Abstract
Aflatoxins are a group of potent foodborne toxicants naturally occurring in maize and groundnuts. Differential species-specific sensitivity to aflatoxins has been documented but cannot be fully explained by the differences in metabolism of these toxicants among animal species. Commensal microbial communities (microbiota) are critical to human and animal health, but few studies have assessed interactions between xenobiotic toxins and those microbiota, and its potential effects to humans and animals. Here, an exploratory dosing experiment was conducted to explore effects of Aflatoxin B1 (AFB1) on the gut microbiota in a commonly used rat model. Male F344 rats were randomly divided into groups and treated with different concentrations of AFB1. Microbial communities in fecal samples were assessed using 16S rRNA sequence analysis. We found that samples from the control group had a phylogenetically diverse community, and that increasing AFB1 doses decreased this diversity but increased evenness of community composition. In addition, the gut microbiota from different samples was clustered according to their dosing regimens. There is no community shift at the phylum level but some lactic acid bacteria were significantly depleted by AFB1. These findings suggested that AFB1 could modify the gut microbiota in a dose-dependent manner.
Keywords: Aflatoxin B1, lactic acid bacteria, microbiome, mycotoxins, next-generation sequencing, 16S rRNA sequencing
Commensal microbial communities (microbiota), especially those that reside in the gastrointestinal tract of animals are critical to the healthy development of some important internal structures, eg, villous structures, immune organs (DuPont and DuPont, 2011; Nicholson et al., 2005). Recently animal and human studies have shown that perturbed gut microbiota are associated with multiple health problems such as obesity and inflammatory bowel disease (Ley et al., 2006; Qin et al., 2010; Turnbaugh et al., 2006). Researchers have become increasingly interested in how xenobiotic exposure would impact these crucial communities.
Aflatoxins are a group of mycotoxins attracting consistent research and public interests because the toxigenic fungus, Aspergillus, contaminates a large fraction of the world’s crops (Lizarraga-Paulin et al., 2011). Williams et al. (2004) estimated that approximately 4.5 billion people in developing countries are chronically exposed to uncontrolled amounts of aflatoxins from foods. Extensive studies have documented that aflatoxins, especially the most potent congener, aflatoxin B1 (AFB1), can be metabolically activated in the liver to cause acute hepatic toxicity and immune toxicity, form DNA adducts, and induce gene mutations (Qian et al., 2014; Wogan et al., 2012). Furthermore, chronic exposure to low levels of aflatoxins is a risk factor for hepatocellular carcinoma and AFB1 has been categorized as a Group 1 human carcinogen by International Agency for Cancer Research (IARC, 2012).
The effects of only a few types of xenobiotic toxicants on gut microbiota have been studied so far: 2 heavy metals (Cd and Pb) (Breton et al., 2013a,b), metal nano-particles (Han et al., 2014; Merrifield et al., 2013), arsenic (Lu et al., 2013, 2014a,b), alcohol (Bull-Otterson et al., 2013; Xie et al., 2013), and a few other toxic organics (Choi and Toborek, 2012; Kish et al., 2013; Kohl et al., 2014; Peterfreund et al., 2012; Swann et al., 2009; Yip et al., 2014). Two major findings have been revealed from these studies: first, gut microbiota contributes to host resistance to xenobiotic toxicants; and second, xenobiotics can modify the composition of microbial communities. Results from these studies begin to shed light on this important issue, but much work remains to be done. Reliable standard experimental protocols are needed to perform such toxicological experiments, including better data analysis procedures to assess the effects and interactions of toxins/toxicants on microbiota.
Differential species-specific sensitivity to AFB1 has been documented but cannot be fully explained by the differences in metabolism of AFB1 among animal species (Rawal et al., 2010). We therefore hypothesize that the gut microbiota plays a role in modulation of species responses to AFB1 toxicity. Accordingly, the rationale underlying this study is that AFB1 could induce changes in gut microbiota of its host and therefore eventually influence the effects of this toxin. This hypothesis was tested via community compositional assessment of gut microbiota in a rat model dosed with different concentrations of AFB1. 16S rRNA gene sequence analysis was used to assess the microbiota. The objectives of this study were designed to: (1) establish reliable experiment protocols for preparing microbial 16S rRNA gene libraries and data preparation/analysis procedures for assessing the effects of AFB1 on commensal microbiota, and (2) use these protocols to determine if there is a dose–response relationship between AFB1 exposure and the observed microbial communities of the exposed rats.
MATERIALS AND METHODS
Chemicals, animals, and experiment design
This study used a design that was similar to a previous toxicity study in which detailed procedures on chemicals and animal preparations were described (Qian et al., 2014). Briefly, 20 male Fischer 344 rats (5-weeks old, 120–130 g) were acclimatized for 1-week before the experiment, randomly allocated into 4 dosing groups: control, low (5 μg AFB1/kg body weight [b.w.]), medium (25 μg/kg b.w.), high (75 μg/kg b.w.); upon starting of the experiment, gavaged with control vehicle (DMSO) or AFB1 in DMSO, 5 days per week for a duration of 4 weeks. DMSO was used due to the hydrophobicity of AFB1 and its minimum effects on animals (<50 µl/animal). Rat feces were collected at the end of the fourth week in metabolic cages and stored at −80°C.
16S rRNA gene sequencing
Genomic DNA of gut microbiota was extracted from fecal samples collected from each individual using QIAamp DNA stool mini kits (QIAGEN, Valencia, California) following manufacture protocols with minor modification. Briefly, immediately after removal from the freezer, about 90 mg of frozen feces was weighed into a 2-ml tube and placed on ice with 500 μl of stool lysis buffer added. A disposable spatula or pipette tip was used to help homogenize the sample. Another 900 μl of stool lysis buffer was added after sufficient homogenization was obtained. We then followed the manufacturer’s protocols, resulting in 200 μl of DNA in buffer from each sample with concentrations ranged from about 70 to 150 ng/μl and stored at −20°C.
A 2-step Quadruple-index PCR method was used to prepare the 16S rRNA gene libraries. Briefly, locus-specific primers to the V3 and V4 regions of 16S rRNA (Klindworth et al., 2013): S-D-Bact-0341-b-S-17 (Bakt_341F) CCTACGGGNGGCWGCAG and S-D-Bact-0785-a-A-21 (Bakt_805R) GACTACHVGGGTATCTAATCC) were fused with Illumina TruSeq Read1 and Read2 sequences. Internal tag sequences 5 bases long (Faircloth and Glenn, 2012) with 0–3 base spacers were inserted between the Illumina and locus-specific portions of the oligonucleotides. The DNA samples were diluted to approximately 20 ng/µl and PCR amplified using Kapa HiFi HotStart PCR kits (Kapa Biosystems, Inc, Boston, Massachusetts). 1 µl of DNA was added to the 25 µl reaction with following thermal cycling conditions: one cycle of pre-denaturation at 95°C for 3 min, 20 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 30 s, elongation at 72°C for 30 s, and 1 cycle of post-elongation at 72°C for 5 min. Following generation of the primary amplicons, another pair of fusion oligonucleotides was used as primers to generate full-length dual-indexed Illumina TruSeqHT style libraries. The resultant amplicons were normalized, purified 1:1 with Speedbeads (Faircloth and Glenn, 2014), and stored at −20°C prior to sequencing. Sequencing of these 16S rRNA fragment libraries was performed in the Georgia Genomic Facility (University of Georgia, Athens, Georgia) using the Illumina MiSeq with v2 500 cycle chemistry, resulting in paired-end 250 base reads to obtain approximately 30 000 reads per sample.
Data preparation
The raw paired-end, demultiplexed sequence reads for each sample from sequencing facility was merged using FLASH 1.2.9 (Magoc and Salzberg, 2011) available through Geneious 8.1 software (Biomatters Inc, San Francisco, California). Internal tags, base spacers, and locus-specific primers from each end of merged sequences were trimmed and sequences outside the length range 400–450 base-pairs were discarded using Geneious 8.1. Outputs from Geneious 8.1 were quality filtered using QIIME pipeline (Quantitative Insights Into Microbial Ecology) (Caporaso et al., 2010b) with following settings: sequences were trimmed from each end to obtain the shortest sequences without 3 consecutive low-quality base calls which were defined as having a Phred quality score lower than 20 and without any ambiguous base calls; the resultant sequences that were shorter than 75% of their original length were discarded (Bokulich et al., 2013). The resultant analyzable sequence reads were deposited into Sequence Read Archive database with the accession number PRJNA291923.
Analyzable sequences from each sample were clustered into Operational Taxonomic Unit (OTU) by uclust algorithm (Edgar, 2010) using a de novo strategy with the similarity threshold of 97%. Representative sequences for each OTU were compared against the Greengene 16S rRNA gene database 13-8 release (DeSantis et al., 2006) using uclust algorithm with the similarity threshold of 90%. The top 3 database hits that matched the above representative sequences for each OTU were selected; the taxonomic information assigned to an OTU was set to be the most detailed lineage description which was shared by at least 51% of these 3 database hits. A phylogenetic tree was constructed by firstly aligning the representative sequences of each OTU using PyNAST algorithm (Caporaso et al., 2010a) against Greengene reference sequences and then using FastTree software with GTR model and Gamma 20 approximation (Price et al., 2010). Above mentioned procedures were implemented through QIIME with customized commands. The OTU table and phylogenetic tree were the major elements for data analysis.
Data analysis
Complementary divergence- and taxon-based methods were used to compare samples among different treatment groups. Divergence-based methods are more powerful in certain circumstances because not all species in one sample are equally related; samples containing species that are phylogenetically closely related should be considered to be more similar as compared with samples containing species that are phylogenetically distant from each other (Lozupone and Knight, 2008). Three methods were employed to facilitate data analysis: α-, β-diversity analysis, and direct comparisons of community compositions in samples from different group. All analysis used 0.05 as significance level.
α-Diversity analysis
α-Diversity describes the diversity within a community (ie, a sample in this study). A divergence-based metric, phylogenetic diversity (PD) (Faith, 1992), and a taxon-based metric, Shannon index (Magurran, 2004) were computed for each sample using QIIME. Mean value and 95% confidence interval were calculated for each dosing group. To avoid sequencing depth bias, the sample sizes from different sample were equalized by randomly jackknifing 24 000 sequences per sample (approximately the number of sequences of the sample with least analyzable reads). One-way ANOVA test were conducted to analyze dosing effect on alpha diversity and Duncan’s test were conducted for post hoc comparisons using R 3.1.2 (R Core Team, 2014).
β-Diversity analysis
β-Diversity describes the relationship between 2 communities (or one community at different time points). The unweighted UniFrac index describes divergence distance between 2 samples, which measures the fraction of branches in a phylogenetic trees, which lead to an OTU that only appears in either 1 of the 2 communities but not both (Lozupone and Knight, 2005). Unweighted Unifrac index was computed for all pairs of samples to construct distance matrix. To avoid sequencing depth bias, calculation of unweighted UniFrac index was performed on subsampled samples as previously described. The principle coordinate analysis (PCoA) was performed using the distance matrix constructed by unweighted UniFrac index to examine sample clustering. In order to measure the uncertainty of β-diversity analysis, each sample was randomly jackknifed 18 000 sequences per sample and analyzed following the above procedures; the jackknifing procedure was repeated 10 times. All above procedures were implemented in QIIME.
Direct comparisons of community compositions
α- and β-Diversity analysis target the gut microbial community. From the perspective of toxicology, the specific gut bacteria that were significantly impacted by exposure to AFB1 may also be important. Consequently, direct comparisons of community compositions were performed using MetagenomeSeq software (Paulson et al., 2013) implemented in R 3.1.2. Briefly, MetagenomeSeq uses normalized OTU counts from each sample as input to avoid bias due to preferentially sampling associated with sequencing yields and a Zero-Inflated Gaussian model to account for the zero OTU counts; a maximum-likelihood model and Empirical Bayes were used to generate significance of dosing effects (Paulson et al., 2013). This analysis compares differentiated abundance of each OTU in different dosing groups. Before performing the analysis, the original datasets were subset to include only OTUs that had counts in at least 6 samples (half the total) to improve statistical capacity. The P-values generated from the Empirical Bayes for each OTU were further corrected to adjust for multiple-comparison using Bonferroni method implemented in R 3.1.2.
RESULTS
16S rRNA Gene Sequencing and Data Preparation
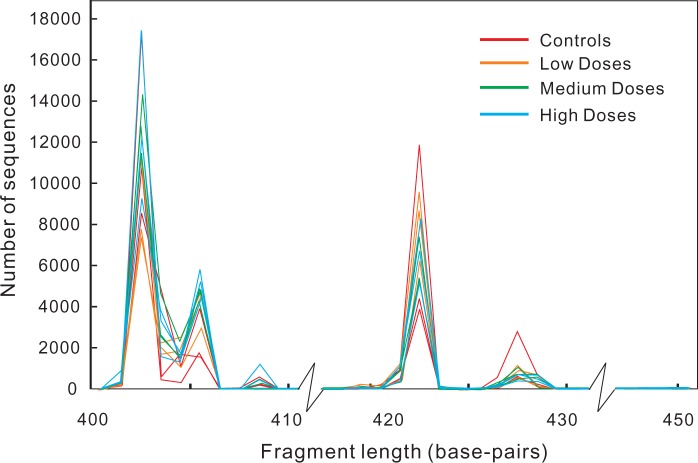
Among the initial 20 rats, fecal samples from 3 rats in each group were available, making a total of 12 samples for analysis in this study. Microbial DNA from fecal samples were extracted, 16S rRNA gene libraries were prepared using a 2-step quadruple-index PCR methods before sending out for sequencing. An average of 32 786 pairs of raw paired-end sequences (ranged from 27 293 to 38 113, Table 1) were obtained from the sequencing facility for each sample. The length distributions of the target 16S rRNA gene fragment from each sample after merging and trimming are plotted in Figure 1. More than 80% of sequences from each sample passed quality filtering, indicating that the sequencing technique is robust and efficient.
TABLE 1.
Sequences obtained from genomic sequencing facility and processing for 12 samples
| Data Processing | Control1 | Control2 | Control3 | Low1 | Low2 | Low3 | Medium1 | Medium2 | Medium3 | High1 | High2 | High3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Original Data (Pairs) | 29 502a | 30 370 | 29 575 | 30 832 | 27 293 | 31 420 | 34 726 | 31 083 | 36 886 | 38 113 | 37 382 | 36 254 |
| Merging and Trimmingb | 27 595 (93.5) | 28 578 (94.1) | 27 390 (92.6) | 28 597 (92.8) | 25 479 (93.4) | 29 470 (93.8) | 31 584 (91.0) | 28 435 (91.5) | 34 618 (93.9) | 34 043 (89.3) | 32 901 (88.0) | 30 750 (84.8) |
| Quality Filteringc | 26 228 (88.9) | 27 127 (89.3) | 25 907 (87.6) | 27 107 (87.9) | 24 199 (88.7) | 28 010 (89.1) | 30 014 (86.4) | 27 145 (87.3) | 32 996 (89.5) | 32 369 (84.9) | 31 328 (83.8) | 29 179 (80.5) |
aData value indicates the number of sequences in respective samples and percentages in original counts in parentheses.
bMerging is a process of combining pair-end Illumina reads of the same fragment to be a single nucleotide sequence. Trimming is a process of cutting inner tags, locus specific primers, and extracting sequences between 400 and 450 base pairs.
cQuality filtering is a process in which merged sequences were cut from each end to obtain the shortest sequences without consecutive 3 bases with a quality score lower than 20 and without any ambiguous base calls. Sequences shorter than 75% the original length after cutting were excluded.
FIG. 1.
Fragment length distribution of sequences from each sample after merging and trimming. The target fragment is 427 base-pairs long in Escherichia coli (from site 358 to 784, excluding primers [Klindworth et al., 2013]).
α-Diversity Analysis
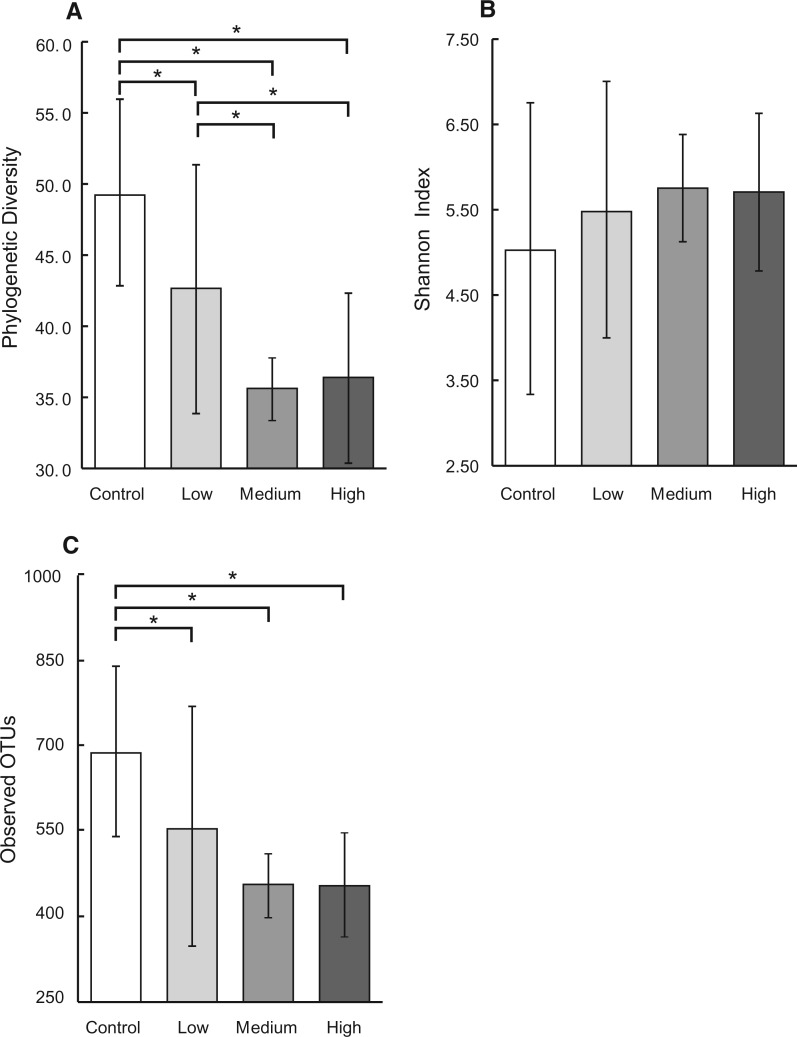
PD reflects the genetic diversity of a microbial community. The more distantly related species a community has, the higher is PD for the community. As illustrated in Figure 2A, increasing AFB1 doses decreased the PD; 1-way ANOVA test suggested a significant AFB1 dosing effect on PD (F-value = 123.24, P-value<0.05). Duncan’s post hoc test was used to adjust for experiment-wise error rate, which showed significant differences in all pairwise comparisons except that between medium and high dosing group.
FIG. 2.
Alpha diversity indexes based on 16S rRNA gene sequence analysis among AFB1 treatment groups with 3 replicate individuals per treatment group and 24 000 sequences per sample. Error bars indicate 95% confidence interval. *indicates significant difference between means of different treatment groups determined by Duncan’s post hoc test. A, Phylogenetic diversity decreased significantly with increasing AFB1 in a dose-dependent manner (1-way ANOVA P < .05). B, Shannon index increased with increasing of AFB1 in a dose-dependent manner, but was not significant (1-way ANOVA P > .05). C, Observed OTUs decreased with increasing of AFB1 in a dose-dependent manner (1-way ANOVA P < .05).
Shannon index simultaneously considers species richness and evenness. The more species (regardless of the genetic relationship between species) a community has and the more even a community is distributed by each species, the higher Shannon index is for the community. As illustrated in Figure 2B, Shannon index had an exact opposite pattern as presented for PD. That is, increasing AFB1 doses increased Shannon index, although the one-way ANOVA test failed to suggest a significant dosing effect. As for comparison, the number of unique OTUs averaged across each dosing group is also plotted in Figure 2C. This measurement directly presents the number of different microbial species within a community. One-way ANOVA test suggested a significant dosing effect on unique OTUs. The control group had an average of 689 OTUs and was significantly higher than the other groups as confirmed by Duncan’s test; the low, medium, and high dose had decreased number of unique OTUs, 556, 453, and 455, respectively.
Detailed information regarding α-diversity, observed OTUs, and taxonomic information for each sample can be found in the supplementary Material.
β-Diversity Analysis
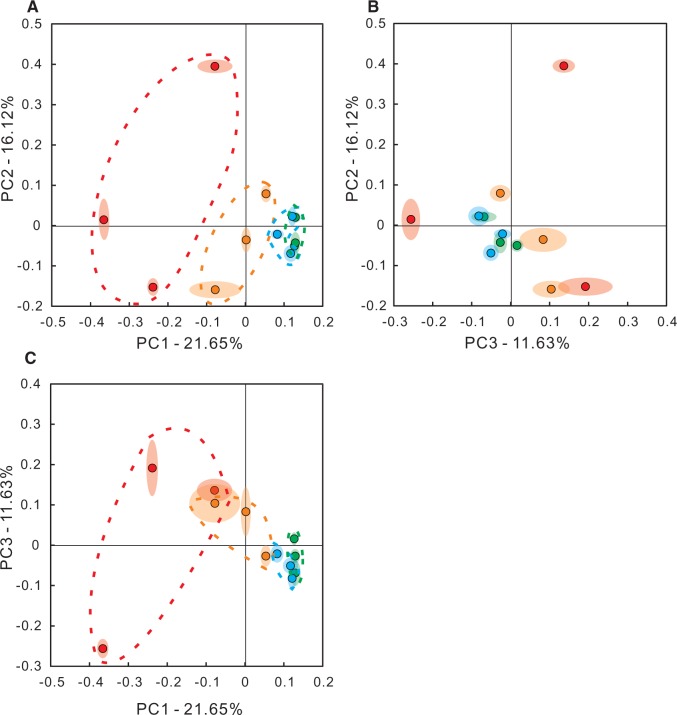
Ranging from 0 to 1, the 2 communities that have closer related species will have a smaller Unweighted UniFrac Index; vice versa. Pair-wise comparisons will produce an N by N Unifrac Index matrix, where N is the number of total samples in all groups. This matrix has the property of a distance matrix and therefore could be analyzed by PCoA. As illustrated in Figure 3, the first 3 principle component could readily cluster the samples according to their dosing regimens. Ellipsoids around each sample point generated via jackknifing analysis suggested low uncertainties. Samples plotted in PCoA also appeared to locate in a dose-dependent manner: samples from low dose group located in between the control samples and higher dose groups. In addition, control samples had relatively large variation, increasing doses brought samples from the same dosing group closer together. It is worth noticing that similar as in α-diversity analysis, samples from medium and high dosing groups had relatively small intragroup variation and cannot be differentiated.
FIG. 3.
Beta diversity analysis based on the unweighted Unifrac index. A–C, 2-dimension plots for the top 3 principle components (PC1, PC2, PC3). Each dot represents a sample point. Ellipsoids around each dot represent the uncertainty of each sample point based on jackknifing analysis. Red, orange, green, and blue dots represent control, low, medium, and high dosing groups, respectively. Coordinates in axis are for illustration purpose only and selected arbitrary and therefore do not have clear biological meanings. Percentage associated with each PC is the proportion of an eigenvalue for the respective PC in the sum of eigenvalues for all PCs. Full color version available online.
Direct Comparison of Community Compositions
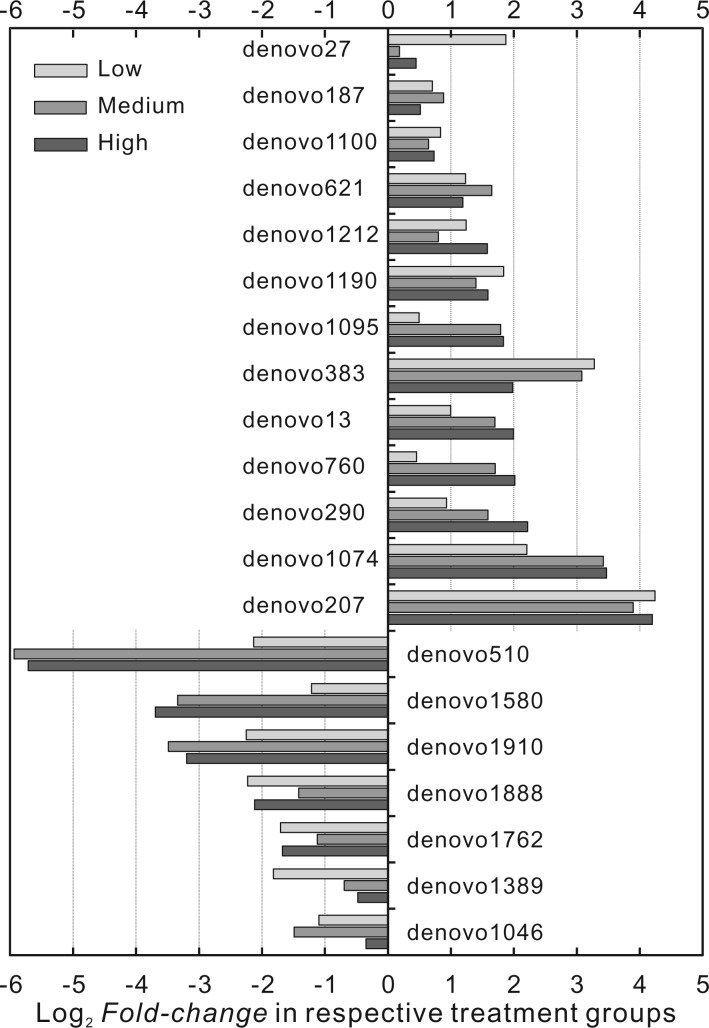
Despite advantages of divergence-based methods, above analyses cannot recognize the species (OTUs) or lineages that drive the clustering in association with different AFB1 doses. Consequently, a direct comparison of community compositions could complement these analyses. Through initial screening, among 2479 unique OTUs in all samples, 253 OTUs appeared in at least 6 samples; following analysis was performed on these 253 OTUs. After fitting the Zero-Inflated Gaussian model, Empirical Bayes testing, and Bonferroni corrections, 191 OTUs appeared to have a corrected P-value below pre-defined significance level, among which the top 20 consistent OTUs were plotted in Figure 4 and listed in Table 2. Figure 4 plotted the model estimated Log2 fold-change of normalized OTU abundance in low, medium, and high dosing groups as compared with the control groups. Among the 20 OTUs, the increases in treatment groups ranged from 1.13-fold (OTU:denovo27) to as high as 19-fold (OTU: denovo207) as compared with controls; the decreases ranged from 78% (OTU: denovo1046) to as low as 2% (OTU: denovo510) of what was in controls. Given taxonomic information of these top 20 OTUs in Table 2, there was no consistent pattern of increase or decrease at phylum level and at least down to order level. Three Clostridiales species had the largest increase while 2 Lactobacillales species had the largest decrease; the 2 species Lactobacillales belongs to the widely known probiotic lactic acid bacteria (LAB) (de Vries et al., 2006).
FIG. 4.
Log2 fold-change in respective treatment groups as compared with control group among top OTUs with highest F-test value, estimated from Zero-Inflated Gaussian model. ‘denovo’ followed with a number represents a specific OTU detected within the gut microbial community.
TABLE 2.
Taxonomic information for the top 20 OTUs with highest F-test values
| OTUa | Phylum | Class | Order | Family | Genus | Species | Control | Low | Medium | High |
|---|---|---|---|---|---|---|---|---|---|---|
| Significantly increased | Average abundance (%)b | |||||||||
| denovo207 | Firmicutes | Clostridia | Clostridiales | 0.53 | 5.28 | 2.28 | 3.14 | |||
| denovo1074 | Firmicutes | Clostridia | Clostridiales | 0.02 | 0.14 | 0.30 | 0.3 | |||
| denovo290 | Firmicutes | Clostridia | Clostridiales | 1.41 | 2.09 | 2.84 | 3.85 | |||
| denovo760 | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | uniformis | 0.82 | 0.6 | 0.52 | 0.68 |
| denovo13 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillospira | 0.05 | 0.11 | 0.19 | 0.23 | |
| denovo383 | Firmicutes | Clostridia | Clostridiales | 0.09 | 0.82 | 1.07 | 0.56 | |||
| denovo1095 | Bacteroidetes | Bacteroidia | Bacteroidales | S24‐7 | 0.89 | 0.94 | 0.55 | 0.56 | ||
| denovo1190 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillospira | 0.06 | 0.36 | 0.38 | 0.35 | |
| denovo1212 | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 3.61 | 5.37 | 2.5 | 4.51 | |
| denovo621 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Coprococcus | 0.08 | 0.47 | 0.87 | 0.59 | |
| denovo1100 | Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 12.21 | 15.36 | 11.6 | 11.36 | |
| denovo187 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillospira | 0.21 | 0.78 | 1.42 | 0.92 | |
| denovo27 | Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | 0.07 | 0.31 | 0.11 | 0.14 | ||
| Significantly decreased | ||||||||||
| denovo510 | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 1.83 | 0.65 | 0.11 | 0.14 | |
| denovo1580 | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Lactococcus | 0.71 | 0.59 | 0.34 | 0.26 | |
| denovo1910 | Bacteroidetes | Bacteroidia | Bacteroidales | 0.18 | 0.11 | 0.07 | 0.09 | |||
| denovo1888 | Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Bilophila | 0.98 | 0.85 | 1.20 | 1.34 | |
| denovo1762 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcus | 0.98 | 0.43 | 1.13 | 0.94 | |
| denovo1389 | Firmicutes | Clostridia | Clostridiales | 0.49 | 0.25 | 0.81 | 0.95 | |||
| denovo1046 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | 0.30 | 0.26 | 0.31 | 0.67 | ||
aOTUs are ordered in descending of estimated fold changes in respective categories (significant increased/decreased).
bData values indicate the percentages of each species in original samples averaged over each group.
DISCUSSION
This study was prompted by the increasing interests on the effects of xenobiotic agent on organism’s commensal gut microbiota. We hypothesized that a xenobiotic toxin/toxicant could induce compositional and/or functional changes in gut microbiota, and these changes will eventually influence the toxic effects of these agents. This study analyzed fecal samples from groups of male F344 rats continuously dosed by different sub-lethal concentrations of AFB1, a Group 1 human carcinogen, using 16S rRNA gene sequencing analysis, and developed a standard protocol for future studies. We found that AFB1 treatment induced the microbial community compositional changes in a dose-dependent manner.
Doses selected in this study were equivalent to 0.03–0.45 mg/kg (30–450 ppb) of AFB1 in food. It is in the range commonly found in contaminated foods consumed daily by several populations especially in developing countries (Daniel et al., 2011; Li et al., 2001; Sharma and Sumbali, 1999) and was in the category of realistic dose as suggested by a recent publication (Grenier and Applegate, 2013). The repeated dosing protocol adopted in this study is the standard protocol we developed for continuously dosing animal experiments. The protocol may have some influence on short-term metabolites, such as urinary metabolites, but no influence on blood and fecal metabolites as well as aflatoxin biomarkers.
To our knowledge, this study is the first study to attempt to establish a dose-response relationship between a toxin/toxicant and responses of microbial community. It was surprising to observe that PD decreased while Shannon index increased with increasing AFB1 doses. We speculate that there were more rare species (in terms of both phylogenetic relationship and abundance) in the control samples, which drove the PD high. Increasing AFB1 exposure depleted low tolerant species (probably those rare species), resulting in a decrease of PD but an increase in evenness of community compositions, and therefore increased the Shannon index. This explanation was further supported by a decrease of unique OTU numbers found in samples while increasing doses as seen in Figure 2C; approximately one-third of unique OTUs was depleted in high dosing group as compared to control samples. Similar findings have been observed in many other studies, eg, a species of fruit fly larvae dosed with Ag nano-particle had a lower diversity in mid-gut microbiota as compared with controls (Han et al., 2014); inflammatory bowel disease patients were documented to have fewer species (Manichanh et al., 2006), fewer genes (Qin et al., 2010) as compared to healthy controls; individuals with obesity was associated with a significant decrease in phylogenetic diversity as compared to lean individuals from a twin-pairs study (Turnbaugh et al., 2009). A number of studies that focused on antibiotics effects on human gut microbiome also documented the depletion of gut microbiota in human volunteers, too, and the effects persisted after the cease of antibiotics treatment for an extended period (Jakobsson et al., 2010; Jernberg et al., 2010). It would be good for future studies to investigate the effects of xenobiotic toxicants on gut microbiome to include a time-course analysis, which may have important implications regarding the temporal role of microbiota in response to exposure to toxicants.
It is also worth noting that samples from medium- and high-dosing groups had similar α-diversity and clustered together in β-diversity analysis despite a large increase in doses (from 25 to 75 µg/kg b.w.). The implications of this finding are unclear because of lack of previous evidences. One explanation may be that increased doses of AFB1 had screened the resistant microbial species; the remaining species were toxic tolerant and could survive this toxin. These explanations may have important implications in that if the gut microbiota contain toxic resistant species, its interaction with this toxin may reduce the toxicity to its host. From another perspective, results from this study showed the potential that the gut microbiota of a host could be intentionally modified to accumulate toxin-tolerant species, a potential natural biological intervention strategy for mitigation of AFB1-induced toxic effects. As previously mentioned, the toxicity resistance functions of gut microbiota have been documented in multiple studies. For example, Kohl et al. (2014) has showed that the gut microbiota was responsible for the toxic-resistance ability to a toxic Creosote resin of a Desert Woodrat strain. Series of studies by Lu et al. (2013, 2014b) reported that perturbed gut microbiota in laboratory mice modified the host response to arsenic toxicity. Breton et al. (2013b) reported that after multiple oral doses of Cadmium and Lead up to 100 ppm, germ-free mice had significantly higher tissue metal concentrations than conventional mice. Swann et al. (2009) reported that toxicity effects, in terms of loss of vitality, of sub-toxic dosing of hydrazine, was more severe in germ-free rats than conventional and control rats.
Direct comparisons on community compositions of samples from different groups may not be informative in some cases because different microbial species may share similar genes and functions, and therefore, might not be appropriate to be treated as different. This idea is evidenced in that different individuals may share the same ‘core microbiome’ which were identifiable at the gene level instead of at the organismal lineage level (Turnbaugh et al., 2009). We did this comparison here, however, to complement divergence-based methods and provide further insights on the effects of AFB1. An overview of community taxonomic compositions at phylum level have showed that Firmicutes and Bacteroidetes constituted >90% of the microbiota from all samples, consistent to what have been found previously in mice and human gut microbiota (Turnbaugh et al., 2006); however the relative abundance of these 2 phylum was not appeared to be impacted by AFB1 dosing (results not shown). The microbial community compositional changes have been documented in all previously mentioned toxicological studies. However, considering the large number of species (500–1000) resides in gut, false positive should be carefully avoided. In this study, control of false positive was accomplished by multiple steps: only OTUs that appeared in more than half of the all samples were included (reducing the comparisons to 253 OTUs); the conservative Bonferroni method was used to correct for the multiple comparisons; only consistent OTUs (OTUs consistently increased or decreased in all 3 treatment groups as compared with control groups) were selected; only the top 20 consistent OTUs were selected. Species whose compositions significantly increased among the top 20 OTUs were from Clostridiales of Firmicutes and Bacteroidales of Bacteroidetes (Table 2). Species whose compositions significantly decreased among the top 20 OTUs were all from Firmicutes except one species of Bacteroidales from Bacteroidetes and one unknown Bilophila species from Proteobacteria. The 2 Lactobacillales from Firmicutes, Streptococcus sp. and Lactococcus sp. had the largest decrease, appearing in a dose-dependent manner. These species are members of LAB (Stiles and Holzapfel, 1997) which have been shown to bind and remove AFB1 via surface protein in several studies (Haskard et al., 2001; Peltonen et al., 2001). It was difficult to speculate the specific functions, the importance of other species which were also significantly changed and their interactions with AFB1, partly due to the lack of previous information, except that antimicrobial activities of AFB1 to gram-negative and -positive bacteria have been revealed decades ago (Arai et al., 1967). In addition, many different microbial species have been previously reported to be impacted by xenobiotic toxicants or by disease status. However, there has not been a consensus on the linkage between specific species with a toxicant. The ratio between Bacteroidetes and Firmicutes has been previously reported in several studies, which was linked to certain disease status such as obesity, although a recent meta-analysis (Walters et al., 2014) suggested that the ratio between these 2 phyla might not be indicative after reviewing all current studies. From the present study, species that were either significantly increased or decreased were from both Firmicutes and Bacteroidetes, suggesting that changes at that phylum level might not be a good indicator to assess compositional changes.
The 16S rRNA fragment amplified in this study is from site 358 to 784 under Escherichia coli system of nomenclature, a 427 base-pairs long fragment (Klindworth et al., 2013). As shown in Figure 1, in the present rat gut microbiota, this gene fragment peaked in 5 different length including 427. The fact of all 12 samples has highly consistent distributions further verified the precision of current data. A recent study (Sinclair et al., 2015) has shown multiple peaks of the same fragment in sediment samples and demonstrated that the peaks were the result of different microbial species. We did not, however, differentiate different species according to the fragment length because it is irrelevant to the scope of this study.
The strength of this study mainly falls in 3 aspects. First, this is the first study as far as we know that successfully established a dose-response relationship between xenobiotic exposure and gut microbiome alternations. Different from traditional toxicological research, study on microbiota generates multi-dimensional data which caused some theoretical difficulties to assess response (effect). This study employed α- and β-diversity matrix, describing the response at the whole community level to conquer the difficulties. Second, this study adopted reliable phylogenetic and statistical methods to assess the community compositional alternations of the model animal using both divergence-based and taxon-based based methods. Last but not the least, this study developed a complete protocol from sample collection, DNA library preparation, data preparation and analysis for conducting a 16S rRNA gene survey based microbiome study in toxicology. This procedure and parameters adopted in this study could serve as a framework for any future similar study.
The study is exploratory, and primarily limited by its small-scale nature. Consequently, the relative small sample size may limit our ability to assess the statistical power of current analysis and to perform more sophisticated analysis. However, results presented in this study did provide solid evidence of AFB1 treatment effect. The fact that increasing doses decreased variations within treatment group as seen in β-diversity analysis suggested that AFB1, not the sample size, may be accounted for the intragroup variation. Regarding the sequence depth, a previous study suggested that 20 000 sequences per individual were sufficient to stabilize individual’s microbiome functional profile (Turnbaugh et al., 2009). In rarefaction analysis, our data showed that α-diversity of an individual sample plateaued after around 30 000 reads (results not shown). Consequently, we deemed targeting at 30,000 reads per sample was sufficient for current study scope. Male F344 rats are more sensitive to AFB1 induced effects compared to female rats (Prince and Campbell, 1982). Consequently, we only included male rats to avoid confounding effects due to rat sex in this exploratory study.
Supplementary Material
ACKNOWLEDGMENTS
Special thanks are due to Dr Guoqing Qian and Kathy S. Xue for animal experiments; Troy Kieran, Bei Gao, and Jesse Thomas for general assistants; Dr. Kun Lu and Dr. Ming Zhang for kindly guidance on sample processing and data analysis; and Interdisciplinary Toxicology Program at the University of Georgia for stipend supports. The authors acknowledge the support of research contract, ECG-A-00-07-00001-00, from the U.S. Agency for International Development via Peanut CRSP at University of Georgia, and the planning grant, 1R24TW009489, from the National Institute of Health Fogarty International Center.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Arai T., Ito T., Koyama Y. (1967). Antimicrobial activity of aflatoxins. J. Bacteriol. 93, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., Mills D. A., Caporaso J. G. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton J., Massart S., Vandamme P., De Brandt E., Pot B., Foligne B. (2013a). Ecotoxicology inside the gut: Impact of heavy metals on the mouse microbiome. BMC Pharmacol. Toxicol. 14, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton J., Daniel C., Dewulf J., Pothion S., Froux N., Sauty M., Thomas P., Pot B., Foligne B. (2013b). Gut microbiota limits heavy metals burden caused by chronic oral exposure. Toxicol. Lett. 222, 132–138. [DOI] [PubMed] [Google Scholar]
- Bull-Otterson L., Feng W., Kirpich I., Wang Y., Qin X., Liu Y., Gobejishvili L., Joshi-Barve S., Ayvaz T., Petrosino J., et al. (2013). Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. Plos One 8, e53028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., Knight R. (2010a). PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. J., Toborek M. (2012). Exercise alters the abundance and composition of gut microflora and attenuates PCB-induced changes in gut microbiome. J. Neuroimmune. Pharmacol. 7, S33–S34. [Google Scholar]
- Daniel J. H., Lewis L. W., Redwood Y. A., Kieszak S., Breiman R. F., Flanders W. D., Bell C., Mwihia J., Ogana G., Likimani S., et al. (2011). Comprehensive assessment of maize aflatoxin levels in Eastern Kenya, 2005–2007. Environ. Health Perspect. 119, 1794–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries M. C., Vaughan E. E., Kleerebezem M., de Vos W. M. (2006). Lactobacillus plantarum-survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16, 1018–1028. [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPont A. W., DuPont H. L. (2011). The intestinal microbiota and chronic disorders of the gut. Nat. Rev. Gastroenterol. Hepatol. 8, 523–531. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Faircloth B. C., Glenn T. C. (2012). Not all sequence tags are created equal: Designing and validating sequence identification tags robust to indels. PLoS One 7, e42543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth B. C., Glenn T. C. (2014). Protocol: Preparation of an AMPure XP substitute (AKA Serapure). doi: 10.6079/J9MW2F26. [Google Scholar]
- Faith D. P. (1992). Conservation Evaluation and Phylogenetic Diversity. Biol. Conserv. 61, 1–10. [Google Scholar]
- Grenier B., Applegate T. J. (2013). Modulation of intestinal functions following mycotoxin ingestion: Meta-analysis of published experiments in animals. Toxins 5, 396–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Geller B., Moniz K., Das P., Chippindale A. K., Walker V. K. (2014). Monitoring the developmental impact of copper and silver nanoparticle exposure in Drosophila and their microbiomes. Sci. Total Environ. 487, 822–829. [DOI] [PubMed] [Google Scholar]
- Haskard C. A., El-Nezami H. S., Kankaanpaa P. E., Salminen S., Ahokas J. T. (2001). Surface binding of aflatoxin B(1) by lactic acid bacteria. Appl. Environ. Microbiol. 67, 3086–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) (2012). Chemical agents and related occupations. IARC Monographs on the Evaluation of Carcinogenic Risks to humans 100F, 225–248. [PMC free article] [PubMed] [Google Scholar]
- Jakobsson H. E., Jernberg C., Andersson A. F., Sjolund-Karlsson M., Jansson J. K., Engstrand L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5, e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C., Lofmark S., Edlund C., Jansson J. K. (2010). Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156, 3216–3223. [DOI] [PubMed] [Google Scholar]
- Kish L., Hotte N., Kaplan G. G., Vincent R., Tso R., Gaenzle M., Rioux K. P., Thiesen A., Barkema H. W., Wine E., et al. (2013). Environmental particulate matter induces murine intestinal inflammatory responses and alters the gut microbiome. PLoS One 8, e62220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glockner F. O. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. D., Weiss R. B., Cox J., Dale C., Dearing M. D. (2014). Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 17, 1238–1246. [DOI] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology - Human gut microbes associated with obesity. Nature 444, 1022–1023. [DOI] [PubMed] [Google Scholar]
- Li F. Q., Yoshizawa T., Kawamura O., Luo X. Y., Li Y. W. (2001). Aflatoxins and fumonisins in corn from the high-incidence area for human hepatocellular carcinoma in Guangxi, China. J. Agric. Food Chem. 49, 4122–4126. [DOI] [PubMed] [Google Scholar]
- Lizarraga-Paulin E. G., Moreno-Martinez E., Miranda-Castro S. P. (2011). Aflatoxins and their impact on human and animal health: An emerging problem. In Aflatoxins - Biochemistry and Molecular Biology (Guevara-González R. G., Ed.), pp. 255–282. InTech, Rijeka, Croatia. [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. A., Knight R. (2008). Species divergence and the measurement of microbial diversity. FEMS Microbiol. Rev. 32, 557–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Cable P. H., Abo R. P., Ru H., Graffam M. E., Schlieper K. A., Parry N. M. A., Levine S., Bodnar W. M., Wishnok J. S., et al. (2013). Gut microbiome perturbations induced by bacterial infection affect arsenic biotransformation. Chem. Res. Toxicol. 26, 1893–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Abo R. P., Schlieper K. A., Graffam M. E., Levine S., Wishnok J. S., Swenberg J. A., Tannenbaum S. R., Fox J. G. (2014a). Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: An integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 122, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Mahbub R., Cable P. H., Ru H., Parry N. M. A., Bodnar W. M., Wishnok J. S., Styblo M., Swenberg J. A., Fox J. G., et al. (2014b). Gut microbiome phenotypes driven by host genetics affect arsenic metabolism. Chem. Res. Toxicol. 27, 172–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran A. E. (2004). Measuring Biological Diversity. Blackwell Publishing, Malden, MA. [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., et al. (2006). Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55, 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield D. L., Shaw B. J., Harper G. M., Saoud I. P., Davies S. J., Handy R. D., Henry T. B. (2013). Ingestion of metal-nanoparticle contaminated food disrupts endogenous microbiota in zebrafish (Danio rerio). Environ. Pollut. 174, 157–163. [DOI] [PubMed] [Google Scholar]
- Nicholson J. K., Holmes E., Wilson I. D. (2005). Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3, 431–438. [DOI] [PubMed] [Google Scholar]
- Paulson J. N., Stine O. C., Bravo H. C., Pop M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K., El-Nezami H., Haskard C., Ahokas J., Salminen S. (2001). Aflatoxin B-1 binding by dairy strains of lactic acid bacteria and bifidobacteria. J. Dairy Sci. 84, 2152–2156. [DOI] [PubMed] [Google Scholar]
- Peterfreund G. L., Vandivier L. E., Sinha R., Marozsan A. J., Olson W. C., Zhu J., Bushman F. D. (2012). Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PLoS One 7, e46966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L. O., Campbell T. C. (1982). Effects of sex difference and dietary-protein level on the binding of Aflatoxin B1 to rat liver chromatin proteins in Vivo. Cancer Res. 42, 5053– 5059. [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G., Tang L., Guo X., Wang F., Massey M. E., Su J., Guo T. L., Williams J. H., Phillips T. D., Wang J. S. (2014). Aflatoxin B-1 modulates the expression of phenotypic markers and cytokines by splenic lymphocytes of male F344 rats. J. Appl. Toxicol. 34, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2014). R: A Language and Environment for Statistical Computing. Vienna, Austria. [Google Scholar]
- Rawal S., Kim J. E., Coulombe R., Jr. (2010). Aflatoxin B1 in poultry: Toxicology, metabolism and prevention. Res. Vet. Sci. 89, 325–331. [DOI] [PubMed] [Google Scholar]
- Sharma Y. P., Sumbali G. (1999). Incidence of aflatoxin producing strains and aflatoxin contamination in dry fruit slices of quinces (Cydonia oblonga Mill.) from the Indian State of Jammu and Kashmir. Mycopathologia 148, 103–107. [DOI] [PubMed] [Google Scholar]
- Sinclair L., Osman O. A., Bertilsson S., Eiler A. (2015). Microbial community composition and diversity via 16S rRNA gene amplicons: Evaluating the illumina platform. Plos One 10, e0116955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles M. E., Holzapfel W. H. (1997). Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36, 1–29. [DOI] [PubMed] [Google Scholar]
- Swann J., Wang Y., Abecia L., Costabile A., Tuohy K., Gibson G., Roberts D., Sidaway J., Jones H., Wilson I. D., Nicholson J., Holmes E. (2009). Gut microbiome modulates the toxicity of hydrazine: A metabonomic study. Mol. Biosyst. 5, 351–355. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters W. A., Xu Z., Knight R. (2014). Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. H., Phillips T. D., Jolly P. E., Stiles J. K., Jolly C. M., Aggarwal D. (2004). Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 80, 1106–1122. [DOI] [PubMed] [Google Scholar]
- Wogan G. N., Kensler T. W., Groopman J. D. (2012). Present and future directions of translational research on aflatoxin and hepatocellular carcinoma. A review. Food Addit. Contam. Part A 29, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G., Zhong W., Zheng X., Li Q., Qiu Y., Li H., Chen H., Zhou Z., Jia W. (2013). Chronic ethanol consumption alters mammalian gastrointestinal content metabolites. J. Proteome Res. 12, 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip L. Y., Ku H. C., Aw C. C., Chen W. S., Browne E., Mahendran R., Lee Y. K., Chan E. C. Y. (2014). Investigation of host-gut microbiome interaction in Tacrine-dosed Lister-hooded rats through fecal metabolic profiling. Drug Metab. Rev. 45, 147–147. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.