Abstract
Peripheral macrophages and resident microglia constitute the dominant glioma-infiltrating cells. The tumor induces an immunosuppressive and tumor-supportive phenotype in these glioma-associated microglia/brain macrophages (GAMs). A subpopulation of glioma cells acts as glioma stem cells (GSCs). We explored the interaction between GSCs and GAMs. Using CD133 as a marker of stemness, we enriched for or deprived the mouse glioma cell line GL261 of GSCs by fluorescence-activated cell sorting (FACS). Over the same period of time, 100 CD133+ GSCs had the capacity to form a tumor of comparable size to the ones formed by 10,000 CD133- GL261 cells. In IL-6-/- mice, only tumors formed by CD133+ cells were smaller compared with wild type. After stimulation of primary cultured microglia with medium from CD133-enriched GL261 glioma cells, we observed an selective upregulation in microglial IL-6 secretion dependent on Toll-like receptor (TLR) 4. Our results show that GSCs, but not the bulk glioma cells, initiate microglial IL-6 secretion via TLR4 signaling and that IL-6 regulates glioma growth by supporting GSCs. Using human glioma tissue, we could confirm the finding that GAMs are the major source of IL-6 in the tumor context.
Keywords: Glioma, Glioma-associated microglia/brain macrophages, Glioma stem cells, IL-6, Toll-like receptor 4.
INTRODUCTION
Glioblastoma (GBM) is the most common and most malignant primary brain tumor in adults with high recurrence rates even after a complete resection. New GBM therapeutic strategies are desperately needed, requiring insights into the biological and molecular mechanisms controlling glioma growth. GBMs are complex tumors that display cellular heterogeneity within the tumor mass. Several studies suggest that GBMs contain a subpopulation with tumorigenic potential and stem cell characteristics (1–5). These glioma stem cells (GSCs) play key roles in the growth, invasion, angiogenesis, and immune evasion of glioma (6, 7). They have also been identified as the major cellular entity for conferring chemo- and radioresistance (3, 8, 9) and have thus emerged as a new therapeutic target. GSCs express neural stem cell markers such as nestin, SOX2, and Musashi-1 (2). In addition, CD133 is an enrichment marker for GSCs; however, several studies have demonstrated its limitations as a specific marker (10, 11). A functional feature of GSCs is their ability to form neurospheres in the culture medium containing B27 supplement and defined growth factors, such as epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF). To identify GSCs, we tested for their ability to form neurospheres in combination with cell sorting using CD133.
A perivascular niche has been proposed for GSCs that determines the characteristics of GSCs and controls the malignant behavior of tumor cells (12). However, there is only limited knowledge about the composition of the GSC niche. Inflammatory mediators and inflammatory cells are indispensable components of the neoplastic microenvironment (13). Glioma-associated microglia (GAMs), the brain-resident macrophages, together with blood-borne monocytes, are the predominant cells, contributing up to 30% of the total tumor mass (14), and their abundance is positively correlated with glioma malignancy (15). Tumor-secreted CXCL12 (stromal cell–derived factor-1, SDF-1) is a potent microglia- and macrophage-recruiting molecule, especially for attracting GAMs to hypoxic areas (16). Activation of SDF-1 and its receptor CXCR4 has been shown to promote macrophage mobilization and tumor revascularization (17). Despite their cytotoxic and phagocytic potential (18), these GAMs rather support tumor growth. Depletion of microglia reduced glioma invasion in organotypic brain slices (19) and also decreased glioma expansion in vivo (20). Microglia release many factors, including extracellular matrix proteases and cytokines, which directly or indirectly influence tumor migration and proliferation (21). We have previously shown that glioma-triggered microglial membrane type 1 matrix metalloprotease (MT1-MMP) and Matrix metallopeptidase 9 (MMP9) expression via Toll-like receptor (TLR) 2 is one of the mechanisms for microglia-induced tumor expansion (20, 22, 23). TLRs are the main members of the pattern recognition receptor (PRR) family that are necessary for the induction of an innate immune response to damage-associated molecular patterns (DAMPs) through the activation and maturation of macrophages and dendritic cells (24). Microglia have been reported to be the predominant TLR-expressing cell type in the normal CNS (25), as well as in the glioma tissue (23). Microglia/brain macrophages freshly isolated from human glioma tissue also express substantial levels of TLR2, TLR3, and especially TLR4 (26). Interleukin (IL)-6 is a cytokine secreted after TLR activation, and its expression has been shown to correlate with glioma invasiveness (27). It also plays a major role in the response to injury or infection and is involved in the immune response, inflammation, and hematopoiesis (28). Glioma cells have been reported to secrete IL-6 (29, 30) and express IL-6 receptors (27). Glioma-derived IL-6, working together with other tumor-secreted factors such as transforming growth factor β (TGF-β) and prostaglandin E2 (PGE2), polarize glioma-infiltrating microglia toward an anti-inflammatory phenotype (21), and microglia-derived IL-6 has been reported to induce glioma cell migration and invasiveness (30).
In this study, we have investigated the link between microglial IL-6 production, TLR expression, and the potential of tumor stem cells to form glioma by utilizing 2 glioma murine models and human samples.
MATERIALS AND METHODS
Animals
All experiments were carried out using C57BL/6J WT mice (Charles River Laboratories, Sulzfeld, Germany) or TLRs 2, 4, 7, 9, and MyD88-/- on a C57BL/6J background. The TLR knockout mice were generated by Dr Shizuo Akira and colleagues from the Osaka University, Japan, and obtained from Oriental BioServices Inc., Kyoto, Japan (22). The generation of IL-6 knockout mice (IL-6-/-) has been described in detail previously (31). Briefly, IL-6–deficient mice had been backcrossed on a C57BL/6J background for >10 generations (32). To broaden the relevance of our findings, we employed another murine model in which the tumor is initiated by the overexpression of platelet-derived growth factor subunit B (PDGFb) in Nestin-expressing cells in vivo: Ntv-a/Ink4a-Arf-/- mice develop high-grade gliomas, which reflect features of the proneural subtype in human GBMs (33–35), 6–8 weeks following intracranial injection of RCAS-PDGFb–producing DF-1 chicken fibroblast cells at 4.5–10 weeks of age (36, 37). The mice were bred and maintained in the animal housing facilities of the Max Delbrück Center for Molecular Medicine and Charité University Hospital (Berlin, Germany) as per rules of the local governmental institutions (LaGeSo, G 0268/10, G 0343/10, G 0438/12). The mice were housed with a 12 h/12 h light-dark cycle and received food and water ad libitum.
Human Materials
All human glioma materials in this study were obtained from the Department of Neurosurgery at Charité University Hospital according to the rules of the Ethical Committee (Charité, EA4/098/11).
Cell Culture
The murine GL261 glioma cell line (which is isogenic to C57BL/6J mice; National Cancer Institute, Frederick, MD), rat glioma cell line C6 (American Type Culture Collection, ATCC, Teddington, UK) and human glioma cell line U87 (ATCC) were grown in DMEM with 10% FCS, 200 mM glutamine, 100 U/mL penicillin, and 100 ng/mL streptomycin (Invitrogen, Darmstadt, Germany). Enhanced green fluorescent protein (EGFP) GL261 cells were generated as previously described (22). NCH421K is a primary human glioblastoma cell line, which is highly enriched in CD133+ GSCs (38). NCH421K cells (CLS Cell Lines Service GmbH, Eppelheim, Germany) were grown in stem cell medium consisting of Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12; Invitrogen, Carlsbad, CA) containing supplement (2% B27; Invitrogen), growth factors (20 ng/mL EGF and bFGF; from PeproTech, Hamburg, Germany), and additives (100 U/mL penicillin and 100 mg/mL streptomycin; both from Invitrogen). Neonatal microglial cells were prepared from WT, MyD88-/-, and TLRs 2, 4, 7, and 9-/- mice according to previously established protocols (22). Microglia were also cultured from adult mice (P49–P56) as previously described in detail (39). Briefly, cortical and midbrain tissue was freed of blood vessels and meninges in Hank’s balanced salt solution (HBSS), mechanically dissociated into 1-mm3 pieces and trypsinized in 1% trypsin and 0.05% deoxyribonuclease for 5 minutes at room temperature, as described for neonatal microglia. Digested tissue was dissociated using a fire-polished pipette and washed twice in HBSS. Cells were then plated on a confluent monolayer of P0 astrocytes in 75-cm2 flasks. The feeder layer of astrocytes was depleted of neonatal microglial cells using clodronate (200 mg/mL) before the adult microglia were added. The adult mixed glial cultures received fresh complete DMEM medium every other day and were treated with 33% L929-conditioned medium after 7 days or once cells became confluent. Microglia were shaken off 1 week later and were used for experiments within 1 day of plating. For analysis of the inflammatory response in vitro, microglia were plated at 3 x 104 cells/96 wells. All cells were maintained in a 37 °C incubator with a 5% CO2 humidified atmosphere. The TLR ligands lipopolysaccharide (LPS) and Poly I:C were obtained from InvivoGen (San Diego, CA).
Isolation and Cell Culture of Glioma Stem Cells
Mouse glioma stem cells were isolated from the GL261 cell line as previously described (40). In summary, bulk cultures of GL261 cells were grown in stem cell medium for at least 4–6 weeks. To broaden the relevance of our findings, we employed the RCAS-PDGFb murine tumor model. RCAS-PDGFb tumors were excised from tumor brains using a scalpel, minced, and incubated with Accutase (eBioscience, San Diego, CA) for 15 minutes at 37 °C. Tissue pieces were mechanically dissociated using a 1-mL pipette and washed in DMEM (Sigma-Aldrich, St Louis, MO). Cells were passed through a 70-μm cell strainer and seeded into a T25 cell-culture flask. Cells were grown in stem cell medium. Glioma stem cells were isolated by fluorescence-activated cell sorting (FACS) using phycoerythrin (PE)-conjugated anti-mouse CD133 antibody (Miltenyi Biotec, Bergisch Gladbach, Germany) and allophycocyanin (APC)-conjugated anti-mouse CD133 (eBioscience) or by magnetic activated cell sorting (MACS) using murine CD133 magnetic beads according to the manufacturer’s instructions (Miltenyi Biotec). CD133 expression level was analyzed by FACS, and the percentage of CD133 cells in the CD133+ population was more than 90%, while in the CD133- population, it was less than 1%, as previously described (40) (Supplementary Data Figures 1 and 2).
Preparation of Glioma-Conditioned Medium
Neurosphere/adhesive cultured GL261 cells or CD133+/CD133- FACS sorted GL261 and RCAS cells were seeded at a density of 0.5 x 106 cells in Greiner CELLSTAR cell culture dishes (diameter × H 100 mm × 20 mm; Sigma-Aldrich, Munich, Germany). The stem cell culture medium was left on the cells for 16–18 hours after seeding before being harvested. The conditioned medium was collected, briefly centrifuged to remove cell debris, filtered using a 0.2-µm filter (Sartorius Stedim Biotech GmbH, Göttingen, Germany), and used for all further experiments. Protein concentrations were measured in the conditioned media using Pierce BCA protein assay kit (Thermo Scientific, Rockford, IL). The protein concentration in the conditioned medium was equal for all conditions (Supplementary Data Figure 3).
Multiple Analyte Detection and ELISA
Multiple analyte detection of cytokines and chemokines in supernatants was performed using FlowCytomix (Bender MedSytems, Burlingame, CA). The immunoassay is a bead-based method to detect the concentrations of up to 20 analytes in 1 sample using a flow cytometer. The mouse/rat basic kit was used in combination with mouse simplex kits. Inflammatory mediators analyzed by the FlowCytomix assay included IL-13, IL-22, IL-2, IL-5, IL-6, IL-1β, IL-23, interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-4, and IL-17. Additionally IL-6, TNF-α, and IL-1β concentrations in cell culture supernatants were measured by enzyme-linked immunosorbent assay (ELISA) using the BD OptEIA Set Mouse IL-6, TNF-α, and IL-1β (BD Biosciences, San Diego, CA) according to the manufacturer’s manual.
TLR4 Antibody Treatment
For the treatment with a TLR4 antibody, anti-mouse MTS510 or isotype control (both from eBioscience) was applied to the medium after seeding primary cultured neonatal microglia to incubate overnight, and new antibody or isotype was added when the medium was changed to stimulate with glioma-conditioned medium (GCM).
Magnetic Cell Separation of Human Brain Tumor Tissue
Glioma-associated brain microglia/macrophages were isolated from the human tumor resected tissues. Fresh tissue was dissociated immediately after resection using the neural tissue dissociation kit (Miltenyi Biotec). Erythrocytes were lysed by adding 5 mL ammonium chloride solution for 10 minutes. Next, cells were resuspended in phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin and 2 mM ethylenediaminetetraacetic acid (EDTA). Magnetic sorting for CD11b+ cells was then performed using a CD11b MicroBead kit (Miltenyi Biotec) following the manufacturer’s instructions. MACS into CD11b--and CD11b+-enriched cell populations was carried out using several MACS columns in a series. Both CD11b- and CD11b+ fractions were collected. A purity check was performed after MACS separation by flow-cytometry analysis of a small fraction of the sorted populations.
Real-Time qPCR
Total RNA was isolated from microglia obtained from wild type (WT) mice, as well as from CD133+ and CD133- FACS-sorted GL261 cells and MACS-purified microglia from human GBM tissue using InviTrap Spin Universal RNA Mini Kit (Invitek GmbH, Berlin, Germany). Quality and yield were determined by NanoDrop ND-1000 (Thermo Scientific, Schwerte, Germany). First strand cDNA synthesis of RNA was done using the Superscript II (Invitrogen) reverse transcriptase according to the manufacturer’s instructions. For mRNA transcription, oligo-dT primers (Invitrogen) were used. Gene amplification was done in duplicates using SYBR Green PCR mix (Applied Biosystems, Foster City, CA) with the following PCR conditions: 95 °C for 10 minutes, 95 °C for 15 seconds, 60 °C for 30 seconds, 72 °C for 15 seconds for 40 cycles using the 7500 Fast Real-Time PCR System (Applied Biosystems). Sequences of primers used were: sense 5′-GCTGGCAGCACCCTGAGACC-3′, antisense 5′-TCCAAGGAGTGCCCGTGACC-3′ (mouse IL-6R); sense 5′-AGAAGGCCAGCAGCATCATT-3′, antisense 5′-TGACAGACCCAGAAACGAGC-3′ (mouse gp130); sense 5′-CCCTGAAGTACCCCATTGAA-3′, antisense 5′-GTGGACAGTGAGGCCAAGAT-3′ (mouse β-actin); sense 5′-GTAGCCGCCCCACACAGA-3′, antisense 5′-CATGTCTCCTTTCTCAGGGCTG3′ (human IL-6); and sense 5′-CACCATTGGCAATGAGCGGTTC-3′, antisense 5′-AGGTCTTTGCGGATGTCCACGT-3′ (human β-actin). Changes in human IL-6, mouse IL-6R, and mouse gp130 gene expressions were analyzed by the comparative 2(-ΔΔCt) method relative to β-actin gene-expression levels. For assessing CD133+-derived TLR4 ligands, gene amplification was performed in triplicate using SYBR Green PCR mix (Applied Biosystems) with the following PCR conditions: 50 °C for 2 minutes, 95 °C for 10 minutes, 95 °C for 15 seconds, 60–64 °C for 30 seconds, 72 °C for 1 minute for 40 cycles using the 7500 Fast Real-Time PCR System (Applied Biosystems). Primer sequences: sense 5′-AGTACGTGGCCCAAGAGTTG-3′, antisense 5′-AGGGCATTTGTGGTTCCAGT-3′ (mouse fibrinogen α-chain); sense 5′-ATGACCATCCACAACGGCAT-3′, antisense 5′-GATCCGTAGTTACCCAGCCG-3′ (mouse fibrinogen β -chain); sense 5′- CAACCCCCAAAGCCAGGTAT-3′, antisense 5′-GCAGCGCTTCGTATTTCACA-3′ (mouse fibrinogen γ–chain); sense 5′-GATGGTGAAGACGACACTGC-3′, antisense 5′-GAATGGCTGTGGACTGGATT-3′ (mouse fibronectin extra domain A [EDA]); sense 5′-GGGAGCCTTGGAAAGTGTGT-3′, antisense 5′-GCTTCTCTTCATGTTTGCCTGA-3′ (mouse Hsp22); sense 5′-AAACAAGCATCGGGATTCCAG-3′, antisense 5′-ACAATGCAGTCTTCCGTGGTG-3′ (mouse lactotransferrin); sense 5′-ATTTCCGGTCAGTGCAGGTAGT-3′, antisense 5′-GGTCAAAGCCATTCTCGAAGAT-3′ (mouse neutrophil elastase); sense 5′-GTAATTGTGTCCACCTTCCAC-3′, antisense 5′-AGTTGCTCATCACCTTCTGGA-3′ (mouse S100A4); sense 5′-CCGTCTTCAAGACATCGTTTGA-3′, antisense 5′-GTAGAGGGCATGGTGATTTCCT-3′ (mouse S100A8); sense 5′-CCCTGACACCCTGAGCAAGAAG-3′, antisense 5′-TTTCCCAGAACAAAGGCCATTGAG-3′ (mouse S100A9); sense 5′-GTTTGGAGACCGCAGAGAAGAA-3′, antisense 5′-TGTCCCCATATCTGCCCATCA-3′ (mouse tenascin-C). Fold changes in CD133+ gene expression compared to respective CD133- samples were analyzed by the comparative 2(-ΔΔCt) method relative to β-actin gene-expression levels.
Flow Cytometry
FACS analysis was carried out on an LSR Fortessa 5Laser (BD Biosciences, Erembodegem, Belgium). Cell sorting was carried out using a FACS-Aria-II (BD Biosciences). Antibodies for fluorescein isothiocyanate (FITC)–conjugated anti-human CD11b, APC-conjugated anti-mouse CD133, eFluor 660–conjugated anti-human/mouse Sox2, Alexa Fluor 488–conjugated anti-mouse anti-glial fibrillary acidic protein (GFAP), PE-conjugated anti-mouse Notch1, and matched isotype controls were all from eBioscience; PE-conjugated anti-mouse CD133 and matched isotype control were from Miltenyi Biotech. Data were analyzed using FlowJo software (Treestar, Ashland, OR).
In Vivo Glioma Implantation
WT and IL-6-/- mice were used for the in vivo studies to investigate glioma expansion. To broaden the relevance of our findings, 4.5–10-week-old mice (Ntv-a/Ink4a-Arf-/- for DF-1 RCAS-PDGFb injection) were used to culture RCAS GSCs. Surgical procedures were performed as described (41, 42). Briefly, mice were anesthetized, immobilized, and mounted onto a stereotactic frame (David Kopf Instruments, Tujunga, CA) in the flat-skull position. After skin incision 1 mm anterior and 1.5 mm lateral to the bregma, the skull was carefully drilled with a dental drill or a 20G needle tip. A 1-µl syringe with a blunt tip (Mikroliterspritze 7001N, Bonaduz, Switzerland) was inserted to a depth of 4 mm and retracted to a depth of 3 mm from the dural surface into the right caudate putamen. Over 2 minutes, 1 µl glioma cell suspension (2 x 104 cells/µl of EGFP-GL261, 1 x 104 CD133- or 100 CD133+ GL261 cells, 4 x 104 transfected DF-1 cells or 5 x 104 RCAS-PDGFb tumor cells) was slowly injected into the brain. Coordinates for injections of DF-1 cells and RCAS-PDGFb tumor cells into Ntva/Ink4a-Arf-/- mice, respectively, were bregma 1.5 mm anterior, lateral 0.5 mm (right of midline) and a depth 2.0 mm from the dural surface. The needle was then carefully retracted from the injection canal, and the skin was sutured with a surgical sewing cone (Johnson & Johnson International, Langhorne, PA). After surgery, the mice were kept warm until awake. Mice were monitored daily for the first 2 weeks and twice a day starting from day 15 postinjection for symptoms of tumor development (lethargy, weight loss, head tilting). The size of resulting tumors ranged from 1.5–2.5 mm (GL261 tumors) and 2.5–3.5 mm (RCAS-PDGFb tumors).
Immunofluorescent Staining and Image Processing
Free-floating 40-µm-thick brain sections from tumor-bearing mice were prepared as previously described (22). Nuclei were visualized using 4,6-diamidino-2-phenylindole (DAPI) from Sigma-Aldrich. The GL261 glioma cells were identified by green fluorescence of the EGFP construct.
Unbiased Stereology for Tumor Volume Estimation
Fourteen days (EGFP-GL261) or 21 days (CD133- and CD133+ GL261 cells) after tumor implantation, mice were anesthetized with pentobarbital (Narcoren, Merial, Hallbergmoos, Germany), brains were perfused and fixed, and resulting brain slices were subsequently used to analyze glioma expansion in vivo. The tumor volume in brain slices of glioma-bearing mice was quantified according to the Cavalieri principle by determining tumor area in every 12th 40-µm-thick brain slice and then multiplying this area by the factor 12 x 40 µm using the Stereoinvestigator software (MBF Bioscience, Williston, VT). Experimental groups were blinded to the investigator performing the analysis.
Statistical Analysis
All data represent the average of at least 3 independent experiments. Datasets were analyzed statistically with SPSS11.5 software and tested for normality with the Shapiro-Wilks test. The Mann-Whitney U test was used for nonparametric analysis. Parametric testing was done with the t test. Comparisons between multiple groups were done using 1-way ANOVA with the Scheffé post hoc test. Statistical significance was determined at p values < 0.05 (*) and < 0.01 (**) while “n.s.” implied a nonsignificant p value.
RESULTS
IL-6–Deficient Mice Show Reduced Glioma Growth
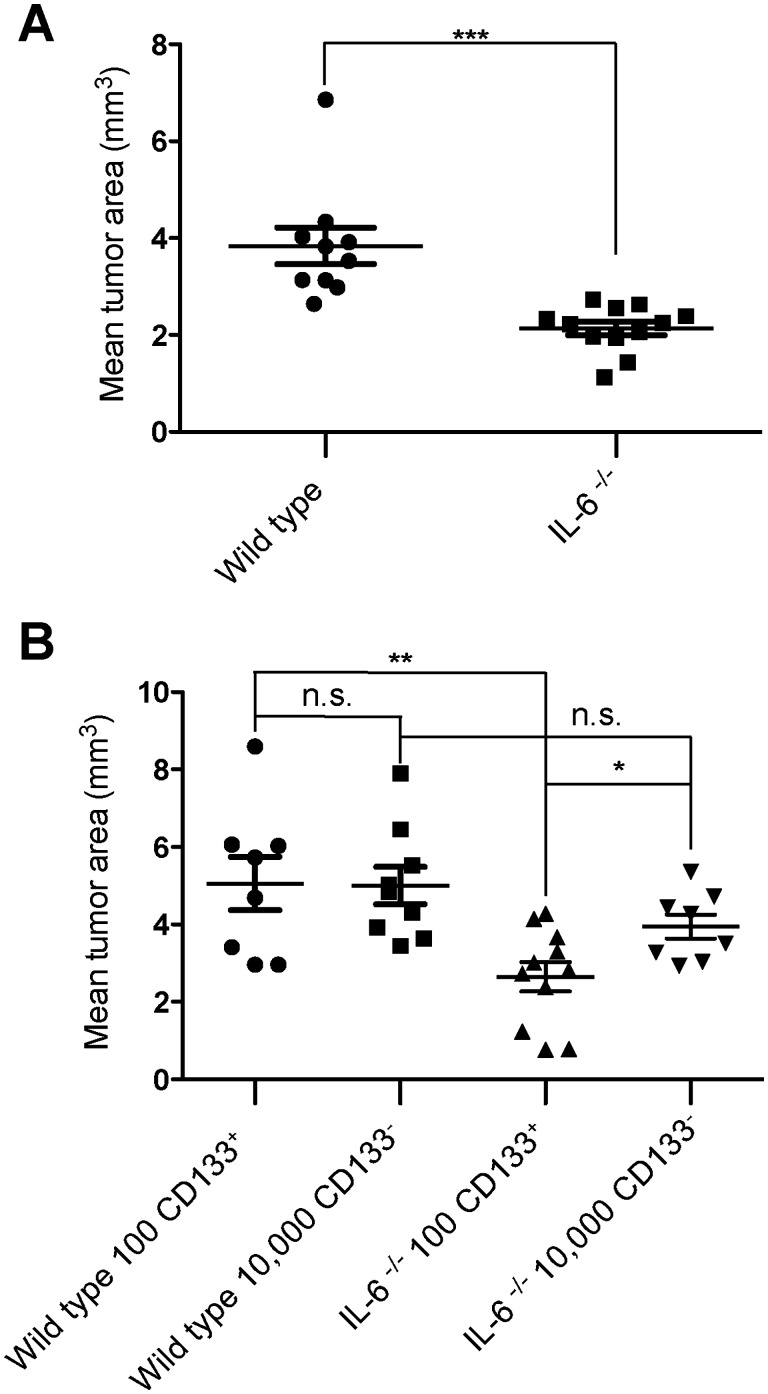
To investigate whether ablation of the IL-6 gene locus in the host interfered with tumor expansion in vivo, we implanted EGFP-GL261 cells into WT and IL-6-/- mice and measured glioma volume by unbiased stereological estimation (Cavalieri method). After 2 weeks of implantation, the tumor volume in IL-6-/- mice was significantly smaller compared with the WT mice (WT 3.84 ± 1.13 mm3, IL-6-/- 2.14 ± 0.46 mm3, p = 0.0002; Fig. 1A). It has been shown by us and others that 100 CD133+ glioma cells have a similar tumor forming capacity as 10,000 CD133- glioma cells (40). To see the impact of host IL-6 on CD133+ cells versus CD133- cells, we injected 100 CD133+ cells or 10,000 CD133- cells into the WT and IL-6-/- mice and analyzed tumor growth. After 3 weeks of tumor growth, we found that, in WT mice, 100 CD133+ cells formed tumors of similar size compared with the 10,000 CD133- cells (WT-CD133+: 5.06 ± 0.69 mm3, WT-CD133-:5.0 ± 0.48 mm3, p = 0.89; Fig. 1B). However, in IL-6-/- animals, inoculation of 100 CD133+ GL261 cells induced significantly smaller tumors (IL-6-/--CD133+: 2.65 ± 0.38 mm3, p = 0.004) compared with WT. Injection of 10,000 CD133- cells into IL-6-/- mice also resulted in smaller tumors, but this decrease was not significant (IL-6-/--CD133-: 3.94 ± 0.31 mm3, p = 0.09). These data indicate that IL-6 from the host cells supports tumor growth by GSCs, but not by bulk glioma cells.
FIGURE 1.
Host IL-6 interferes with glioma expansion by influencing GSCs. (A) EGFP-GL261 cells were intracerebrally implanted into WT and IL-6-/- mice; tumor volume in WT versus IL-6-/- animals was evaluated based on unbiased stereology. (B) WT and IL-6-/- mice were intracerebrally implanted with 100 CD133+ or 10,000 CD133- GL261 cells, and after 3 weeks, tumor volume was evaluated based on unbiased stereology.
Microglial IL-6 Is Upregulated by Supernatant From Glioma Stem Cells but Not From Bulk Glioma Cells
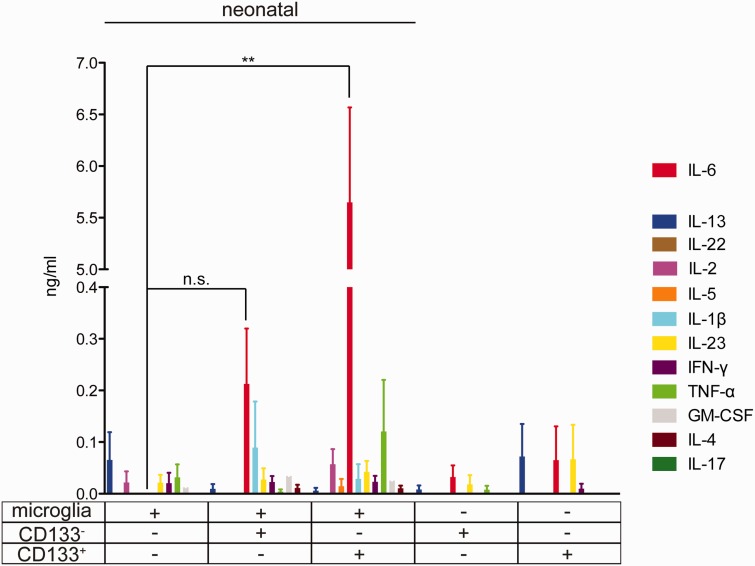
To investigate the potential of GSCs versus bulk cells to induce microglial cytokine release, mouse primary neonatal microglia cultures were treated with control medium (stem cell culture medium) or supernatant medium from GL261 cells (GCM) either enriched for CD133 or deprived of CD133. After 24 hours of stimulation, cell supernatant was collected to measure expression levels of 12 cytokines. As shown in Figure 2, the level of IL-6 in supernatant from microglial cells that were stimulated with CD133+-conditioned medium was higher than the levels in supernatant from microglial cells that were stimulated with CD133--conditioned medium (control: undetectable, CD133+: 6.28 ± 1.16 ng/mL, CD133-: 0.32 ± 0.03 ng/mL, p = 0.004). However, levels of IL-1β, TNF-α, IL-13, IL-22, IL-2, IL-5, IL-23, IFN-γ, GM-CSF, IL-4, and IL-17 in supernatant from microglia did not change between treatment groups.
FIGURE 2.
Cytokine release by multiple analyte detection in microglia stimulated with conditioned medium from GSCs and non-GSCs. Neonatal primary cultured microglia were stimulated with conditioned medium from CD133- and CD133+ GL261 cells for 24 hours, and the release of the cytokines IL-13, IL-22, IL-2, IL-5, IL-6, IL-1β, IL-23, IFN-γ, TNF-α, GM-CSF, IL-4, and IL-17 were analyzed by FlowCytomix. We also measured the cytokine levels of the conditioned medium from CD133- and CD133+ GL261 before application to microglia.
Supernatant From Glioma Stem Cells Induced IL-6 Release in Both Neonatal and Adult Microglia
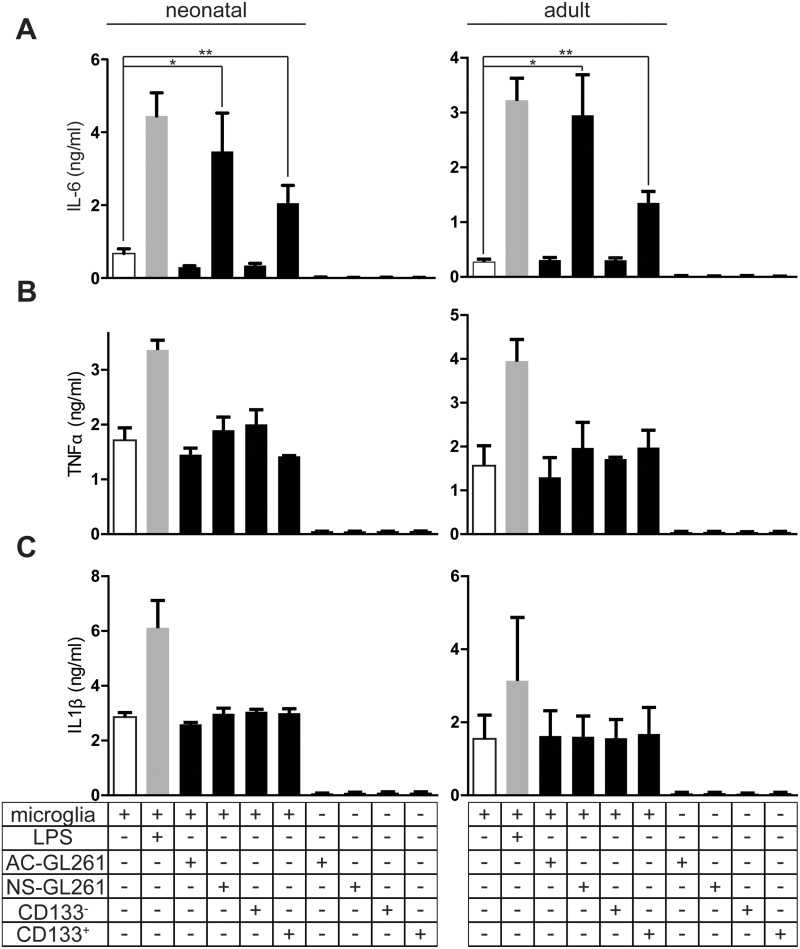
To further investigate the potential of GSCs versus bulk cells to induce cytokines, mouse primary neonatal and adult microglial cultures were treated with medium only (as controls) or GCM from CD133+, CD133-, neurosphere GL261 (NS-GL261), or adhesive GL261 (AC-GL261) cells. The percentage of CD133+ cells in NS-GL261 cells is 30.1%, while in AC-GL261, it is less than 1%, as described elsewhere (43) (Supplementary Data Figures 1 and 2). Treatment with LPS served as a positive control. After 24 hours of treatment, cell supernatant was collected to perform ELISA for 3 proinflammatory cytokines: IL-6, TNF-α, and IL-1β. As shown in Figure 3, the level of IL-6 in supernatant from microglial cells that were stimulated with CD133+– or NS-GL261–conditioned medium was higher than the levels in supernatant from microglial cells that were stimulated with CD133-– or AC-GL261–conditioned medium (control: 0.67 ± 0.22 ng/mL, CD133+: 2.05 ± 0.85 ng/mL, CD133-: 0.34 ± 0.11 ng/mL, NS-GL261: 3.47 ± 1.83 ng/mL, AC-GL261: 0.3 ± 0.06 ng/mL, p = 0.04). The difference between CD133+ cell populations (enriched from NS-GL261 cells) and NS-GL261 cells in Figure 3A was not significant: neonatal microglia (NS-GL261: 3.47 ± 1.83, CD133+: 2.05 ± 0.85, p = 0.27) and adult microglia (NS-GL261: 2.95 ± 1.28, CD133+: 1.35 ± 0.29, p = 0.13). However, when we analyzed TNF-α and IL-1β levels in microglia treated in different conditions, there was no significant difference. LPS always triggered an induction of these cytokines. IL-6, IL-1β, and TNF-α were not detectable in GCM from all glioma cells.
FIGURE 3.
Factors released from glioma stem cells induced IL-6 but not TNF-α and IL-1β secretion from both neonatal and adult primary microglia. (A) Neonatal and adult primary cultured microglia were stimulated with conditioned medium from adhesive (AC-), neurosphere (NS-), CD133-, and CD133+ GL261 cells for 24 hours, and IL-6 release was analyzed by ELISA. TNF-α (B) and IL-1β (C) were also analyzed in a similar way. LPS was used as a positive control. We also determined the cytokine levels of the conditioned media from different cells before application to microglia.
Factors Released From Glioma Stem Cells Induced Microglial IL-6 Secretion Through MyD88-TLR4 Signaling
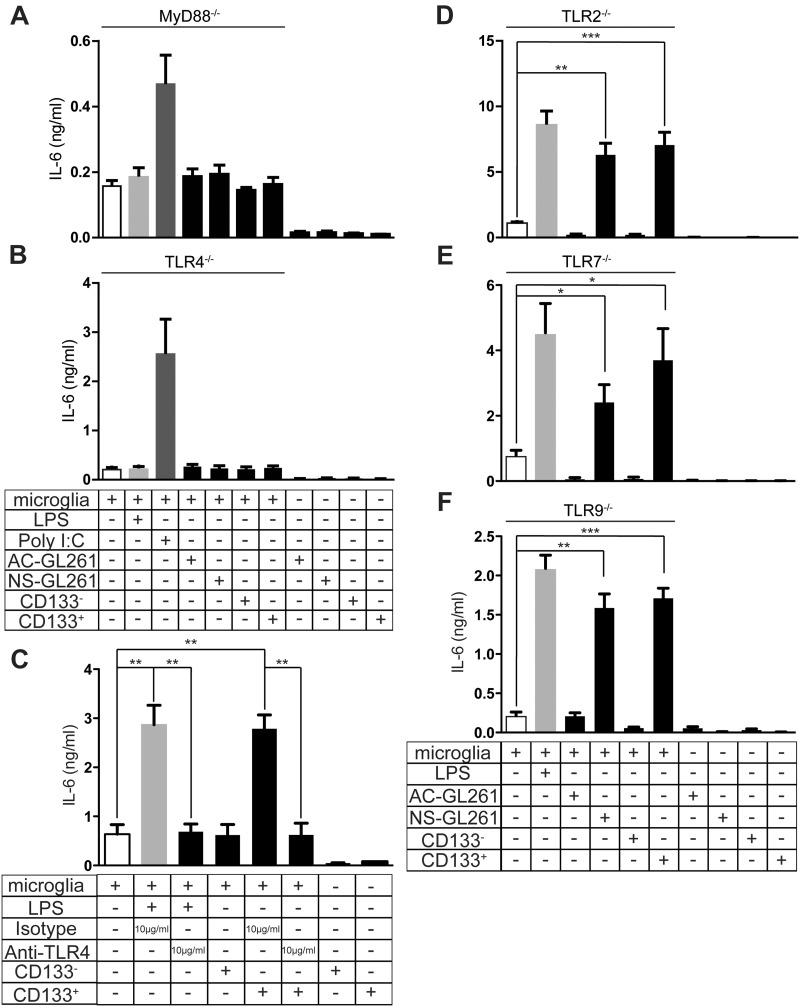
We have previously shown that glioma-released versican induced microglial MT1-MMP production through the TLR2 signaling pathway (23). We therefore tested whether IL-6 release is also regulated by TLR signaling. Since MyD88 is the adapter protein for all TLRs except TLR3 (44), we stimulated microglia from MyD88-/- animals with GSC supernatant. The TLR3 agonist Poly I:C was used as a positive control. As shown in Figure 4A, in microglial cells deficient for MyD88, IL-6 induction was completely abolished, indicating that GSCs triggered microglial IL-6 induction through TLR signaling. We then screened GSC-triggered microglial IL-6 induction in TLR2, TLR4, TLR7, and TLR9-/- animals. Interestingly, only the knockout of TLR4 impaired microglial IL-6 induction (control: 0.2 ± 0.08 ng/mL, CD133+: 0.24 ± 0.07 ng/mL, NS-GL261: 0.23 ± 0.04 ng/mL; Fig. 4B) while IL-6 upregulation was similar to WT levels in microglial cells from TLR2-, TLR7-, or TLR9-deficient animals (Fig. 4D–F). The TLR4 monoclonal neutralizing antibody MTS510 has previously been shown to functionally block TLR4 (45). We first verified that MTS510 blocks microglial TLR4 functionality in vitro. MTS510 attenuated the TLR4 agonist LPS-induced microglial IL-6 induction (Fig. 4C). To test whether MTS510 impairs microglial IL-6 secretion induced by supernatant from GSCs, microglia were stimulated with either GSC supernatant in combination with MTS510 or GSC supernatant with isotype control antibody for 24 hours. GSC supernatant treatment with isotype control resulted in an increase (control: 0.64 ± 0.27 ng/mL, CD133+/Isotype: 2.78 ± 0.4 ng/mL, p = 0.003; Fig. 4C) of microglial IL-6 secretion compared with the untreated cells. Treatment with GSC supernatant in combination with MTS510 did not trigger IL-6 secretion (CD133+/Anti-TLR4: 0.62 ± 0.34 ng/mL, p = 0.004; Fig. 4C), which indicates that MTS510 blocked microglial TLR4 signaling. These data indicate that induction of IL-6 by stimulation with GSCs-conditioned medium was fully dependent on MyD88-TLR4 signaling but not on TLR2, TLR7, and TLR9. In an attempt to narrow down possible TLR4-specific ligands as candidate factors released by the glioma cells, we compared the level of eleven TLR4 specific known endogenous ligands in CD133+ versus CD133- cells (Supplementary Data Figure 4). Fibrinogen alpha, beta and gamma chain, lactotransferrin, and neutrophil elastase, as well as the S100 proteins S100A8 and S100A9, are not expressed in either cell population (respective controls with liver and bone marrow revealed primer specificity and validity; data not shown). Fibronectin EDA and HSP22 are expressed by both CD133+ and CD133- at the same level (fold change CD133+ vs CD133-, respectively, 0.76 ± 0.35 and 1.26 ± 0.66). The specific TLR4 ligand tenascin-C is, on average, 3.93 ± 2.06-fold significantly higher expressed in CD133+ compared with CD133- cells. Interestingly, we also identified S100A4 as a TLR4-specific ligand that is significantly 64-fold (64.16 ± 20.71; data not shown) lower expressed in CD133+ versus CD133-. However, from these data, tenascin-C is the best candidate to be investigated further to verify its role in IL-6 upregulation after TLR4 stimulation in microglia cells by soluble factor derived from CD133+ cells.
FIGURE 4.
IL-6 release induced by GCM in microglia from MyD88- and TLR-deficient mice. (A) Neonatal microglia from MyD88-/- mice were stimulated with conditioned medium from adhesive (AC-), neurosphere (NS-), CD133-, and CD133+ GL261 cells for 24 hours, and IL-6 release was analyzed by ELISA and compared to an unstimulated control. Similarly, IL-6 release was analyzed in MyD88-/- (A), TLR4-/- (B), TLR2-/- (D), TLR7-/- (E), and TLR9-/- mice (F). For MyD88-/- and TLR4-/-, Poly I:C was used as a positive control, while LPS was used as a positive control in other groups. (C) Monoclonal antibody MTS510 inhibited GSCs-conditioned medium-driven microglial IL-6 secretion. Primary microglial cells were stimulated with CD133--and CD133+-conditioned medium together with 10 µg/mL isotype and 10 µg/mL MTS510 for 24 hours. We also determined the cytokine levels of the conditioned media from different cells before application to microglia.
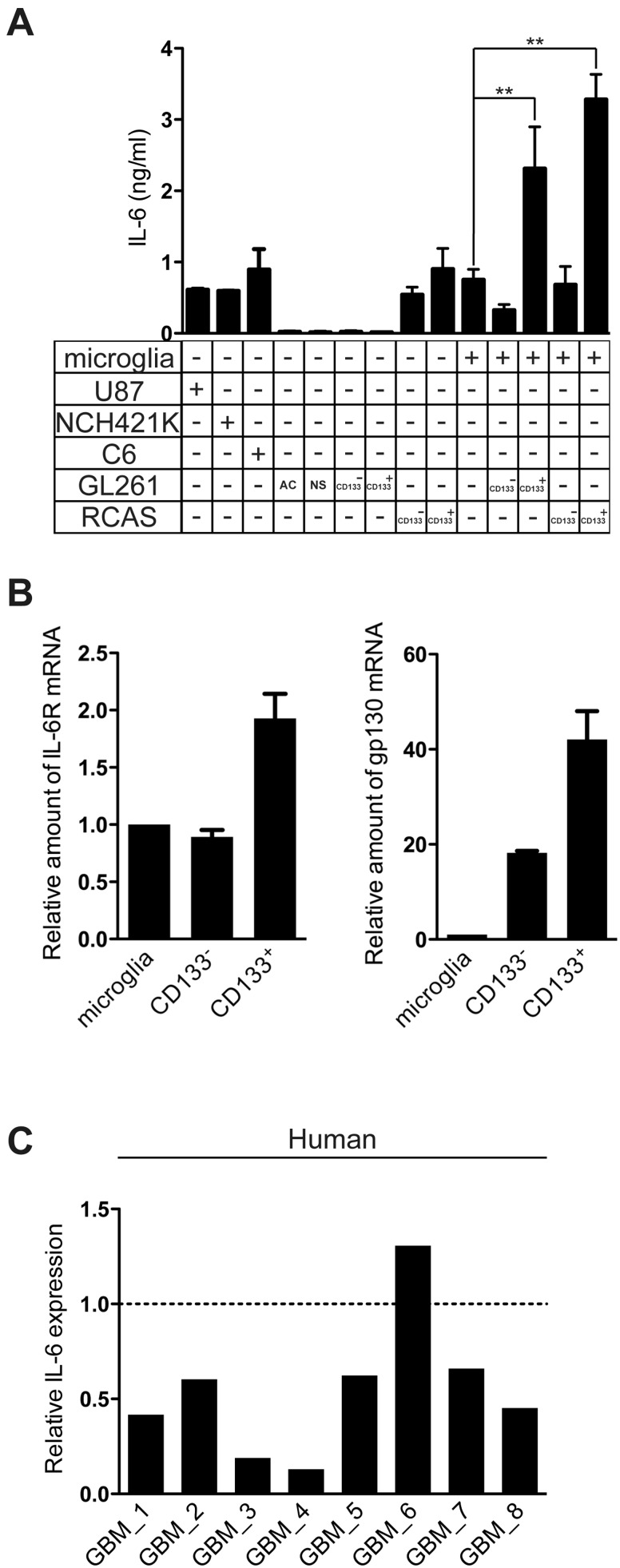
GAMs Are Predominating IL-6 Expressing Population in Gliomas
To evaluate the potential contribution of IL-6 signaling to the glioma microenvironment, we measured IL-6 release in a series of glioma cell lines as well as mouse microglia and microglia stimulated from conditioned medium of CD133- and CD133+ GL261 cells. While some of the gliomas do secrete IL-6, both CD133- and CD133+ GL261 cells express very low levels of IL-6 compared with naïve microglia and microglia primed by CD133+ GL261–conditioned medium (Fig. 5A). To broaden the relevance of our findings, we employed the RCAS-PDGFb murine tumor model. Both CD133- and CD133+ RCAS cells do secrete low levels of IL-6 (CD133-: 0.54 ± 0.15 ng/mL, CD133+: 0.9 ± 0.41 ng/mL) compared with microglia primed by CD133+ RCAS–conditioned medium (Fig. 5A). To investigate the potential of GSCs versus bulk cells to induce microglial cytokine release in another murine tumor model, mouse primary neonatal microglia cultures were treated with control medium (stem cell medium) or supernatant medium from RCAS cells either enriched for CD133 or deprived of CD133. After 24 hours of stimulation, cell supernatant was collected to measure expression levels of IL-6. As seen in Figure 5A, the level of IL-6 in supernatant from microglial cells that were stimulated with CD133+ RCAS–conditioned medium was higher than the levels in supernatant from control microglial cells or microglial cells that were stimulated with CD133--conditioned medium (control: 0.64 ± 0.27 ng/mL, CD133+: 3.28 ± 0.5 ng/mL, p = 0.03). To verify that the IL-6 receptor is expressed on the target cell, we measured IL-6 receptor expression in primary cultured neonatal microglia and freshly FACS-isolated CD133+ and CD133- GL261 cells by qRT-PCR (Fig. 5B). CD133+ cells expressed higher IL-6 receptor and gp130 mRNA levels than CD133- cells or microglial cells. These data demonstrate that the expression of IL-6 receptors was elevated on CD133+ in comparison with CD133- cells and supports the concept of paracrine signaling between GSCs and microglia (Fig. 6). We also purified microglia/brain macrophages from human GBM tissue by MACS (the purity of CD11b+ cells was described previously [46]) and tested for the expression of IL-6 by qRT-PCR. In 7 out of 8 samples, the CD11b+ cells (ie, GAMs) expressed higher IL-6 than the CD11b- cells (mainly tumor cells) (Fig. 5C). These data suggest that GAMs are the main source of IL-6 in gliomas.
FIGURE 5.
IL-6 expression in GAMs and gliomas. (A) IL-6 release was analyzed in conditioned medium from U87, C6, NCH421K, AC-, NS-, CD133-, and CD133+ from GL261 and RCAS cells; primary cultured microglia and microglia treated with GSCs-conditioned medium were used as controls. (B) IL-6 receptor and gp130 expression were analyzed in microglia; FACS-sorted CD133- and CD133+ cells from GL261 by qRT-PCR. (C) MACS freshly isolated CD11b+ cells and CD11b- (ie, mainly glioma cells) from 8 human GBM (Supplementary Data Table 1) samples were analyzed for IL-6 by qRT-PCR. Dashed line represents IL-6 expression in CD11b+ cells in each sample, solid bars represent fold changes of IL-6 expression in flow through cells compared to CD11b + cells.
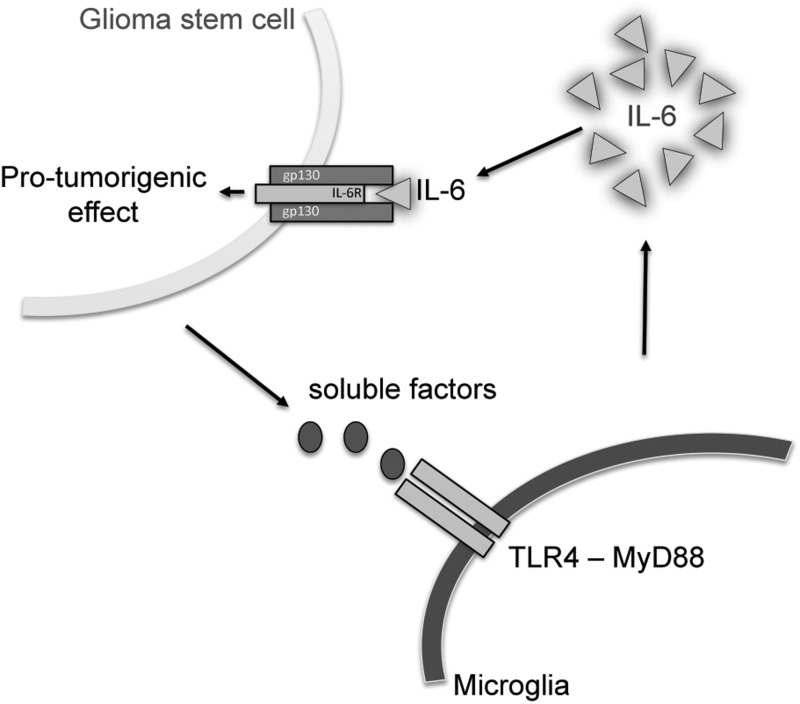
FIGURE 6.
Scheme of the paracrine relationship between glioma stem cells and microglial cells. GSCs, but not the bulk glioma cells, initiate microglial IL-6 secretion via TLR4 signaling, and IL-6 regulates glioma growth by supporting GSCs.
DISCUSSION
The interaction between tumor cells and their microenvironment has attracted increasing attention over the last few years. Microglia, as the immune competent cells of the brain, are the key resident cells interacting with glioma. We, along with others, have demonstrated that glioma attract microglia/macrophages and educate them to develop a tumor-supportive phenotype (20, 23, 46–48). Glioma cells in the tumor tissue are a heterogeneous population containing different subgroups. GSCs are the minority among the glioma cells, but have a strong impact on the disease progression since they are believed to be responsible for glioma relapse and therapy resistance. So far, there have only been a few studies addressing the crosstalk between GSCs and microglia/macrophages. GSCs recruit more GAMs than the bulk glioma cells (non-GSCs) by releasing higher levels of chemoattractants, including CCL2 and VEGF-A. In both primary human gliomas and orthotopical transplanted syngeneic glioma, the density of GAMs at the invasive front is increased by the presence of CD133+ GSCs. The interdependence of these 2 cell types became evident since GAMs release TGF-ß1, which promotes the upregulation of MMP-9 in GSCs and thus tumor invasion (49). In the present study, we found that CD133+ GSCs trigger IL-6 release from microglia, which promotes glioma growth.
Microglia associated with glioma have been assigned the M2 phenotype since they are tumor-supportive and immune-suppressive (50). However, in a recent microarray study, we demonstrated that GAMs share markers of both the M1 and M2 phenotype, and thus, the genetic profile defines them as a unique phenotype (36). The simple view of separation into the M1 and M2 category has also recently been challenged for macrophages, and a more differentiated scheme has been proposed (51). Indeed, IL-6, as a proinflammatory cytokine, is not upregulated in microglia associated with both mouse and human gliomas compared with naive microglia (26, 52). We confirmed this finding by stimulating primary microglia with conditioned medium from GL261 cells, and we also did not detect any induction of proinflammatory cytokines. GSCs, however, lead to a selective upregulation of IL-6 but not of a battery of other cytokines including TNF-α. Thus, microglial cells associated with GSCs acquire a phenotype, which is distinct from other glioma-associated microglia. GAMs are thus a heterogeneous population imposed by glioma heterogeneity. Recently, it was shown by a single-cell sequencing approach that glioma cells are heterogenic also with respect to the classical diagnostic categories (53).
We found that TLR4 signaling in the microglia/macrophages is essential for IL-6 secretion since release was abolished in TLR4-deficient mice and in a mouse line deficient for the TLR-adaptor protein MyD88, which is an essential component for TLR signaling. Moreover, the anti-TLR4 monoclonal antibody MTS510 inhibited GSC-conditioned medium-driven microglial IL-6 secretion. We thus propose that glioma cells release ligands that activate TLR4 signaling. We measured the levels of 11 TLR4-specific ligands in CD133+ and CD133- cells and identified tenascin-C (TNC) as a possible candidate mediating the IL-6 release in microglia via TLR4 signaling. This is in line with recent findings where TNC was reported as a stem cell marker in human glioma samples (54). Whereas TNC is produced by stromal fibroblasts in the majority of solid tumors, brain tumor cells themselves are the main source of extracellular matrix TNC in glial malignancies (55,56). The intensity of TNC expression is shown to correlate with glioma grade and patient prognosis (55,57). Due to its expression in solid tumors, TNC has been used as a tumor-associated antigen to deliver antibody-conjugated radiotherapeutic agents to GBM (58). While the classic ligand for TLR4, LPS, triggers the release of several proinflammatory cytokines including IL-6 and TNF-α, we thus assume that the ligand released from glioma CD133+ cells triggers a selective release of IL-6. It is possible that other factors such as epigenetic regulations like miRNA deregulation, histone modification, or DNA methylation specifically suppress the transcription of other proinflammatory cytokines (59). Recently, it was shown that an miR-142-3p–driven autocrine and paracrine positive loop epigenetically regulates the progression and cancer stem-like property of glioblastoma by targeting the secretion of the proinflammatory cytokine IL-6 (60). The exact mechanism of how tumor cells educate bulk cells to serve in favor of tumor growth and invasiveness remains to be elucidated. At this point we like to stress that, due to the global deletion of IL-6 in our mouse model, we cannot exclude primary or secondary effects of IL-6 on tumor growth apart from the microglia/macrophage population. However, our in vitro data strongly support the importance of the CD133-TLR4-IL6 pathway.
Zhang et al describe another mode of interaction between glioma and microglia mediated by IL-6 independent of TLR4. They used human glioma lines and nontransformed human microglia isolated from surgically resected epileptic brain tissue and described that gliomas release the chemokine CCL2. Overexpression of CCL2 in the U87 glioma line stimulated microglia to release IL-6 (30).
Our results are in line with a study where it was demonstrated that interfering with IL-6 signaling in GSCs led to reduced growth and neurosphere formation capacity and to an increase in the apoptosis rate (27). From a clinical perspective, the expression of IL-6 and its receptors in the human glioma tissues is inversely correlated to patient survival. The median survival times were 16 months in patients with negative IL-6 expression and 7 months in those with positive IL-6 expression (61). An IL-6 neutralizing antibody attenuated microglia-stimulated glioma invasiveness and reduced glioma growth in vivo (27, 30). Our data further corroborate the important role of microglial IL-6 in glioma growth. Specifically, we propose that TLR4 signaling is an important component of GSC-microglia crosstalk and propose tenascin-C released by glioma CD133+ cells as a regulator of this pathway (Fig. 6).
Supplementary Material
ACKNOWLEDGMENTS
The authors sincerely thank Christina Krüger for the extensive help in providing MyD88, TLRs 4, 7, and 9-/- mice from Charité, Berlin. Many thanks to Irene Haupt, Regina Piske, Hanna Schmidt, and Nadine Scharek for excellent technical assistance. We appreciate the support of Maria Pannell for manuscript proofreading.
REFERENCES
- 1.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396–401 [DOI] [PubMed] [Google Scholar]
- 2.Ignatova TN, Kukekov VG, Laywell ED, et al. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002;39:193–206 [DOI] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756–60 [DOI] [PubMed] [Google Scholar]
- 4.Hambardzumyan D, Squatrito M, Holland EC. Radiation resistance and stem-like cells in brain tumors. Cancer Cell 2006;10:454–6 [DOI] [PubMed] [Google Scholar]
- 5.Bleau AM, Hambardzumyan D, Ozawa T, et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell 2009;4:226–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao S, Wu Q, Sathornsumetee S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res 2006;66:7843–8 [DOI] [PubMed] [Google Scholar]
- 7.Wei J, Barr J, Kong LY, et al. Glioma-associated cancer-initiating cells induce immunosuppression. Clin Cancer Res 2010;16:461–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.McCord AM, Jamal M, Williams ES, et al. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clin Cancer Res 2009;15:5145–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dey M, Ulasov IV, Lesniak MS. Virotherapy against malignant glioma stem cells. Cancer Lett 2010;289:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res 2007;67:4010–5 [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Sakariassen PO, Tsinkalovsky O, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer 2008;122:761–8 [DOI] [PubMed] [Google Scholar]
- 12.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell 2007;11:69–82 [DOI] [PubMed] [Google Scholar]
- 13.Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature 2008;454:436–44 [DOI] [PubMed] [Google Scholar]
- 14.Morimura T, Neuchrist C, Kitz K, et al. Monocyte subpopulations in human gliomas: Expression of Fc and complement receptors and correlation with tumor proliferation. Acta Neuropathol 1990;80:287–94 [DOI] [PubMed] [Google Scholar]
- 15.Yi L, Xiao H, Xu M, et al. Glioma-initiating cells: A predominant role in microglia/macrophages tropism to glioma. J Neuroimmunol 2011;232:75–82 [DOI] [PubMed] [Google Scholar]
- 16.Wang SC, Hong JH, Hsueh C, et al. Tumor-secreted SDF-1 promotes glioma invasiveness and TAM tropism toward hypoxia in a murine astrocytoma model. Lab Invest 2012;92:151–62 [DOI] [PubMed] [Google Scholar]
- 17.Wang SC, Yu CF, Hong JH, et al. Radiation therapy-induced tumor invasiveness is associated with SDF-1-regulated macrophage mobilization and vasculogenesis. PLoS One 2013;8:e69182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreutzberg GW. Microglia: A sensor for pathological events in the CNS. Trends Neurosci 1996;19:312–8 [DOI] [PubMed] [Google Scholar]
- 19.Markovic DS, Glass R, Synowitz M, et al. Microglia stimulate the invasiveness of glioma cells by increasing the activity of metalloprotease-2. J Neuropathol Exp Neurol 2005;64:754–62 [DOI] [PubMed] [Google Scholar]
- 20.Markovic DS, Vinnakota K, Chirasani S, et al. Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion. Proc Natl Acad Sci U S A 2009;106:12530–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charles NA, Holland EC, Gilbertson R, et al. The brain tumor microenvironment. Glia 2012;60:502–14 [DOI] [PubMed] [Google Scholar]
- 22.Vinnakota K, Hu F, Ku MC, et al. Toll-like receptor 2 mediates microglia/brain macrophage MT1-MMP expression and glioma expansion. Neuro Oncol 2013;15:1457–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu F, a Dzaye OD, Hahn A, et al. Glioma-derived versican promotes tumor expansion via glioma-associated microglial/macrophages Toll-like receptor 2 signaling. Neuro Oncol 2015;17:200–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou B, Reizis B, DeFranco AL. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity 2008;29:272–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson JK, Miller SD. Microglia initiate central nervous system innate and adaptive immune responses through multiple TLRs. J Immunol 2004;173:3916–24 [DOI] [PubMed] [Google Scholar]
- 26.Hussain SF, Yang D, Suki D, et al. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol 2006;8:261–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Lathia JD, Wu Q, et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells 2009;27:2393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sehgal PB, Wang L, Rayanade R, et al. Interleukin-6-type cytokines. Ann N Y Acad Sci 1995;762:1–14 [PubMed] [Google Scholar]
- 29.Van Meir E, Sawamura Y, Diserens AC, et al. Human glioblastoma cells release interleukin 6 in vivo and in vitro. Cancer Res 1990;50:6683–8 [PubMed] [Google Scholar]
- 30.Zhang J, Sarkar S, Cua R, et al. A dialog between glioma and microglia that promotes tumor invasiveness through the CCL2/CCR2/interleukin-6 axis. Carcinogenesis 2012;33:312–9 [DOI] [PubMed] [Google Scholar]
- 31.Kopf M, Baumann H, Freer G, et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature 1994;368:339–42 [DOI] [PubMed] [Google Scholar]
- 32.Chourbaji S, Urani A, Inta I, et al. IL-6 knockout mice exhibit resistance to stress-induced development of depression-like behaviors. Neurobiol Dis 2006;23:587–94 [DOI] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brennan C, Momota H, Hambardzumyan D, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One 2009;4:e7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006;9:157–73 [DOI] [PubMed] [Google Scholar]
- 36.Szulzewsky F, Pelz A, Feng X, et al. Glioma-associated microglia/macrophages display an expression profile different from m1 and m2 polarization and highly express gpnmb and spp1. PLoS One 2015;10:e0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hambardzumyan D, Amankulor NM, Helmy KY, et al. Modeling adult gliomas using RCAS/t-va technology. Transl Oncol 2009;2:89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campos B, Wan F, Farhadi M, et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer Res 2010;16:2715–28 [DOI] [PubMed] [Google Scholar]
- 39.Scheffel J, Regen T, Van Rossum D, et al. Toll-like receptor activation reveals developmental reorganization and unmasks responder subsets of microglia. Glia 2012;60:1930–43 [DOI] [PubMed] [Google Scholar]
- 40.Chirasani SR, Sternjak A, Wend P, et al. Bone morphogenetic protein-7 release from endogenous neural precursor cells suppresses the tumourigenicity of stem-like glioblastoma cells. Brain 2010;133:1961–72 [DOI] [PubMed] [Google Scholar]
- 41.Walzlein JH, Synowitz M, Engels B, et al. The antitumorigenic response of neural precursors depends on subventricular proliferation and age. Stem Cells 2008;26:2945–54 [DOI] [PubMed] [Google Scholar]
- 42.Glass R, Synowitz M, Kronenberg G, et al. Glioblastoma-induced attraction of endogenous neural precursor cells is associated with improved survival. J Neurosci 2005;25:2637–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi L, Zhou C, Wang B, et al. Implantation of GL261 neurospheres into C57/BL6 mice: A more reliable syngeneic graft model for research on glioma-initiating cells. Int J Oncol 2013;43:477–84 [DOI] [PubMed] [Google Scholar]
- 44.Beutler B, Hoebe K, Shamel L. Forward genetic dissection of afferent immunity: The role of TIR adapter proteins in innate and adaptive immune responses. C R Biol 2004;327:571–80 [DOI] [PubMed] [Google Scholar]
- 45.Mortaz E, Redegeld FA, Nijkamp FP, et al. Acetylsalicylic acid-induced release of HSP70 from mast cells results in cell activation through TLR pathway. Exp Hematol 2006;34:8–18 [DOI] [PubMed] [Google Scholar]
- 46.Hu F, Ku MC, Markovic D, et al. Glioma-associated microglial MMP9 expression is upregulated by TLR2 signaling and sensitive to minocycline. Int J Cancer 2014;135:2569–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ku MC, Wolf SA, Respondek D, et al. GDNF mediates glioblastoma-induced microglia attraction but not astrogliosis. Acta Neuropathol 2013;125:609–20 [DOI] [PubMed] [Google Scholar]
- 48.Ellert-Miklaszewska A, Dabrowski M, Lipko M, et al. Molecular definition of the pro-tumorigenic phenotype of glioma-activated microglia. Glia 2013;61:1178–90 [DOI] [PubMed] [Google Scholar]
- 49.Ye XZ, Xu SL, Xin YH, et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-beta1 signaling pathway. J Immunol 2012;189:444–53 [DOI] [PubMed] [Google Scholar]
- 50.Li W, Graeber MB. The molecular profile of microglia under the influence of glioma. Neuro Oncol 2012;14:958–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murray PJ, Allen JE, Biswas SK, et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014;41:14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sliwa M, Markovic D, Gabrusiewicz K, et al. The invasion promoting effect of microglia on glioblastoma cells is inhibited by cyclosporin A. Brain 2007;130:476–89 [DOI] [PubMed] [Google Scholar]
- 53.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jachetti E, Caputo S, Mazzoleni S, et al. Tenascin-C protects cancer stem-like cells from immune surveillance by arresting T-cell activation. Cancer Res 2015;75:2095–108 [DOI] [PubMed] [Google Scholar]
- 55.Behrem S, Zarkovic K, Eskinja N, et al. Distribution pattern of tenascin-C in glioblastoma: Correlation with angiogenesis and tumor cell proliferation. Pathol Oncol Res 2005;11:229–35 [DOI] [PubMed] [Google Scholar]
- 56.Leins A, Riva P, Lindstedt R, et al. Expression of tenascin-C in various human brain tumors and its relevance for survival in patients with astrocytoma. Cancer 2003;98:2430–9 [DOI] [PubMed] [Google Scholar]
- 57.Brosicke N, van Landeghem FK, Scheffler B, et al. Tenascin-C is expressed by human glioma in vivo and shows a strong association with tumor blood vessels. Cell Tissue Res 2013;354:409–30 [DOI] [PubMed] [Google Scholar]
- 58.Spenle C, Saupe F, Midwood K, Burckel H, Noel G, Orend G. Tenascin-C: Exploitation and collateral damage in cancer management. Cell Adh Migr 2015;9:141–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubuc AM, Mack S, Unterberger A, et al. The epigenetics of brain tumors. Methods Mol Biol 2012;863:139–53 [DOI] [PubMed] [Google Scholar]
- 60.Chiou GY, Chien CS, Wang ML, et al. Epigenetic regulation of the miR142-3p/interleukin-6 circuit in glioblastoma. Mol Cell 2013;52:693–706 [DOI] [PubMed] [Google Scholar]
- 61.Chang CY, Li MC, Liao SL, et al. Prognostic and clinical implication of IL-6 expression in glioblastoma multiforme. J Clin Neurosci 2005;12:930–3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.