Abstract
Chemical exposure of cells may damage biomolecules, cellular structures, and organelles thereby jeopardizing cellular homeostasis. A multitude of defense mechanisms have evolved that can recognize specific types of damaged molecules and will initiate distinct cellular programs aiming to remove the damage inflicted and prevent cellular havoc. As a consequence, quantitative assessment of the activity of the cellular stress responses may serve as a sensitive reporter for the induction of specific types of damage. We have previously developed the ToxTracker assay, a mammalian stem cell-based genotoxicity assay employing two green fluorescent protein reporters specific for DNA damage and oxidative stress. We have now expanded the ToxTracker assay with an additional four reporter cell lines to include monitoring of additional stress signaling pathways. This panel of six green fluorescent protein reporters is able to discriminate between different primary reactivity of chemicals being their ability to react with DNA and block DNA replication, induce oxidative stress, activate the unfolded protein response, or cause a general P53-dependent cellular stress response. Extensive validation using the compound library suggested by the European Centre for the Validation of Alternative Methods (ECVAM) and a large panel of reference chemicals shows that the ToxTracker assay has an outstanding sensitivity and specificity. In addition, we developed Toxplot, a dedicated software tool for automated data analysis and graphical representation of the test results. Rapid and reliable identification by the ToxTracker assay of specific biological reactivity can significantly improve in vitro human hazard assessment of chemicals.
Keywords: genotoxicity, DNA damage response, oxidative stress, reporter cell lines, mechanisms of toxicity
Pharmaceutical, chemical, and cosmetics industries yearly develop increasing numbers of novel chemical compounds for a wide range of applications that benefit society. With the increasing production of new chemicals comes a strong demand for novel approaches that can reliably assess the adverse and potentially carcinogenic properties of chemicals already during the early phases of the development process. Currently, in vitro genotoxicity prediction of chemicals generally relies on the classical Ames bacterial mutation test, followed by a mammalian mutation test and chromosome damage assay (Kirkland et al., 2011). Unfortunately, this battery of genotoxicity assays has a relative low specificity and importantly provides only limited insight into the mechanisms of genotoxicity. Currently, various efforts are ongoing to include novel technologies, adverse outcome pathways, and physiological relevant cell models to improve safety assessment of novel chemicals (Lynch et al., 2010).
Interaction of newly developed materials, chemicals, and drugs with biomolecules may disrupt cellular homeostasis and can ultimately lead to severe tissue damage or induction of cancer (Kidane et al., 2014; Ray et al., 2012). Quantitative assessment of the activation status of damage-specific cellular stress response pathways upon exposure to chemicals or xenobiotics provides insight into the type and extent of cellular damage that has been induced and thus the biological reactivity of compounds (Hendriks et al., 2013; Jennings et al., 2013). For this purpose, a cellular system is required in which all these pathways are unimpaired. Mouse embryonic stem (mES) cells are, in contrast to cancer-derived cell lines, eg, TK6 or HepG2 that are often used for in vitro genotoxicity testing, genetically stable and proficient in all cellular pathways necessary for accurate detection of potentially carcinogenic properties of compounds (Giachino et al., 2013). In addition, stem cells have a high rate of cell proliferation making them highly sensitive to DNA damage. Furthermore, stem cells are considered to be the cells of origin for most cancers and therefore a highly relevant cell type for genotoxicity studies.
Although cancer is a complex, multifactorial disease, only for limited types of cellular damage a direct association has been established (Jennings et al., 2013). DNA damage can lead to gene mutations that are a hallmark for tumor induction and promotion. Reactive oxygen species (ROS), often originating from dysfunctional mitochondria, can not only lead to oxidative DNA damage but also affect cell metabolism. Furthermore, upregulation of the unfolded protein response (UPR) and various heat shock proteins (HSPs) has been associated with the development of various tumor types. To determine the activity of these different cancer-associated cellular stress responses, we have previously identified a panel of biomarker genes that are preferentially activated upon exposure to different classes of genotoxic carcinogens, nongenotoxic carcinogens, and noncarcinogenic chemicals by extensive whole-genome transcription profiling of mES cells (Hendriks et al., 2011; Schaap et al., 2014). Interestingly, the majority of genes that were induced by the various carcinogens was associated with the DNA damage, anti-oxidant, and unfolded protein responses. Based on the identified biomarker genes, we have generated various green fluorescent protein (GFP) mES reporter cell lines (Hendriks et al., 2012). The original ToxTracker assay consisted of the DNA damage-associated Berardinelli-Seip congenital lipodystrophy 2 (BSCL2)-GFP reporter and the SRXN1 (sulfiredoxin 1)-GFP reporter that is induced by oxidative stress. Bscl2 encodes the SEIPIN protein and was identified in patients suffering from Berardinelli–Seip congenital lipodystrophy who completely lack adipocyte differentiation (Magré et al., 2001). SEIPIN accumulates in the endoplasmic reticulum (ER) and associates with lipids (Szymanski et al., 2007). We previously found that in mES cells, induction of BSCL2 in response to DNA damage is associated with the ATR (ataxia telangiectasia and Rad3-related)/Checkpoint Kinase 1 (CHK1) DNA damage signaling pathway (Hendriks et al., 2012). ATR together with the CHK1 kinase activates multiple DNA repair mechanisms and checkpoint responses to delay cell cycle progression, modulates DNA replication, and induces apoptosis (Smith et al., 2010). The SRXN1-GFP reporter is strongly activated upon cellular oxidative stress and is directly controlled by the nuclear factor (erythroid-derived 2)-like 2 (NFE2L2/NRF2) transcription factor (Soriano et al., 2008). NRF2 plays a key role in the oxidative stress response and is involved in the activation of various antioxidant gene networks (Itoh et al., 1999).
ATM (ataxia telangiectasia mutated) is another major kinase involved in the cellular DNA damage response. ATM is rapidly activated upon induction of DNA double-strand breaks (Lecona and Fernandez-Capetillo, 2014). ATM directly phosphorylates both the CHK2 checkpoint kinase and the P53 tumor suppressor, thereby inhibiting cell cycle progression, activating DNA repair or inducing apoptosis (Shiloh and Ziv, 2013). Also the cytokine signaling-associated nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) transcription factor, which plays a pivotal role in cell proliferation, cell survival and apoptosis, is rapidly activated in response to DNA damage (Sabatel et al., 2011). Activation of the ATR, ATM, and NF-κB signaling cascades is associated with different types of DNA damage and their relative activation can therefore indicate the primary mechanism of genotoxicity of DNA reactive compounds. Expansion of the ToxTracker assay with reporters for the ATM and NF-κB signaling pathways could therefore significantly improve its ability to identify hazardous properties of chemicals.
Although NRF2 plays a central role in the antioxidant response upon cellular oxidative stress (Kensler et al., 2007), also other cellular signaling pathways are activated by increased ROS levels. Various MAPK and PI3K signaling pathways are associated with oxidative stress and control cell proliferation, differentiation, and apoptosis in response to increased ROS levels (Ray et al., 2012). Chemical exposure will also often elicit protein damage. Accumulation of damaged and unfolded proteins triggers a signal transduction cascade referred to as the UPR and originates from the ER (Hetz, 2012). Accumulation and aggregation of misfolded proteins in the cytoplasm will activate an HSF1 (heat shock factor 1)-associated cellular response (Vihervaara and Sistonen, 2014). Induction of protein misfolding and subsequent activation of the UPR and HSF1 signaling pathways, P53 activation, cytokine secretion, and apoptosis has been associated with various cancers (Wang and Kaufman, 2014). Biomarkers that would be able to identify activation of different oxidative stress and protein unfolding signaling pathways could further improve the in vitro chemical hazard assessment.
Because there is extensive cross-talk between the different cellular signaling pathways in response to different cellular damages and because compounds often induce multiple types of biological damage, integration of the activated cellular pathways is essential to interpret the biological reactivity of compounds.
Here, we report on the expansion of the original ToxTracker assay into a panel of six different mES GFP reporter cell lines representing four distinct biological responses, ie, general cellular stress, DNA damage, oxidative stress, and the UPR. The specificity and sensitivity of the ToxTracker reporter cell lines were validated using the ECVAM-recommended compound library (Kirkland et al., 2008), supplemented with a library of Organisation for Economic Co-operation and Development (OECD)-recognized nongenotoxic carcinogens and an additional selection of reference genotoxins.
MATERIALS AND METHODS
ES cell culture and treatments
C57/Bl6 B4418 wild-type mES cells were cultured in mES knockout medium (Gibco) containing 10% fetal calf serum (FCS), 2 mM glutamax, 1 mM sodium pyruvate, 100 µM β-mercaptoethanol and leukemia inhibitory factor and were propagated on irradiated primary mouse embryonic fibroblasts as feeders according to established protocols (Hendriks et al., 2012). For chemical exposure, cells were seeded 24 h prior to exposure on gelatin-coated plates in Buffalo rat liver (BRL)-conditioned mES cell medium in the absence of feeder cells, and subsequently exposed to the test compounds for 24 h. For analysis of compounds that required metabolic activation, cells were exposed for 3 h in the presence of 1% S9 rat liver extract in 3.2 mM KCl, 0.8 mM MgCl2, 0.5 mM glucose-6-phosphate, and 0.4 mM NADP. After 3 h cells were washed with phosphate-buffered saline (PBS) and cultured for 24 h in BRL-conditioned medium without the tested compounds and S9.
Generation of GFP reporter cell lines
The biomarker genes that were used to create GFP-based reporters were selected from earlier whole-genome transcription profiling studies of mES cells following exposure to 40 genotoxic and nongenotoxic carcinogens (Hendriks et al., 2011; Schaap et al., 2014). The GFP reporters were generated by bacterial artificial chromosome (BAC) recombineering as described previously (Hendriks et al., 2012). Bacterial strains with a BAC containing the biomarker gene were selected using mouse BAC finder ( http://www.mitocheck.org/cgi-bin/BACfinder) and ordered from BACPAC (http://bacpac.chori.org). The BACs used in the ToxTracker assay contain apart from the biomarker gene on average 70-100 kb of upstream sequence and 50-70 kb of downstream sequence encompassing all regulatory sequences that are in close proximity to the gene. The putative biomarker genes on the BAC were modified with a C-terminal GFP marker (Poser et al., 2008) using the Quick & Easy BAC modification Kit (Gene Bridges) as described previously (Hendriks et al., 2012). PCR fragments that contain a GFP-IRES-neomycin/kanamycin reporter cassette and on both ends 50 nucleotides of sequence homology to the 3′-sequence of the biomarker gene on the BAC were generated using the KAPA HiFi PCR system (Kapa Biosystems). BAC strains were transformed with the GFP-IRES-Neo PCR fragment by electroporation, incubated at 37°C for 2 h to allow recombination of the PCR fragment with the BAC and plated on kanamycin selection plates. Individual clones were analyzed for proper integration of the GFP cassette by PCR. Modified BACs were isolated using the Nucleobond PC100 DNA isolation kit (Macherey-Nagel). Current modification of the BAC was confirmed by sequence analysis.
Mouse ES cells were seeded on gelatin-coated culture dishes 24 h prior to transfection. Modified BACs were transfected into mES cells using Lipofectamine 2000 (Invitrogen) as described previously (Hendriks et al., 2012). Monoclonal mES cell lines were selected based on the level of induction of the GFP reporters after exposure to cisplatin, diethyl maleate (DEM), or tunicamycin.
Detection of GFP expression
GPF reporter expression was determined by flow cytometry (Guava easyCyte 6HT, Millipore). Cells were exposed to genotoxic agents in gelatin-coated 96 wells. All tested compounds were dissolved in Dimethyl sulfoxide (DMSO) or phosphate buffered saline (PBS) and diluted in fresh BRL-conditioned mES cell medium just before incubation with the cells. After 24-h exposure, cells were washed with PBS, trypsinized, and resuspended into PBS supplemented with 2% FCS, immediately followed by flow cytometry analysis. Reporter activity was determined as the mean fluorescence intensity (MFI) of 5000 intact cells.
Test criteria
Activation of a reporter cell line was considered positive when at any applied dose exposure to a compound resulted in > 1.5-fold induction of GFP expression. This 1.5-fold increase cut-off for a positive test result is based on extensive validation of the ToxTracker assay using various libraries of reference compounds (Hendriks et al., 2012, this study, and unpublished data G. Hendriks). Statistical support for this threshold comes from the fact that a 1.5-fold induction is at least five times higher than the standard deviation of background fluorescence in DMSO-exposed cells. Measurements at concentrations that induce > 75% cytotoxicity were not considered for data analysis. Application of the 1.5-fold induction cut-off threshold provides positive test results with a confidence of > 99.9%. In a representative experiment, reporter cells were exposed to five different concentrations of a compound, generally at 2-fold dilutions, starting with a concentration that shows no cytotoxicity up to a concentration that results in 50%-75% cell killing. The relative cell survival after 24 h of treatment was calculated as the ratio in concentration of intact cells for treated versus untreated samples as determined by the flow cytometer (Guava, Millipore). In case of no or limited induction of cytotoxicity, the highest dose applied was 1 mM. All presented data are the summary of at least three independent biological replicates. All shown error bars represent the standard error.
Toxplot data analysis
Toxplot is a collection of custom scripts written in the R statistical analysis language (http://www.r-project.org). Toxplot imports the raw data files in CSV format that are generated by the flow cytometer and that contain for each exposure data on the MFI and cell concentration. Following calculation of GFP induction as well as cytotoxicity levels, GFP fold induction is plotted against cytotoxicity. The extent of GFP induction at a particular level of cytotoxicity is calculated by linear regression between the two adjacent cytotoxicity measurements. Next, GFP induction levels for each of the ToxTracker reporters are used for hierarchical clustering of the analyzed compounds based on similarity of reporter activation. Results are visualized in a heat map. Toxplot can be run as a Windows, Linux or MacOS shell script and uses the R-library shiny for a web browser-based graphical user interface.
Validation of GFP reporter cell lines
GFP reporter cells were exposed to 50 genotoxic and nongenotoxic compounds at five or more concentrations. The selection of compounds was largely based on the ECVAM suggested list of chemicals for validation of in vitro genotoxicity test assays (Kirkland et al., 2008). Compound concentrations were based on cytotoxicity with the highest concentration inducing significant cell death (25%-50% viable cells after 24-h treatment).
RESULTS
The carcinogenicity hazard of novel chemicals, xenobiotics, and biologicals is often assessed by testing their ability to induce DNA damage. DNA damage perturbs gene transcription and DNA replication, affects cell cycle progression, induces mutations and genome instability, and is strongly correlated with carcinogenesis (Figure 1A). However, also damage to other cellular biomolecules, structures and organelles has been associated with carcinogenicity. The relative ability of compounds to induce various types of cellular damage and their potency to perturb cellular homeostasis are important determinants in carcinogenicity hazard. Current in vitro carcinogenicity hazard assays are primarily focused on assessing genotoxic potential, but generally do not consider alternative types of biological damage and fail to integrate the various reactive properties of compounds for human hazard prediction.
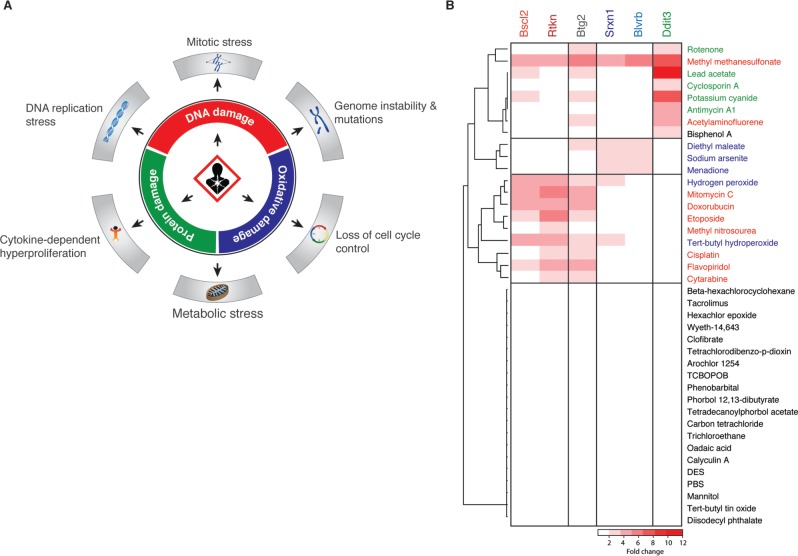
FIG. 1.
Biomarkers for cancer-associated biological damages. A, Overview of the three major types of biological damage and the respective cellular consequences that have been associated with increased carcinogenicity hazard. B, Heatmap indicating the preferential induction of six selected biomarker genes at 8 h after exposure to various genotoxic and nongenotoxic carcinogens as determined by whole-genome transcription profiling of mouse embryonic stem cells.
Biomarker identification
We previously described our extensive toxicogenomics approaches to identify biomarker genes in mES cells that are preferentially activated upon cellular exposure to different classes of carcinogenic compounds (Hendriks et al., 2011; Schaap et al., 2014). We identified two highly specific biomarker genes for DNA damage and oxidative stress that were used to generate fluorescent reporter cell lines which together we named ToxTracker. Utilizing these transcription profiling data we aimed to expand the panel of biomarker genes to allow monitoring of a broader spectrum of cellular stress responses following exposure. In addition to the previously described BSCL2 reporter for DNA replication stress, we identified RTKN (Figure 1B) as a second biomarker to monitor activation of DNA damage responses. Rhotekin is encoded by the Rtkn gene which is strongly upregulated following exposure to a wide variety of DNA damaging agents. RTKN modulates Rho activity and is involved in cytokinesis, cell growth, and transformation (Thumkeo et al., 2013).
We previously described the NRF2-associated SRXN1 reporter for detection of oxidative stress. To extend our ability to detect induction of oxidative stress, we searched for a biomarker gene that was preferentially induced by multiple oxidative treatments but was not regulated by NRF2. BLVRB (biliverdin reductase B) has been shown to play an important role in heme metabolism and has been associated with the cellular antioxidant response (Smith et al., 2008). Importantly, the BLVRB promoter sequence does not contain any NRF2 binding motif.
Finally, we searched for a marker that could be used to monitor activation of the UPR. The UPR is in addition to genotoxicity and oxidative stress the third biological stress pathway that has been directly associated with carcinogenicity. DDIT3 (DNA damage-inducible transcript 3) is a multifunctional transcription factor that has been associated with cell cycle arrest, apoptosis, and the ER stress response (Tabas and Ron, 2011).
Whole-genome transcription data of mES cells after exposure to various genotoxic and nongenotoxic carcinogens clearly demonstrate the differential responsiveness of the selected biomarker genes (Figure 1B). Bscl2 and Rtkn are preferentially induced by DNA damaging agents, Srxn1 and Blvrb are induced upon oxidative stress. Upregulated B-cell translocation gene 2 (BTG2) expression reflects activation of the P53 response and is therefore induced upon DNA damage and oxidative stress. Finally, enhanced DDIT3 expression represents activation of the UPR.
GFP-based reporter cell lines
Next, we created GFP-based reporters for RTKN, BTG2, BLVRB, and DDIT3 in a similar way as the initial ToxTracker cell reporters (Hendriks et al., 2012). In short, GFP was tagged to the C-terminus of the biomarker genes by the Red/ET-mediated BAC recombineering system, reporters were introduced into mES cells by transfection, and stable cell lines were generated. A single cell line was selected for each biomarker gene where the specificity of the induction of GFP expression after carcinogen treatment closely resembled the observed transcriptional response of the endogenous biomarker gene as was verified by Quantitative Reverse Transcription PCR (Hendriks et al., 2012 and results not shown).
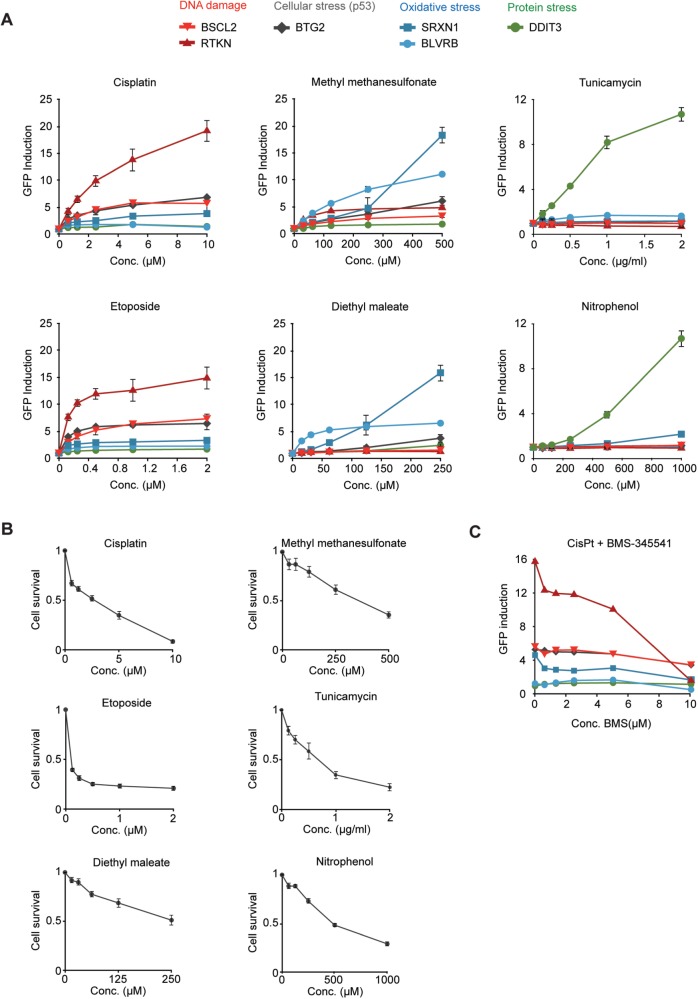
To demonstrate their differential responsiveness, we exposed the six ToxTracker reporter cell lines to the DNA damaging agents cisplatin and etoposide, the genotoxin methyl methanesulfonate (MMS) that was previously shown to strongly activate the cellular antioxidant response (Lee et al., 2007), the pro-oxidant DEM and the UPR-inducing compounds tunicamycin and nitrophenol. For each exposure, the fold-change in GFP reporter induction is determined by flow cytometry. It is important to note that although the same fold-induction threshold (> 1.5) is used for all reporters to indicate a positive response, the reporters differ considerably in the maximum fold-induction that has historically been observed by us. Maximum fold-inductions range between 6× (BSCL2), 6× (BTG2), 8× (DDIT3), 12× (BLVRB), 20× (RTKN), and 50× (SRXN1). Differences in fold-induction levels between the reporters are caused by variation in background levels and extent of changes in gene expression upon exposure. All reporter cell lines show a uniform response where increased GFP expression is observed in all cells (Supplementary Figs. 1 and 2). After 24 h of exposure to cisplatin and etoposide, the BSCL2 and RTKN genotoxicity reporters (red) as well as the P53-responsive BTG2 reporter (gray) were strongly induced (Figs. 2A and B). Although the NRF2-dependent SRXN1 oxidative stress reporter was slightly induced, the BLVRB and DDIT3 reporters for oxidative and protein stress were not activated indicating that the genotoxins cisplatin and etoposide either induce only limited levels of oxidative stress or that activation of the NRF2 antioxidant response is secondary to P53 activation as has been reported previously (Wakabayashi et al., 2010). Exposure to the genotoxin MMS induced both DNA damage reporters, BSCL2 and RTKN, but resulted in a much stronger response for the SRXN1 and BLVRB oxidative stress reporters as observed previously (Hendriks et al., 2011, 2012). The preferential induction of the SRXN1 and BLVRB reporters following oxidative stress was also detected after exposure to the pro-oxidant DEM. Here, no induction of the DNA damage reporters BSCL2 or RTKN was observed even at higher cytotoxicity levels (Figure 2B), indicating that oxidative stress is a poor inducer of the DDR. Increased levels of oxidative stress following DEM exposure did induce activation of the P53 response pathway as indicated by the activation of the BTG2-GFP reporter. Finally, exposure to tunicamycin, a strong inducer of the ER-associated UPR or to nitrophenol, which induces protein unfolding, selectively activated the DDIT3-GFP protein stress reporter. Even at high levels of cytotoxicity, none of the DNA damage or oxidative stress reporters was activated, confirming their specificity. GFP reporter induction levels for these reference chemicals, as well as assessment of their statistical significance, are summarized in Supplementary Table 1.
FIG. 2.
Selective activation of the ToxTracker reporter cell lines. A, green fluorescent protein (GFP) reporter cells were exposed to increasing concentrations of the DNA damaging agents cisplatin and etoposide, the alkylating agent methyl methanesulfonate (MMS), the oxidative stress-inducing agent diethyl maleate (DEM), and the unfolded protein response-activating compounds tunicamycin and nitrophenol. GFP induction levels in intact cells were determined by flow cytometry at 24 h after initiation of the exposure. B, Cell survival was determined by flow cytometry after 24-h exposure as the relative decrease in cell concentration compared with untreated controls. C, Reporter cell lines were exposed to 10 μM cisplatin in the presence of an increasing concentration of the NF-κB inhibitor BMS-345541.
As expression of RTKN has been associated with the NF-κB cytokine signaling pathway, we tested whether addition of the synthetic NF-κB inhibitor BMS-345541 during cisplatin treatment could selectively abolish the induction of RTKN. Indeed, although expression of RTKN was strongly reduced, the responsiveness of the BSCL2 genotoxicity biomarker was largely unaffected (Figure 2C). This result suggests that BSCL2 and RTKN function in distinct subpathways of the DNA damage response.
Metabolic activation
Occasionally, compounds are not directly reactive but can become genotoxic following metabolic activation during detoxification reactions in the liver, kidney, and lung (Nebert and Dalton, 2006). The major enzymes that are involved in the metabolic activation of pro-genotoxins, eg, cytochrome P450s and epoxide hydrolyses, are scarcely or not expressed in mESC. We tested whether addition of S9 rat liver extract during pro-carcinogen treatment could supplement the ToxTracker assay with the necessary enzymatic activities to allow detection of the genotoxic properties of compounds that required metabolic activation. Reporter cell lines were exposed to six reference pro-genotoxins in the presence of S9 rat liver extract for 3 h. After removal of S9 and compounds, fresh medium was added and after 24-h recovery, GFP reporter induction was measured by flow cytometry. All tested pro-genotoxins induced both DNA damage reporters BSCL2 and RTKN, consistent with the reported DNA damage inducing properties of these pro-genotoxins after metabolic activation (Supplementary Figure 3). In addition to activation of the DNA damage reporters, exposure to the pro-genotoxins in the presence of S9 extract frequently led to activation of the oxidative stress reporters. Only after treatment with benzo[a]pyrene induction of the UPR was detected, as observed previously in other studies (Boysen and Hecht, 2003).
ToxTracker data integration
In order to allow comparison of induction levels of the ToxTracker reporter cell lines for large number of compounds, we developed Toxplot, a dedicated data analysis software package. Toxplot is a collection of R scripts that imports fluorescence data and cell concentration information from the flow cytometer, calculates GFP induction levels and cytotoxicity, performs statistical analysis of the data, hierarchically clusters tested compounds based on similarity in reporter activation, and visualizes reporter activities in a heatmap allowing convenient interpretation of obtained test results. To compare the induction of the six GFP reporters for a collection of compounds, each with different biological reactivity, dose–response relationships and kinetics, Toxplot calculates for every reporter and for each compound the extent of GFP induction at a specified level of cytotoxicity (Figure 3A). In this way, the relative induction of the various GFP reporters by different compounds can be compared at equitoxic doses. As GFP induction by flow cytometry is determined exclusively in intact, viable cells, we generally determine the extent of GFP reporter activation at compound concentrations that induce 50% cytotoxicity after 24-h exposure.
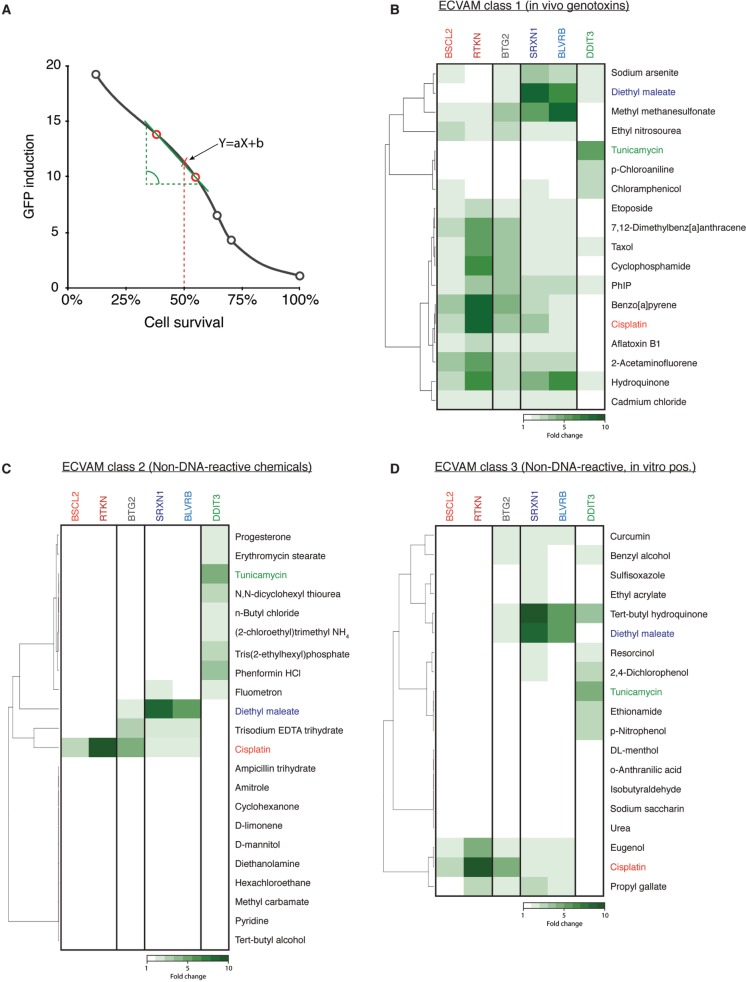
FIG. 3.
Validation of the differential activation of the ToxTracker reporter cell lines. A, Induction levels of the different GFP reporters were calculated for all compounds at an equitoxic concentration that induced 50% cytotoxicity. GFP levels were determined by linear regression of the GFP induction data points of the two doses encompassing 50% cytotoxicity. B, Induction of the GFP reporter upon exposure to a selection of ECVAM-recommended carcinogens that are established positives in an in vitro genotoxicity assay. Cisplatin (red), diethyl maleate (blue), and tunicamycin (green) are considered as positive controls for induction of DNA damage, oxidative stress and activation of the unfolded protein response and color-coded in all heatmaps and graphs. C, Induction of ToxTracker reporters following exposure to a selection of ECVAM-recommended noncarcinogens that are established negatives in an in vitro genotoxicity assay. D, Activation of ToxTracker reporters after exposure to ECVAM-recommended compounds that are noncarcinogens or nongenotoxic carcinogens but that occasionally scored positive in a conventional in vitro/in vivo genotoxicity test.
Validation of the ToxTracker assay
The sensitivity and specificity of the ToxTracker reporter assay were further validated using the reference compound library as suggested by ECVAM (Kirkland et al., 2008). The library consists of genotoxic carcinogens (class 1) that should be positive in an in vitro genotoxicity test, a collection of nongenotoxic carcinogens and noncarcinogens (class 2) that should give negative results in a genotoxicity test and a set of noncarcinogenic compounds that are occasionally found positive in an in vitro genotoxicity assay (class 3). All compounds were tested either up to a concentration that induces 75% cytotoxicity or up to a maximum concentration of 1 mM. For compounds that required metabolic activation, exposures were performed in the presence of S9 rat liver extract.
Almost all class 1 ECVAM compounds induced the DNA damage-associated GFP reporters (Figure 3B). Only chloroaniline failed to activate the genotoxicity reporters. Of note, chloroaniline is generally found negative in other in vitro genotoxicity assays (Birrell et al., 2010). Many of the tested carcinogenic compounds not only activated the genotoxicity reporters but also displayed additional biological reactivities. For MMS, hydroquinone and sodium arsenite, induction of oxidative stress, rather than induction of DNA damage, appear to be the primary mode of reactivity. These data underscore the often complex reactive properties of compounds and subsequent cellular responses upon exposure. None of the class 2 ECVAM-suggested compounds induced the BSCL2 or RTKN reporters, correctly classifying these compounds as being nongenotoxic (Figure 3C). A number of these compounds did induce the oxidative stress or protein stress reporters. Results for the genotoxicity reporters were generally also negative for the class 3 ECVAM noncarcinogens with the exception of eugenol and propyl gallate (Figure 3D). Although eugenol is negative in carcinogenicity studies in rats and mice, it has been reported positive in both the in vitro and in vivo micronucleus assay and the in vivo chromosome aberration test, indicative for genotoxic properties of eugenol (Maralhas et al., 2006). Also propyl gallate is a rodent noncarcinogen but has occasionally been found positive in an in vivo mutation assay and chromosome aberration test (Fowler et al., 2012). Various class 3 compounds strongly induced the ToxTracker reporters for oxidative stress (eg, curcumin, ethyl acrylate, tert-butyl hydroquinone, resorcinol) or the UPR (eg, dinitrophenol, chlorophenol). This reactivity may be related to their occasional positive score in conventional genotoxicity assays.
Extended validation
To extend the validation of the ToxTracker assay, we tested a collection of 13 nongenotoxic carcinogens that were previously used for whole-genome transcription profiling in mESC (Schaap et al., 2014). As anticipated, none of the tested nongenotoxic carcinogens activated the BSCL2 or RTKN DNA damage reporters (Figure 4A). However, various nongenotoxic carcinogens induced the oxidative stress response and the UPR.
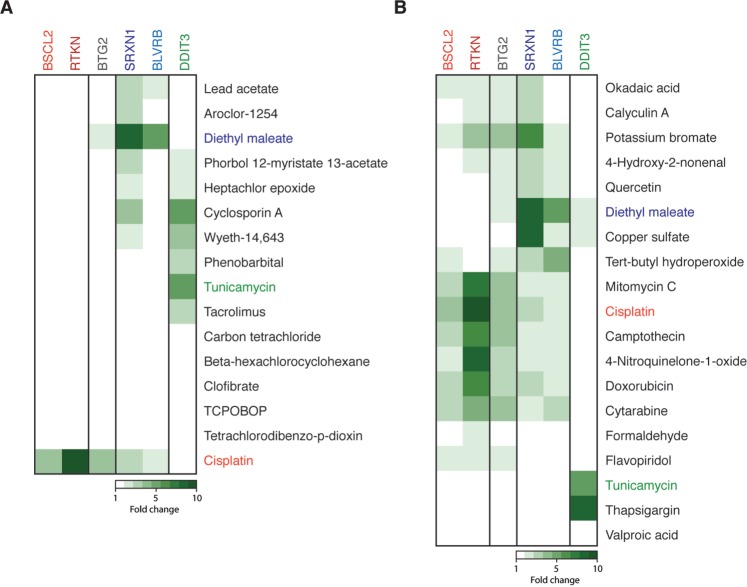
FIG. 4.
Extended validation of the GFP reporter cell lines. A, ToxTracker reporter cell lines were exposed to a collection of reference nongenotoxic carcinogens. GFP induction levels were calculated for a compound concentration that induce 50% cytotoxicity. Cisplatin, DEM, and tunicamycin were used as positive controls for the induction of DNA damage, cellular oxidative stress, and protein damage, respectively. B, Induction of the GFP reporters following exposure to a selection of established genotoxic and nongenotoxic chemicals.
Next, we tested a collection of compounds which have been positive in conventional in vitro genotoxicity tests. All well-established DNA damaging agents including mitomycin C, camptothecin, and doxorubicin strongly induced both the BSCL2 and RTKN genotoxicity reporters (Figure 4B). Interestingly, also the nucleoside analog and DNA replication chain inhibitor cytosine arabinoside (cytarabine) and the cyclin-dependent kinase inhibitor flavopiridol were identified as genotoxin. In addition, we observed a slight induction of the two genotoxicity reporters by the protein phosphatase inhibitors calyculin A and okadaic acid. The protein phosphatases PP2A and PP2B are broad-range serine/threonine phosphatases and their inhibition has been shown to affect various DNA-associated processes including transcription, DNA replication, cell growth, and apoptosis (Wurzenberger and Gerlich, 2011). As a consequence inhibition of these phosphatases is likely to indirectly induce a DNA damage response. In addition to induction of the DNA damage reporters, many of the tested compounds activated the oxidative stress-responsive reporters SRXN1 and BLVRB. For some of these compounds, eg, potassium bromate and tert-butyl hydroperoxide, oxidative stress induction appears to be the primary mode of toxicity. The flavonoid quercetin and copper sulfate did not display DNA damaging properties. The HDAC inhibitor valproic acid, which promotes chromatin relaxation and stimulates general gene transcription, did not induce any reporter of the ToxTracker assay under the tested conditions. The specificity of the DDIT3 reporter for activation of the UPR was confirmed after exposure to the well-established UPR-activator thapsigargin.
All ToxTracker results are summarized and compared with test results in the conventional genotoxicity assays (Table 1 and Supplementary Table 2). A compound was classified as genotoxic in the ToxTracker assay when either the BSCL2-GFP or the RTKN-GFP DNA damage reporter was induced more than 1.5-fold. This validation indicates that the ToxTracker reporter assay displays a much higher sensitivity (95%) and specificity (94%) than the classical genotoxicity tests. The ability to discriminate between induction of DNA damage, oxidative stress, and protein unfolding allows the ToxTracker genotoxicity assay to discern between genuine and false-positive genotoxicity measurements.
TABLE 1.
Sensitivity and Specificity of the Conventional and ToxTracker in vitro Genotoxicity Tests
| Sensitivity (%) | Specificity (%) | |
|---|---|---|
| Ames testa | 46b | 81 |
| In vitro micronucleus testa | 96 | 47 |
| Chromosome aberration testa | 94 | 50 |
| ToxTracker assay | 95 | 94 |
aData obtained from Kirkland et al. (2008, 2011, 2014).
bSensitivity increases to 62% when microtubule disrupting compounds were omitted from the calculations.
Reporter specificity independent of cytotoxicity
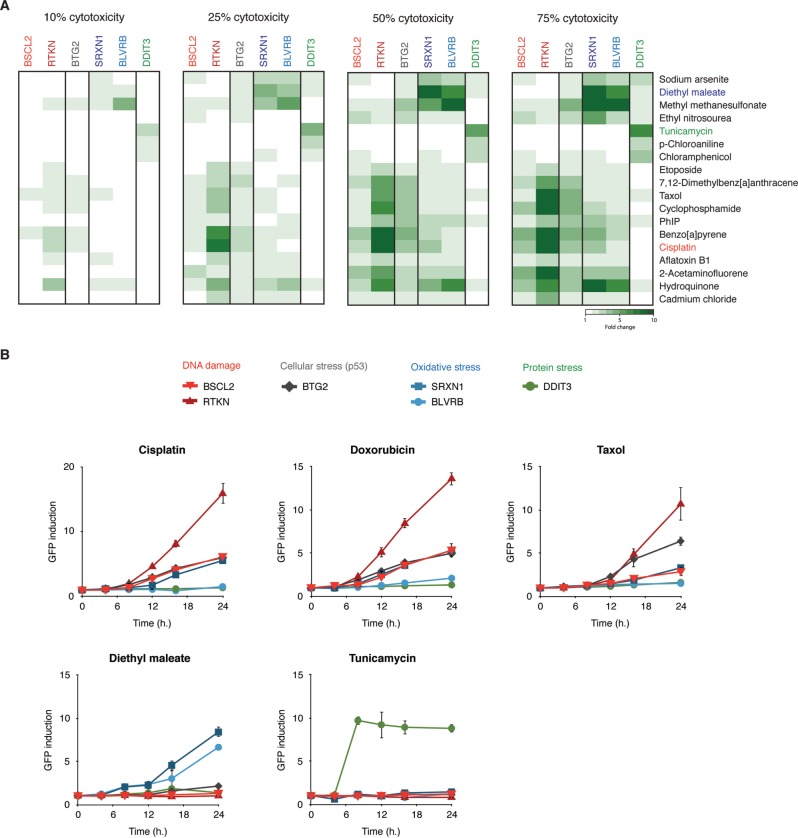
Although we quantify GFP measurements by flow cytometry at a single cell level exclusively in intact cells, we questioned whether the sensitivity or specificity of the different GFP reporters may be affected by the level of cytotoxicity. We therefore calculated the extent of induction for each of the ToxTracker reporters at four different cytotoxicity levels (10%, 25%, 50%, and 75%) for the collection of genotoxins suggested by ECVAM (Figure 5). Nearly all compounds that were classified as genotoxic at high levels of cytotoxicity (75%) were already positive at low cytotoxicity levels (25%). Furthermore, the identification of the primary mode of toxicity, ie, DNA damage, oxidative stress of protein damage, was not affected by the level of cytotoxicity. The only exception is chloramphenicol, which at weak cytotoxic concentrations already induced the DDIT3 protein damage reporter, but at high levels of cytotoxicity in addition activated the BSCL2 genotoxicity reporter. As described before, no reactivity was observed for chloroaniline. Only at very low levels of cytotoxicity (< 10%) induction of the ToxTracker reporters does not for all genotoxic compounds meet the 1.5-fold increase threshold any more, although often a significant induction of the reporters was observed.
FIG. 5.
Specificity of the GFP reporters is largely unaffected by cytotoxicity. A, GFP reporter induction levels were calculated at compound concentrations that induce 10%, 25%, 50%, or 75% cytotoxicity using the panel of ECVAM-selected genotoxic carcinogens. Clustering of compounds was based on the similarity in reporter activation at 50% cytotoxicity. B, Kinetics of GFP induction was determined by flow cytometric analysis of intact cells following 4-, 8-, 12-, 16-, and 24-h exposure to the DNA damaging agents cisplatin and doxorubicin, the microtubule disrupting agent taxol and the control compounds DEM and tunicamycin for induction of oxidative stress and protein unfolding, respectively.
ToxTracker reporter kinetics
We previously established 24 h as an optimal exposure time for reliable assessment of GFP reporters activation in mESC for both DNA damaging agents and pro-oxidants (Hendriks et al., 2011, 2012). To gain more insight into the response kinetics of the different GFP reporter genes, we exposed the ToxTracker cell lines to the genotoxins cisplatin and doxorubicin, the microtubule disruptor taxol, the oxidative stress-inducing agent DEM and the UPR inducer tunicamycin, and determined at various time points GFP induction. For BSCL2-GFP, RTKN-GFP, BLVRB-GFP, SRXN1-GFP, and BTG2-GFP, the earliest time point at which a response could be detected was at 8 h after which induction levels increased steadily over time (Figure 5B). The kinetics of induction for DDIT3-GFP by the UPR-inducing compound tunicamycin was clearly deviant from those of the other reporters by their respective potentiating agents. The DDIT3 reporter was strongly induced after 8 h of exposure to tunicamycin, after which the levels of induction remained constant over time. We did not observe any indications for the appearance of a secondary wave of stress response induction for any of the tested compounds that may occur at late exposure times when significant cytotoxicity is expected to occur. We conclude that the specificity of the ToxTracker reporter assay is not affected by the time of exposure although the GFP levels increase over time.
Detection of aneugenic compounds
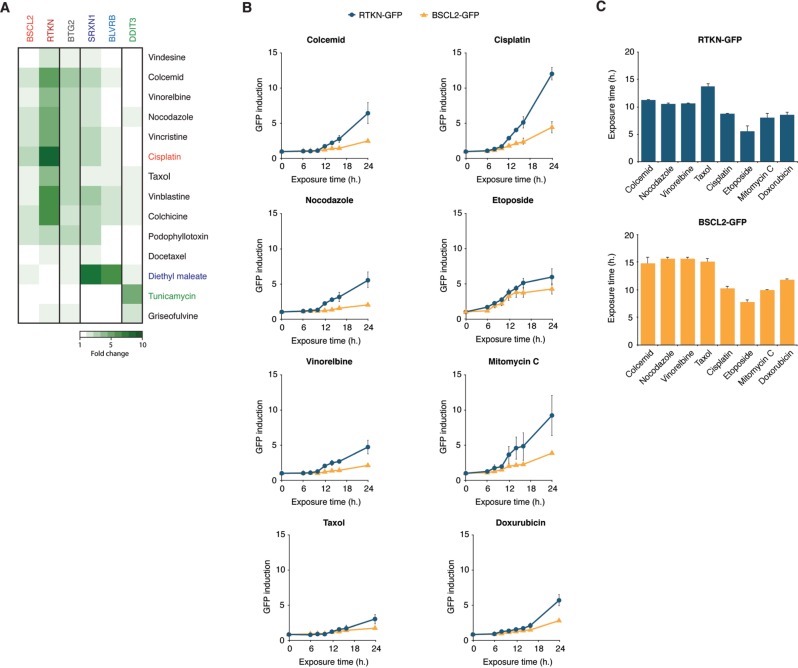
A distinct class of compounds that are notoriously difficult to classify by the conventional genotoxicity assays are microtubule disrupting agents. Microtubule disruption leads to missegregation of chromosomes during mitosis, resulting in chromosome breakage and aneuploidy. Microtubule disruptors are generally noncarcinogenic but give a positive score in the micronucleus test (Kirsch-Volders et al., 2011). To determine the reactivity of such aneugenic compounds in the ToxTracker assay, we exposed the reporter cell lines to a collection of the well-established microtubule disrupting agents colcemid, nocodazole, vinorelbine, and taxol. In contrast to the direct DNA damaging agents cisplatin, etoposide, mitomycin C, and doxorubicin that induce both BSCL2- and RTKN-GFP DNA reporters, the tested mitotic spindle poisons predominantly activated only the RTKN-GFP reporter (Figure 6A). In addition, exposure to the microtubule disruptors also induced the P53- and NRF2-associated cellular signaling response.
FIG. 6.
Differential responses of the BSCL2 and RTKN-GFP reporters discriminate between clastogenic and aneugenic compounds. A, ToxTracker reporter cell lines were exposed to a selection of 11 established microtubule disrupting agents. GFP induction levels were determined after 24-h exposure by flow cytometry. B, Kinetics of BSCL2-GFP and RTKN-GFP following exposure to the DNA damaging compounds cisplatin, etoposide, mitomycin C, and doxorubicin or the mitotic spindle poisons colcemid, nocodazole, vinorelbine and taxol. GFP induction was determined after 4-, 8-, 12-, 16-, and 24-h exposure. C, Exposure times that resulted in a 1.5-fold increase in GFP signal for the BSCL2-GFP and RTKN-GFP reporters after exposure to the tested clastogenic and aneugenic compounds were calculated by linear regression of the GFP induction data points of the two time points encompassing 1.5-fold induction.
Next, we investigated the kinetics of reporter activation by the microtubule disruptors. Although induction of the DNA damage reporters BSCL2 and RTKN after exposure to the DNA damaging agents cisplatin, etoposide, mitomycin C, and doxorubicin could already be observed after approximately 8 h, activation of the RTKN-GFP reporter by the microtubule disruptors took significantly longer (12 h) (Figs. 6B and C). Interestingly, the BSCL2-GFP reporter that is associated with DNA replication inhibition was hardly induced within 24-h exposure to the microtubule disrupting agents. Together, these data indicate that the ToxTracker assay is able to identify mitotic spindle poisons as genotoxic agents. By comparing the differential induction and kinetics of the BSCL2 and RTKN DNA damage reporters, ToxTracker is able to discriminate between direct DNA damaging agents and non-DNA reactive aneugenic agents.
DISCUSSION
The current standard battery of in vitro genotoxicity assays generally includes the bacterial Ames gene mutation test and a micronucleus test using mammalian cell line, often TK6 human lymphoblastoid cells, occasionally expanded with a mammalian gene mutation assay (Kirkland et al., 2011). Extensive evaluations indicate that this battery of in vitro genotoxicity assay can identify genotoxic carcinogens and in vivo genotoxins with a high accuracy (Kirkland et al., 2011). However, the relative high frequency of false positive results in the in vitro micronucleus test occasionally identifies nongenotoxic carcinogens and noncarcinogens as potentially genotoxic. Recently, it was shown that misleading “false positive” test results could dramatically be reduced by using p53 proficient human cell lines (Fowler et al., 2014). In addition to the regulatory in vitro genotoxicity assays, the Comet assay is occasionally included for detection of DNA strand breaks (Witte et al., 2007).
A profound limitation of the conventional in vitro genotoxicity assays is that they often lack the ability to provide insight into the mechanisms of toxicity that are exerted by a compound. Understanding the mechanisms of toxicity of compounds is important to identify false-positive genotoxicity findings and to better estimate the relevance of obtained test results for human hazard assessment. An attractive approach to obtain mechanistic insight into toxicity is to visualize the cellular stress response pathways that are activated after exposure. Various in vitro systems have been developed that use quantitation of the activation of a specific cellular stress response for the assessment of genotoxicity of compounds. The Vitotox test visualizes activation of the bacterial S.O.S. DNA damage response (van der Lelie et al., 1997), and the yeast RadarScreen employs a Rad54-beta galactosidase reporter for detection of double-strand DNA breaks (Westerink et al., 2011). Mammalian reporter cell lines have been described in which the promoters of rad51c P53, cystatin, and NRF2 are fused to a luciferase gene to detect induction of DNA damage, apoptosis, and cellular oxidative stress (Westerink et al., 2010). More recently a HepG2 liver cell assay has been reported that uses phosphorylation of histone variant H2AX for detection of DNA damage (Khoury et al., 2013). Phosphorylation of H2AX is a hallmark for induction of double-strand DNA breaks and plays an essential role in initiating repair of these breaks (Scully and Xie, 2013). Validation of the H2AX-based assay indicates a high sensitivity and specificity for detection of genotoxic carcinogens. However, various noncanonical functions of H2AX phosphorylation, including chromatin remodeling have been described which could complicate interpretation of the H2AX-based genotoxicity assay (Scully and Xie, 2013).
The most extensively validated in vitro cell signaling-based genotoxicity test is the GreenScreen HC assay, a fluorescent reporter assay that uses the GADD45α (growth arrest and DNA damage inducible alpha) gene promoter fused to a GFP marker for the detection of genotoxic agents (Hastwell et al., 2006). GADD45α is a member of the growth arrest and DNA damage (GADD) proteins that are induced upon various cellular stresses including nutrient deprivation, oxidative stress, and DNA damage (Rosemary Siafakas and Richardson, 2009). GADD45α expression is directly activated by P53 upon induction of DNA damage (Zhan, 2005). However, GADD45α is also induced by NF-κB and the NRF2 antioxidant pathway (Han et al., 2008). The GreenScreen HC assay has been validated using large libraries of reference chemicals as well as proprietary compounds (Birrell et al., 2010; Olaharski et al., 2009). GreenScreen HC shows an excellent sensitivity and specificity compared with the conventional genotoxicity tests. More recently, the BlueScreen HC, a luciferase-based version of the GADD45α reporter assay, has been described (Hughes et al., 2012).
To gain understanding of the often complex biological reactivity and interactions of chemicals, biologics and (micro/nano)materials, monitoring of various cellular damage response pathways will be required. For the development of the ToxTracker assay, we employed whole-genome transcription analysis of mES cells to identify biomarker genes that were selectively activated upon exposure to different classes of chemicals and biologics (Hendriks et al., 2011). The original ToxTracker assay consisted of the DNA damage-associated BSCL2-GFP reporter, associated with perturbed DNA replication, and controlled by ATR and the oxidative stress SRXN1-GFP reporter being part of the NRF2 antioxidant response pathway.
We identified Rhotekin (Rtkn) as another gene that is preferentially induced in response to DNA damaging agents. RTKN is an effector of the RHO GTPase with relative unknown function (Reid et al., 1996) and has been implicated in neural stem cell differentiation and apoptosis regulation (Collier et al., 2009). Activation of RHO/RTKN has been associated with the NF-κB cytokine signaling pathway and its overexpression has been implemented in the activation of antiapoptotic responses leading to increased chemoresistance of gastric cancer cells (Liu et al., 2004). In this study, we confirmed the association between RTKN induction and NF-κB signaling (Figure 2C).
Combining the BSCL2 and RTKN biomarkers in a single genotoxicity test further increased the high sensitivity and specificity of the ToxTracker assay. More importantly, the different reporters allow assessment of the mechanisms of genotoxicity. The BSCL2-GFP reporter identifies compounds that induce DNA replication blocking lesions that are particularly hazardous in proliferating cells and tissues. Microtubule disruption agents that can induce chromosome breakage during mitosis selectively activate the RTKN-GFP reporter. Activation of NF-κB by these mitotic spindle poisons has previously been associated with activation of the ATM DNA damage kinase and its effector NF-κB essential modulator (NEMO) (McCool and Miyamoto, 2012).
We previously identified SRXN1 as biomarker for cellular oxidative stress. SRXN1 is involved in the reduction of hyperoxidized peroxiredoxin (Prx) after oxidative stress (Chang et al., 2004). We extended the ToxTracker assay with a second reporter for oxidative stress. The Blvrb gene encodes an oxidoreductase that catalyzes the NADPH-dependent reduction of a variety of flavins (Komuro et al., 1996). Overexpression of BLVRB has been associated with prostate cancer (Pallua et al., 2013). We showed that in mES cells, BLVRB expression is specifically induced upon oxidative stress although the specificity and kinetics of the BLVRB-GFP reporter clearly deviate from the NRF2-controlled SRXN1-GFP reporter (Figs. 3 and 4). Currently, the cellular signaling pathways that control expression of BLVRB are largely unknown.
In addition to DNA damage and induction of ROS production, also protein unfolding has been associated with increased carcinogenicity. The UPR is mediated by the ER protein chaperone GRP78 (BiP) (78 kDa glucose regulated protein), three ER transmembrane receptors PERK, ATF6 and IRE1, and the transcription factor X-box binding protein 1 (XBP1) (Hetz, 2012). Accumulation of misfolded proteins in the cytosol is recognized by various members of the HSP family including HSP70 and HSP90 and the HSF1 transcription factor (Frydman, 2001; Whitesell and Lindquist, 2005). Various UPR and HSPs have been associated with cancer (Wang et al., 2014). We expanded the ToxTracker assay with the DDIT3-GFP reporter. DDIT3 encodes the transcription factor CHOP, which plays an essential role in the response to a wide variety of cell stresses and induces cell cycle arrest and apoptosis following ER stress but is not activated by DNA damage in mES cells.
The sixth reporter in the ToxTracker assay is P53-controlled BTG2-GFP. P53 is a transcription factor that plays a pivotal role in multiple cellular stress responses including the DNA damage, oxidative stress, and the UPRs (Meek, 2009). BTG2 is a component of the P53-dependent DNA damage response, is involved in regulation of the G1/S cell cycle checkpoint (Winkler, 2010), and is induced by various cellular stressors.
In general, the fold-induction of the various ToxTracker reporters is determined at exposure levels with a relatively high level of cytotoxicity (i.e. 50%) which in theory might affect their specificity. However, assessment of the response of the various reporters over a wide dose range showed that the identification of the primary mode of toxicity, ie, DNA damage, oxidative stress of protein damage, was not affected by the level of cytotoxicity.
One might wonder to what extent the stress response specificity of the ToxTracker biomarkers is specific for mouse ES cells. Both SRXN1 and DDIT3 have recently been used as stress reporters in human HepG2 cells (Wink, 2014) and behave with a similar preferentiality as in mouse ES cells. In addition, we found BSCL2 expression to be highly upregulated in mouse lymphoma which suffered from high levels of replication stress (unpublished results), whereas Btg2 has been reported also in human cells to be induced in response to DNA damage through a p53-dependent pathway (Cortes 2000). Conservation of the stress response specificity of the remaining two biomarker genes (Blvrb and Rtkn) in human cells and tissues is unclear.
Here, we report on the extended ToxTracker assay for mechanistic genotoxicity screening. By utilizing a panel of reporter genes that are preferentially induced by either DNA damage, oxidative stress or protein damage, the ToxTracker assay provides toxicologists with a valuable tool to gain understanding of the principal types of biological damage induced by chemicals, biologicals, and other materials. Detailed dissection of the signaling cascades that underlie induction of the various reporters is required to fully unravel the cellular reactivity of compounds.
Supplementary Material
ACKNOWLEDGMENTS
We thank all colleagues and former colleagues at the Department of Human Genetics at the Leiden University Medical Center (LUMC) and Professor Bob van de Water at the Division of Toxicology of the LACDR at Leiden University for their help and useful discussions and Dr Tahar van Straaten (LUMC) for technical support.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NWO, project 050-060-510); the Technology Program of the Ministry of Economic Affairs (project LCG.6935).
REFERENCES
- Birrell L., Cahill P., Hughes C., Tate M., Walmsley R. M. (2010). GADD45a-GFP GreenScreen HC assay results for the ECVAM recommended lists of genotoxic and non-genotoxic chemicals for assessment of new genotoxicity tests. Mutat. Res. 695, 87–95. [DOI] [PubMed] [Google Scholar]
- Boysen G., Hecht S. S. (2003). Analysis of DNA and protein adducts of benzo[a]pyrene in human tissues using structure-specific methods. Mutat. Res. 543, 17–30. [DOI] [PubMed] [Google Scholar]
- Chang T. S., Jeong W., Woo H. A., Lee S. M., Park S., Rhee S. G. (2004). Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem. 279, 50994–51001. [DOI] [PubMed] [Google Scholar]
- Collier F. M., Loving A., Baker A. J., McLeod J., Walder K., Kirkland M. A. (2009). RTKN2 induces NF-KappaB dependent resistance to intrinsic apoptosis in HEK cells and regulates BCL-2 genes in human CD4(+) lymphocytes. J. Cell Death 2, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes U., Moyret-Lalle C., Falette N., Duriez C., Ghissassi F. E., Barnas C., Morel A. P., Hainaut P., Magaud J. P., Puisieux A. (2000). BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol Carcinog 27, 57–64. [PubMed] [Google Scholar]
- Fowler P., Smith K., Young J., Jeffrey L., Kirkland D., Pfuhler S., Carmichael P. (2012). Reduction of misleading (“false”) positive results in mammalian cell genotoxicity assays. I. Choice of cell type. Mutat. Res. 742, 11–25. [DOI] [PubMed] [Google Scholar]
- Frydman J. (2001). Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647. [DOI] [PubMed] [Google Scholar]
- Giachino C., Orlando L., Turinetto V. (2013). Maintenance of genomic stability in mouse embryonic stem cells: Relevance in aging and disease. Int. J. Mol. Sci. 14, 2617–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han E.-S., Muller F. L., Pérez V. I., Qi W., Liang H., Xi L., Fu C., Doyle E., Hickey M., Cornell J. (2008). The in vivo gene expression signature of oxidative stress. Physiol. Genomics 34, 112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastwell P. W., Chai L. L., Roberts K. J., Webster T. W., Harvey J. S., Rees R. W., Walmsley R.M. (2006). High-specificity and high-sensitivity genotoxicity assessment in a human cell line: Validation of the GreenScreen HC GADD45a-GFP genotoxicity assay. Mutat. Res. 607, 160–175. [DOI] [PubMed] [Google Scholar]
- Hendriks G., Atallah M., Raamsman M., Morolli B., van der Putten H., Jaadar H., Tijdens I., Esveldt-van Lange R., Mullenders L., van de Water B., et al. (2011). Sensitive DsRed fluorescence-based reporter cell systems for genotoxicity and oxidative stress assessment. Mutat. Res. 709-710, 49–59. [DOI] [PubMed] [Google Scholar]
- Hendriks G., Atallah M., Morolli B., Calléja F., Ras-Verloop N., Huijskens I., Raamsman M., van de Water B., Vrieling H. (2012). The ToxTracker assay: Novel GFP reporter systems that provide mechanistic insight into the genotoxic properties of chemicals. Toxicol. Sci. 125, 285–298. [DOI] [PubMed] [Google Scholar]
- Hendriks G., van de Water B., Schoonen W., Vrieling H. (2013). Cellular-signaling pathways unveil the carcinogenic potential of chemicals. J. Appl. Toxicol. 33, 399–409. [DOI] [PubMed] [Google Scholar]
- Hetz C. (2012). The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell. Biol. 13, 89–102. [DOI] [PubMed] [Google Scholar]
- Hughes C., Rabinowitz A., Tate M., Birrell L., Allsup J., Billinton N., Walmsley R. M. (2012). Development of a high-throughput Gaussia luciferase reporter assay for the activation of the GADD45a gene by mutagens, promutagens, clastogens, and aneugens. J. Biomol. Screen. 17, 1302–1315. [DOI] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999). Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P., Limonciel A., Felice L., Leonard M. O. (2013). An overview of transcriptional regulation in response to toxicological insult. Arch. Toxicol. 87, 49–72. [DOI] [PubMed] [Google Scholar]
- Kensler T. W., Wakabayashi N., Biswal S. (2007). Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47, 89–116. [DOI] [PubMed] [Google Scholar]
- Khoury L., Zalko D., Audebert M. (2013). Validation of high-throughput genotoxicity assay screening using γH2AX in-cell western assay on HepG2 cells. Environ. Mol. Mutagen. 54, 737–746. [DOI] [PubMed] [Google Scholar]
- Kidane D., Chae W. J., Czochor J., Eckert K. A., Glazer P. M., Bothwell AL. M., Sweasy J. B. (2014). Interplay between DNA repair and inflammation, and the link to cancer. Crit. Rev. Biochem. Mol. Biol. 49, 116–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland D., Kasper P., Müller L., Corvi R., Speit G. (2008). Recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests: A follow-up to an ECVAM workshop. Mutat. Res. 653, 99–108. [DOI] [PubMed] [Google Scholar]
- Kirkland D., Reeve L., Gatehouse D., Vanparys P. (2011). A core in vitro genotoxicity battery comprising the Ames test plus the in vitro micronucleus test is sufficient to detect rodent carcinogens and in vivo genotoxins. Mutat. Res. 721, 27–73. [DOI] [PubMed] [Google Scholar]
- Kirkland D., Zeiger E., Madia F., Corvi R. (2014). Can in vitro mammalian cell genotoxicity test results be used to complement positive results in the Ames test and help predict carcinogenic or in vivo genotoxic activity? II. Construction and analysis of a consolidated database. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 775, 69–80. [DOI] [PubMed] [Google Scholar]
- Kirsch-Volders M., Plas G., Elhajouji A., Lukamowicz M., Gonzalez L., Vande Loock K., Decordier I. (2011). The in vitro MN assay in 2011: Origin and fate, biological significance, protocols, high throughput methodologies and toxicological relevance. Arch. Toxicol. 85, 873–899. [DOI] [PubMed] [Google Scholar]
- Komuro A., Tobe T., Hashimoto K., Nakano Y., Yamaguchi T., Nakajima H., Tomita M. (1996). Molecular cloning and expression of human liver biliverdin-IX beta reductase. Biol. Pharm. Bull. 19, 796–804. [DOI] [PubMed] [Google Scholar]
- Lecona E., Fernandez-Capetillo O. (2014). Replication stress and cancer: It takes two to tango. Exp. Cell Res. 329, 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-Y, Kim M-A, Kim H-J, Bae Y-S, Park J-I, Kwak J-Y, Chung J. H., Yun J. (2007). Alkylating agent methyl methanesulfonate (MMS) induces a wave of global protein hyperacetylation: Implications in cancer cell death. Biochem. Biophys. Res. Commun. 360, 483–489. [DOI] [PubMed] [Google Scholar]
- Liu C-A, Wang M-J, Chi C-W, Wu C-W, Chen J-Y. (2004). Rho/Rhotekin-mediated NF-kappaB activation confers resistance to apoptosis. Oncogene 23, 8731–8742. [DOI] [PubMed] [Google Scholar]
- Lynch A. M., Sasaki J. C., Elespuru R., Jacobson Kram D., Thybaud V., De Boeck M., Aardema M. J., Aubrecht J., Benz R. D., Dertinger S. D., et al. (2010). New and emerging technologies for genetic toxicity testing. Environ. Mol. Mutagen. 52, 205–223. [DOI] [PubMed] [Google Scholar]
- Magré J., Delépine M., Khallouf E., Gedde-Dahl T., Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A., et al. (2001). Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28, 365–370. [DOI] [PubMed] [Google Scholar]
- Maralhas A., Monteiro A., Martins C., Kranendonk M., Laires A., Rueff J., Rodrigues A. S. (2006). Genotoxicity and endoreduplication inducing activity of the food flavouring eugenol. Mutagenesis 21, 199–204. [DOI] [PubMed] [Google Scholar]
- McCool K. W., Miyamoto S. (2012). DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out. Immunol. Rev. 246, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D. W. (2009). Tumour suppression by p53: A role for the DNA damage response? Nat. Rev. Cancer 9, 714–723. [DOI] [PubMed] [Google Scholar]
- Nebert D., Dalton T. (2006). The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 6, 947–960. [DOI] [PubMed] [Google Scholar]
- Olaharski A., Albertini S., Kirchner S., Platz S., Uppal H., Lin H., Kolaja K. (2009). Evaluation of the GreenScreen GADD45α-GFP indicator assay with non-proprietary and proprietary compounds. Mutat. Res. 672, 10–16. [DOI] [PubMed] [Google Scholar]
- Pallua J. D., Schaefer G., Seifarth C., Becker M., Meding S., Rauser S., Walch A., Handler M., Netzer M., Popovscaia M., et al. (2013). MALDI-MS tissue imaging identification of biliverdin reductase B overexpression in prostate cancer. J. Proteomics 91, 500–514. [DOI] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J R. A., Hériché J-K, Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A. W., et al. (2008). BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P. D., Huang B. W., Tsuji Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 24, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T., Furuyashiki T., Ishizaki T., Watanabe G., Watanabe N., Fujisawa K., Morii N., Madaule P., Narumiya S. (1996). Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J. Biol. Chem. 271, 13556–13560. [DOI] [PubMed] [Google Scholar]
- Rosemary Siafakas A., Richardson D. R. (2009). Growth arrest and DNA damage-45 alpha (GADD45α). Int. J. Biochem. Cell Biol. 41, 986–989. [DOI] [PubMed] [Google Scholar]
- Sabatel H., Pirlot C., Piette J., Habraken Y. (2011). Importance of PIKKs in NF-κB activation by genotoxic stress. Biochem. Pharmacol. 82, 1371–1383. [DOI] [PubMed] [Google Scholar]
- Schaap M. M., Wackers PF. K., Zwart E. P., Huijskens I., Jonker M. J., Hendriks G., Breit T. M., van Steeg H., van de Water B., Luijten M. (2014). A novel toxicogenomics-based approach to categorize (non-)genotoxic carcinogens. Arch. Toxicol. [DOI] [PubMed] [Google Scholar]
- Scully R., Xie A. (2013). Double strand break repair functions of histone H2AX. Mutat. Res. 750, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y., Ziv Y. (2013). The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell. Biol. 14, 197–210. [PubMed] [Google Scholar]
- Smith J., Tho L. M., Xu N., Gillespie D. A. (2010). The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108, 73–112. [DOI] [PubMed] [Google Scholar]
- Smith L. J., Browne S., Mulholland A. J., Mantle T. J. (2008). Computational and experimental studies on the catalytic mechanism of biliverdin-IXbeta reductase. Biochem. J. 411, 475–484. [DOI] [PubMed] [Google Scholar]
- Soriano F. X., Léveillé F., Papadia S., Higgins L. G., Varley J., Baxter P., Hayes J. D., Hardingham G. E. (2008). Induction of sulfiredoxin expression and reduction of peroxiredoxin hyperoxidation by the neuroprotective Nrf2 activator 3H-1,2-dithiole-3-thione. J. Neurochem. 107, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W-P, Agarwal A. K., Garg A., Anderson RG. W., Goodman J. M. (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U.S.A. 104, 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., Ron D. (2011). Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumkeo D., Watanabe S., Narumiya S. (2013). Physiological roles of Rho and Rho effectors in mammals. Eur. J. Cell Biol. 92, 303–315. [DOI] [PubMed] [Google Scholar]
- van der Lelie D., Regniers L., Borremans B., Provoost A., Verschaeve L. (1997). The VITOTOX test, an SOS bioluminescence Salmonella typhimurium test to measure genotoxicity kinetics. Mutat. Res. 389, 279–290. [DOI] [PubMed] [Google Scholar]
- Vihervaara A., Sistonen L. (2014). HSF1 at a glance. 127(Pt 2), 261–266. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N., Slocum S. L., Skoko J. J., Shin S., Kensler T. W. (2010). When NRF2 talks, who's listening? Antioxid. Redox. Signal. 13, 1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Kaufman R. J. (2014). The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 14, 581–597. [DOI] [PubMed] [Google Scholar]
- Wang W.-A., Groenendyk J., Michalak M. (2014). Endoplasmic reticulum stress associated responses in cancer. Biochim. Biophys. Acta 1843, 2143–2149. [DOI] [PubMed] [Google Scholar]
- Westerink W M. A., Stevenson J. C. R., Horbach G. J., Schoonen W. G. E. J. (2010). The development of RAD51C, Cystatin A, p53 and Nrf2 luciferase-reporter assays in metabolically competent HepG2 cells for the assessment of mechanism-based genotoxicity and of oxidative stress in the early research phase of drug development. Mutat. Res. 696, 21–40. [DOI] [PubMed] [Google Scholar]
- Wink S., Hiemstra S., Huppelschoten S., Danen E., Niemeijer M., Hendriks G., Vrieling H., Herpers B., van de Water B. (2014). Quantitative high content imaging of cellular adaptive stress response pathways in toxicity for chemical safety assessment. Chem Res Toxicol. 27, 338–355. [DOI] [PubMed] [Google Scholar]
- Witte I., Plappert U., de Wall H., Hartmann A. (2007). Genetic toxicity assessment: employing the best science for human safety evaluation part III: the comet assay as an alternative to in vitro clastogenicity tests for early drug candidate selection. Toxicological Sciences. 97, 21–26. [DOI] [PubMed] [Google Scholar]
- Westerink W. M. A., Schirris T. J. J., Horbach G. J., Schoonen W. G. E. J. (2011). Development and validation of a high-content screening in vitro micronucleus assay in CHO-k1 and HepG2 cells. Mutat. Res. 724, 7–21. [DOI] [PubMed] [Google Scholar]
- Whitesell L. L., Lindquist S. L. S. (2005). HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772. [DOI] [PubMed] [Google Scholar]
- Winkler G. S. (2010). The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 222, 66–72. [DOI] [PubMed] [Google Scholar]
- Wurzenberger C., Gerlich D. W. (2011). Phosphatases: Providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 12, 469–482. [DOI] [PubMed] [Google Scholar]
- Zhan Q. (2005). Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat. Res. 569, 133–143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.