Abstract
Angiotensin Converting Enzyme Inhibitors (ACEI) and Angiotensin II Receptor Blockers (ARB) are two common medication classes used for heart failure treatment. The ADAHF (Automated Data Acquisition for Heart Failure) project aimed at automatically extracting heart failure treatment performance metrics from clinical narrative documents, and these medications are an important component of the performance metrics. We developed two different systems to detect these medications, rule-based and machine learning-based. The rule-based system uses dictionary lookups with fuzzy string searching and showed successful performance even if our corpus contains various misspelled medications. The machine learning-based system uses lexical and morphological features and produced similar results. The best performance was achieved when combining the two methods, reaching 99.3% recall and 98.8% precision. To determine the prescription status of each medication (i.e., active, discontinued, or negative), we implemented a SVM classifier with lexical features and achieved good performance, reaching 95.49% accuracy, in a five-fold cross validation evaluation.
Keywords: Heart Failure, Angiotensin Converting Enzyme Inhibitors, Angiotensin II Receptor Blockers, Biomedical Informatics, Natural Language Processing
Introduction
Heart Failure (HF) is characterized by the inability of the heart to pump blood at a rate sufficient to answer metabolizing tissues needs. It is a frequent condition in the U.S. adult population, causing more hospitalizations than all forms of cancer combined.[1] HF treatment can include dietary and physical activity therapies and invasive therapies, but pharmacologic therapies are the most common. Among pharmacologic therapies, Angiotensin converting enzyme inhibitors (ACEI) are a mainstay of treatment in patients who can tolerate them; for patients who cannot take these drugs, Angiotensin receptor blockers (ARB) agents offer an alternative. Assessment of the care HF patients benefit from relies on detecting evidence of current treatment with these medications in clinical notes, along with analysis of the status of these medications. This information, combined with the functional assessment of the left ventricular function, is a key component of HF treatment performance measures.
This study was realized in the context of the ADAHF (Automated Data Acquisition for Heart Failure) project, a U.S. Department of Veterans Administration (VA) project which aims to automatically extract data for HF treatment performance measures from clinical notes. These performance measures include left ventricular ejection fraction (LVEF) assessments (their mention and measured values), medications (ACEIs and ARBs), or reasons not to administer these medications, as part of the Joint Commission National Hospital Quality Measures Heart Failure Core Measure Set that the VA has adopted for use in Veteran care.
Previous efforts have focused on detecting LVEF mentions and values in clinical notes and determining their abnormality (LVEF < 40%).[2–4]
However, to assess whether each patient’s clinical record substantiates proper treatment for HF, active medications should be considered as well. In this study, we focused on ACEIs and ARBs detection and classification of the status of these detected medications.
Clinical notes are mostly composed of unstructured text and have multiple different formats depending on specialties and institutions. Misspelled words and non-narrative text formats can be found in clinical notes, especially when directly typed into the system by healthcare providers. For example, one of the medications included in our study, “Losartan”, was found with the following issues:
“Lorsartan” (misspelled word)
“Losartan30 mg” (tokenization problem: missing whitespace)
“L [newline] osartan” (word wrapping problem: word wrongly split)
These problems make medication detection based on a simple dictionary lookup more difficult.
Medications can be mentioned in multiple contexts in clinical notes. Most are currently taken by the patient (i.e., active), but some can be mentioned in the patient medical history as discontinued, or even mentioned as specifically not taken by the patient, because of an allergic reaction for example. This contextual information, the status of the medication, therefore needs accurate analysis, and we designated three different categories: active (the patient currently takes the medication), discontinued (the patient remains off the medication or is temporarily taken off the medication), and negative (the medication does not pertain to the patient or is negated).
The automatic extraction of medication information from clinical notes was the main task of the 2009 i2b2 NLP challenge.[5] It focused on the identification of medications and attributes including dosage, frequency, treatment duration, mode of administration, and reason for the administration of the medication. Almost twenty teams participated in this challenge, and Meystre and colleagues built a system called Textractor that combined dictionary lookups with machine learning approaches and reached a performance of about 72% recall and 83% precision.[6]
Patrick and Li trained a sequence-tagging model using conditional random fields (CRF) to detect medications with various lexical, morphological, and gazetteer features. Their tagger reached about 86% recall and 91% precision (the highest score in the challenge).[7]
No published research attempted prescription status classification, but some developed systems to recognize the context or assertions of medical concepts. Chapman and colleagues created the NegEx algorithm, a simple rule-based system that uses regular expressions with trigger terms to determine whether a medical term is negated. They reported 77.8% recall and 84.5% precision for medical problems in discharge summaries.[8] Chapman and colleagues also introduced the ConText algorithm, which extended the NegEx algorithm to detect four context categories: negated, hypothetical, historical, and not associated with the patient.[9]
Kim and colleagues [10] developed a Support Vector Machines (SVM [11])-based assertion classifier for the 2010 i2b2 NLP challenge [12] and their system reached 94.17% accuracy by regulating un-balanced class probabilities and adding features designed to improve performance recognizing minority classes. Our system for medication status classification expanded this system with a simplified feature set.
In the following sections, we will describe the methods we used for medication detection and prescription status classification and present our experimental results.
Methods
Materials
The ADAHF project included development of a large annotated corpus of clinical narrative text notes from patients with HF treated in a group of VHA medical centers in 2008. Each document in this corpus was annotated by two reviewers independently, and a third reviewer adjudicated their disagreements.
For this study, we randomly sampled 3,000 clinical notes from our training corpus. The most common clinical note types were progress notes, discharge summaries, history and physical notes, cardiology consultation notes, and echocardiogram reports. These 3,000 notes included 6,007 medication annotations (4,911 ACEIs and 1,096 ARBs). Medications annotated in our project included all ACEI and ARB preparations available in the U.S. The distribution of the most common medications annotated in our corpus is presented in Table 1.
Table 1.
Most Frequent Medications with Term Variants
| Medication | Type | % | Frequent term variants |
|---|---|---|---|
| Lisinopril | ACEI | 39.9 | Lisinopril |
| ACEI | ACEI | 16.7 | ACE, ACEi, ACE inhibitor |
| Benazepril | ACEI | 16.2 | Benazepril, Lotensin |
| Losartan | ARB | 8.2 | Losartan |
| Fosinopril | ACEI | 6.0 | Fosinopril, Fos |
| Valsartan | ARB | 5.0 | Valsartan, Diovan |
| ARB | ARB | 4.5 | ARB, Angiotensin receptor blocker |
| Captopril | ACEI | 1.6 | Captopril |
| Enalapril | ACEI | 0.8 | Enalapril |
| Irbesartan | ARB | 0.4 | Irbesartan |
| Others | 0.7 |
Each annotated medication was also assigned a status category: active, discontinued, or negative. Among annotated medications, 4,491 (74.76%) were active, 1,191 (19.83%) were discontinued, and 325 (5.41%) were negative. Even though the discontinued or negative status was not common, they have to be classified accurately.
Rule-Based Medication Detection
Our first approach was based on a dictionary lookup with pre-defined medication entries. We used a modified version of ConceptMapper,[13] a highly configurable UIMA [14] dictionary annotator.
To build this baseline system (DL1 system), the dictionary of medication terms has to be manually built. For each ACEI and ARB medication, we included generic names, brand names, and other frequently used name variations (from RxNorm and clinical experts’ experience with clinical text).
Misspelled medication names are common in our corpus. For example, “Lisinopril”, the most frequent I medication had 21 different misspellings in our corpus:
| Lisinipril | Lisinoppril | Lisnopril |
| Lasinopril | Lisiniprli | Lisinopiril |
| Lisinorpril | Linsinopril | Liinopril |
| Linsopril | Lisiniopril | Lisiniprol |
| Lisinoril | Lisinorpil | Lisinorpirl |
| Lisinpril | Lisionpril | Lisniopril |
| Lisnoril | Lisonopril | Loisinopril |
To improve the sensitivity of our detection and include misspellings, we added fuzzy string searching for spelling variant replacement (DL1 + fuzzy searching system). We used the edit distance (or Levenshtein distance),[15] the minimum number of single-character edits needed to transform one word into another, to check whether each text token was a spelling variant of our pre-defined medication terms.
To reduce the number of false positive matches in a second version of our system (DL2 system), we only considered matched word tokens as medication name candidates when they met the following criteria: 1) the first character was matched or the last four characters matched one of our dictionary terms, and 2) the edit distance between the word token and one of our dictionary terms was less than 2.
In addition to fuzzy searching for each word token, we also analyzed all tokens separated by a newline to reconstruct wrongly split words like “L [newline] osartan.” After removal of the newline character between two word tokens, treating them as one word token, we considered them as a correctly reconstructed word if they met the following criteria: 1) the first character was matched and the last four characters matched with one of our dictionary terms, and 2) the edit distance between the word token and one of our dictionary terms was less than 2.
These fuzzy searching strategies allowed detection of more medications even when they were misspelled or erroneously split by some newline character.
Machine Learning-Based Medication Detection
The second approach for medication detection was based on machine learning methods. We built a token-based classifier without sequential learning. A sequential tagger requires sentences as inputs and predicts the sequence of labeled tags with probabilities. In a Hidden Markov Model,[16] a popular choice for sequence tagging, transition probabilities from one tag to the next tag are considered when learning a model with a dynamic learning algorithm like Viterbi path.[17]
In token-based learning, the tagger only predicts a label for each token, independently from the previous tags. We used LIBLINEAR[18] with a linear SVM classifier to train our token-based model (SVM system). We used lexical features (the word itself, two words preceding it, and two words following it) and morphological features (prefix and suffix up to length of five) with B-I-O tags (B denotes the beginning of a term, I a token inside a term, and O a token outside a term). Because the classifier predicts each tag independently from the previous tags, we did some post-processing (using a few heuristic rules) to avoid undesirable tag sequences like a “B-ACEI” (token at the beginning of an ACEI name) followed by “I-ARB” (token inside an ARB name).
Finally, we combined the rule-based and machine learning-based methods by using DL2 system’s predictions as new features for the SVM classifier (SVM + DL2 system). The feature vector was augmented with the DL-II system predictions for the current token, the two previous tokens, and the two following tokens.
Medication Status Classification
To classify medications as active, discontinued, or negative (details in the Introduction), pre-processing included tokenization with a modified version of the cTAKES [19] tokenizer. The prescription status classifier only used lexical features from the tokenizer (i.e., all word tokens). We also used LIBLINEAR for this task, with a wider context window than for medication detection (five preceding and five following words) and no morphological features.
Metrics and Statistical Analysis
Accuracy of the detection of medications is reported using typical metrics for information extraction or retrieval: Recall (equivalent to sensitivity in this context; equals true positives/(true positives + false negatives)), Precision (equivalent to positive predictive value on this context; equals true positives/(true positives + false positives), and the F1-measure (harmonic mean of recall and precision; equals (2*recall*precision)/(recall+precision) when giving equal weight to recall and precision). These metrics were macro-averaged to obtain average values for each system (i.e., each metric was calculated for each document, and then averaged across all 3,000 documents).
Descriptive statistics are reported with 95% confidence intervals. Statistical analysis to compare our different approaches to detect medications was realized using the Student’s t-test as well as the Mann-Whitney U test for its higher efficiency with non-normal distributions.
Results
Medication Detection
As an easily accessible baseline system for our evaluation, we used eHOST,[20] the Extensible Human Oracle Suite of Tools, an open source text annotation tool, to detect medications with a pre-compiled dictionary of medication terms, as specified in our annotation guideline.
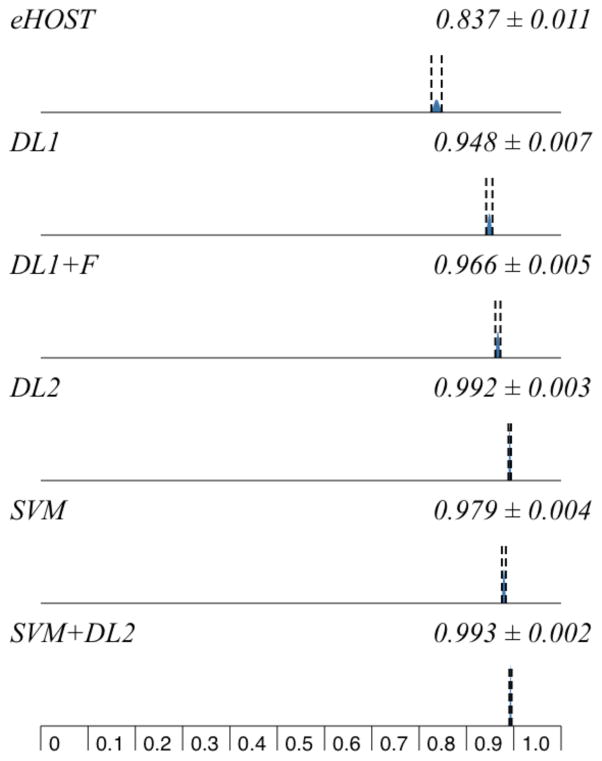
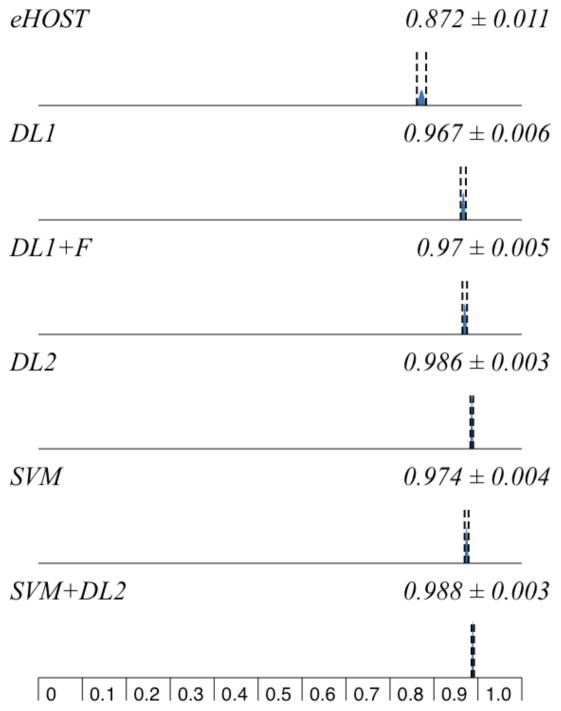
This dictionary listed multiple terms for 44 different medications and general categories. eHOST reached moderate performance (Table 2, Figures 1 and 2).
Table 2.
Five-fold Cross Validation Results for Medication Detection (macro-averaged percentages)
| System | R | P | F |
|---|---|---|---|
| eHOST | 83.7 ±1.1 | 87.2 ±1.1 | 84.8 ±1.1 |
| DL1 | 94.8 ±0.7 | 96.7 ±0.6 | 95.3 ±0.6 |
| DL1+fuzzy searching | 96.6 ±0.5 | 97.0 ±0.5 | 96.5 ±0.5 |
| DL2 | 99.2 ±0.3 | 98.6 ±0.3 | 98.7 ±0.3 |
| SVM | 97.9 ±0.4 | 97.4 ±0.4 | 97.5 ±0.4 |
| SVM+DL2 | 99.3 ±0.2 | 98.8 ±0.3 | 98.9 ±0.2 |
R=Recall, P=Precision, F=F1-measure. Percentages reported with 95% confidence intervals
Figure 1.
Systems Recall Comparison
Figure 2.
Systems Precision Comparison
However, many medications detected by eHOST would be considered true positives if partial span matches were counted. For example, medication names are sometimes attached to punctuation in our corpus (e.g., “Losartan;”, “Losartan)” ) and punctuation (; or ) in the same examples) should be excluded for exact matches, but eHOST detected these medications with the punctuation characters.
Our first dictionary lookup system (DL1) reached good performance, especially for precision (96.7%). Adding fuzzy string searching to DL1 increased recall to 96.6%. When adding wrongly split words correction (DL2), recall reached more than 99% by detecting medication names that contained newline characters.
When evaluating the machine learning-based system with a five-fold cross validation, the SVM classifier achieved lower recall and precision than the DL2 system, but closer to the DL1 system with fuzzy searching. SVM with DL2output allowed for the best performance, with the highest recall at 99.3% and a 98.9% F1-measure.
As indicated in Table 3, pairwise comparion and statistical analysis of the results reported in Table 2 and Figures 1 and 2 demonstrated that all differences were significant (p<0.001) except all metrics between DL2 and SVM+DL2 (p=0.306–368), and precision between DL1 and DL1+fuzzy searching (p=0.411), and between DL1+fuzzy searching and SVM (p=0.233).
Table 3.
Systems Recall Pairwise Comparison
| Precision | eHOST | DL1 | DL1+F | DL2 | SVM |
|---|---|---|---|---|---|
| DL1 | * | ||||
| DL1+F | * | 0.411 | |||
| DL2 | * | * | * | ||
| SVM | * | 0.047 | 0.233 | * | |
| SVM+DL2 | * | * | * | 0.306 | * |
<0.001
Medication Status Classification
We also used a five-fold cross validation with the 6,007 medication annotations to measure performance of medication status classification (Table 4). The overall accuracy was 95.49%. Precision of each status was above 90%, and recall of the discontinued status was 86.23%. Interestingly, recall was higher than precision with the negative status, even though they were associated with only 5.41% of the annotated medications in our corpus.
Table 4.
Five-fold Cross Validation Results for Medication Status Classification
| Medication Status | R | P | F |
|---|---|---|---|
| Active | 98.0 | 96.3 | 97.1 |
| Discontinued | 86.2 | 92.9 | 89.5 |
| Negative | 94.2 | 93.3 | 93.7 |
| Overall | 95.5 | 95.5 | 95.5 |
R=Recall, P=Precision, F=F1-measure.
Discussion
The dictionary lookup approach for medications detection was very efficient and the fuzzy string searching boosted performance, especially recall. The main advantages of this method are that there is no need for manually annotated training examples and processing is fast. With the SVM classifier, morphological features helped detect misspelled medication names but didn’t help with erroneously split medication names. We observed that a machine learning-based method could be applied successfully to this task without an external medical knowledge base, but it didn’t add any significant performance improvement when compared to the improved dictionary-based and rule-based system (DL2). Overall, it and was probably not a worthy effort in our case, considering the requirement for annotated training examples to train the SVM classifier.
Medication status classification was satisfactory, and even though performance with active and negative cases was quite good, there is ample room for improvement with the discontinued status. A total of 230 (71+159) active or discontinued cases were misclassified as the other class (Table 5).
Table 5.
Medication Status Classification Confusion Matrix
| Active | Classified as Discontinued | Negative | |
|---|---|---|---|
| Active | 4403 | 71 | 17 |
| Discontinued | 159 | 1027 | 5 |
| Negative | 12 | 7 | 306 |
One possible avenue for future work is to develop specific patterns or lexicons for this discontinued status, including terms like ‘hold’, ‘discontinue’, or ‘d/c’. Recognizing clinical document sections or detecting phrases mentioning why the patient was not on the medication might play an important role as classifier.
Our experimentation with machine learning-based approaches to detect specific medications was limited to one method: SVMs. Other machine learning algorithms such as Conditional Random Fields have been successfully applied to similar tasks and could also be applied to detect ACEIs and ARBs.
Conclusion
This study showed that information extraction methods using rule-based or machine learning-based approaches could be successfully applied to the detection of ACEI and ARB medications in unstructured and somewhat messy clinical notes. We boosted medication detection performance with fuzzy string searching and combining the two approaches. The preliminary work to classify the status of each medication showed that the words surrounding medication names were the most beneficial features.
Acknowledgments
This publication is based upon work supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, HSR&D, grant numbers HSR&D IBE 09-069. The views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veterans Affairs or the University of Utah School of Medicine.
References
- 1.Chen J, Normand S-LT, Wang Y, Krumholz HM. National and regional trends in heart failure hospitalization and mortality rates for Medicare beneficiaries, 1998–2008. JAMA. 2011;306:1669–1678. doi: 10.1001/jama.2011.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garvin JH, DuVall SL, South BR, Bray BE, Bolton D, Heavirland J, Pickard S, Heidenreich P, Shen S, Weir C, Samore M, Goldstein MK. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc. 2012;19:859–866. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meystre SM, Kim J, Garvin J. Comparing Methods for Left Ventricular Ejection Fraction Clinical Information Extraction. AMIA Summits Transl Sci Proc, CRI. 2012:138. [Google Scholar]
- 4.Kim Y, Garvin JH, Heavirland J, Meystre SM. Relatedness Analysis of LVEF Qualitative Assessments and Quantitative Values. AMIA Summits Transl Sci Proc, CRI. 2013:123. [Google Scholar]
- 5.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17:514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meystre SM, Thibault J, Shen S, Hurdle JF, South BR. Textractor: a hybrid system for medications and reason for their prescription extraction from clinical text documents. J Am Med Inform Assoc. 2010;17:559–562. doi: 10.1136/jamia.2010.004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc. 2010;17:524–527. doi: 10.1136/jamia.2010.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman W, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 9.Harkema H, Dowling JN, Thornblade T, Chapman W. ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42:839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Riloff E, Meystre SM. Improving classification of medical assertions in clinical notes. HLT '11 Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies. 2011:311–316. [Google Scholar]
- 11.Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. New York, New York, USA: ACM Press; 1992. pp. 144–152. [Google Scholar]
- 12.Uzuner O, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18:552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ConceptMapper. Available at: http://uima.apache.org/d/uima-addons-current/ConceptMapper/ConceptMapperAnnotatorUserGuide.html.
- 14.Apache UIMA. Available at http://uima.apache.org.
- 15.Levenshtein VI. Binary Codes Capable of Correcting Deletions, Insertions and Reversals. Soviet Physics Doklady. 1966;10:707. [Google Scholar]
- 16.Baum LE, Petrie T. Statistical inference for probabilistic functions of finite state Markov chains. The Annals of Mathematical Statistics. 1966 [Google Scholar]
- 17.Viterbi AJ. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. Information Theory, IEEE Transactions on. 1967;13:260–269. [Google Scholar]
- 18.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. 2001 Available at: http://www.csie.ntu.edu.tw/~cjlin/libsvm.
- 19.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.South BR, Shen S, Leng J, Forbush TB, DuVall SL. A Prototype Tool Set to Support Machine-Assisted Annotation. BioNLP '12 Proceedings of the 2012 Workshop on Biomedical Natural Language Processing. 2012:130–139. [Google Scholar]