Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Atg7 expression is associated with shorter remission duration in AML.
Atg7 inhibition is a proapoptotic phenotype and enhances sensitivity to chemotherapy.
Abstract
Autophagy is a cellular adaptive mechanism to stress, including that induced by chemotherapeutic agents. Reverse phase protein array suggested that high expression of the essential autophagy-related protein, Atg7, was associated with shorter remission in newly diagnosed acute myeloid leukemia (AML) patient samples, indicating a role in chemoresistance. Knockdown of Atg7 in AML cells using short hairpin RNA markedly increased apoptosis and DNA damage following treatment with cytarabine and idarubicin. Interestingly, coculture of AML cells with stromal cells increased autophagy and chemoresistance in the AML cells exposed to chemotherapeutic agents, and this was reversed following Atg7 knockdown. This effect was further enhanced by concomitant knockdown of Atg7 in both AML and stromal cells. These findings strongly suggest that Atg7, and likely microenvironment autophagy in general, plays an important role in AML chemoresistance. Mechanistic studies revealed that Atg7 knockdown induced a proapoptotic phenotype in AML cells, which was manifested by an increased NOXA expression at the transcriptional level. Finally, in a mouse model of human leukemia, Atg7 knockdown extended overall survival after chemotherapy. Thus, the inhibition of Atg7 appears to be a valid strategy to enhance chemosensitivity, and it could indeed improve outcomes in AML therapy.
Introduction
Combinations of cytarabine (Ara-C) and an anthracycline (eg, either idarubicin [Ida] or daunorubicin) are used as frontline induction chemotherapy for patients with acute myeloid leukemia (AML), particularly those younger than 65 years of age.1 Even though the majority of these patients achieve remission, disease relapse is frequent within the first year following treatment.2 As notion of that, the persistence of minimal residual disease (MRD) is involved in disease relapse.3 Hence, a better understanding of survival mechanisms of AML cells, including the protection rendered by their microenvironment during chemotherapy, is critical for improving the depth and the quality of response to agents used in frontline therapy.
Autophagy is an evolutionally conserved pathway that is used by cells to recycle damaged cellular components as a mechanism of adaption to adverse environmental stimuli, including that triggered by exposure to chemotherapeutic agents.4 Indeed, chemotherapy can upregulate autophagy in tumors, which in turn can protect transformed cells from organelle damage and nutrient deprivation.5-7 Moreover, the profoundly hypoxic bone marrow niche is believed to induce autophagy in AML cells.8 Therefore, autophagy induction occurring within this niche, as well as that occurring as a secondary response to chemotherapy, could protect leukemic cells during therapy and contribute to the persistence of MRD.9,10 Finally, autophagy in stromal cells has been implicated in tumor-stroma interdependence.11 Indeed, our proteomic analysis of several autophagy-related proteins (eg, LKB1, Beclin1, and Atg7) in AML blasts and stromal cells showed that the abnormal expression of several autophagy proteins was associated with poor disease outcome.12
Autophagy-related E1 ligase 7 (Atg7) protein is a key molecule in autophagy vesicle elongation, and it is involved in 2 essential ubiquitin-like reactions (ie, LC3 lipidation and Atg5/12 conjugation).13 In carboplatin-treated breast cancer cells, transcriptional upregulation of Atg7 was associated with chemoresistance.14 Conversely, in Down syndrome associated–AML, autophagy is suppressed by mTOR activation.15 In this context, further Atg7 knockdown resulted in an accumulation of defective mitochondria, DNA damage, and enhanced apoptosis.
To gain mechanistic insights into the consequences of autophagy induction/inhibition in response to frontline AML chemotherapy, we conducted experiments testing how the inhibition of Atg7 by genetic silencing could affect the sensitivity of AML cells to chemotherapeutic agents. Second, to study the role of the protein in stroma-mediated AML chemoresistance, we assessed Atg7 in a stromal coculture system to determine the effects of suppression of Atg7 in leukemia cells alone, or concomitant suppression in stromal and AML cells, on the sensitivity of AML cells to chemotherapy. Mechanistically, our results indicate that Atg7 knockdown impairs autophagy, and this then can tilt the survival axis in AML cells to a proapoptotic state, which sensitizes these cells to Ara-C- and Ida-induced apoptosis. In vivo, Atg74 knockdown in AML cells translated into prolonged overall survival of mice in a systemic engraftment human leukemia model. Collectively, our studies support targeting of autophagy regulator Atg7 in the treatment of AML.
Methods
Reagents and antibodies
Ara-C, Ida, and dimethyl sulfoxide (DMSO) were purchased from APP Pharmaceuticals (Schaumberg, IL) and Pfizer (New York, NY), respectively. 3-Methyladenine (3-MA) was obtained from Sigma-Aldrich (St. Louis, MO). Antibodies to Atg5 (A2859) from Sigma-Aldrich Atg7 (2010), Bax (5023), Beclin 1 (3738), CHOP (2895), Cleaved Caspase 3 (9664), LC3-B (2775), PE-conjugated LC3II (8899), p53 (2524), cleaved PARP (9541), and P-PERK (3179) were obtained from Cell Signaling (Danvers, MA). Similarly, antibodies to β-actin (sc-47778), p62 (sc-28359), Bcl-2 (sc-7382), Bcl-XL (sc-56021), PARP (sc-365315), and α-tubulin (sc-53646) were purchased from Santa Cruz (Dallas, TX). Antibodies to Mcl-1 (BD 559027), Noxa (ab13654), γ H2AX (phospho S139) (ab11174), and H2AX antibody (ab11175) were purchased from BD Bioscience and Abcam, respectively.
Reverse phase protein array (RPPA)
Proteomic profiling was performed after fresh samples were enriched for leukemic cells by performing Ficoll-Hypaque (Sigma-Aldrich) density-gradient separation followed by CD3/CD19 depletion. The details of the methodology and validation of the technique have been published earlier.16,17 Briefly, lysates were printed in 5 serial dilutions onto slides along with normalization and expression controls. Slides were probed with 232 strictly validated primary antibodies including the one against Atg7 and a secondary antibody to amplify the signal. The stained slides were analyzed using MicroVigene software (Vigene Tech) to produce quantified data. Normal bone marrow–derived CD34+ cells (N = 11) were used to define normal range of Atg7 expression.
AML cell lines and primary samples
Human leukemic cell lines CCRF-CEM, HL60, K562, MOLM-13, MOLM-16, MOLT-4, OCI-AML2, OCI-AML3, and THP-1 (purchased from American Type Culture Collection [ATCC], Manassas, VA) cells were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal calf serum with penicillin and streptomycin (all from Sigma-Aldrich) at 37°C with 5% CO2 in humidified incubator.
Peripheral blood (PB) primary AML specimens were collected from patients during routine diagnostic assessments in the MD Anderson Cancer Center. The study was conducted in accordance with the Declaration of Helsinki and under a protocol approved by the MD Anderson Cancer Center Investigational Review Board. Peripheral blood mononuclear cells (PBMC) were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal calf serum supplemented with penicillin and streptomycin at 37°C with 5% CO2 in humidified incubator.
Generation of stable Atg7 knockdown AML cell lines using lentiviral transfection system
Atg7 was knocked down by lentiviral transduction using the following shRNAmir transfer vectors (Open Biosystems, Huntsville, AL): V3LHS_405877 for MSCs and V3LHS_342077 for OCI-AML3 and PBMC cells, which target residues 2418 to 2436 and 961 to 979 on RefSeq NM_006395.2, respectively. As a negative control, we used a nonsilencing GIPZ lenti shRNAmir control (Open Biosystems). We generated lentivirus by transfecting HEK 293T cells (ATCC) with an equimolar mixture of the GIPZ plasmid with packaging plasmids pMD2.G and psPAX2 from the laboratory of Didier Trono (plasmids 12259 and 12260, respectively; Addgene, Cambridge, MA) using JetPrime Reagent (Polyplus, Illkirch, France) as directed by the manufacturer. Lentiviral supernatants were harvested 48 hours after transfection and passed through 0.45-μm surfactant-free cellulose acetate membranes. Polybrene (Chemicon, Temecula, CA) was added to 8 μm/mL, and the virus stock was used at once to spinoculate OCI-AML3 cells as described.18 Infected cells were selected with puromycin (Invivogen, San Diego, CA), starting at 1.0 μm/mL. Decreased expression of ATG7 was verified by immunoblot analysis.
The transfection of PBMC cells with small interfering RNA (siRNA) targeting human Atg7 (Sigma SASI_Hs01_00077649; a generous gift from Phil Lorenzi, Department of Bioinformatics and Computational Biology, MD Anderson Cancer Center) or nontarget siRNA was performed using INTERFERin polyplus with minor modification according to manufacturer’s instructions. After a 36-hour transfection, cells were used for experiments outlined below.
Transient transfection with pCDNA-myc-Atg7 (Addgene plasmid 24921) or empty vector was carried out on AML cells using X-treme GENE HP DNA Transfection Reagent (Roche Diagnostic, Indianapolis, IN) following the manufacturer’s instructions with minor modification. Forty-eight hours after transfection, the cells were treated with Ara-C or Ida for different time intervals.
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-Cl [pH 7.5], and 150 mM NaCl) in the presence of 1× protease cocktail inhibitor. Soluble lysates were subjected to SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA). Membranes were probed with specific antibodies. Signals were visualized using Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) and quantitated using Image Studio Lite (LI-COR Biosciences). β-Actin or α-tubulin was used as a loading control.
Apoptosis quantitation by flow cytometry
Apoptosis was analyzed by flow cytometry (Gallios Flow Cytometer; Beckman Coulter, Inc, Brea, CA) following double staining with either annexin V–fluorescein isothiocyanate (FITC) or antigen-presenting cell (APC) and 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) following standard protocols. Briefly, 3 × 105 cells were either treated with DMSO or Ara-C or Ida for specific durations. The cells were collected and washed once with phosphate-buffered saline (PBS) and once with annexin V binding buffer. The resulting pellets were then stained with APC/FITC conjugated annexin V (Roche Diagnostic) antibody in 100 µL annexin-binding buffer followed by incubation for 15 minutes. Cells were then washed twice in annexin binding buffer and resuspended in 300 µL annexin binding buffer and analyzed by flow cytometry. The cells were analyzed in a GALLIOS analytical flow cytometer, with the APC on FL-6 or FITC on FL-1 on the log scale. CountBright Beads (Molecular Probes, Grand Island, NY) and 4′,6-diamidino-2-phenylindole were used to quantitate the number of positive cells as well as the absolute cell number.
Autophagy detection by flow cytometry
Phycoerythrin-conjugated specific LC3-II antibody was used for quantification of autophagy following standard protocol for flow cytometry. Briefly, healthy cells of log phase were treated with drug for specific time points. The cells were pelleted by centrifugation and washed with ice-cold PBS followed by permeabilization by adding ice-cold 100% methanol slowly to prechilled cells, while gently vortexing, to a final concentration of 90% methanol. After incubation for 30 minutes on ice, the cells were washed with ice-cold PBS. The cells were resuspended in 100 µL of incubation buffer (PBS containing 0.5% bovine serum albumin) with 1 µL specific antibody and then incubated at 37°C in the dark for 1 hour. After incubation, cells were collected by centrifugation and washed twice in incubation buffer. Finally, the cell pellets were resuspended in 300 µL of PBS and analyzed by flow cytometry.
Autophagy detection by Cyto-ID autophagy detection kit
The Cyto-ID autophagy dye was used for quantification of autophagy following instructions provided in the manufacturer’s manual. Briefly, 5 × 105 cells were collected after treatment with DMSO, Ara-C, or Ida. The 10× assay buffer was allowed to warm to room temperature and then diluted to 1× (with deionized H2O). The working concentration of Cyto-ID Green autophagy dye solution was prepared by mixing 8 µL of the dye and 4 mL of 1× assay buffer. Cells were washed twice and adjusted to a final concentration of ∼5 × 105 cells/mL. After centrifugation for 5 minutes at 1400 rpm, 500 µL of the working dye was added to each cell pellet, resuspended, and incubated for 30 minutes at 37°C, followed by a wash and resuspension with 500 µL of 1× assay buffer before flow cytometry analysis.
In vivo model of human AML
All animal studies were conducted in accordance with the guidelines approved by the Institutional Animal Care and Use Committees at the University of Texas MD Anderson Cancer Center. Forty male NOD/SCID/IL-2rγnull (NSG) mice (7-weeks-old; Jackson Laboratory, Bar Harbor, MA) were intravenously injected with OCI-AML3 cells (1 × 106 cells/100 µL) transduced with a nontargeted short hairpin RNA (shRNA; OCIAML3: ShControl, n = 20 mice) or ATG7 shRNA (OCIAML3: ShAtg7, n = 20 mice). The mice were monitored daily for evidence of leukemia. After 7 days, the mice with each cell type were chosen randomly for treatment with vehicle or Ara-C (100 mg/kg, intraperitoneally 3 times/wk) for 2 weeks, n = 10 each. On day 29, PB was collected through intraorbital route and processed to measure leukemic burden as green fluorescent protein (GFP)/hCD45-positive cells by flow cytometry as described previously.18 The spleen size from the euthanized mice was measured. The survival of mice is also followed and presented with a Kaplan-Meier survival plot.
Statistical analyses
All data are expressed as the mean ± standard deviation and are representative of triplicate samples. Statistics were prepared using Graphpad Prism 6 software. Kaplan-Meier survival curve was employed to compare the in vivo survival data. Normally distributed groups were compared by 2-tailed Student t test. P ≤ .05 was considered statistically significant. For RPPA data, Super curve algorithms were used to generate a single value from the 5 serial dilutions. Loading control and topographical normalization procedures accounted for protein concentration and background staining variations. The Kaplan-Meier method was used to generate the relapse-free survival curve.
Results
High Atg7 levels in AML blasts are associated with shorter remission duration
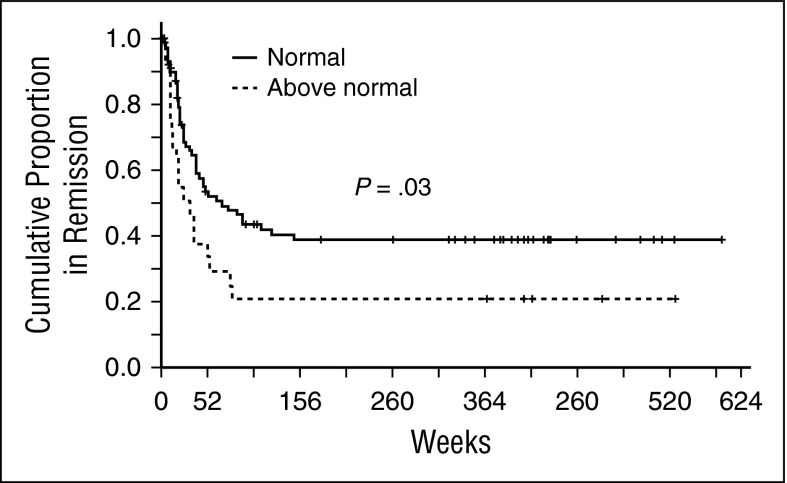
We hypothesized that Atg7 related chemoresistance and allowed for persistence of MRD, which would be clinically relevant in remission duration. To avoid sample heterogeneity, we used only protein samples prepared from fresh blasts on the day of collection (excluding samples prepared from cryopreserved cells) for RPPA analysis. Fresh samples were available from 185 patients with newly diagnosed AML treated with frontline regimens (82% Ara-C-based regimens and 18% low-intensity regimens). Distribution of regimen intensity and remission rates were balanced among Atg7 expressing groups (both P = .97). Among 106 patients achieving complete remission, duration in patients with higher Atg7 levels (N = 26) was shorter than those expressing levels comparable (N = 80) to normal marrow–derived CD34+ cells (median 23 versus 49 weeks, P = .03) (Figure 1). ATG7 high versus normal expression groups were balanced for age, performance status, FLT3-ITD mutation, white blood cell count, and blast percentage at presentation. The number of patients with unfavorable cytogenetics (P = .02) is lower and NPM1 mutation is higher (P = .01) in the ATG7 high group.
Figure 1.
High Atg7 levels in leukemic AML blasts are associated with shorter remissions. Fresh samples were available from patients with newly diagnosed AML treated with age-appropriate frontline regimens. Among 106 patients achieving complete remission, duration in patients with higher Atg7 (dashed line) levels was shorter than those expressing levels comparable to normal marrow–derived CD34+ cells (continuous line) (median 23 versus 49 weeks, P = . 03).
Enhancement of Ara-C- and Ida-induced apoptosis in AML cells by Atg7 suppression
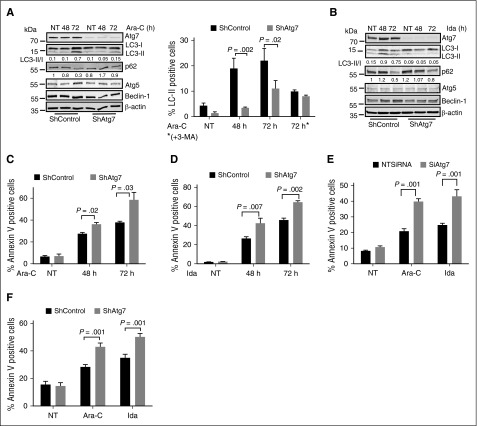
Autophagy can be induced in cancer cells by treatment with purine analogs or anthracyclines, and it can decrease the cytotoxic response to these agents.19 The purine analog Ara-C and anthracycline-like agent Ida are frontline therapies for AML. We confirmed by immunoblotting that both Ara-C and Ida individually induced autophagy in AML cells. Autophagosome maturation and lysosomal degradation were robustly induced in a time-dependent manner by these agents, as indicated by changes in LC3-II/-I ratio and reduced p62 levels, respectively (Figure 2A-B lanes 2 and 3). In agreement with the western blot data, treatment with Ara-C and Ida increased the number of LC3-II-positive cells shown by flow cytometry. However, this was reversed by Atg7 knockdown (Figure 2A right panel). 3-MA is an inhibitor of type III phosphatidylinositol 3-kinases that are involved in autophagy.20 3-MA treatment of AML cells reduced the number of LC3-II-positive cells after exposure to Ara-C, but did not add to the effect associated with the Atg7 knockdown (Figure 2A right panel), suggesting that Atg7 knockdown alone adequately disable autophagy.
Figure 2.
Atg7 knockdown enhances chemosensitivity in AML cells. (A) Atg7 knockdown (ShAtg7) or nonsilencing scrambled control (ShControl) OCI-AML3 AML cells were treated with Ara-C (2 μM) or (B) Ida (5 ng/mL) for different time intervals, and total lysates were immunoblotted to detect autophagy. Duplicate samples were stained with autophagy marker LC3-II to quantify autophagy using flow-based assay (A right panel). Similarly, apoptosis was measured using annexin V binding after treatment with (C) Ara-C (2 μM) and (D) Ida (5 ng/mL) for different time intervals, respectively. AML patient samples were either transfected or transduced with siRNA (E) or shRNA (F) targeting Atg7 for 48 hours, and then cells were further treated with Ara-C (2 μM) and Ida (5 ng/mL) for 72 hours. The apoptosis was measured using flow cytometry (annexin binding). Lysates were immunoblotted with Atg7 antibody to confirm knockdown of Atg7 (data not shown). The statistical significance of percent apoptosis in ShAtg7 compared with ShControl was calculated by the Student t test.
To determine the functional significance of autophagy inhibition in AML cells, apoptosis (annexin V-positive cells) was assessed in OCI-AML3 cells after treatment with Ara-C (Figure 2C) or Ida (Figure 2D) and was higher in Atg7 knockdown cells (P = .03 and .002 at 72 hours, respectively). To confirm the role of autophagy inhibition in AML cells’ survival and proliferation, we further performed clonogenic assays in chemotherapy-treated AML cells with wild-type (WT) or Atg7 knockdown. Interestingly, clonogenic growth was markedly suppressed in Atg7 knockdown cells compared with the WT counterpart (supplemental Figure 1A-B, available on the Blood Web site). Therefore, Atg7 knockdown enhanced the toxic effect of both drugs, indicating that autophagy inhibition enhances the cytotoxic effects of chemotherapeutic agents in AML cells. In corollary to this, treatment of 3-MA that blocks autophagosome formation by inhibition of class III PI3K3 recapitulates the effect of Atg7 knockdown in OCI-AML3 (supplemental Figure 1C).
Confirming relevance of these findings in primary AML cells, Atg7 knockdown with siRNA or lentiviral shRNA enhanced the apoptotic effect of both Ara-C and Ida (Figure 2E-F; supplemental Figure 2A-B).
Furthermore, we tested the effect of Ara-C and Ida on ATG7WT, and ATG7−/− mouse embryonic fibroblast (MEF) cell proliferation and clonogenic growth. Although cell proliferation did not appear to be substantially different in ATG7−/− MEFs, there was a reduction, albeit not a statistically significant one, in clonogenic growth in the treated ATG7−/− MEFs compared with their WT counterparts (supplemental Figure 3A-B).
Atg7 expression protects leukemic cells from chemotherapy-induced cell death
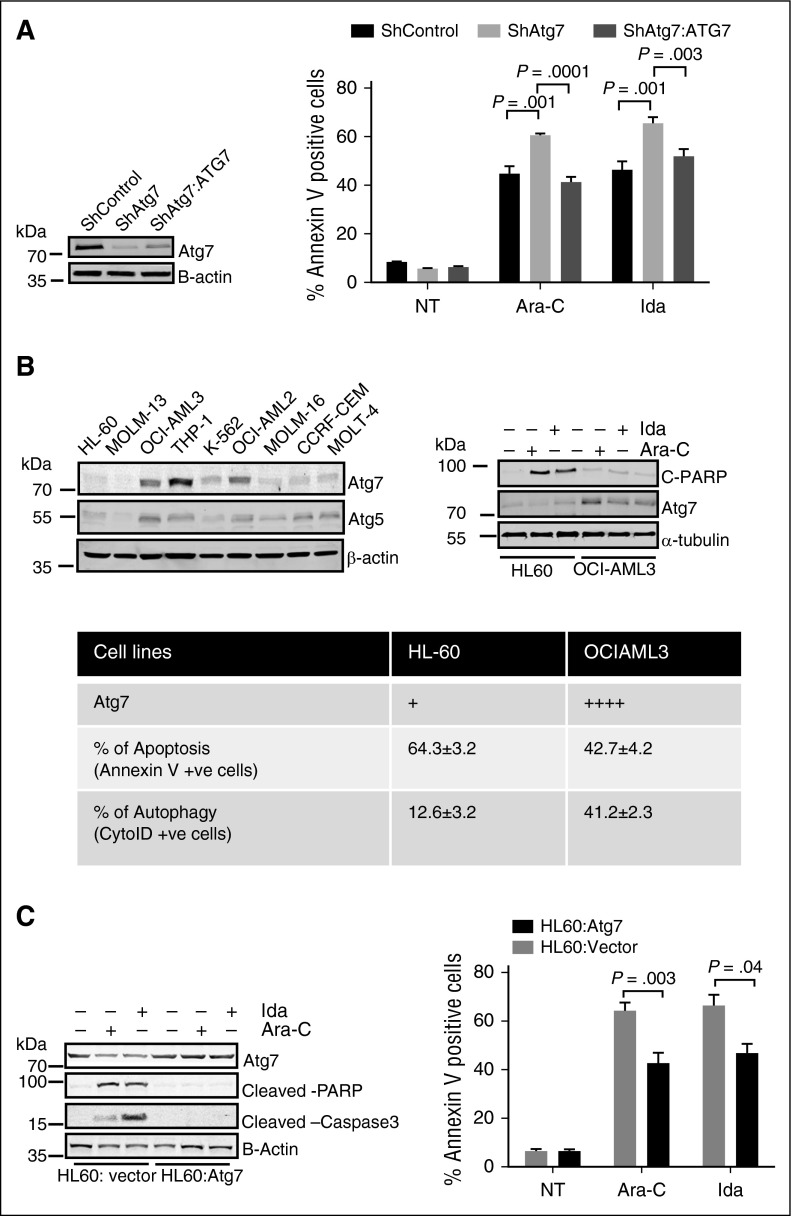
To verify the role of Atg7 and autophagy in chemoresistance, we performed complementation experiments in which we transiently reintroduced a form of Atg7, unaffected by the Atg7-targeting shRNA, in stable shAtg7-expressing cells. Restoration of Atg7, attenuated apoptosis, and cell growth inhibition were caused by treatment with Ara-C or Ida (Figure 3A).
Figure 3.
Atg7 expression is correlated with associated chemoresistance in AML cells. (A) ShControl, stable Atg7 knockdown, and stable Atg7 knockdown transiently overexpressing Atg7 cells were treated with Ara-C (2 μM) and Ida (5 ng/mL) for 72 hours. The flow cytometry was used to measure apoptosis. Whole cell lysate was subjected to immunoblotting for Atg7 (box). β-Actin was used as loading control. (B) Whole cell lysates of AML cell lines (THP1, OCI-AML2 and 3, HL-60, MOLT-4, MOLM 13 and 16) separated on SDS gel electrophoresis and immunoblotted with Atg7 (left panel). HL60 and OCI-AML3 cells were treated with Ara-C (2 μM) and Ida (5 ng/mL) for indicated time periods. The whole cell lysates were immunoblotted with cleaved PARP and cleaved caspase-3 (right panel). Tubulin was used as loading control. Apoptosis was measured using flow cytometry, and autophagy was measured by CytoID kit (bottom table). (C) Ectopic Atg7-expressed HL60 cells were treated with Ara-C (2 μM) and Ida (5 ng/mL) for 48 hours. Lysates were immunoblotted with Atg7, cleaved caspase-3, and cleaved PARP-specific antibody (left panel). β-Actin was used as the loading control. The apoptosis (right panel) was measured using flow cytometry. The statistical significance between the 2 compare groups was calculated by a standard Student t test.
To assess further whether Atg7 expression, and presumably autophagy in general, protects AML cells from chemotherapy, we first semiquantitatively assessed Atg7 protein levels by immunoblot in different AML cell lines (Figure 3B left top panel). We then compared sensitivity to, and autophagy induction by, chemotherapeutic agents in lines with high (OCI-AML3) or low (HL-60) levels of Atg7. As shown in Figure 3B (bottom/top right panel), Atg7-high OCIAML3 cells showed more autophagy induction (ie, increased CytoID-positive cells) and less apoptosis (less cleaved PARP and annexin V-positive cells) in response to Ara-C. Overexpression of Atg7 in HL-60 cells reduced sensitivity to Ara-C and Ida, as indicated by decreases in cleaved PARP, cleaved Caspase 3 (Figure 3C left panel), and annexin V staining (Figure 3C right panel). Finally, to confirm the role of autophagy in adaption to nutrient deprivation, Atg7 knockdown OCI-AML3 cells underwent apoptosis at a higher rate than their control counterparts upon glutamine and serum deprivation (supplemental Figure 4A-B). Together, these data confirm that Atg7 plays an important role in AML cell survival as well as resistance to Ara-C- and Ida-induced cytotoxicity.
Atg7 plays a critical role in stroma-mediated protection of AML cells
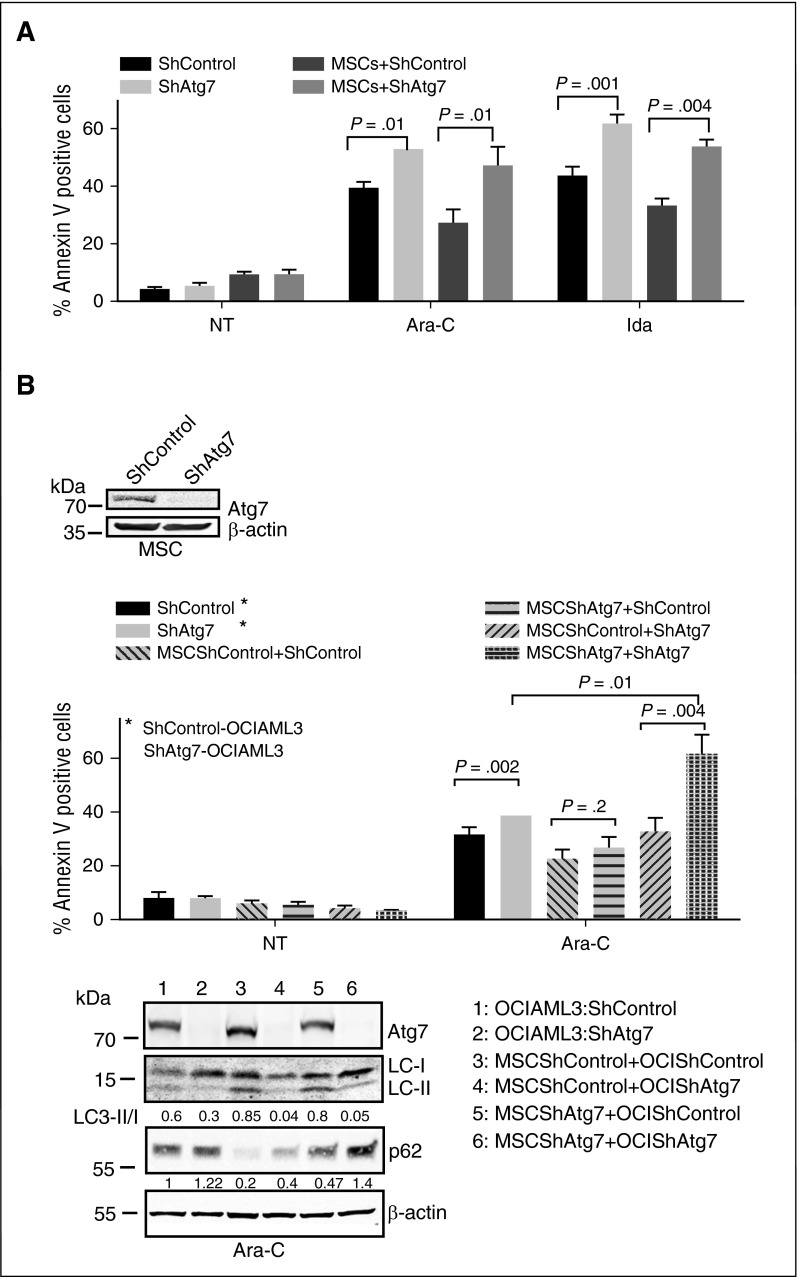
Persistent AML MRD in the bone marrow niche can result in relapse after frontline chemotherapy.21 The bone marrow microenvironmental niche contributes to chemoresistance by providing prosurvival signals and critical nutrients and assisting in the adaption to hypoxia.22,23 To better understand the potential role of autophagy in the AML microenvironment and its possible contribution to MRD, specifically that of Atg7 in stroma-mediated chemoresistance, we cocultured Atg7 or control knockdown OCI-AML3 cells with normal bone marrow–derived mesenchymal stromal cells (MSCs) to mimic the bone marrow microenvironment. Under these conditions, shControl OCI-ML3 cells were rendered resistant to Ara-C or Ida, and this resistance was partially abrogated in shAtg7 cells (Figure 4A).
Figure 4.
Atg7 knockdown abrogates marrow/stroma-induced chemoprotection and enhances susceptibility to chemotherapeutic stress. OCI-AML3 cells (ShControl or ShAtg7) were cultured with (A) monolayer of normal bone marrow–derived stromal cells or (B) stromal cells (ShControl or ShAtg7) and treated with Ara-C (2 μM) and Ida (5 ng/mL) for 72 hours, and apoptosis was assessed by annexin V binding (flow cytometry). Lysates were immunoblotted to Atg7 and β-actin to confirm knockdown of Atg7 and loading control, respectively (box). Duplicate whole cell lysates were separated on SDS-polyacrylamide gel electrophoresis and probed with LC3 and p62. β-Actin was used as loading control (bottom panel).
We confirmed by immunoblot that coculture of AML cells with MSCs indeed induces autophagy in OCI-AML3 cells (ie, inducing a higher LC3 II/I ratio) (Figure 4B lane 3 bottom panel), which is abrogated by Atg7 knockdown in both OCI-AML3 and MSCs (Figure 4B lane 6 bottom panel). When Atg7 was knocked down individually in MSCs or OCI-AML3 cells, or in both, it was interesting to observe that while Atg7 knockdown in the MSCs alone did not overcome the chemoprotection provided to OCI-AML3 cells, Atg7 knockdown in both cell types maximized the apoptotic activity of the chemotherapeutic agents (Figure 4B top panel bar 12; supplemental Figure 5), suggesting that autophagy was a critical element of chemoresistance that perhaps arises from a bidirectional stroma-leukemia cell interaction. These findings are clinically relevant because inhibitors of autophagy/Atg7 are likely to inhibit this process in both AML and stroma cells in the bone marrow microenvironment.
Atg7 knockdown alters expression of Bcl-2 family proteins and results in sustained DNA damage following treatment with chemotherapeutic agents
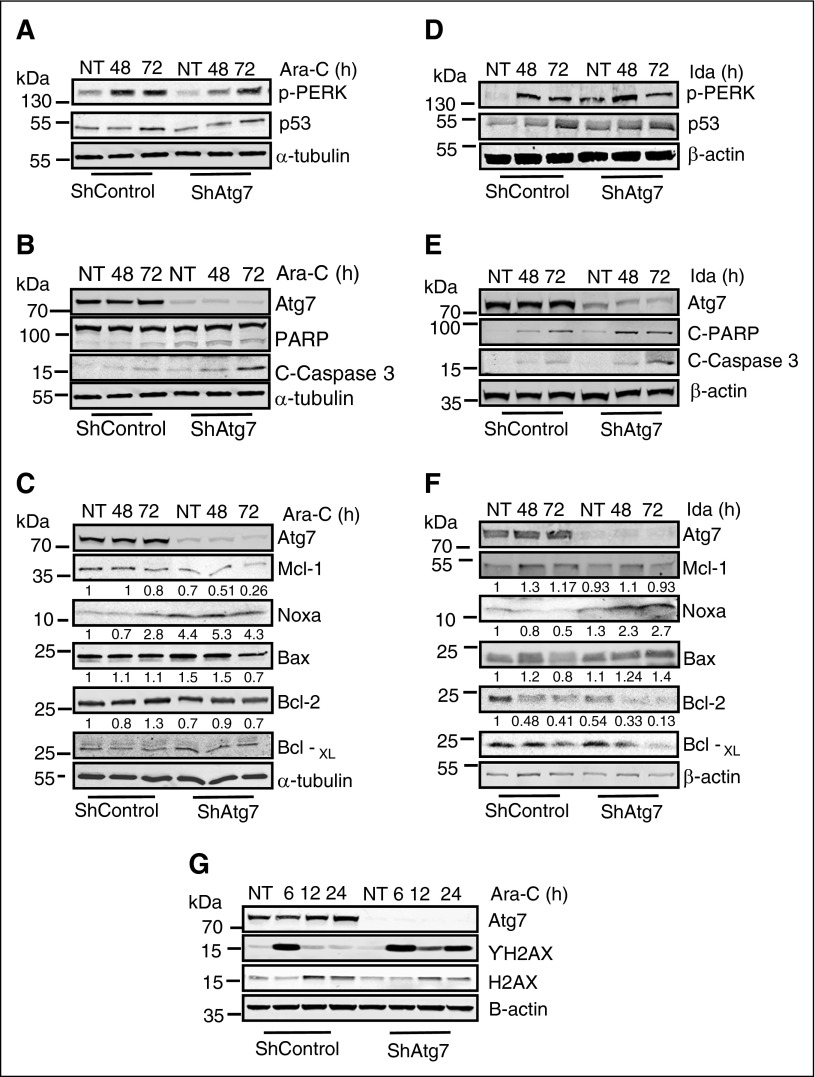
To gain mechanistic insight into possible effector molecules involved in the increased chemosensitivity resulting from Atg7 ablation, we studied changes in the Bcl-2 family and endoplasmic reticulum stress proteins in OCI-AML3 cells. Induction of p-PERK (a marker of ER stress) by Ara-C or Ida was largely unaffected by Atg7 knockdown (Figure 5A,D). Conversely, both baseline and posttreatment levels of the proapoptotic Bcl-2 family member NOXA were increased in Atg7 knockdown cells. In addition, levels of antiapoptotic Mcl-1 were lower in shAtg7 cells after treatment with Ara-C and Ida (Figure 5C,F, respectively). Thus, with the shift in the ratio between these 2 apoptosis regulators, it appears that Atg7 knockdown cells are primed to undergo apoptosis. This theory is supported by higher levels of cleaved PARP and caspase 3 observed in the shAtg7 cells after treatment with Ara-C or Ida (Figure 5B,E). These cells were also seemingly primed for enhanced DNA damage or suppression of DNA repair because ablation of Atg7 resulted in a sustained phosphorylation of histone H2AX (Figure 5G), indicating double-stranded DNA damage.24 Thus, Atg7 silencing appears to create an AML phenotype that is more primed toward apoptosis and DNA damage.
Figure 5.
Examination of changes in Bcl-2 family proteins and ER stress response proteins after treatment with Ara-C and Ida. Atg7 knockdown cells (ShAtg7) or nonsilencing scrambled control (ShControl) OCI-AML3 AML cell lines were treated with Ara-C (2 μM) or Ida (5 ng/mL) for indicated time intervals, and total lysates were immunoblotted to detect biochemical markers of ER stress (A and D), regulation of apoptosis (B and E), and alteration of Bcl-2 family proteins (C and F). Similarly, immunoblotting detected DNA damage (G) in indicated time intervals in response to Ara-C and Ida. β-Actin was used as a loading control.
We confirmed the increase in transcript levels of NOXA by quantitative polymerase chain reaction at baseline and after exposure to Ara-C in OCI-AML3 shAtg7 cells. Compared with scramble controls, transcript levels of Bcl-2, Mcl-1, and Bcl-XL were comparable (supplemental Figure 6). To confirm the critical role of NOXA, we knocked down NOXA in OCI-AML3 Atg7 knockdown cells using siRNA. NOXA knockdown reverses the apoptotic phenotype in Ara-C- and Ida-treated cells (supplemental Figure 7A-B).
Silencing of Atg7 improves overall survival in a systemic human AML model
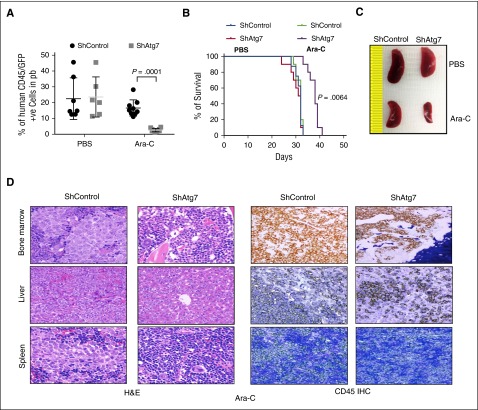
To investigate the in vivo effect of Atg7 silencing in AML chemotherapy, we established a systemic human AML mouse model using OCI-AML3 cells transduced with either a nontargeted shRNA OCI-AML3 (control) or ATG7 shRNA introduced to NSG mice via tail vein injection.25 On the seventh day after injection, the mice were treated with Ara-C at 100 mg/kg 3 times/wk for 2 weeks (PBS was used as the untreated control). Leukemia burden was followed by flow cytometry detection of human CD45+GFP+ circulating cells in the PB of the mice. On day 29 after treatment with Ara-C, the leukemia burden in the mice bearing OCI-AML3 shAtg7 cells was significantly lower than the control mice (P = .0001, Figure 6A). In addition, a Kaplan-Meier plot in Figure 6B demonstrates that mice bearing OCI-AML3 shAtg7 survived longer after Ara-C therapy compared with control mice treated with Ara-C (P = .0064). The leukemia burden was further characterized by spleen size and immunohistochemistry. As shown in Figure 6C, spleen enlargement was significantly less in shAtg7 AML mice compared with the control mice. Immunohistochemical studies confirmed a lower leukemia burden in the femurs, spleens, and livers of mice bearing OCI-AML3 shAtg7 cells compared with controls after Ara-C treatment (Figure 6D). These results strongly suggest that the suppression of Atg7 in leukemic cells can improve outcome during chemotherapy of human AML.
Figure 6.
Atg7 knockdown improves antileukemic activity of Ara-C in OCI AML3 in NSG model of engrafting leukemia. NOD-SCID-IL2Rϒ null mice were injected with Atg7 knockdown or ShControl OCI-AML3 (1 × 106) through tail vein and Ara-C (100 mg/kg) administered on days 7, 9, 11, 14, 16, 18, and 24 through intraperitoneal route. At day 29, 50 μL of PB was collected from intraorbital bleeding and tumor burden was measured by detection of double-positive (GFP/human CD45) cells (A). Two mice from each group were euthanized to assess splenomegaly (C) and infiltration of tumor in other organs (D). A plot of overall survival probability, estimated with the Kaplan-Meier method, is shown in (B). Original magnification ×40 for panel D.
Discussion
With few exceptions, intensification of frontline AML therapy has not improved treatment outcomes. Mutational profiling can provide potential therapeutic targets (eg, FLT3, RAS, DNMT3A, and IDH).26-28 However, multiple mutations can coexist in different clones; new mutations can be acquired, and many epigenetic mutations cannot currently be targeted.29 Thus, additional cell survival pathways need to be identified that can potentially overcome to potentially prevent the development of chemoresistance and MRD. Several lines of evidence have identified autophagy as such a pathway, and it contributes to resistance even in a setting of mutation-targeted therapy.30 For example, autophagy induced in stem cells in response to tyrosine kinase inhibitors such as imatinib, dasatinib, or nilotinib promotes persistence of chronic myeloid leukemia stem cells.31 Preclinical data suggest a role of autophagy inhibitors in combination with tyrosine kinase inhibitors, for the elimination of leukemic stem cells in chronic myeloid leukemia.32 Similarly, chemotherapy-induced autophagy in AML and acute lymphocytic leukemia cell lines9 has been shown to be cytoprotective.19
Atg7, being an E1 ligase, is potentially targetable, and targeting Atg7 largely disables the autophagic machinery in AML cells.33 From a drug development perspective, it is exciting to see that recently several small molecules have been developed that specifically target components of autophagy network. These targets include vps34, a class III PI3Kinase specific for autophagy, ULK1, the apical kinase for autophagy, and AMPK1, a kinase that activates ULK1.34,35 However, because of the current lack of a small-molecule inhibitor of Atg7, we used RNA interference to suppress Atg7 and to illustrate proof of concept for this study. Our results show that Atg7 knockdown in AML cells results in a proapoptotic phenotype with increased chemosensitivity that translates to improved overall survival in vivo. In addition, Atg7 expression levels trended with chemoresistance in AML cells, and complementation or ectopic expression of Atg7 in low Atg7 expressing HL60 cells confirmed the effect on chemoresistance mediated by Atg7. This data parallel our RPPA data, suggesting the negative impact of high Atg7 expression on remission duration in patients with newly diagnosed AML. We hope to expand this analysis to a larger cohort of AML patients in future studies.
Because autophagy can have prosurvival and prodeath effects, it is not surprising that autophagy intersects at various points with apoptosis. Phosphorylation of Beclin 1 or Bcl-2 dissociates Beclin 1 from its inhibition by Bcl-2 and allows autophagy to proceed.36 Apart from its role as an E1 ligase, Atg7 binds to p53 to influence p53-mediated transcription. In the absence of Atg7, p53-mediated transcription in response to prolonged metabolic stress induced DNA damage, shifts to a proapoptotic pattern rather than cell-cycle arrest, resulting in lower p21 and higher levels of NOXA, BAX, and PUMA.37 This mechanism could potentially explain our finding that levels of NOXA (both protein and RNA) were higher even at baseline in Atg7 knockdown cells and increased further after treatment with chemotherapy. Our NOXA knockdown experiments confirm the key role of NOXA transcription in this context.
An important and novel aspect of our study is in defining the role of Atg7 in leukemia-stroma interaction.23 Although chemokine or adhesion molecules have been studied for their role in stroma-mediated chemoresistance in leukemia, the role played by autophagy is underinvestigated. In the present study, we demonstrated that coculture with stroma enhanced autophagy in AML cells and that concomitant inhibition of autophagy in both leukemia and stroma, a scenario potentially achievable with Atg7 inhibitors in clinic, reduced chemoresistance and holds great promise for improving response to chemotherapy. Our data also confirm that Atg7 knockdown puts AML cells at a disadvantage in adapting to nutrient stress that is prevalent in the bone marrows of AML patients.38
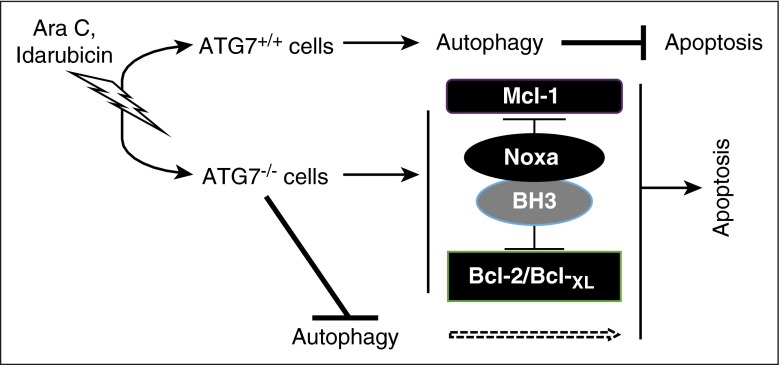
In conclusion, our data support the strategy of targeting Atg7 to overcome chemoresistance in AML, including that provided by the stromal microenvironment. Based on our findings, we propose the apoptotic response to genotoxic stress caused by Ara-C and Ida, at least part, is dependent on Atg7/autophagy (Figure 7). Suppression of Atg7 is accompanied by a modulation of Bcl-2 family proteins that prime the cell for apoptosis. Atg7 knockdown also enhances DNA damage after exposure to chemotherapy. In agreement with this observation, recent studies have demonstrated that inhibition of chaperon-mediated autophagy renders cells susceptible to genotoxic agents via nuclear retention of CHK1 protein and increases DNA damage.39 Thus, inhibition of Atg7, a potentially druggable target in the autophagy network, merits further evaluation for the treatment of AML.
Figure 7.
Model of Atg7-mediated AML chemoresistance. Chemotherapy exerts ER stress/genotoxic stress, and cell fate depends on the type of cells. If they are Atg7+/+ cells, stress might be mitigated with autophagy to inhibit apoptosis. However, if they are Atg7−/− cells, stress will lead to an increase in Noxa and a decrease in its counterpart Mcl-1, resetting apoptosis regulation at the mitochondrial outer membrane to induce the mitochondrial-mediated pathway of cell death.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (National Cancer Institute P01 CA055164 and MD Anderson’s Cancer Center Support grant CA016672) and the Paul and Mary Haas Chair in Genetics to M.A.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.P. and G.B. designed experiments, analyzed data, and wrote the paper. S.P. performed experiments. H.M., V.R.R., P.P.R., and R.E.D. supported the study. N.H. edited the paper. S.M.K. generated and analyzed RPPA data and edited the paper. M.A. supported the study. T.M. managed the stromal and primary AML cells. H.K. reviewed the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gautam Borthakur, Department of Leukemia, University of Texas, MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 0448, Houston, TX 77030; e-mail: gborthak@mdanderson.org.
References
- 1.Padron E, Fernandez H. Anthracycline dose intensification in young adults with acute myeloid leukemia. Ther Adv Hematol. 2012;3(1):17–27. doi: 10.1177/2040620711427069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El Rassi F, Arellano M. Update on optimal management of acute myeloid leukemia. Clin Med Insights Oncol. 2013;7:181–197. doi: 10.4137/CMO.S8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paietta E. Minimal residual disease in acute myeloid leukemia: coming of age. Hematology Am Soc Hematol Educ Program. 2012;2012:35-42. [DOI] [PubMed]
- 4.White EJ, Martin V, Liu JL, et al. Autophagy regulation in cancer development and therapy. Am J Cancer Res. 2011;1(3):362–372. [PMC free article] [PubMed] [Google Scholar]
- 5.Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24(1):42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu YL, DeLay M, Jahangiri A, et al. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res. 2012;72(7):1773–1783. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui X, Chen R, Wang Z, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan JL, Simon AK, Prescott M, et al. Autophagy in stem cells. Autophagy. 2013;9(6):830–849. doi: 10.4161/auto.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabe Y, Konopleva M. Advances in understanding the leukaemia microenvironment. Br J Haematol. 2014;164(6):767–778. doi: 10.1111/bjh.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabe Y, Konopleva M. Role of microenvironment in resistance to therapy in AML. Curr Hematol Malig Rep. 2015;10(2):96–103. doi: 10.1007/s11899-015-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borthakur G, Andreeff M, Coombes KR, et al. High expression of autophagy related proteins negatively impacts clinical outcomes in acute myelogenous leukemia—time to target autophagy to improve therapy outcomes? [abstract]. Blood. 2011;118(21). Abstract 2513. [Google Scholar]
- 13.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20(1):21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai S, Liu Z, Yao J, et al. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J Biol Chem. 2013;288(13):9165–9176. doi: 10.1074/jbc.M112.422071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stankov MV, El Khatib M, Kumar Thakur B, et al. Histone deacetylase inhibitors induce apoptosis in myeloid leukemia by suppressing autophagy. Leukemia. 2014;28(3):577–588. doi: 10.1038/leu.2013.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornblau SM, Coombes KR. Use of reverse phase protein microarrays to study protein expression in leukemia: technical and methodological lessons learned. Methods Mol Biol. 2011;785:141–155. doi: 10.1007/978-1-61779-286-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5(10):2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 18.Ruvolo VR, Karanjeet KB, Schuster TF, et al. Role for PKC delta in fenretinide-mediated apoptosis in lymphoid leukemia cells. J Signal Transduct. 2010;2010:584657. [DOI] [PMC free article] [PubMed]
- 19.Bosnjak M, Ristic B, Arsikin K, et al. Inhibition of mTOR-dependent autophagy sensitizes leukemic cells to cytarabine-induced apoptotic death. PLoS One. 2014;9(4):e94374. doi: 10.1371/journal.pone.0094374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar D, Das B, Sen R, et al. Andrographolide Analogue Induces Apoptosis and Autophagy Mediated Cell Death in U937 Cells by Inhibition of PI3K/Akt/mTOR Pathway. PLoS One. 2015;10(10):e0139657. doi: 10.1371/journal.pone.0139657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyerinas B, Zafrir M, Yesilkanal AE, Price TT, Hyjek EM, Sipkins DA. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood. 2013;121(24):4821–4831. doi: 10.1182/blood-2012-12-475483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weekes CD, Kuszynski CA, Sharp JG. VLA-4 mediated adhesion to bone marrow stromal cells confers chemoresistance to adherent lymphoma cells. Leuk Lymphoma. 2001;40(5-6):631–645. doi: 10.3109/10428190109097661. [DOI] [PubMed] [Google Scholar]
- 23.Nauta AJ, Fibbe WE. Immunomodulatory properties of mesenchymal stromal cells. Blood. 2007;110(10):3499–3506. doi: 10.1182/blood-2007-02-069716. [DOI] [PubMed] [Google Scholar]
- 24.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 25.Carter BZ, Mak PY, Mak DH, et al. Synergistic targeting of AML stem/progenitor cells with IAP antagonist birinapant and demethylating agents. J Natl Cancer Inst. 2014;106(2):djt440. doi: 10.1093/jnci/djt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Konopleva M, Shi YX, et al. Mutant FLT3: a direct target of sorafenib in acute myelogenous leukemia. J Natl Cancer Inst. 2008;100(3):184–198. doi: 10.1093/jnci/djm328. [DOI] [PubMed] [Google Scholar]
- 27.Bowen DT, Frew ME, Hills R, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106(6):2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 28.Im AP, Sehgal AR, Carroll MP, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014;28(9):1774–1783. doi: 10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar CC. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer. 2011;2(2):95–107. doi: 10.1177/1947601911408076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Bhatia R. Role of SIRT1 in the growth and regulation of normal hematopoietic and leukemia stem cells. Curr Opin Hematol. 2015;22(4):324–329. doi: 10.1097/MOH.0000000000000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamad A, Sahli Z, El Sabban M, Mouteirik M, Nasr R. Emerging therapeutic strategies for targeting chronic myeloid leukemia stem cells. Stem Cells Int. 2013;2013:724360. [DOI] [PMC free article] [PubMed]
- 32.Bellodi C, Lidonnici MR, Hamilton A, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119(5):1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 34.Dowdle WE, Nyfeler B, Nagel J, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16(11):1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 35.Egan DF, Chun MG, Vamos M, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell. 2015;59(2):285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiuri MC, Criollo A, Kroemer G. Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J. 2010;29(3):515–516. doi: 10.1038/emboj.2009.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee IH, Kawai Y, Fergusson MM, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336(6078):225–228. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiegl M, Samudio I, Clise-Dwyer K, Burks JK, Mnjoyan Z, Andreeff M. CXCR4 expression and biologic activity in acute myeloid leukemia are dependent on oxygen partial pressure. Blood. 2009;113(7):1504–1512. doi: 10.1182/blood-2008-06-161539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun. 2015;6:6823. doi: 10.1038/ncomms7823. [DOI] [PMC free article] [PubMed] [Google Scholar]