Abstract
The activation of naïve B lymphocyte involves rapid and major changes in chromatin organization and gene expression; however, the complete repertoire of nuclear factors affecting these genomic changes is not known. We report that HMGN proteins, which bind to nucleosomes and affect chromatin structure and function, co-localize with, and maintain the intensity of DNase I hypersensitive sites genome wide, in resting but not in activated B cells. Transcription analyses of resting and activated B cells from wild-type and Hmgn−/− mice, show that loss of HMGNs dampens the magnitude of the transcriptional response and alters the pattern of gene expression during the course of B-cell activation; defense response genes are most affected at the onset of activation. Our study provides insights into the biological function of the ubiquitous HMGN chromatin binding proteins and into epigenetic processes that affect the fidelity of the transcriptional response during the activation of B cell lymphocytes.
INTRODUCTION
The dynamic structure of the chromatin fiber and the organization of regulatory sites in chromatin play a major role in the transcriptional response of cells to various internal or external stimuli. Among the numerous factors known to affect chromatin compaction and dynamics are architectural chromatin-binding proteins such as the H1 linker histone variants (1–4), and all the members of the three high mobility group (HMG) protein families (5,6). H1s and HMGs are among the most abundant chromatin binding proteins; they are ubiquitously found in the nuclei of vertebrate cells, bind dynamically to chromatin without obvious specificity for the underlying DNA sequence and have been shown to affect chromatin compaction and genomic functions, including transcription (2–6). Yet, despite numerous studies on these ubiquitous architectural proteins, their biological function and effects on gene expression are not fully understood. The HMG proteins consist of three families named HMGA, HMGB and HMGN, each characterized by a distinct structure and a unique chromatin-binding motif (5–8). Here we focus on the role of the high mobility groups N (HMGN) protein family in regulating chromatin structure and gene expression during the activation of mouse B cells.
The HMGN protein family consists of five variants (9,10), all of which contain a conserved, positively charged region that facilitates their specific binding to the 147 base pair nucleosome core particle, the building block of the chromatin fiber (11,12). HMGN variants bind to chromatin dynamically with a short residence time (13,14), compete with each other for nucleosome binding sites, decrease the interaction of H1 variants with chromatin (14) and reduce chromatin compaction (15). Genomic profiling revealed that the binding sites of the major variants, HMGN1 and HMGN2, overlap with DNase I hypersensitive sites, the hallmark of regulatory sites in chromatin, such as gene promoters and enhancers (16). Given the known effects of genome organization on gene expression, it could be expected that HMGNs affect the cellular transcriptome thereby impacting the cellular phenotype. Indeed, genetically altered Hmgn−/− mice show distinct phenotypes and transcriptional analyses of tissues taken from these mice revealed HMGN variant-specific effects on gene expression (16,17). A major unresolved question is whether HMGNs affect the ability of a cell to adequately respond to biological stimuli which involve rapid and major changes in chromatin organization and gene expression. Here we address this question in a biological relevant setting, by examining the role of HMGNs in the lipopolysaccharide and interleukin-4 (LPS + IL4) induced activation of naïve splenic B cells.
Naïve B cell lymphocytes reside in the mouse spleen in the quiescent G0 state, when transcription and protein synthesis are at basal levels (18). Upon antigen stimulation, the resting lymphocytes drastically increase their transcriptional output and undergo additional changes that play a major role in the immune response (19). Lymphocyte activation has been extensively studied ex vivo by stimulating quiescent CD43− B cells isolated from spleen with LPS + IL4 (20,21). LPS + IL4 stimulation rapidly leads to major chromatin de-condensation, significant changes in epigenetic marks, promoter melting and RNA polymerase recruitment, resulting in more than 10-fold increase in RNA output (22,23). Transcriptional responses have been detected as early as 30 min after activation; however it can last as long as 72 h for the full response to develop (24). Given the biological significance of the immune response, it is important to understand the full spectrum of factors that affect the transcriptional response elicited during B cell activation.
Here we use lymphocytes prepared from the spleens of WT and Hmgn−/− mice, to study the effects of HMGN on the organization of chromatin regulatory site and to examine the role of HMGN proteins in the transcriptional response during the course of activation of naïve B cells. We found that stimulation of B cells leads to genome wide changes in the binding of HMGN to chromatin, that HMGN proteins co-localize with and maintain the intensity of DNase I hypersensitive sites genome wide in resting but not in activated B cells, and that loss of HMGNs dampens the magnitude of the transcriptional response and alters the pattern of gene expression during the course of B-cell activation. Our results provide new insights on the biological function of a ubiquitous family of nucleosome binding proteins and identify additional epigenetic factors that affect gene expression during B cell activation.
MATERIALS AND METHODS
Mouse strains
Hmgn1−/− mice, in which the exons 2–4 were deleted, Hmgn2−/− mice which lack the entire Hmgn2 gene, and Hmgn1−/−n2−/−, double knockout mice (DKO) generated by crossing Hmgn1−/− mice and Hmgn2−/− mice were previously described (16,25).
Preparation of resting and activated B cells
B lymphocytes were prepared as described (26). Briefly, B lymphocytes were isolated from spleens of 8–12 week old male mice by immunomagnetic depletion using anti-CD43 MicroBeads (Miltenyi Biotech). Purified B cells were cultured at 37°C for up to 72 h in RPMI 1640 media supplemented with 10% fetal calf serum, 1 × antibiotic-antimycotic, 1% glutamine, 53 μM 2-mercaptoethanol and 10 mM Hepes. Activation of B cells was achieved by incubation with 50 μg/ml lipopolysaccharide LPS (Escherichia coli 0111:B4; Sigma-Aldrich) and 2.5 ng/ml mouse IL-4 (Sigma-Aldrich) for IgG1 switching.
Chromatin immunoprecipitation, illumina library construction and sequencing
Resting or LPS + IL4 stimulated B cells were harvested and cross-linked with 1% formaldehyde (W/V) for 10 min at room temperature on a rocking platform, followed by quenching with 125 mM glycine. Cells were washed twice with ice-cold phosphate-buffered saline and incubated in 1 ml RIPA buffer (10 mM Tris–HCl, PH 7.6, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.1% sodium dodecyl sulphate (SDS), 0.1% sodium deoxycholate, 1% triton X100) containing protease inhibitor (Complete, Mini, EDTA-free Cocktail Tablets from Roche Applied Science) for 45 min. Lysates were sonicated to result in DNA fragments of 200–500 bp in length. Sonicated chromatin was pre-cleared with 20 μl Dynabeads Protein G (Invitrogen) for 30 min at 4°C. A total of 40 μl Dynabeads Protein G and 5–10 μg of specific antibodies were then added to the rest of chromatin samples and incubated for overnight at 4°C with rotation. The antibodies used in chromatin immunoprecipitation include: rabbit anti mouse HMGN1 polyclonal antibody, rabbit anti HMGN2 polyclonal antibody (25,27). The DNA immunoprecipitated with HMGN1 antibody from Hmgn1−/− cells was used as control.
The beads were washed sequentially with the following buffers at 4°C: twice with RIPA buffer, twice with RIPA buffer plus 0.3 M NaCl, twice with LiCl buffer (0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), once with Tris–EDTA buffer (pH 8.0) plus 0.2% Triton X-100 and once with Tris-EDTA buffer (pH8.0) alone. Cross-links were reversed at 65°C for overnight in the presence of 3 μl of 10% SDS and 5 μl of proteinase K (20 mg/ml). The DNA samples were eluted in 21 μl of elution buffer using MiniElute kit (Qiagen). ChIP-seq library was prepared following the manufacturer's instructions (Illumina). Briefly, immunoprecipitated DNA were blunt ended, ligated to adapters, amplified with polymerase chain reaction and size selected. The ChIP templates were sequenced at 36 bp read length with Illumina GAIIx by CCR-sequencing facility at NIH. Sequence reads were aligned to the Build 37 assembly of the National Center for Biotechnology Information mouse genome data (NCBI37/mm9).
Genome-wide DNase I hypersensitivity assay
Isolated B cells were washed twice with phosphate buffered saline (PBS), then pelleted and resuspended in buffer A (15 mM Tris–Cl, pH 8.0, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, pH 8.0, 0.5 mM EGTA, pH 8.0) with freshly added 0.5 mM spermidine and 1 × protease inhibitor (Roche Applied Science). Cell membranes were disrupted by addition of equal volume of buffer A containing 0.01% IGEPAL, followed by incubation on ice for 10 min. Following centrifuge at 1500 rpm for 5 min, 2 × 107 pelleted nuclei were resuspended in 2.5 ml of DNase I digestion buffer (15 mM Tris–Cl, pH 8.0, 90 mM NaCl, 60 mM KCl, 6 mM CaCl2 1 mM EDTA, pH 8.0, 0.5 mM ethylene glycol tetraacetic acid (EGTA), pH 8.0, 0.5 mM spermidine). DNase I digestion was carried out with 80 U/ml and 40 U/ml DNase I, for resting nuclei and activated nuclei, respectively, at 37°C for 3 min followed by termination of reaction with addition of equal volume of stop buffer (50 mM Tris–Cl, pH 8.0, 100 mM NaCl, 0.10% SDS, 100 mM EDTA, pH 8.0, 10 µg/ml RNAse A) and incubated for 4 h at 55°C. After addition of proteinase K (50 µg/ml), samples were further incubated for overnight at 55°C. On the following day, DNA was recovered by phenol/chloroform extraction. DNA fragments with size between100 and 500 bp were isolated by sucrose gradient centrifugation. DNaseI library construction was performed according to Illumina's protocol and sequenced with HiSeq 2000.
MNase digestion of chromatin followed by paired-end sequencing
MNase digestion of nuclei isolated from resting and LPS + IL4 activated B cells were performed with EZ Nucleosomal DNA Prep Kit (ZYMO RESEARCH) according to manufacturer's instruction. Briefly, 2 × 107 cells were washed twice in 1 ml PBS, cell pellet were resuspended in 1 ml Nuclei Prep Buffer and incubated on ice for 5 min. After washing twice with MN Digestion Buffer, 1.5 × 106 nuclei were treated with 0.001–0.2 units of micrococcal nuclease in 100 µl MN Digestion Buffer at room temperature for 5 min. Reaction was terminated with addition of 5× Stop Buffer. Mono-nucleosomal DNA was purified and processed for library construction (Illumina). The sequencing was performed on a NextSeq instrument (Illumina), with single-end read lengths of 76 nucleotides.
Total RNA extraction and mRNA sequencing
Total RNA was extracted from resting or LPS + IL4 stimulated B cells with TRIzol reagent (Invitrogen). Purified RNA was cleaned up with RNeasy kit (Qiagen). Potential genomic DNA contamination was removed with on-column DNaseI treatment. mRNA-seq assay was performed using HiSeq2000 (Illumina) at the Laboratory of Molecular Technology (LMT), NCI, Frederick, MD, USA.
Bioinformatics, DNase I-Seq, ChIP-seq, MNase-Seq and mRNA-seq analyses
Unfiltered sequencing reads were aligned to the mouse reference genome (NCBI build 37, mm9) using BoWTie (28). Up to two mismatch was allowed for each aligned read. Only uniquely aligned reads were collected for further analysis. Binding regions were identified using SICER (29)with the following parameters: effective genome size, 0.787 (78.7% of the mouse genome is mappable); E-value, 0.1; window size, 100 bp for HMGN ChIP-Seq and 50 bp for DNase I-Seq and histone marker ChIP-seq; gap size, 100 bp for HMGN ChIP-Seq and 50 bp for DNase I-Seq and histone marker ChIP-seq. Calculation of coverage and identification of overlapping binding regions were performed with the ‘chipseq’ and ‘GenomicRanges’ packages in BioConductor (30). Specifically, in analyzing MNase-seq data, since the reads are from the ends of the mononucleosome particle, to best visualize the nucleosome positions, all the reads were shifted to the middle of the corresponding nucleosome core. The average sizes of mononucleosome fragments purified from a 2% agarose gel were determined by Bioanalyzer (Agilent). For normalization, the calculation of coverage at any regions and the comparisons between different datasets were preceded by library size normalization. Control subtraction was carried out in the following way: coverage (exp)/N1—coverage (control)/N2, in which ‘exp’ is the dataset (in .bam format) to be examined, N1 is the library size of the experimental data (‘exp’), and N2 is the library size of the control. Samples obtained from DKO mice were used as a control. The function coverage that calculates genome coverage from .bam files is from the ‘chipseq’ package in BioConductor (30). RNAseq analysis was conducted with R package EdgeR, with expression level cutoff set as 0.5 RPKM (31).
RESULTS
Activation of splenic B lymphocytes alters the binding of HMGN1 and HMGN2 to chromatin
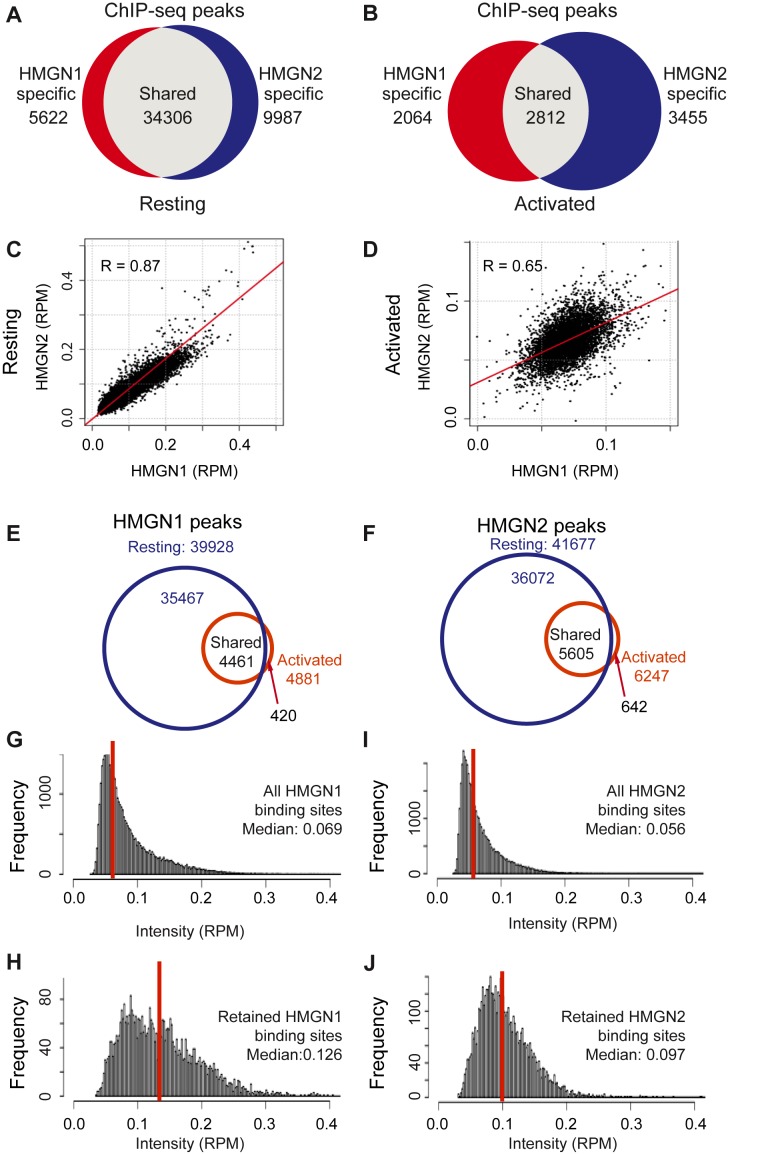
To test whether potentially HMGNs could affect the course of B cell activation we mapped the genome wide organization of HMGN1 and HMGN2, in both resting and stimulated cells, i.e. before and after 72 h exposure to LPS + IL4. In resting B cells we identified a total of 49 915 HMGN binding sites; of these, 34 306 (69%) were common to both HMGN1 and HMGN2 showing a Pearson correlation coefficient of 0.87, 5622 sites were specific to HMGN1 and 9987 sites were bound only by HMGN2 (Figure 1A and C). Transcriptional activation by LPS + IL4 led to drastic changes in the HMGN chromatin occupancy, without noticeable changes in the level of these proteins (Supplementary Figure S1A). The total number of binding sites was reduced by over 80%, to 8331. The sites shared by HMGN1 and HMGN2 were reduced to 2812, and the two HMGN variants bind to these sites with different intensities, as indicated by the large decrease in the Pearson correlation coefficient. The HMGN1and the HMGN2 specific sites decreased by over 60%, to 2064 and 3455, respectively (Figure 1B and D).
Figure 1.
LPS + IL4 induced activation of B cells alters the binding of HMGN proteins to chromatin. (A and B) Venn diagrams showing the overlap between HMGN1 and HMGN2 binding sites in resting (A) and activated (B) B cells, as determined by ChIP-Seq. Binding sites were identified by SICER, with an E-value cutoff of 0.1. (C and D) Scatter plots showing similar binding intensities of HMGN1 and HMGN2 in resting (C), but not in activated (D) B cells. R = Pearson correlation coefficient. (E and F) Venn diagrams showing the overlap in HMGN1 and HMGN2 occupancy between resting and activated cells. (G and H) Histogram showing signal intensities (RPM) in resting B cells at all HMGN1 binding sites (G) or only at HMGN1 binding sites retained after activation (H). (I and J) Histogram showing signal intensity (RPM) in resting B cells at all HMGN2 binding sites (I) or only at HMGN2 binding sites retained after activation (J).
Over 90% of the HMGN binding sites seen in activated B cells overlapped with sites that were seen in resting cells, an indication that the activation did not generate a significant number of new HMGN binding sites (Figure 1E and F). In activated cells, the median intensity of the new sites is of 0.023 and 0.025 for HMGN1 and HMGN2, respectively. In the overlapping, retained sites, the median density for both HMGN1 and HMGN2 is 0.031. Thus, the new peaks in the activated cells are weaker than the retained sites. In resting cells, the median intensity of HMGN1 and HMGN2 binding sites was 0.069 and 0.056 respectively, measured in reads per million (RPM) (Figure 1G and I). The HMGN1 and HMGN2 sites that were retained in activated cells were the sites that in resting cells showed respectively, a median intensity of 0.126 RPM and 0.097 RPM (Figure 1H and J) indicating that the stronger HMGN binding sites were retained in the activated cells.
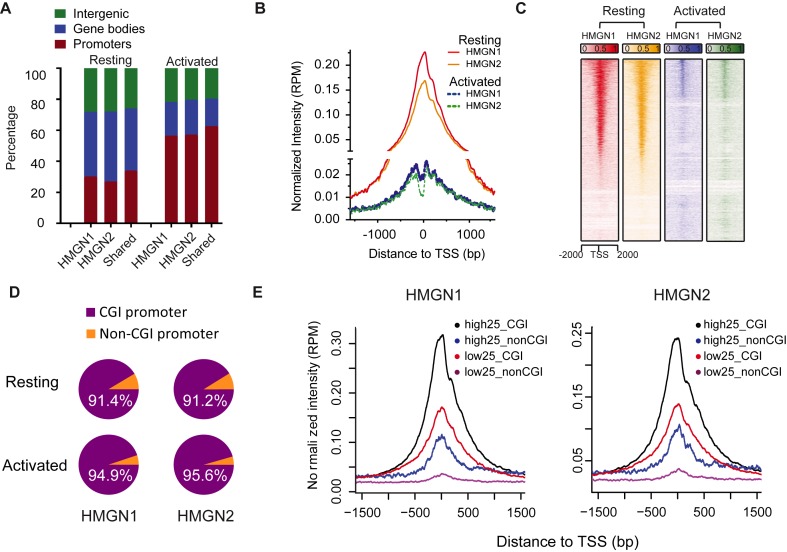
Further analysis of HMGN1 and HMGN2 binding in resting cells shows that 30% are located at promoters, 40% at gene bodies and 30% in intergenic regions (Figure 2A). In activated cells, the proportion of HMGNs associated with promoters was double of that of resting cells (Figure 2A). Occupancy profiles (Figure 2B) and heat maps (Figure 2C) indicated that in both resting and activated cells, HMGN proteins are markedly enriched at transcriptional start sites (TSS), but in resting B cells the average occupancy of HMGN at TSS is 10× higher than that in activated B cells. Thus, although the HMGN binding sites detected in activated cells were preferentially located at promoters, their chromatin binding was significantly altered as compared to their binding in resting cells. In sum, the results indicate that LPS + IL4 activation causes significant changes in the chromatin interactions of HMGN proteins.
Figure 2.
HMGN proteins are enriched at CGI promoters. (A) Global distribution of HMGN1 and HMGN2 throughout the genome. (B and C) Enrichment profile and heat map representation of relative occupancy of HMGN1 and HMGN2 at TSS regions. (D) Most of HMGN1 and HMGN2 binding promoters are CGI containing promoters. (E) HMGN1 and HMGN2 are most enriched at CGI containing promoters of highly expressed genes. Shown are average HMGN signals at promoters that either do, or do not, contain CGI sorted by high and low expression levels. High25 and low25 indicate top 25% genes with highest and lowest expression level, in resting B cells, as measured by RNA-seq. Genes with expression level below 0.5 RPKM were excluded from the analysis.
Of the 24 303 non-redundant RefSeq promoters annotated in the NCBI Mouse mm9 refseq data base (http://hgdownload.soe.ucsc.edu/goldenPath/mm9/database/), 15 112 (62%) contain CpG islands (CGI). We find that both HMGN1 and HMGN2 preferentially bind to CGI promoters, since over 90% of the promoters bound by either of these HMGN variants are classified as CGI promoters, in both resting and activated B cells (Figure 2D). CGI promoters are often found in housekeeping gene and may affect transcription levels (32). Therefore, we examined the relationship between CGI, HMGN occupancy and transcription levels in resting B cells, as determined by RNAseq of three biological replicates, which showed very low variability among the samples (Supplementary Figure S1). We identified the 25% most actively transcribed genes (high25) and the 25% least actively transcribed genes (low25) and examined the HMGN occupancies and the CGI content at the promoters of these genes. We find that the average occupancy of both HMGN1 and HMGN2 proteins at the high25 promoters containing CGI is 3-fold higher than at high25 promoters that do not contain CGI (compare black and blue lines in Figure 2E). Furthermore, the occupancy of HMGNs at the low25 CGI promoters is higher than at the high25 promoters that do not contain CGI (Figure 2E). Together, these results indicate that the major factor affecting HMGN binding to promoters is the presence of a CGI, rather than gene expression levels.
HMGN proteins modulate the landscape of DNase I hypersensitive sites in the chromatin of B cells
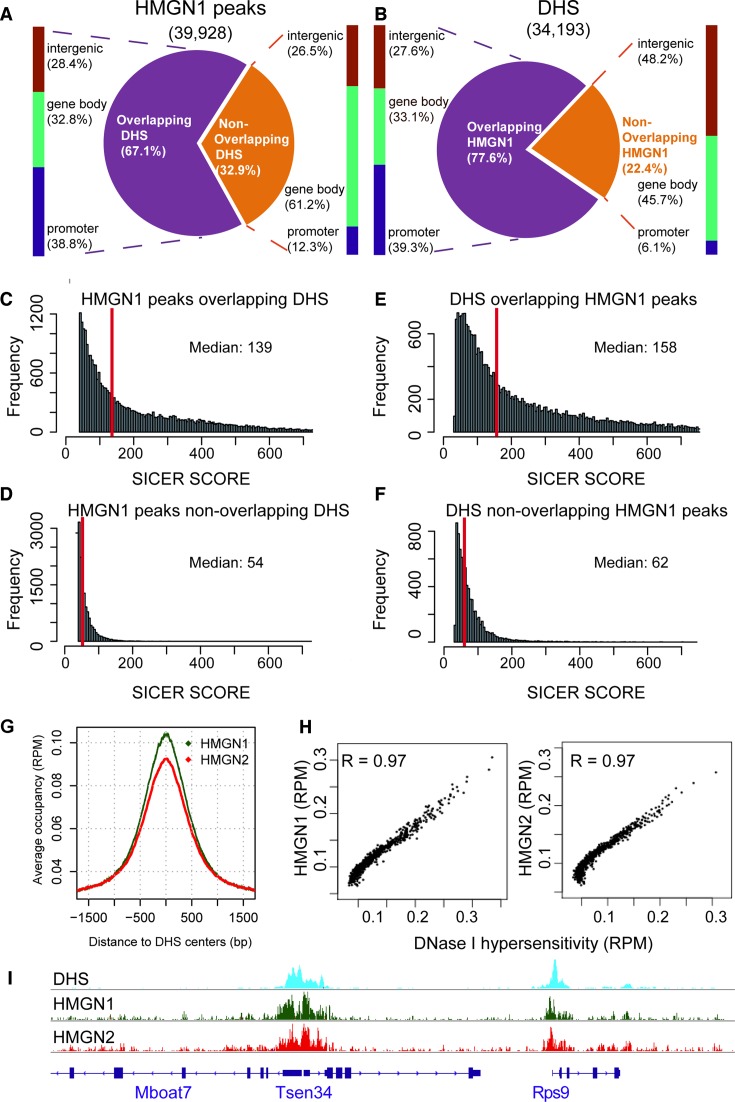
Enhancers and promoters are known to play a key role in gene expression; the location of these regulatory elements in chromatin can be identified and mapped by their hypersensitivity to digestion by DNase I (33). In view of the high occupancy of HMGN1 and HMGN2 at CGI promoters we mapped the genome wide association of these HMGN variants with chromatin regulatory sites (Figure 3, Supplementary Figure S2). In resting cells, we identified 34 193 DHSs; 78% of these co-localized with HMGN1 (Figure 3B). In agreement, of the 39 928 HMGN1 binding sites that we identified in resting cells 67% co-localize with DHSs (Figure 3A). Similar relationships were seen for HMGN2 (Supplemental Figure S2). Thus, most of DHSs co-localize with HMGNs.
Figure 3.
Co-localization of DHSs and HMGN1 binding sites in resting B cells. (A) Distributions of HMGN1 peaks that either do or do not overlap DHSs. (B) Distribution of DHSs that either do, or do not overlap HMGN1 peaks. (C–F) HMGN1 and DHSs overlapping regions have stronger signal intensities than the non-overlapping regions. (G) Enrichment of HMGN1 and HMGN2 at DHSs. (H) Scatter plot showing co-localization of DHSs with HMGN1 (left) and HMGN2 (right). Normalized coverage depths for HMGNs and DHSs were sorted by DHSs coverage depth, grouped into 100 data point bins and averaged. The Pearson correlation coefficient (R) was calculated for the binned data. (I) Genome browser snapshot visualizes the co-localization of HMGN1 and HMGN2 binding sites with DHSs.
Of the HMGN1 binding sites that overlapped DHSs, 39% mapped to promoter regions. In contrast, only 12% of the HMGN1 peaks that did not overlap with DHSs located to promoters (Figure 3A). Likewise, 39% of the DHSs that did, but only 6% that did not, overlap with HMGN1 localized to promoters (Figure 3B). The median value of the Sicer scores indicated that the binding of HMGN1 at sites overlapping DHSs was significantly stronger than at sites not overlapping DHSs (Figure 3C and D). Likewise, the signal strength of DHSs at sites overlapping HMGNs was significantly stronger than at sites that did not overlap HMGNs (Figure 3E and F). Similar results were noted with HMGN2 (Supplemental Figure S2). The strongest HMGN occupancies localized to the center of the DHSs, and the Pearson correlation coefficient between the DHSs signal intensity and HMGN occupancy was 0.97 for either HMGN1 or HMGN2 (Figure 3G–I). Taken together, the results indicate that across the genome of resting B cells, DHSs co-localize with HMGN variants, and that the most prominent DHSs correspond to the strongest HMGN binding sites.
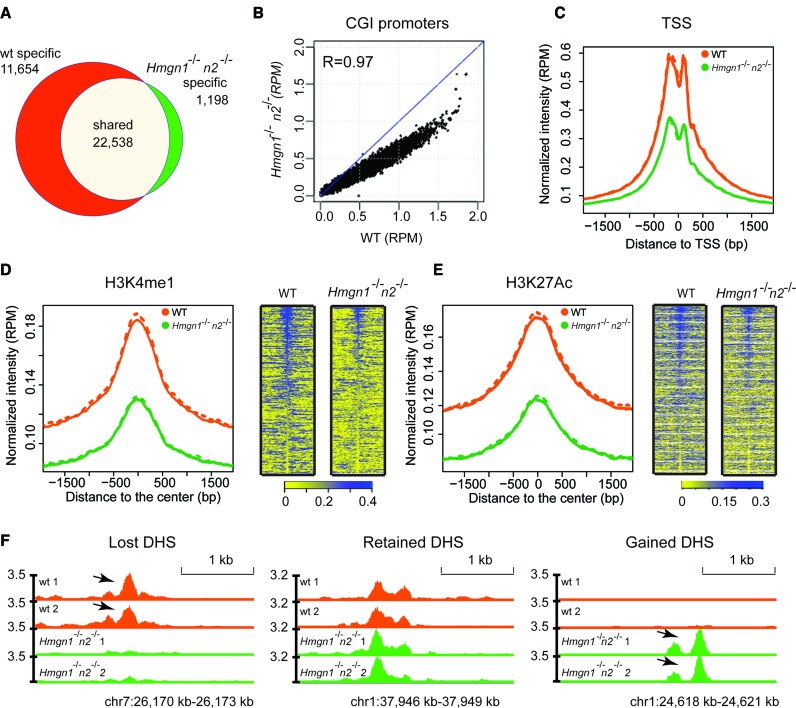
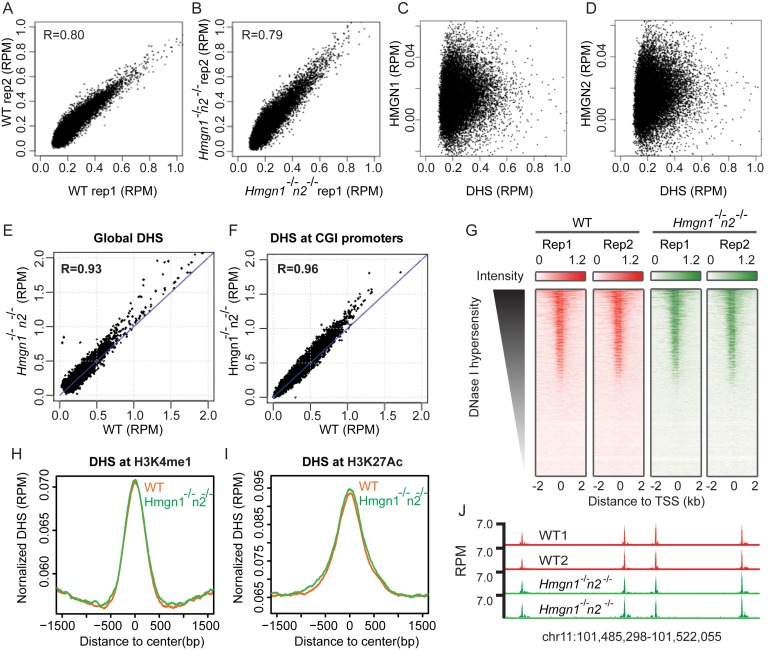
A major question is whether HMGNs just co-localize, or whether they also affect the genome wide organization of the DHSs. To examine whether HMGN variants affects the landscape of these regulatory sites we mapped the DHSs site in the chromatin of resting B cells obtained from wild-type (WT) or Hmgn1−/−n2−/− DKO B cells. We used DKO cells because HMGNs bind redundantly to chromatin (16) and because we found that the chromatin binding of HMGN2 in B cells lacking HMGN1 was higher than in WT cells (Supplementary Figure S3).
We identified 34 192 DHSs in WT cells but only 23 736 DHS sites in DKO cells, a 30% loss of DHSs (Figure 4A). Of the DHSs present in DKO cells, 22 538 overlapped with the DHSs seen in WT cells and 1198 sites were newly formed, i.e. not mapped in WT cells (Figure 4A and F). Thus, the number of lost DHSs is approximately 10× larger than the number of gained DHSs sites, indicating that loss of HMGNs leads to a global reduction in DNase I hypersensitivity. Almost 95% of the DHSs observed in DKO cells were also detected in WT cells. The scatter plot of the DHSs at the CGI promoters shows a strong correlation between the intensities of the DHSs in WT and DKO cells, with a correlation coefficient of 0.97; however, the intensities of most DHSs, as measured by the reads per million (RPM) at any specific site in the DKO cells is reduced compared to the same sites in WT cells (Figure 4B). Likewise, although in both WT and DKO cells the DHSs seem to center at the TSS, the intensities of these sites in DKO is significantly reduced, showing only 50% of the intensities seen in WT cells (Figure 4C). A similar level of reduction in DNase I hypersensitivity is observed at chromatin regions enriched in H3K4me1 and H3K27ac (34), histone modifications that mark enhancers (35,36). However, both the enrichment profiles and the heat maps indicate that the loss of DNase I hypersensitivity occurred not only at these specific marks but also in the regions flanking these marks (Figure 4D and E). Taken together, these results indicate that loss of HMGN proteins reduces the DNase I hypersensitivity throughout the genome of resting B cells.
Figure 4.
Loss of HMGN1 and HMGN2 proteins reduces DHSs intensity in the chromatin of resting B cells. (A) Venn diagrams showing the DHSs overlap between WT and Hmgn1−/−n2−/- resting B cells. (B) Scatter plot showing the difference in the intensity of DHSs between WT and Hmgn1−/−n2−/- cells. DHS signals are normalized by library size and averaged over two biological replicates over each site, in reads per million of reads (RPM). (C–E) Loss of HMGNs reduces the DHSs intensity at TSS and at the chromatin regions with the indicated histone marks. Solid line and dotted line represent two biological replicates. (F) Genomic browser snapshot visualizing lost, retained and gained DHSs in Hmgn1−/−n2−/- cells. Two biological replicates are shown for the DHSs maps. Numbers next to Y axis indicate the scales in RPM.
Since in activated B cells the chromatin binding of HMGN variants is reduced (Figure 1A and B) we examined whether HMGNs affect the DHSs landscape in these cells, using two biological replicates for each genotype. The high correlation between the biological replicates verified the reproducibility of the data (Figure 5A and B). Alignment of these DHSs sites with the chromatin binding sites of either HMGN1 (Figure 5C) or HMGN2 (Figure 5D) indicated that there is no correlation between HMGNs and DHSs, raising the possibility that in activated B cells HMGNs do not significantly affect the DHSs landscape. Indeed, the DHSs scatter plot revealed very little difference between WT and DKO cells for all DHSs (Figure 5E) and for the DHSs located at CGI promoters (Figure 5F). Likewise, loss of HMGNs did not affect the DHSs at transcription start sites (Figure 5G), or at regions marked by either H3K4me1 or H3K27ac where the enrichment profiles of the DHSs obtained from WT and mutant cells almost overlap (Figure 5H–J). Thus, loss of HMGNs affects the landscape of DHSs in the chromatin of resting, but not of activated B cells.
Figure 5.
Loss of HMGN1 and HMGN2 does not significantly change DHSs landscape in activated B cells. (A and B) Scatter plot showing similar intensity of DHSs between two biological replicates. (C and D) Scatter plot showing poor correlation between HMGN binding sites and DHSs in activated B cells. (E and F) Scatter plot showing similar intensity of DHSs between WT and Hmgn1−/−n2−/− cells at global DHS sites and CGI promoters. DHSs signals are averaged over two biological replicates. (G) Heat maps of DHSs intensities at TSS region in two biological replicates of WT and Hmgn1−/−n2−/− cells. (H and I) Normalized average intensity of DHSs in WT and Hmgn1−/−n2−/− cells at H3K4me1 and H3K27ac marked regions. (J) Genomic browser snapshot visualizing the DHSs in WT and Hmgn1−/−n2−/− cells.
Loss of HMGN proteins increases nucleosome occupancy at promoters
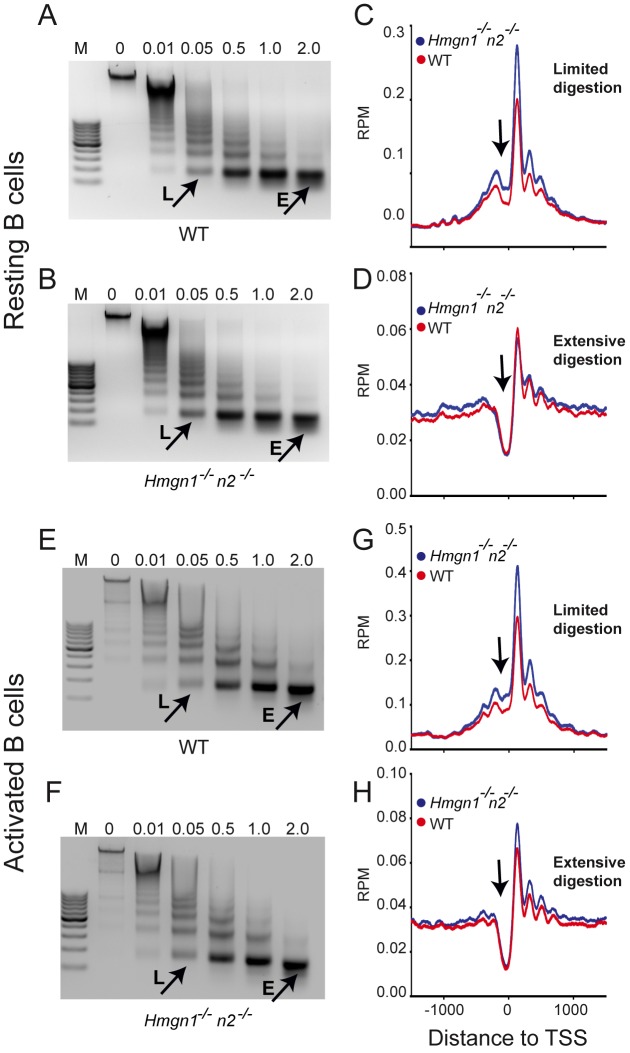
Altered DNase I hypersensitivity at regulatory sites reflects changes in nucleosome occupancy. Since HMGNs are known to bind specifically to nucleosomes (6) we mapped the nucleosome positions in resting and activated B cells obtained from WT and DKO mice, by deep sequencing the DNA purified from mononucleosomes isolated from micrococcal nuclease (MNase) digests. In either resting or activated cells, the MNase digestion kinetics and nucleosome pattern of WT and DKO cells are indistinguishable, suggesting that loss of HMGN proteins did not significantly affect the global chromatin compaction of the cells (Figure 6A, B, E and F).
Figure 6.
Loss of HMGNs enhances the occupancy of unstable nucleosomes at TSS in both resting and activated B cells. (A and B) Loss of HMGNs does not affect the kinetics of MNase digestion of resting cells. (C and D) Loss of HMGNs enhances the occupancy of unstable (C) but not of stable (D) nucleosomes at the TSS of resting cells. (E and F) Loss of HMGNs does not affect the kinetics of MNase digestion of activated cells. (G and H) Loss of HMGNs enhances occupancy of unstable (G) but not of stable (H) nucleosome at the TSS of activated cells. Resting and activated B cells were digested with increasing concentration of MNase (0.01–2 U/ml). Mononucleosomes from limited (L) and extensive (E) digestions were isolated, their DNA sequenced and aligned to the TSS of top 3000 expressed genes. Arrow points to the ‘nucleosome free’ region near the TSS.
During MNase digestion, the linker DNA flanking of the most accessible nucleosomes, which are often located at regulatory sites, is digested first thereby releasing these nucleosomes from the chromatin fiber during the early phases of digestion. The released nucleosomes are further digested and their DNA may not be included in the mononucleosomal fraction of an extensive digest. Therefore, we isolated and sequenced the nucleosomes obtained from both limited (0.05 U/ml) and extensive (2 U/ml) MNase digestion where respectively, either less than 5% or more than 90% of the chromatin was converted to mononucleosomes (Figure 6). Mapping of these nucleosomes with the TSS of the top 3000 expressed genes revealed differences between the nucleosomes isolated from limited and extensive digests. The nucleosome occupancy profile obtained from extensive digests show a nucleosome free region at the TSS, flanked by nucleosomes both down- and upstream. The nucleosome at position +1 is best positioned, as seen by the highest peak at that site (Figure 6D and H). In contrast, the limited digests show the presence of a nucleosome positioned at the TSS, in the ‘nucleosome free’ region seen in extensive digests (Figure 6C and G). The nucleosome positions of WT and DKO cells are similar in both resting and activated cells; however, in the limited digests of both resting and activated cells, and to a lesser degree in the extensive digests of activated cells, the nucleosomes obtained from DKO cells have higher occupancy than those obtained from WT cells, as indicated by the height of the peaks surrounding the TSS. The increased nucleosome occupancy, may be due to increased H1 binding (14), changes in histone marks (37,38) or reduced nucleosome remodeling activity (39).
In contrast to the differences noticed at promoter regions, we did not notice differences between WT and DKO cells in nucleosome occupancy at enhancer regions, as identified by enrichment in both H3K4me1 and H3K27ac peaks (not shown).
Loss of HMGN proteins affects gene expression during B-cell activation
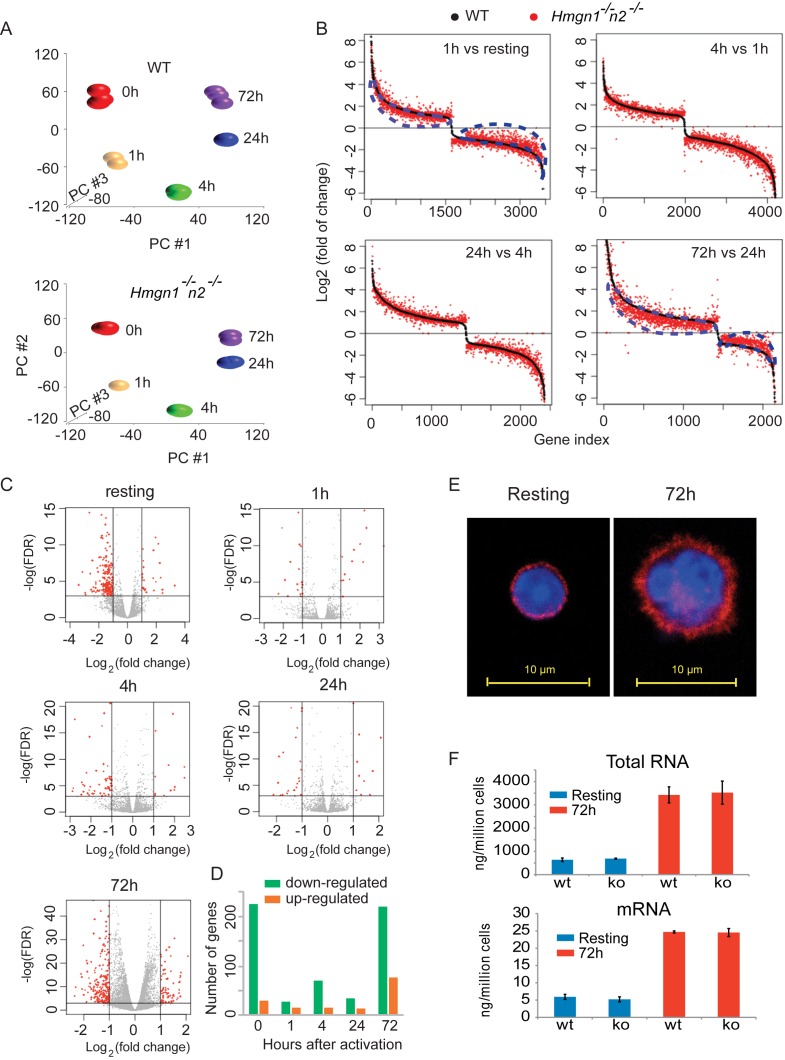
Stimulation of isolated mouse CD43-splenic B cells with LPS + IL4 is known to lead to a significant increase in the size of the cell and the nucleus, to major alterations in chromatin compaction, and to substantial increase in transcription (22). In view of our finding that loss of HMGNs altered the landscape of chromatin regulatory sites, we tested whether loss of HMGN1 and HMGN2 affects the course of gene expression during the LPS + IL4 induced activation of primary B spleenocytes. To this end we used RNA-seq to analyze the transcriptome of WT and DKO B cells prior to, and at several time points after LPS + IL4 stimulation, using three3 biological replicates for each point. The scatter plots of the RNA-seq among the three biological replicates show high overlap with Pearson correlation coefficients of over 0.99 for all time points, in both WT and DKO (Supplemental Figure S4).
Principal component analysis of the transcriptomes at different activation time points shows distinct clustering in both WT and DKO cells, indicating significant changes in the transcription profile during the course of LPS + IL4 activation (Figure 7A). The transcriptional changes involved numerous genes with the great majority of the changes involving the same genes, in WT and DKO cells. We selected the genes whose expression changed 2-fold or more between two consecutive time points in either WT or DKO cells and plotted these changes in Figure 7B, with black dots indicating WT and red dots indicating DKO cells. Sorting of the fold changes of all genes based on their values in WT cells, showed subtle but obvious differences between the two genotypes. The scatter of the red dots around the black line indicates that loss of HMGN1 and HMGN2 altered the magnitude of the transcriptional changes in most genes. Most noticeable differences are seen during the time course from 24 to 72 h after LPS + IL4 stimulation, where the overall magnitude of both upregulation and downregulation in gene expression was lower in DKO cells, since the great majority of the red dots cluster below the black lines in the upregulated genes, but above the black line in the downregulated genes (Figure 7B lower right panel). A similar, but somewhat less pronounced trend is seen in the first hour after activation (Figure 7B, upper left panel). Thus, loss of HMGNs dampens the magnitude of gene expression changes during the last and the first stages of B-cell activation.
Figure 7.
Loss of HMGNs affects the transcriptional response of B cells to LPS + IL4 activation. (A) Principal component analysis of transcription profiles following activation of WT (top) and Hmgn1−/−n2−/− (bottom) B cells. Data from RNA-seq of three biological replicates at different activation time points. (B) Gene expression fold changes between consecutive time points during B-cell activation in WT and Hmgn1−/−n2−/−cells. The difference between red and black visualizes the effects of HMGN loss on the gene expression during B-cell activation. Blue dotted lines highlight the marked difference between WT and Hmgn1−/−n2−/− in the transcriptional changes occurring from resting to 1 h activation (upper left) and from 24 to 72 h after LPS + IL4 activation (lower right). (C) Transcription differences between WT and Hmgn1−/−n2−/− B cells at the indicated times following LPS + IL4 activation. (D) The total number of genes that were either down (green) or upregulated (orange), with a fold change >2, at each time point shown in panel C. Data derived from RNA-seq of three biological replicates for each point. (E) Confocal images showing mouse B cells before (left) and 72 h after (right) LPS + IL4-induced activation. Blue: DAPI; red: Anti-β-Actin. (F). Bar graph showing the amount of total RNA (upper panel) or mRNA (lower panel) in resting (blue) or 72 h after LPS + IL4-activation (red) of wild-type (WT) or Hmgn1−/−n2−/− (KO) cells. Error bars show the standard deviations, n = 3 for all groups.
In a further test, we identified the differentially expressed genes between DKO and WT (fold change ≥ 2 and FDR ≤ 0.05) at several time points following LPS + IL4 activation (Figure 7C). The largest number of significantly affected genes due to loss of HMGNs is seen in resting cells and during the last period of LPS + IL4 activation, with significantly more genes downregulated than upregulated (Figure 7D). The differences between WT and DKO cells in the number of changed genes seen 1, 4 and 24 h after LPS + IL4 addition, i.e. during the course of activation, were significantly lower (Figure 7D). Conceivably, the significant effects of LPS + IL4 stimulation on chromatin structure and gene expression mask HMGN-dependent effects on transcription during the progression of activation to a larger degree than at the onset, or in the latest stages of activation, when the cells adjust to a new steady state of transcription. However globally, loss of HMGNs did not alter the ability of the splenic B cells to respond to LPS + IL4, since WT and DKO cells showed similar increases in cell size and in transcription, as measured by the amount of either total RNA, or mRNA produced 72 h after LPS + IL4 stimulation (Figure 7E and F).
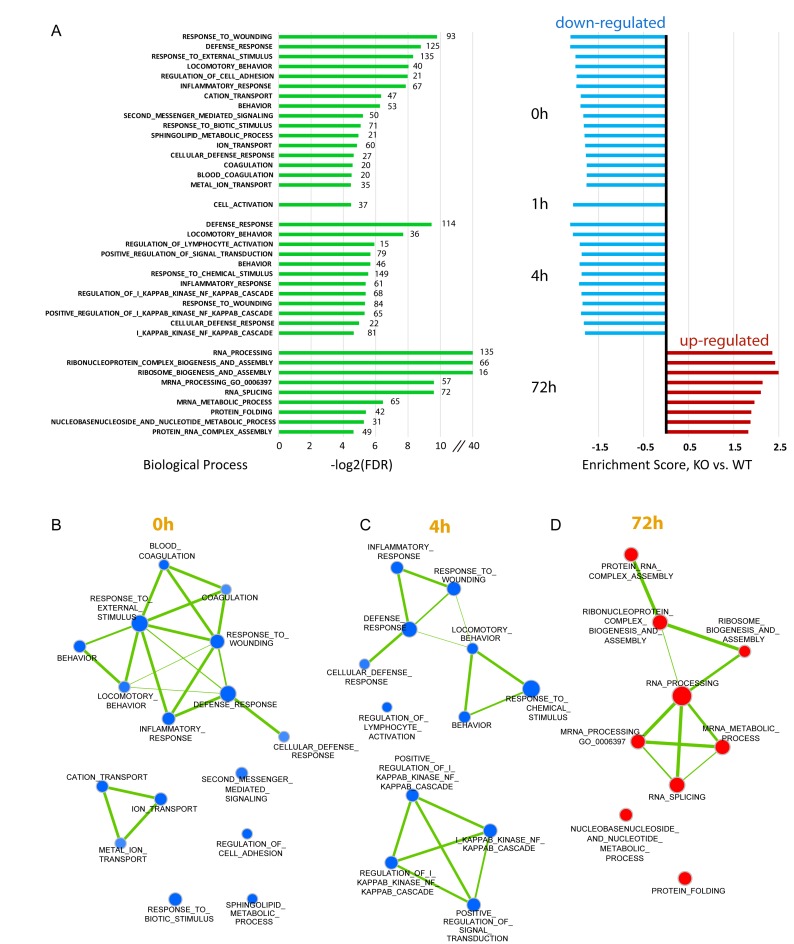
To identify possible biological pathways affected by the loss of HMGNs, we performed gene set enrichment analysis using Gene Set Enrichment Analysis (GSEA) software (40) which analyze transcriptome data by focusing on gene sets, rather than on individual genes and identifies statistically significant differences between gene sets of two biological states. The biological processes altered in DKO compared to WT cells at a high confidence level, at each of the time points tested, are listed in Figure 8A with blue indicating downregulated, and red indicating upregulated pathway. In resting cells (0 h) and at early activation time points, i.e. 1 and 4 h after LPS + IL4 stimulation, all the identified processes are downregulated in DKO cells (Figure 8A). Several of the processes significantly altered, such as ‘response to wounding’, ‘defense responses’, ‘inflammatory responses’, ‘regulation of lymphocyte activation’ and ‘NF-KappaB’ cascades are related to immune functions. In addition pathways related to signal transduction such as ‘response to biotic stimulus’, ‘cation transport’, ‘second messenger mediated signaling’, ‘ion transport’ and ‘positive regulation of signal transduction’ are also down regulated. Indeed, examination of the proteins significantly downregulated at the first three time points (0, 1 and 4 h), indicated that the majority are located in the extracellular space, or are integral to the plasma membrane, or are components of typical signal transduction pathways, which is consistent with the identified biological processes affected (supplemental Table S1). The derived interaction maps generated with Cytoscape (41), in which the node size represents the relative gene set size and the line width represents similarity coefficient of the two connected nodes, visualizes the interconnection between the biological pathways affected by loss of HMGNs (Figure 8B). At the onset (0 h) and 4 h after LPS + IL4 stimulation, all the interconnected pathways are downregulated and with very few exemptions affect cellular responses related to immune or defense processes. However at the later stages of activation (72 h after stimulation), when the expression of over 200 genes were downregulated (Figure 7D), no significantly under-represented biological pathways were identified in DKO cells; all the identified pathways were actually upregulated and related to ‘RNA processing’ (Figure 8A). Thus, in resting cells and during initial stages of activation, the pathways most affected by loss of HMGNs are downregulated and related to immune system processes while in the terminal stages of activation the pathways affected by loss of HMGNs are upregulated and not related to obvious specific B cell functions.
Figure 8.
Loss of HMGN1 and HMGN2 affects the expression of genes involved in immune response and signal transduction. (A) Gene enrichment analysis by GSEA. The transcriptome data of WT and Hmgn1−/−n2−/− cells (with three replicates) at each time point were submitted as input and GS biological process sets (c5.all.v5.0.symbols.gmt) were used as reference. FDR threshold is 0.05. All the identified processes are shown. Green bars on the left side show the log2 (FDR) for the process identified with the numbers at the end of the green bars indicating the number of genes in that process. Bars on right side are the normalized enrichment fold with blue indicating down regulated processes and red showing upregulated processes. (B–D) Interaction maps between the identified pathways at the time point 0, 4 and 72 h were made with Cytoscape based on results in panel A. Node size represents the gene set size and line width represents the similarity coefficient of two connected nodes.
DISCUSSION
Our ChIP-seq data demonstrates a significant decrease of HMGN chromatin binding in activated B cells, which could be partially due to chromatin de-condensation and ∼4-fold expansion in the volume of the B cell nucleus due to the LPS treatment. The dynamic and non-sequence specific nature of HMGN binding makes it harder to capture their chromatin binding through immunoprecipitation. Nonetheless, the strong binding signals detected in the resting states and their close localization with DNase I hypersensitivity suggests that one of the potential major roles of HMGNs is to poise some of the genes for activation.
We find that the HMGN nucleosome binding proteins affect gene expression during the activation of B lymphocytes, a process that involves major changes in chromatin organization and plays a key role in immune responses. The genes most significantly affected during the early stages of activation are related to processes involved in immune response and signaling. The effects of HMGNs on gene expression correlate with their interaction with chromatin. In resting cells, loss of HMGNs affects significantly the expression of over 200 genes, with most of the genes being downregulated. The transcriptional changes affect pathways involved in cellular defense responses suggesting that, HMGNs modulates the innate, predetermined cellular transcription program, and are not regulating HMGN-specific cellular processes. Likely, HMGNs modulate the transcriptional profile of resting cells by interacting with, and modulating the intensity of chromatin regulatory site, as indicated by the genome wide co-localization of both HMGN1 and HMGN2 with DNase I hypersensitive sites, especially at promoters containing CpG islands, and at enhancer regions marked by H3K4me1 and H3K27ac. HMGNs may affect DNase I hypersensitivity by modulating the binding of linker histone H1 to chromatin (14), by interfering with the action of certain nucleosome remodeling complexes (39), by altering the levels of specific epigenetic marks in the tail of core histones (37,38), by changing the interaction of regulatory factors with chromatin (42), or by a combination of these effects.
We find that the DHSs at promoters are universally decreased across the whole genome and therefore we did not find a direct quantitative correlation between gene expression level changes and the DHSs changes. Nevertheless, the reduced DNase I hypersensitivity seen in resting DKO cells could still be the indirect underlying cause for the dampened transcriptional response seen one hour after activation (Figure 7B). Likely, in the initial stages of activation, the HMGN-induced changes in chromatin organization reduced the ability of transcriptional regulators to effectively alter the cellular transcription profile. However, during the progression of lymphocyte activation, the transcription profiles of WT and DKO cells are very similar with very few significant differences in gene expression, most likely because the LPS + IL4 activation leads to a major changes in the interaction of HMGN with chromatin. In addition, the induction of transcription regulators and the high levels of gene expression during the course of activation may mask the effects of HMGNs on chromatin structure and gene expression. Nevertheless, in fully activated cells, 72 h after LPS + IL4 addition, the gene expression profile of WT differs from that of DKO, with over 200 genes showing significant changes in gene expression levels. Interestingly, at this stage, when 10 times more RNA is produced compared with resting cells, the genes changed and the pathways affected are related to basic cellular functions such as ‘RNA processing’ and ‘ribosome assembly’ rather than to B-cell specific functions. Thus, although the interaction of HMGNs with chromatin is significantly decreased, their presence optimizes the expression of a subset of genes, some of which are involved in housekeeping functions, a possibility consistent with their high occupancy at promoters containing CpG islands.
In summary, our present studies indicate that HMGNs fine tune the fidelity of the cellular transcription profile during B cell activation. Likely, the impact of HMGN on transcription is not major because unlike most transcription factors, they bind to chromatin with low affinity and without specificity for DNA sequence and their chromatin interactions are affected by the presence of other chromatin-binding architectural proteins, including other HMG proteins and H1 variants (13,43). Nevertheless, Hmgn−/− mice and cells do show phenotypes and gene expression changes especially when exposed to stress (16,25,44,45). Thus, the changes in gene expression may be the underlying cause for the minor changes in the frequency of B cell subpopulation previously seen in a comprehensive phenotypic analysis of the DKO mice (16). Moreover, changes in HMGN levels can lead to changes in B cell properties since the significant increased risk of B cell acute lymphoblastic leukemia seen in Down syndrome has been linked directly to overexpression of HMGN1 (46). Thus, dysregulation of HMGNs levels may weaken the ability of B cells to cope with biological challenges.
ACCESSION NUMBERS
The data from this study has been submitted to the NCBI Sequence Read Archive (SRA; http://ncbi.nlm.nih.gov/sra/) under accession number SAMN04327545-SAMN04327548
Supplementary Material
Acknowledgments
Author contributions: S.Z., M.R. and M.B. conceived the experiments, I.Z. and D.L. performed bioinformatics analysis, S.Z., T.D., T.F., M.S.V. and M.R. performed experiments and analyzed data, R.C., D.L., R.B. and M.B. analyzed data, S.Z. and M.B. wrote the manuscript, M.B. coordinated the research.
Footnotes
Present address: Mark Rochman, Cincinnati Children's Hospital, Division of Allergy and Immunology, Cincinnati, OH 45229, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Center for Cancer Research; Intramural Research Programs of the National Cancer Institute, NIH; National Center for Biotechnology Information; National Library of Medicine, NIH. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Millan-Arino L., Izquierdo-Bouldstridge A., Jordan A. Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochim. Biophys. Acta. 2015;1859:510–519. doi: 10.1016/j.bbagrm.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Hergeth S.P., Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16:1439–1453. doi: 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catez F., Ueda T., Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 5.Reeves R. Nuclear functions of the HMG proteins. Biochim. Biophys. Acta. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustin M. High mobility group proteins. Biochim. Biophys. Acta. 2010;1799:1–2. doi: 10.1016/j.bbagrm.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Bianchi M.E., Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Postnikov Y., Bustin M. Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta. 2010;1799:62–68. doi: 10.1016/j.bbagrm.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Romero R., Eirin-Lopez J.M., Ausio J. Evolution of high mobility group nucleosome-binding proteins and its implications for vertebrate chromatin specialization. Mol. Biol. Evol. 2015;32:121–131. doi: 10.1093/molbev/msu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda T., Catez F., Gerlitz G., Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol. Cell. Biol. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato H., van Ingen H., Zhou B.R., Feng H., Bustin M., Kay L.E., Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catez F., Hock R. Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim. Biophys. Acta. 2010;1799:15–27. doi: 10.1016/j.bbagrm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Catez F., Brown D.T., Misteli T., Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochman M., Postnikov Y., Correll S., Malicet C., Wincovitch S., Karpova T.S., McNally J.G., Wu X., Bubunenko N.A., Grigoryev S., et al. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng T., Zhu Z.I., Zhang S., Postnikov Y., Huang D., Horsch M., Furusawa T., Beckers J., Rozman J., Klingenspor M., et al. Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers. Genome Res. 2015;25:1295–1308. doi: 10.1101/gr.192229.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kugler J.E., Horsch M., Huang D., Furusawa T., Rochman M., Garrett L., Becker L., Bohla A., Holter S.M., Prehn C., et al. High mobility group N proteins modulate the fidelity of the cellular transcriptional profile in a tissue- and variant-specific manner. J. Biol. Chem. 2013;288:16690–16703. doi: 10.1074/jbc.M113.463315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprent J. Lifespans of naive, memory and effector lymphocytes. Curr. Opin. Immunol. 1993;5:433–438. doi: 10.1016/0952-7915(93)90065-z. [DOI] [PubMed] [Google Scholar]
- 19.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 20.Bryant C.E., Spring D.R., Gangloff M., Gay N.J. The molecular basis of the host response to lipopolysaccharide. Nat. Rev. Microbiol. 2010;8:8–14. doi: 10.1038/nrmicro2266. [DOI] [PubMed] [Google Scholar]
- 21.Major J., Fletcher J.E., Hamilton T.A. IL-4 pretreatment selectively enhances cytokine and chemokine production in lipopolysaccharide-stimulated mouse peritoneal macrophages. J. Immunol. 2002;168:2456–2463. doi: 10.4049/jimmunol.168.5.2456. [DOI] [PubMed] [Google Scholar]
- 22.Kouzine F., Wojtowicz D., Yamane A., Resch W., Kieffer-Kwon K.R., Bandle R., Nelson S., Nakahashi H., Awasthi P., Feigenbaum L., et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaehning J.A., Stewart C.C., Roeder R.G. DNA-dependent RNA polymerase levels during the response of human peripheral lymphocytes to phytohemagglutinin. Cell. 1975;4:51–57. doi: 10.1016/0092-8674(75)90133-6. [DOI] [PubMed] [Google Scholar]
- 24.Fowler T., Garruss A.S., Ghosh A., De S., Becker K.G., Wood W.H., Weirauch M.T., Smale S.T., Aronow B., Sen R., et al. Divergence of transcriptional landscape occurs early in B cell activation. Epigenetics Chromatin. 2015;8 doi: 10.1186/s13072-015-0012-x. doi:10.1186/s13072-015-0012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birger Y., West K.L., Postnikov Y.V., Lim J.H., Furusawa T., Wagner J.P., Laufer C.S., Kraemer K.H., Bustin M. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003;22:1665–1675. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovalchuk A.L., Ansarah-Sobrinho C., Hakim O., Resch W., Tolarova H., Dubois W., Yamane A., Takizawa M., Klein I., Hager G.L., et al. Mouse model of endemic Burkitt translocations reveals the long-range boundaries of Ig-mediated oncogene deregulation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10972–10977. doi: 10.1073/pnas.1200106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postnikov Y.V., Trieschmann L., Rickers A., Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J. Mol. Biol. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 28.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-10-r80. doi:10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchen S., Resch W., Yamane A., Kuo N., Li Z., Chakraborty T., Wei L., Laurence A., Yasuda T., Peng S., et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 36.Bogdanovic O., Fernandez-Minan A., Tena J.J., de la Calle-Mustienes E., Hidalgo C., van Kruysbergen I., van Heeringen S.J., Veenstra G.J., Gomez-Skarmeta J.L. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 2012;22:2043–2053. doi: 10.1101/gr.134833.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim J.H., West K.L., Rubinstein Y., Bergel M., Postnikov Y.V., Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim J.H., Catez F., Birger Y., West K.L., Prymakowska-Bosak M., Postnikov Y.V., Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Rattner B.P., Yusufzai T., Kadonaga J.T. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol. Cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amen M., Espinoza H.M., Cox C., Liang X., Wang J., Link T.M., Brennan R.G., Martin J.F., Amendt B.A. Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/beta-catenin signaling. Nucleic Acids Res. 2008;36:462–476. doi: 10.1093/nar/gkm1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bustin M., Catez F., Lim J.H. The dynamics of histone H1 function in chromatin. Mol. Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Birger Y., Catez F., Furusawa T., Lim J.H., Prymakowska-Bosak M., West K.L., Postnikov Y.V., Haines D.C., Bustin M. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 2005;65:6711–6718. doi: 10.1158/0008-5472.CAN-05-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Postnikov Y.V., Furusawa T., Haines D.C., Factor V.M., Bustin M. Loss of the nucleosome-binding protein HMGN1 affects the rate of N-nitrosodiethylamine-induced hepatocarcinogenesis in mice. Mol. Cancer Res. 2014;12:82–90. doi: 10.1158/1541-7786.MCR-13-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane A.A., Chapuy B., Lin C.Y., Tivey T., Li H., Townsend E.C., van Bodegom D., Day T.A., Wu S.C., Liu H., et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 2014;46:618–623. doi: 10.1038/ng.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.