With their efforts,various sophisticated nanoparticle platforms have been devised that have demonstrated high delivery efficiency and specificity. Material-oriented innovation has been the driving force of the field. However, researchers have started to realize that it is highly unlikely that one material is capable of dealing with all the barriers present in gene delivery. Failure to overcome anyone of the barriers will significantly compromise the delivery efficiency. In order to resolve this problem, distinct functional materials are chosen and incorporated into one nanoparticle to overcome these barriers. This modular-based design approach has turned out to be very effective in engineering nanoparticles for efficient gene delivery. There are a large number of publications on these composite nanoparticles and their sophisticated design to overcome the critical barriers to gene delivery. The barriers to gene delivery are: (1) The adsorption of serum protein compromises the colloidal stability of nanoparticles after systemic administration; (2) the reticuloendothelial system takes up the nanoparticles in the blood circulation; (3) the cytoplasmic membrane prevents the nanoparticles from entering the cells; (4) the nanoparticles are trapped in the endo/lysosome compartment; (5) the nuclear envelope blocks the DNA from entering the nucleus for transcription. We will review the recent advances of composite nanoparticles in this section.
4.1 Lipid-Coated Composite Nanoparticles
Lipids have been extensively used in the fabrication of composite nanoparticles because they can be easily formulated into other nanoformulations through electrostatic or hydrophobic interaction. Lipid coating allows the nanoparticles to acquire liposomal characteristics. The surface of the lipid-coated nanoparticles can be easily manipulated with simple and well-established protocols for liposome modification. For example, PEGylation of nanoparticles can be achieved by postinsertion of lipid-PEG conjugates. Targeting ligands or other functional motifs can be incorporated in the same way. These nanoparticles are PEGylated with lipid-PEG conjugates by simultaneous or postinsertion, giving the nanoparticles a stealth property after systemic administration. The cores of these lipid-coated composite nanoparticles are diverse and can be classified into several categories, which include self-assembled cores such as polyplexes or polymeric nanoparticles, as well as inorganic cores such as gold, silica nanoparticle, or calcium phosphate nanoparticles (details Haynes et al., this book).
4.1.1 Polyplex-Based Cores
Polyplexes refer to nanosized complexes between negatively charged polyanions such as nucleic acids and positively charged polycations including synthetic polymers (polyethyleneimine, polylysine), polypeptides (protamine, spermidine, or histone), and natural polymers (chitosan). These nanoparticles are widely used as gene delivery carriers for siRNA and DNA in vitro. However, due to serum instability and nonspecific uptake by reticuloendothelial system (RES) after systemic administration, their applications in vivo are limited. One of the most exploited approaches to address this issue is coating the polyplexes with lipids. A lipid bilayer spontaneously forms on the surface of the polyplexes when mixing the polyplexes with preformed liposomes or rehydrating the dry lipid membrane with the polyplex solution. This approach takes advantage of the low toxicity and immunogenicity of liposomes along with their ability to be PEGylated for extended circulation.
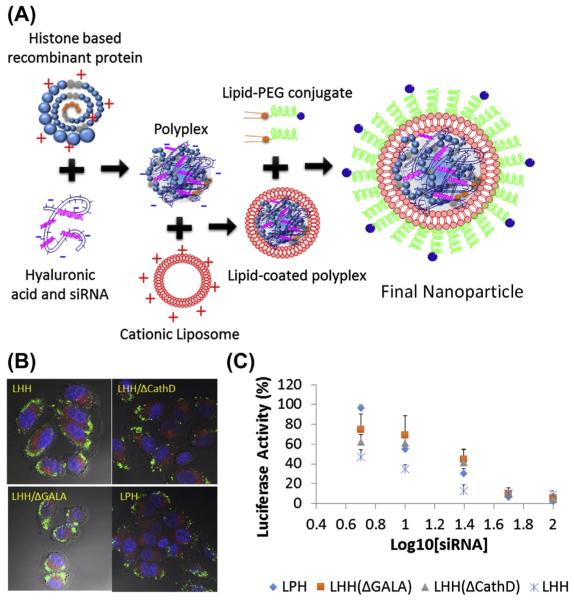
Most representative examples of these core-membrane nanoparticles have been developed in Dr Leaf Huang’s lab. As early as the mid-1990s, Li et al. formulated these core-membrane nanoparticles using poly(l-lysine) and protamine (Li & Huang, 1997) to condense pDNA into negatively charged polyplexes. The cores were then coated with a preformed cationic liposome containing 3β-[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol (DC-Chol) and dioleoyl-phosphatidylethanolamine (DOPE) for lipid coating. These core-membrane-structured composite nanoparticles were named lipid–protamine–DNA (LPD) nanoparticles. Compared with lipoplexes formed by cationic liposomes and pDNA, the size of LPD nanoparticles dramatically decreased and effectively protected DNA from nuclease degradation (Gao & Huang, 1996). This lipid coating enhanced the transfection efficiency of the lipid nanoparticles by up to 28-fold in vitro compared with lipoplexes (Gao & Huang, 1996). LPD was also able to transfect the lungs in vivo with minor modifications; possibly due to the excessively positive charge carried by the nanoparticles (Li & Huang, 1997). Later, Li et al. optimized LPD nanoparticles by incubating the lipid-coated LPD with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)] (DSPE-PEG) micelles to PEGylate the nanoparticle. PEGylation reduced the serum protein adsorption and increased the colloidal stability of the nanoparticles. Therefore, less nanoparticles were taken up by the RES system and a higher tumor accumulation was achieved due to the enhanced permeation and retention effect (Li, Chen, Hackett, & Huang, 2008; Li, Chono, & Huang, 2008). This PEGylated LPD substantially increased in vivo siRNA delivery efficiency by fourfold and the gene silencing effect by two- to threefold (Li & Huang, 2006). It is worth noting that the negatively charged protamine/calf thymus DNA/siRNA core played a critical role in supporting the lipid bilayer, allowing more surfactant-like DSPE-PEG conjugates to be inserted on the membrane without detaching the lipid membrane (Li & Huang, 2009). This high density of sheddable PEG significantly improved the PK of LPD nanoparticles in the blood stream (Li & Huang, 2010). An outstanding issue that remained for LPD nanoparticles was the high binding affinity between nucleic acid and protamine, which was so strong that cytoplasmic release of the cargos became insufficient. This could compromise delivery efficiency as well. To address this issue,Wang et al. engineered a histone-derived fusion protein to replace protamine for nucleic acid condensation. The fusion protein is composed of four repeats of histone H2 peptide derived from N-terminal of histone H2 protein as the condensing element. The tandem repeats were linked with the enzyme-responsive degradation element, cathepsin D cleavage substrate. The exposure of the fusion protein to the corresponding enzyme in the endosome led to the degradation of the fusion protein. This degradation mechanism engineered in the condensation agent facilitated the cargos release and efficiently increased the in vivo target gene knock down efficiency by twofold (Wang, Zhang, Guo, Hatefi, & Huang, 2013) (Figure 5.3).
Figure 5.3.
(A) Schematic illustration of fabrication of polyplex core-based composite nanoparticle for siRNA delivery using histone-based recombinant protein as nucleic acid condensing agent. (B) Intracellular release profiles of oligonucleotides using the different histone-based recombinant proteins and mutants. Lack of degradation element (LHH/ΔCathD) prohibited the dissociation of oligonucleotides and the punctate forms were observed. (C) IC50 of luciferase knockdown with antiluciferase siRNA delivered by different nanoparticles. (See the color plate.) Reprinted with permission from Wang et al. (2013).
A similar approach has been employed by Harashima et al., who have developed a series of lipid-based core-membrane-structured nanoparticles, namely multifunctional envelope-type nanodevice (MEND) (Akita et al., 2009;El-Sayed,Masuda,Khalil,Akita,& Harashima,2009;Hatakeyama,Akita, & Harashima, 2011; Hatakeyama, Akita, Ito, et al., 2011; Hatakeyama et al., 2007, 2009; Ishitsuka, Akita, & Harashima, 2011; Khalil, Hayashi, Mizuno, & Harashima, 2011; Khalil et al., 2007; Kogure et al., 2004; Kuramoto et al., 2008; Masuda et al., 2008; Moriguchi et al., 2005; Mudhakir, Akita,Tan, & Harashima, 2008; Nakamura, Kogure, Futaki, & Harashima, 2007; Nakamura, Kogure, Yamada, Futaki, & Harashima, 2006; Sasaki et al., 2005; Shaheen et al., 2011). However, the lipid-coating procedure for MEND nanoparticles is slightly different from LPD nanoparticles. Instead of using preformed liposomes, the polyplex solution is used to rehydrate a lipid film, which had been dried down from an organic solvent solution. The hydration initiates the formation of the liposome, which subsequently coats the polyplex condensates. In those formulations, stearyl-octaarginine and cholesteryl-GALA (WEAALAEALAEALAEHLAEALAEALEALAA) are incorporated into the membrane for efficient cellular uptake and endosomal escape as a pH responsive fusogenic peptide (Hatakeyama et al., 2009; Khalil et al., 2007, 2011; Nakamura et al., 2006, 2007). The final nanoparticle is sonicated for self-assembly and homogeneity. MEND has been employed to deliver genes to the liver (Khalil et al., 2011) or the lung (Ishitsuka et al., 2011) with surface modifications of octaarginine or IRQ peptide (IRQRRRR), respectively. The incorporation of octaarginine and IRQ peptide facilitate the internalization of MEND nanoparticles through macropinocytosis or caveolar endocytosis, respectively.
4.1.2 Mesoporous Silica Nanoparticle Cores
Mesoporous silica nanoparticles (MSNs) are one of the most well-studied inorganic nanoparticles for the delivery of drugs and contrast agents. These nanoparticles possess some unique characteristics such as high drug-loading capacity due to large surface area and high pore volume as well as tunable pore and particle size. The surfaces of MSNs are also subject to a variety of modifications. In order to deliver nucleic acid agents, the silica surface of the MSNs is converted to carry positive charges for the binding of DNA or siRNA. This is realized by grafting an amine group on the surface (Kneuer et al., 2000) or coating the MSNs with polycations such as PEI (Meng et al., 2010; Shen et al., 2014) and polyamidoamine (Radu et al., 2004). Due to the relatively small pore sizes of MSNs, nucleic acid therapeutics such as DNA and siRNA are usually immobilized on the external surface of the MSNs. Recent advances have engineered MSNs with large pores, which make encapsulation of genes possible. Min et al. synthesized 250 nm monodispersed MSNs with large cationic pores (>15 nm) to load plasmid DNA (Kim et al., 2011). This strategy held DNA inside and provided better protection against nuclease degradation. Qiao et al. fabricated 100–200 nm MSNs that contain 28 nm cage-like pores and a large entry size of 13.4 nm. This is large enough to encapsulate siRNA with a polylysine surface functionalization (Hartono et al., 2012).
The Brinker lab is a pioneer for the engineering of MSN-based composite nanoparticles, referred to as “protocells” (Liu, Stace-Naughton, Jiang, & Brinker, 2009). The protocells are fabricated by fusing liposomes on MSNs and simultaneously loading and sealing the cargos inside the particles with the lipid membrane. Compared to the traditional liposomes, protocells have demonstrated higher drug-loading capacity and stability (Ashley et al., 2011). They are able to encapsulate a variety of cargos including chemodrugs, proteins, quantum dots, and siRNAs (Ashley et al., 2011). Ashley et al. exploited the emulsion processing technique (Carroll, Pylypenko,Atanassov, & Petsev, 2009) to prepare 165 nm MSNs with ultralarge pores (23–30 nm) (Ashley et al., 2012). These MSNs were characterized by a Brunauer–Emmett–Teller surface area of 850 m2/g and a pore volume fraction of 65% with 3–13 interconnecting surface accessible pores. Surface modification of MSNs with 3-[2-(2-aminoethylamino)ethylamino]propyltrimethyoxysilane resulted in a siRNA-loading capacity of about 1 nmol per 1010 particles. Cationic 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP)- or zwitterionic 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)-based lipid coating did not cause much of a difference in siRNA loading, although both types of lipid-coated MSNs showed 10–100 times higher loading than the corresponding lipid nanoparticles. These results confirmed that high surface area cores confer a higher intrinsic loading capacity. To facilitate the targeted cellular uptake and endosomal escape of the cargos, the protocells were surface modified with targeting peptide and lipidated endosomolytic peptide (H5WYG) using the postinsertion method. In vitro studies have shown that the delivery of siRNA using lipid-coated MSNs is able to silence the expression of the target protein by 90% in 72 h (Ashley et al., 2012). In the same group, Dengler et al. reported the delivery of pDNA using a similar platform. The MSNs in this study were prepared by the surfactant template aerosol-assisted self-assembly method (Liu et al., 2009) and surface modified with ATPES to carry a positive charge. These 230 nm MSNs are characterized by a surface area of 935 m2/g and a small pore size of 2–5 nm. During the pDNA loading procedure, the positively charged MSNs adsorbed the pDNA to the external surface of the nanoparticles and was then followed by lipid coating. Due to the extralarge size of the pDNA, the cargo was not meant to be loaded inside the MSNs. Hypothetically, the pDNA were trapped between the nanoparticle surface and lipid layer, although this was not confirmed experimentally. Without a targeting ligand, DOTAP coating of the MSNs resulted in a much higher transgene expression level (Interleukin 10) than DOPC-coated MSNs in HEK cells. Presumably, this is due to enhanced cellular uptake and endosomal escape caused by the cationic lipid. Although the composite MSNs particles have shown capacity for siRNA and pDNA delivery, the loading mechanism is better characterized for siRNA. In contrast, the loading of pDNA primarily depends on surface adsorption to the positively charged nanoparticles, which provides negative charges for the cationic liposomes to bind and fuse. However, whether the cationic liposomes have a chance to compete and rip off the pDNA from the surface of the MSNs, is not well investigated. With advances in MSN fabrication technology, the ultralarge pore size could be engineered to accommodate the loading of pDNA inside the particles.
4.1.3 Polymeric Nanoparticle-Based Core
Biodegradable and biocompatible polymers are a dominant class of nanomaterials for drug delivery. This can be seen through an increasing number of publications and clinical trials. Polymeric nanoparticles (PNPs) exhibit high structural integrity, stability, and controlled release. They are fabricated with distinct synthetic polymers or modified natural polymers through a variety of preparation methods (Hadinoto, Sundaresan, & Cheow, 2013). Therefore rational design of the building block polymers allows fine-tuning of the size, charge, shape, and functionality of the nanoparticles (Panyam & Labhasetwar, 2003). For the aforementioned reasons, lipid-coated polymeric nanoparticles have been developed in an effort to mitigate the shortcomings of PNPs such as in vivo instability, nonspecific toxicity, and early drug release. The lipid-coated PNPs are engineered to load chemodrugs, nucleic acid therapeutics, proteins and peptides by entrapment, adsorption or covalent conjugation. The nanoparticles have a well-defined core–shell nanostructure, which is composed of three parts: (1) the polymeric core where the drugs are loaded; (2) the lipid membrane, which confers the liposomal characteristics to the core and prevents the leakage of the drugs from the core; and (3) the PEG layer, which can also be functionalized for targeting, membrane penetration, or endosomal escape. The most commonly studied polymers are PLGA and polycarprolactone due to their low cost of synthesis and easy functionalization.
In order to load nucleic acids into PLGA nanoparticles, a polycation-based condensing agent is usually used to condense and increase the hydrophobicity of the cargos so that they can be loaded into the hydrophobic core. Zhong et al. (2010) have characterized three methods to encapsulate pDNA in a lipid-PLGA-based composite nanoparticle formulation prepared with a conventional double emulsion method (W/O/W). These pDNA loading methods were designated as: (1) “OUT,” pDNA is adsorbed at the surface of the nanoparticles; (2) “IN,” pDNA is encapsulated in the PLGA core of the nanoparticles; (3) “BOTH,” pDNA is adsorbed and encapsulated in the nanoparticles. This was realized by adding protamine-condensed pDNA before and/or after the formation of PLGA core. In vitro release and transfection studies using luciferase reporter gene in human embryonic kidney cells indicated that the “OUT” loading method tended to cause a burst effect of transgene expression with shorter expression longevity.“IN” and “BOTH” loading methods could generate a sustained expression profile, which indicated a slow release of the cargo due to the hydrolysis of PLGA (Zhong et al., 2010).
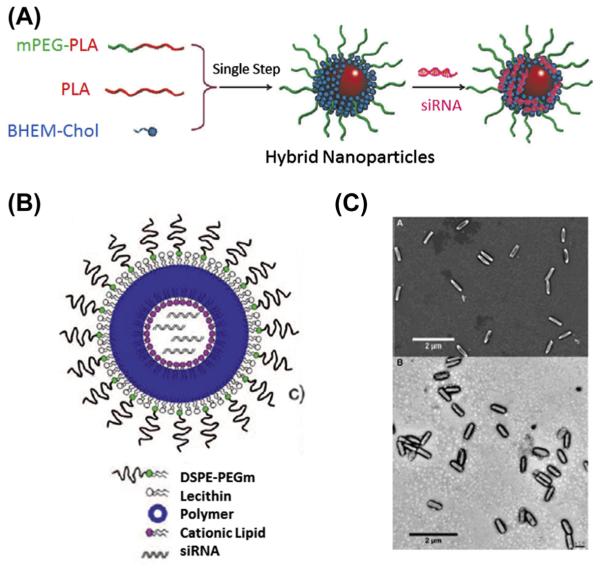
Numerous formulations have been developed for the delivery of siRNA using lipid/polymer composite nanoparticles (Desai et al., 2013; Gao et al., 2014; Shi, Xiao,Votruba,Vilos, & Farokhzad, 2011;Yang et al., 2012) (Figure 5.4). Desai et al. (2013) exploited the OUT loading method to codeliver anti-TNFα siRNA and antinociception agent Capsaicin with lipid-coated PLGA nanoparticles prepared through the one-step double emulsion method. Capsaicin was encapsulated into the core and siRNA was adsorbed on the surface of cationic lipid membrane. The topical administration of the nanoparticles significantly reduced inflammatory cytokines, such as TNF-α, NK-κB, and IL-17 and effectively treated chronic skin inflammation on imiquimod-induced psoriatic plague-like model (Desai et al., 2013). Shi et al. (2011) have reported hollow-structured lipid-PLGA composite nanoparticles that encapsulated siRNA in the cavity of the nanoparticle. The nanoparticle was formulated by adding a siRNA solution to an organic solvent in the presence of ethylphosphocholine (EPC) and PLGA. The amphiphilic EPC stabilized the water-in-oil microemulsion with the hydrophilic head in the aqueous phase and hydrophobic chain in the PLGA phase. This allows the effective encapsulation of siRNA in the aqueous phase. In a second emulsion and subsequent solvent evaporation, DSPE-PEG coated on the surface of PLGA and formed an outer lipid-PEG layer. The lipid bilayer was separated by ester-terminated PLGA and the hollow structure densely loaded siRNA for delivery (Shi et al., 2011).
Figure 5.4.
(A) Schematic illustration of fabrication of lipid-coated Polylactic acid (PLA) nanoparticles with siRNA adsorbed on the surface of carrier. (Reprinted with permission from Yang et al. (2012).) (B) Schematic representation of lipid–polymer nanoparticle composed of outer lipid-PEG, a middle polymer layer, and an inner cationic lipid hollow core for siRNA entrapment. (Reprinted with permission from Shi et al. (2011).) (C) SEM (top) and TEM (bottom) images of lipid-coated PRINT nanoparticles composed of PLGA and siRNA. (See the color plate.) (Reprinted with permission from Hasan et al. (2012).)
Gao et al. reported a composite nanoparticle platform composed of a cholesterol-grafted poly(amidoamine) (rPAA-Chol) core and a lipid coating (Gao et al., 2014). The amphiphilic polymer self-assembled into a nanoparticle in aqueous solution using the thin-film hydration method, followed by the addition of siRNA molecules. The anionic core was then coated with DOTAP-based cationic liposomes for lipid coating. The lipid–polymer nanoparticles were then subject to PEGylation by postinsertion of a lipid-PEG conjugate. An endosomolytic peptide, T7 (HAIYPRH), or transferrin, was conjugated to the distal end of the PEG chain for either enhanced endosomal escape or targeting purposes. The lipid-rPAA-Chol composite nanoparticles were relatively small in size (100 nm) compared to lipid-PLGA nanoparticles and showed a significant tumor inhibition effect when delivering anti-EGFR siRNA to an MCF-7 bearing tumor model (Gao et al., 2014).
Recently, Hasan et al. reported the fabrication of lipid-PLGA nanoparticles using PRINT technology to generate a PLGA/siRNA core with high siRNA loading (Hasan et al., 2012). The siRNA and PLGA (85:15 lactic/glycolic acid) were prepared in Dimethyl sulfoxide/Dimethylformamide/water and molded using PRINT technology. The 80 × 320 nm monodispersed needle-shaped core was fabricated for better cellular uptake. The core was then subject to cationic lipid coating and PEGylation. These particles have been tested in multiple cell lines and target genes knockdown has been demonstrated. This study was a hallmark for the development of composite nanoparticle-based siRNA delivery. The nanoparticles showed great translational potential due to their high reproducibility from batch to batch and a relatively easy scale-up production for clinical evaluation (Hasan et al., 2012).
4.1.4 Gold Nanoparticle-Based Cores
The gold nanoparticles (AuNPs) draw much attention as their applications in bimolecular sensing, computed tomography imaging, as well as photothermal therapy. Over the past decades,AuNPs have also been investigated as efficient nucleic acid delivery systems (Ding et al., 2014) due to their biocompatibility, versatility, and facile surface modification through gold–thiol linkages.
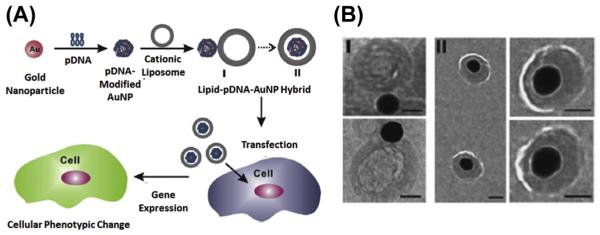
Chad Mirkin and his colleagues have been dedicated to the investigation of such nucleic acid-gold nanostructures as well as their biomedical applications (Barnaby, Lee, & Mirkin, 2014; Choi, Hao, Narayan, Auyeung, & Mirkin, 2013; Giljohann, Seferos, Prigodich, Patel, & Mirkin, 2009; Jensen et al., 2013). In their studies, citrate-stabilized AuNPs were mixed with thiolated siRNA, where siRNAs duplexes were allowed to chemisorb via thio–gold bonds (Giljohann et al., 2009). The dense surface functionalization of nucleic acids increases the stability of the bound cargos, while maintaining the biological activity of the siRNAs. However, negatively charged, nucleic acid-adsorbed AuNPs showed compromised cellular uptake due to the weak interactions between the cellular membrane and nanoparticles. To address this issue, Rhim, Kim, & Nam (2008) coated nucleic acid-adsorbed AuNPs with cationic liposomes to enhance the cellular uptake and protect cargos from nuclease-mediated degradation (Figure 5.5). In this study, they used AuNPs of various sizes (15, 30, 50, and 80 nm) to load pDNA for lipid coating. Two types of lipid–gold nanoparticles were observed under cryo-TEM observation. Type I hybrids were believed to be the intermediate status, which showed the adsorption of AuNPs to the surface of liposomes. Type II hybrids showed that AuNPs were encapsulated in the liposomes, and the diameters of the lipid membrane were much larger than those of AuNPs. The cryo-TEM images suggested that the lipid membrane was not supported by the AuNP core. Although the author did not give the explanation for the structural discrepancy between lipid-AuNPs and other lipid-coated nanoparticles, it could be speculated that the small AuNPs with high curvature made it difficult for the lipid to form bilayer on them. The in vitro transfection studies showed that lipid-coated AuNPs resulted in significantly higher transfection efficiency compared with liposome-mediated or naked AuNPs-mediated DNA delivery due to the high DNA loading capacity and higher cellular uptake. Recently, Kong et al. reported a distinct method for the fabrication of lipid-coated AuNP for siRNA delivery using 5 nm AuNPs. The AuNPs were dissolved in chloroform in the presence of DC-Chol, DOPE, and cholesterol. The system was then emulsified followed by the evaporation of the solvent. Final particles contained cationic lipid membrane with cluster of gold nanoparticle cores. siRNAs were adsorbed on the surface of such nanoparticles via electrostatic interaction (Kong, Bae, Jo, Kim, & Park, 2012). Recently, Alhasan et al. have introduced an approach to take advantage of cells for the fabrication of lipid–gold composite nanoparticle (Alhasan, Patel, Choi, & Mirkin, 2014). In their study, it was discovered that when endocytosed into PC-3 prostate cancer cells, a small percent of the nucleic acid-adsorbed gold nanoparticles could be naturally sorted into exosomes. These AuNPs-containing exosomes could be isolated and enriched for in vivo miRNA delivery. Although the coating and loading efficiency for the nanoparticles fabricated with this method was not efficient, the approach represented engineered nanoparticle of biogenesis, which is a trend regarding the fabrication of the nanoparticles.
Figure 5.5.
(A) Schematic illustration of lipid-coated AuNP for the delivery of plasmid DNA. (B) Cryo-TEM images of two types of lipid-coated AuNPs loaded with pDNA. Scale bars: 50 nm. Reprinted with permission from Rhim et al. (2008).
4.1.5 Magnetic Nanoparticle-Based Core
Magnetic field has been exploited as an external energy to enhance the transfection efficiency of nonviral-mediated gene delivery (Duan et al., 2014; Hao et al., 2010; Miao et al., 2014; Scherer et al., 2002). For efficient in vivo delivery of siRNA,Namiki et al. have prepared lipid-coated magnetic nanocrystals termed “LipoMag.” LipoMags were fabricated by dispersing oleic acid-coated magnetite nanocrystal cores with cationic lipid in chloroform and removing solvent afterward. Final nanoparticles were generated by spontaneously coating of monolayer cationic lipid on top of the nanocrystal cores via hydrophobic interactions. The siRNAs were then adsorbed on the surface of the LipoMag through electrostatic interaction. Under a magnetic field, heavy accumulation of siRNA delivered by LipoMag was observed in the tumor lesions compared with other organs, which resulted in target gene silencing (Namiki et al., 2009). Recently, Jiang, Eltoukhy, Love, Langer, and Anderson (2013) reported a simple method to coat iron oxide nanoparticles with lipid or lipid-like materials and form uniform nanoparticles. In their method, the solvent N-methyl-2-pyrrolidone (NMP) was added to the chloroform where oleic acid-coated iron oxide nanoparticles and lipids were dispersed. NMP serves as adhesive between the lipids and nanoparticle surface, which could be removed by dialyzing nanoparticles against water since it is miscible with both chloroform and water. Introduction of NMP promoted the full arrangement and assembly of outer leaflet lipid into a more complete layer and prevented the aggregation from happening. The nanoparticles generated were highly homogenous and showed high efficiency in delivering pDNA and siRNA in vitro (Jiang et al., 2013).
5. CONCLUSION
The chapter highlights core–shell-structured composite nanoparticles for gene therapy with a focus on lipid-coated systems. The fusion of lipid with other nanomaterials renders colloidal stability as well as other liposome-like properties to the composite nanoparticles, which makes them suitable carriers for in vitro and in vivo delivery. In addition, the well-defined nanostructure allows modular design of multifunctional nanoparticle, with each component overcoming designated extracellular or intracellular barriers to efficient gene delivery in a spatiotemporal manner. The fabrication of composite nanoparticles lowers the demand for the development of omnipotent nanomaterials for nonviral gene delivery and accelerates the advances in clinical translation of the nanomedicine. Moreover, some inorganic components such as gold or magnetic could be used for thermal activation and as contrast agent for noninvasive diagnosis of the cancers as well as monitoring of drug distribution. Therefore, lipid-based composite nanoparticle clearly affords new options for gene therapy and hold great potential for clinical translation.
ACKNOWLEDGMENTS
This work was supported by NIH grants CA149363, CA151652, DK100664. We appreciate Mr Andrew Mackenzie Blair for manuscript editing.
REFERENCES
- Akita H, Kudo A, Minoura A, Yamaguti M, Khalil IA, Moriguchi R, et al. Multi-layered nanoparticles for penetrating the endosome and nuclear membrane via a step-wise membrane fusion process. Biomaterials. 2009;30(15):2940–2949. doi: 10.1016/j.biomaterials.2009.02.009. http://dx.doi.org/10.1016/j.biomaterials.2009.02.009.pii:S0142-9612(09)00157-4. [DOI] [PubMed] [Google Scholar]
- Alhasan AH, Patel PC, Choi CH, Mirkin CA. Exosome encased spherical nucleic acid gold nanoparticle conjugates as potent microRNA regulation agents. Small. 2014;10(1):186–192. doi: 10.1002/smll.201302143. http://dx.doi.org/10.1002/smll.201302143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CE, Carnes EC, Epler KE, Padilla DP, Phillips GK, Castillo RE, et al. Delivery of small interfering RNA by peptide-targeted mesoporous silica nanoparticle-supported lipid bilayers. American Chemical Society Nano. 2012;6(3):2174–2188. doi: 10.1021/nn204102q. http://dx.doi.org/10.1021/nn204102q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, et al. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nature Materials. 2011;10(5):389–397. doi: 10.1038/nmat2992. http://dx.doi.org/10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao G, Mitragotri S, Tong S. Multifunctional nanoparticles for drug delivery and molecular imaging. Annual Review of Biomedical Engineering. 2013;15:253–282. doi: 10.1146/annurev-bioeng-071812-152409. http://dx.doi.org/10.1146/annurev-bioeng-071812-152409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnaby SN, Lee A, Mirkin CA. Probing the inherent stability of siRNA immobilized on nanoparticle constructs. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(27):9739–9744. doi: 10.1073/pnas.1409431111. http://dx.doi.org/10.1073/pnas.1409431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capretto L, Cheng W, Hill M, Zhang X. Micromixing within microfluidic devices. Topics in Current Chemistry. 2011;304:27–68. doi: 10.1007/128_2011_150. http://dx.doi.org/10.1007/128_2011_150. [DOI] [PubMed] [Google Scholar]
- Carroll NJ, Pylypenko S, Atanassov PB, Petsev DN. Microparticles with bimodal nanoporosity derived by microemulsion templating. Langmuir. 2009;25(23):13540–13544. doi: 10.1021/la900988j. http://dx.doi.org/10.1021/la900988j. [DOI] [PubMed] [Google Scholar]
- Choi CH, Hao L, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(19):7625–7630. doi: 10.1073/pnas.1305804110. http://dx.doi.org/10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Shoushan SY. Magnetic nanoparticles-based diagnostics and theranostics. Current Opinion in Biotechnology. 2013;24(4):672–681. doi: 10.1016/j.copbio.2013.01.006. http://dx.doi.org/10.1016/j.copbio.2013.01.006. [DOI] [PubMed] [Google Scholar]
- de Villiers MM, Otto DP, Strydom SJ, Lvov YM. Introduction to nano-coatings produced by layer-by-layer (LbL) self-assembly. Advanced Drug Delivery Reviews. 2011;63(9):701–715. doi: 10.1016/j.addr.2011.05.011. http://dx.doi.org/10.1016/j.addr.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Desai PR, Marepally S, Patel AR, Voshavar C, Chaudhuri A, Singh M. Topical delivery of anti-TNFalpha siRNA and capsaicin via novel lipid-polymer hybrid nanoparticles efficiently inhibits skin inflammation in vivo. Journal of Controlled Release. 2013;170(1):51–63. doi: 10.1016/j.jconrel.2013.04.021. http://dx.doi.org/10.1016/j.jconrel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh PK, Ramani KP, Singh SS, Tekade AR, Chatap VK, Patil GB, et al. Stimuli-sensitive layer-by-layer (LbL) self-assembly systems: targeting and biosensory applications. Journal of Controlled Release. 2013;166(3):294–306. doi: 10.1016/j.jconrel.2012.12.033. http://dx.doi.org/10.1016/j.jconrel.2012.12.033. [DOI] [PubMed] [Google Scholar]
- Ding Y, Jiang Z, Saha K, Kim CS, Kim ST, Landis RF, et al. Gold nanoparticles for nucleic acid delivery. MolecularTherapy. 2014;22(6):1075–1083. doi: 10.1038/mt.2014.30. http://dx.doi.org/10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Dong J, Zhang T, Su Z, Ding J, Zhang Y, et al. Polyethyleneimine-functionalized iron oxide nanoparticles for systemic siRNA delivery in experimental arthritis. Nanomedicine (London) 2014;9(6):789–801. doi: 10.2217/nnm.13.217. http://dx.doi.org/10.2217/nnm.13.217. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, Masuda T, Khalil I, Akita H, Harashima H. Enhanced gene expression by a novel stearylated INF7 peptide derivative through fusion independent endosomal escape. Journal of Controlled Release. 2009;138(2):160–167. doi: 10.1016/j.jconrel.2009.05.018. http://dx.doi.org/10.1016/j.jconrel.2009.05.018.pii:S0168-3659(09)00354-X. [DOI] [PubMed] [Google Scholar]
- Feng L, Zhu C, Yuan H, Liu L, Lv F, Wang S. Conjugated polymer nanoparticles: preparation, properties, functionalization and biological applications. Chemical Society Reviews. 2013;42(16):6620–6633. doi: 10.1039/c3cs60036j. http://dx.doi.org/10.1039/c3cs60036j. [DOI] [PubMed] [Google Scholar]
- Fine D, Grattoni A, Goodall R, Bansal SS, Chiappini C, Hosali S, et al. Silicon micro- and nanofabrication for medicine. Advanced Healthcare Materials. 2013;2(5):632–666. doi: 10.1002/adhm.201200214. http://dx.doi.org/10.1002/adhm.201200214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. http://dx.doi.org/10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gao X, Huang L. Potentiation of cationic liposome-mediated gene delivery by polycations. Biochemistry. 1996;35(3):1027–1036. doi: 10.1021/bi952436a. http://dx.doi.org/10.1021/bi952436a pii:bi952436a. [DOI] [PubMed] [Google Scholar]
- Gao LY, Liu XY, Chen CJ, Wang JC, Feng Q, Yu MZ, et al. Core-shell type lipid/rPAA-Chol polymer hybrid nanoparticles for in vivo siRNA delivery. Biomaterials. 2014;35(6):2066–2078. doi: 10.1016/j.biomaterials.2013.11.046. http://dx.doi.org/10.1016/j.biomaterials.2013.11.046. [DOI] [PubMed] [Google Scholar]
- Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. Journal of the American Chemical Society. 2009;131(6):2072–2073. doi: 10.1021/ja808719p. http://dx.doi.org/10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton SE, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, et al. The effect of particle design on cellular internalization pathways. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(33):11613–11618. doi: 10.1073/pnas.0801763105. http://dx.doi.org/10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadinoto K, Sundaresan A, Cheow WS. Lipid-polymer hybrid nanoparticles as a new generation therapeutic delivery platform: a review. European Journal of Pharmaceutics and Biopharmaceutics. 2013;85(3):427–443. doi: 10.1016/j.ejpb.2013.07.002. Pt A. http://dx.doi.org/10.1016/j.ejpb.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Hammond PT. Recent explorations in electrostatic multilayer thin film assembly. Current Opinion in Colloid & Interface Science. 1999;4(6):430–442. http://dx.doi.org/10.1016/S1359-0294(00)00022-4. [Google Scholar]
- Hao R, Xing R, Xu Z, Hou Y, Gao S, Sun S. Synthesis, functionalization, and biomedical applications of multifunctional magnetic nanoparticles. Advanced Materials. 2010;22(25):2729–2742. doi: 10.1002/adma.201000260. http://dx.doi.org/10.1002/adma.201000260. [DOI] [PubMed] [Google Scholar]
- Hartono SB, Gu W, Kleitz F, Liu J, He L, Middelberg AP, et al. Poly-llysine functionalized large pore cubic mesostructured silica nanoparticles as biocompatible carriers for gene delivery. American Chemical Society Nano. 2012;6(3):2104–2117. doi: 10.1021/nn2039643. http://dx.doi.org/10.1021/nn2039643. [DOI] [PubMed] [Google Scholar]
- Hasan W, Chu K, Gullapalli A, Dunn SS, Enlow EM, Luft JC, et al. Delivery of multiple siRNAs using lipid-coated PLGA nanoparticles for treatment of prostate cancer. Nano Letters. 2012;12(1):287–292. doi: 10.1021/nl2035354. http://dx.doi.org/10.1021/nl2035354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama H, Akita H, Harashima H. A multifunctional envelope type nano device (MEND) for gene delivery to tumours based on the EPR effect: a strategy for overcoming the PEG dilemma. Advanced Drug Delivery Reviews. 2011;63(3):152–160. doi: 10.1016/j.addr.2010.09.001. http://dx.doi.org/10.1016/j.addr.2010.09.001.pii:S0169-409X(10)00179-1. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Akita H, Ito E, Hayashi Y, Oishi M, Nagasaki Y, et al. Systemic delivery of siRNA to tumors using a lipid nanoparticle containing a tumor-specific cleavable PEG-lipid. Biomaterials. 2011;32(18):4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. http://dx.doi.org/10.1016/j.biomaterials.2011.02.045 pii:S0142-9612(11)00208-0. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, kita H, Kogure K, Oishi M, Nagasaki Y, Kihira Y, et al. Development of a novel systemic gene delivery system for cancer therapy with a tumor-specific cleavable PEG-lipid. Gene Therapy. 2007;14(1):68–77. doi: 10.1038/sj.gt.3302843. http://dx.doi.org/10.1038/sj.gt.3302843pii:3302843. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Ito E, Akita H, Oishi M, Nagasaki Y, Futaki S, et al. A pH-sensitive fusogenic peptide facilitates endosomal escape and greatly enhances the gene silencing of siRNA-containing nanoparticles in vitro and in vivo. Journal of Controlled Release. 2009;139(2):127–132. doi: 10.1016/j.jconrel.2009.06.008. http://dx.doi.org/10.1016/j.jconrel.2009.06.008. pii:S0168-3659(09)00418-0. [DOI] [PubMed] [Google Scholar]
- Ishitsuka T, Akita H, Harashima H. Functional improvement of an IRQ-PEG-MEND for delivering genes to the lung. Journal of Controlled Release. 2011;154(1):77–83. doi: 10.1016/j.jconrel.2011.05.012. http://dx.doi.org/10.1016/j.jconrel.2011.05.012pii:S0168-3659(11)00307-5. [DOI] [PubMed] [Google Scholar]
- Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Science Translational Medicine. 2013;5(209) doi: 10.1126/scitranslmed.3006839. http://dx.doi.org/10.1126/scitranslmed.3006839209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Eltoukhy AA, Love KT, Langer R, Anderson DG. Lipidoid-coated iron oxide nanoparticles for efficient DNA and siRNA delivery. Nano Letters. 2013;13(3):1059–1064. doi: 10.1021/nl304287a. http://dx.doi.org/10.1021/nl304287a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BK, Prud’homme RK. Mechanism for rapid self-assembly of block copolymer nanoparticles. Physical Review Letters. 2003;91(11):118302. doi: 10.1103/PhysRevLett.91.118302. [DOI] [PubMed] [Google Scholar]
- Karnik R, Gu F, Basto P, Cannizzaro C, Dean L, Kyei-Manu W, et al. Microfluidic platform for controlled synthesis of polymeric nanoparticles. Nano Letters. 2008;8(9):2906–2912. doi: 10.1021/nl801736q. http://dx.doi.org/10.1021/nl801736q. [DOI] [PubMed] [Google Scholar]
- Kelly JY, DeSimone JM. Shape-specific, monodisperse nano-molding of protein particles. Journal of American Chemical Society. 2008;130(16):5438–5439. doi: 10.1021/ja8014428. http://dx.doi.org/10.1021/ja8014428. [DOI] [PubMed] [Google Scholar]
- Khalil IA, Hayashi Y, Mizuno R, Harashima H. Octaarginine- and pH sensitive fusogenic peptide-modified nanoparticles for liver gene delivery. Journal of Controlled Release. 2011;156(3):374–380. doi: 10.1016/j.jconrel.2011.08.012. http://dx.doi.org/10.1016/j.jconrel.2011.08.012.pii:S0168-3659(11)00580-3. [DOI] [PubMed] [Google Scholar]
- Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M, et al. Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery. Gene Therapy. 2007;14(8):682–689. doi: 10.1038/sj.gt.3302910. http://dx.doi.org/10.1038/sj.gt.3302910.pii:3302910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Na HK, Kim YK, Ryoo SR, Cho HS, Lee KE, et al. Facile synthesis of monodispersed mesoporous silica nanoparticles with ultralarge pores and their application in gene delivery. American Chemical Society Nano. 2011;5(5):3568–3576. doi: 10.1021/nn103130q. http://dx.doi.org/10.1021/nn103130q. [DOI] [PubMed] [Google Scholar]
- Kneuer C, Sameti M, Bakowsky U, Schiestel T, Schirra H, Schmidt H, et al. A nonviral DNA delivery system based on surface modified silica-nanoparticles can efficiently transfect cells in vitro. Bioconjugate Chemistry. 2000;11(6):926–932. doi: 10.1021/bc0000637. [DOI] [PubMed] [Google Scholar]
- Kogure K, Moriguchi R, Sasaki K, Ueno M, Futaki S, Harashima H. Development of a non-viral multifunctional envelope-type nano device by a novel lipid film hydration method. Journal of Controlled Release. 2004;98(2):317–323. doi: 10.1016/j.jconrel.2004.04.024. http://dx.doi.org/10.1016/j.jconrel.2004.04.024.pii:S0168365904002159. [DOI] [PubMed] [Google Scholar]
- Kong WH, Bae KH, Jo SD, Kim JS, Park TG. Cationic lipid-coated gold nanoparticles as efficient and non-cytotoxic intracellular siRNA delivery vehicles. Pharmaceutical Research. 2012;29(2):362–374. doi: 10.1007/s11095-011-0554-y. http://dx.doi.org/10.1007/s11095-011-0554-y. [DOI] [PubMed] [Google Scholar]
- Kuramoto H, Park YS, Kaji N, Tokeshi M, Kogure K, Shinohara Y, et al. On-chip fabrication of mutifunctional envelope-type nanodevices for gene delivery. Analytical and Bioanalytical Chemistry. 2008;391(8):2729–2733. doi: 10.1007/s00216-008-2124-7. http://dx.doi.org/10.1007/s00216-008-2124-7. [DOI] [PubMed] [Google Scholar]
- Lee JN, Park C, Whitesides GM. Solvent compatibility of poly(dimethylsiloxane)-based microfluidic devices. Analytical Chemistry. 2003;75(23):6544–6554. doi: 10.1021/ac0346712. http://dx.doi.org/10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Molecular Therapy. 2008;16(1):163–169. doi: 10.1038/sj.mt.6300323. http://dx.doi.org/10.1038/sj.mt.6300323.pii:6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. Journal of Controlled Release. 2008;126(1):77–84. doi: 10.1016/j.jconrel.2007.11.002. http://dx.doi.org/10.1016/j.jconrel.2007.11.002.pii:S0168-3659(07)00623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Therapy. 1997;4(9):891–900. doi: 10.1038/sj.gt.3300482. http://dx.doi.org/10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- Li SD, Huang L. Surface-modified LPD nanoparticles for tumor targeting. Annals of New York Academy of Sciences. 2006;1082:1–8. doi: 10.1196/annals.1348.001. http://dx.doi.org/10.1196/annals.1348.001.pii:1082/1/1. [DOI] [PubMed] [Google Scholar]
- Li SD, Huang L. Nanoparticles evading the reticuloendothelial system: role of the supported bilayer. Biochimica et Biophysica Acta. 2009;1788(10):2259–2266. doi: 10.1016/j.bbamem.2009.06.022. http://dx.doi.org/10.1016/j.bbamem.2009.06.022.pii:S0005-2736(09)00222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SD, Huang L. Stealth nanoparticles: high density but sheddable PEG is a key for tumor targeting. Journal of Controlled Release. 2010;145(3):178–181. doi: 10.1016/j.jconrel.2010.03.016. http://dx.doi.org/10.1016/j.jconrel.2010.03.016.pii:S0168-3659(10)00221-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stace-Naughton A, Jiang X, Brinker CJ. Porous nanoparticle supported lipid bilayers (protocells) as delivery vehicles. Journal of the American Chemical Society. 2009;131(4):1354–1355. doi: 10.1021/ja808018y. http://dx.doi.org/10.1021/ja808018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Wang H, Chen KJ, Guo F, Lin WY, Chen YC, et al. A digital microfluidic droplet generator produces self-assembled supramolecular nanoparticles for targeted cell imaging. Nanotechnology. 2010;21(44):445603. doi: 10.1088/0957-4484/21/44/445603. http://dx.doi.org/10.1088/0957-4484/21/44/445603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lvov Y, Ariga K, Onda M, Ichinose I, Kunitake T. A careful examination of the adsorption step in the alternate layer-by-layer assembly of linear polyanion and polycation. Colloids and Surfaces A-Physicochemical Engineering Aspects. 1999;146(1–3):337–346. http://dx.doi.org/10.1016/S0927-7757(98)00789-4. [Google Scholar]
- Masuda T, Akita H, Nishio T, Niikura K, Kogure K, Ijiro K, et al. Development of lipid particles targeted via sugar-lipid conjugates as novel nuclear gene delivery system. Biomaterials. 2008;29(6):709–723. doi: 10.1016/j.biomaterials.2007.09.039. http://dx.doi.org/10.1016/j.biomaterials.2007.09.039.pii:S0142-9612(07)00785-5. [DOI] [PubMed] [Google Scholar]
- McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21(1):27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. http://dx.doi.org/10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Meng H, Liong M, Xia T, Li Z, Ji Z, Zink JI, et al. Engineered design of mesoporous silica nanoparticles to deliver doxorubicin and P-glycoprotein siRNA to overcome drug resistance in a cancer cell line. American Chemical Society Nano. 2010;4(8):4539–4550. doi: 10.1021/nn100690m. http://dx.doi.org/10.1021/nn100690m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel TJ, Herlihy KP, Nunes J, Orgel RM, Rolland JP, DeSimone JM. Scalable, shape-specific, top-down fabrication methods for the synthesis of engineered colloidal particles. Langmuir. 2010;26(16):13086–13096. doi: 10.1021/la903890h. http://dx.doi.org/10.1021/la903890h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Liu C, Ge J, Yang W, Liu J, Sun W, et al. Antitumor effect ofTRAIL on oral squamous cell carcinoma using magnetic nanoparticle-mediated gene expression. Cell Biochemistry and Biophysics. 2014;69(3):663–672. doi: 10.1007/s12013-014-9849-z. http://dx.doi.org/10.1007/s12013-014-9849-z. [DOI] [PubMed] [Google Scholar]
- Moriguchi R, Kogure K, Akita H, Futaki S, Miyagishi M, Taira K, et al. A multifunctional envelope-type nano device for novel gene delivery of siRNA plasmids. International Journal of Pharmaceutics. 2005;301(1–2):277–285. doi: 10.1016/j.ijpharm.2005.05.021. http://dx.doi.org/10.1016/j.ijpharm.2005.05.021.pii:S0378-5173(05)00328-5. [DOI] [PubMed] [Google Scholar]
- Mudhakir D, Akita H, Tan E, Harashima H. A novel IRQ ligand-modified nano-carrier targeted to a unique pathway of caveolar endocytic pathway. Journal of Controlled Release. 2008;125(2):164–173. doi: 10.1016/j.jconrel.2007.10.020. http://dx.doi.org/10.1016/j.jconrel.2007.10.020.pii:S0168-3659(07)00572-X. [DOI] [PubMed] [Google Scholar]
- Mulligan RC. The basic science of gene therapy. Science. 1993;260(5110):926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- Murday JS, Siegel RW, Stein J, Wright JF. Translational nanomedicine: status assessment and opportunities. Nanomedicine (London) 2009;5(3):251–273. doi: 10.1016/j.nano.2009.06.001. http://dx.doi.org/10.1016/j.nano.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kogure K, Futaki S, Harashima H. Octaarginine-modified multifunctional envelope-type nano device for siRNA. Journal of Controlled Release. 2007;119(3):360–367. doi: 10.1016/j.jconrel.2007.03.010. http://dx.doi.org/10.1016/j.jconrel.2007.03.010.pii:S0168-3659(07)00144-7. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kogure K, Yamada Y, Futaki S, Harashima H. Significant and prolonged antisense effect of a multifunctional envelope-type nano device encapsulating antisense oligodeoxynucleotide. Journal of Pharmacology and Pharmacotherapeutics. 2006;58(4):431–437. doi: 10.1211/jpp.58.4.0002. http://dx.doi.org/10.1211/jpp.58.4.0002. [DOI] [PubMed] [Google Scholar]
- Namiki Y, Namiki T, Yoshida H, Ishii Y, Tsubota A, Koido S, et al. A novel magnetic crystal-lipid nanostructure for magnetically guided in vivo gene delivery. Nature Nanotechnology. 2009;4(9):598–606. doi: 10.1038/nnano.2009.202. http://dx.doi.org/10.1038/nnano.2009.202. [DOI] [PubMed] [Google Scholar]
- Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Advanced Drug Delivery Reviews. 2003;55(3):329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- Probst CE, Zrazhevskiy P, Bagalkot V, Gao X. Quantum dots as a platform for nanoparticle drug delivery vehicle design. Advanced Drug Delivery Reviews. 2013;65(5):703–718. doi: 10.1016/j.addr.2012.09.036. http://dx.doi.org/10.1016/j.addr.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nature Protocols. 2010;5(3):491–502. doi: 10.1038/nprot.2009.234. http://dx.doi.org/10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- Qiu XP, Leporatti S, Donath E, Mohwald H. Studies on the drug release properties of polysaccharide multilayers encapsulated ibuprofen microparticles. Langmuir. 2001;17(17):5375–5380. http://dx.doi.org/10.1021/La010201w. [Google Scholar]
- Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VS. A poly-amidoamine dendrimer-capped mesoporous silica nanosphere-based gene transfection reagent. Journal of the American Chemical Society. 2004;126(41):13216–13217. doi: 10.1021/ja046275m. http://dx.doi.org/10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- Rhee M, Valencia PM, Rodriguez MI, Langer R, Farokhzad OC, Karnik R. Synthesis of size-tunable polymeric nanoparticles enabled by 3D hydrodynamic flow focusing in single-layer microchannels. Advanced Materials. 2011;23(12):H79–H83. doi: 10.1002/adma.201004333. http://dx.doi.org/10.1002/adma.201004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim WK, Kim JS, Nam JM. Lipid-gold-nanoparticle hybrid-based gene delivery. Small. 2008;4(10):1651–1655. doi: 10.1002/smll.200800628. http://dx.doi.org/10.1002/smll.200800628. [DOI] [PubMed] [Google Scholar]
- Rolland JP, Hagberg EC, Denison GM, Carter KR, De Simone JM. High-resolution soft lithography: enabling materials for nanotechnologies. Angewandte Chemie International Edition in English. 2004;43(43):5796–5799. doi: 10.1002/anie.200461122. http://dx.doi.org/10.1002/anie.200461122. [DOI] [PubMed] [Google Scholar]
- Rolland JP, Maynor BW, Euliss LE, Exner AE, Denison GM, DeSimone JM. Direct fabrication and harvesting of monodisperse, shape-specific nanobiomaterials. Journal of the American Chemical Society. 2005;127(28):10096–10100. doi: 10.1021/ja051977c. http://dx.doi.org/10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]
- Rolland JP, Van Dam RM, Schorzman DA, Quake SR, DeSimone JM. Solvent-resistant photocurable liquid fluoropolymers for microfluidic device fabrication (corrected) Journal of the American Chemical Society. 2004;126(8):2322–2323. doi: 10.1021/ja031657y. http://dx.doi.org/10.1021/ja031657y. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kogure K, Chaki S, Kihira Y, Ueno M, Harashima H. Construction of a multifunctional envelope-type nano device by a SUV*-fusion method. International Journal of Pharmaceutics. 2005;296(1–2):142–150. doi: 10.1016/j.ijpharm.2005.02.020. http://dx.doi.org/10.1016/j.ijpharm.2005.02.020.pii:S0378-5173(05)00152-3. [DOI] [PubMed] [Google Scholar]
- Scherer F, Anton M, Schillinger U, Henke J, Bergemann C, Kruger A, et al. Magnetofection: enhancing and targeting gene delivery by magnetic force in vitro and in vivo. Gene Therapy. 2002;9(2):102–109. doi: 10.1038/sj.gt.3301624. http://dx.doi.org/10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- Shaheen SM, Akita H, Nakamura T, Takayama S, Futaki S, Yamashita A, et al. KALA-modified multi-layered nanoparticles as gene carriers for MHC class-I mediated antigen presentation for a DNA vaccine. Biomaterials. 2011;32(26):6342–6350. doi: 10.1016/j.biomaterials.2011.05.014. http://dx.doi.org/10.1016/j.biomaterials.2011.05.014.pii:S0142-9612(11)00533-3. [DOI] [PubMed] [Google Scholar]
- Shen J, Kim HC, Su H, Wang F, Wolfram J, Kirui D, et al. Cyclodextrin and Polyethylenimine functionalized mesoporous silica nanoparticles for delivery of siRNA cancer therapeutics. Theranostics. 2014;4(5):487–497. doi: 10.7150/thno.8263. http://dx.doi.org/10.7150/thno.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Xiao Z, Votruba AR, Vilos C, Farokhzad OC. Differentially charged hollow core/shell lipid-polymer-lipid hybrid nanoparticles for small interfering RNA delivery. Angewandte Chemie International Edition in English. 2011;50(31):7027–7031. doi: 10.1002/anie.201101554. http://dx.doi.org/10.1002/anie.201101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298(5593):580–584. doi: 10.1126/science.1076996. http://dx.doi.org/10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- Valencia PM, Basto PA, Zhang L, Rhee M, Langer R, Farokhzad OC, et al. Single-step assembly of homogenous lipid-polymeric and lipid-quantum dot nanoparticles enabled by microfluidic rapid mixing. American Chemical Society Nano. 2010;4(3):1671–1679. doi: 10.1021/nn901433u. http://dx.doi.org/10.1021/nn901433u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia PM, Farokhzad OC, Karnik R, Langer R. Microfluidic technologies for accelerating the clinical translation of nanoparticles. Nature Nanotechnology. 2012;7(10):623–629. doi: 10.1038/nnano.2012.168. http://dx.doi.org/10.1038/nnano.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang L, Guo S, Hatefi A, Huang L. Incorporation of histone derived recombinant protein for enhanced disassembly of core-membrane structured liposomal nanoparticles for efficient siRNA delivery. Journal of Controlled Release. 2013;172(1):179–189. doi: 10.1016/j.jconrel.2013.08.015. http://dx.doi.org/10.1016/j.jconrel.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. http://dx.doi.org/10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Yang XZ, Dou S, Wang YC, Long HY, Xiong MH, Mao CQ, et al. Single-step assembly of cationic lipid-polymer hybrid nanoparticles for systemic delivery of siRNA. American Chemical Society Nano. 2012;6(6):4955–4965. doi: 10.1021/nn300500u. http://dx.doi.org/10.1021/nn300500u. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Chinta DM, Pamujula S, Wang H, Yao X, Mandal TK, et al. Optimization of DNA delivery by three classes of hybrid nanoparticle/DNA complexes. Journal of Nanobiotechnology. 2010;8:6. doi: 10.1186/1477-3155-8-6. http://dx.doi.org/10.1186/1477-3155-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]